Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Composition, phosphorylation and dynamic organization of photosynthetic protein complexes in plant thylakoid membrane
Marjaana
Rantala†
 ,
Sanna
Rantala†
,
Sanna
Rantala†
 and
Eva-Mari
Aro
and
Eva-Mari
Aro
 *
*
Molecular Plant Biology, Department of Biochemistry, University of Turku, FI-20520, Turku, Finland. E-mail: evaaro@utu.fi
First published on 16th April 2020
Abstract
The photosystems (PS), catalyzing the photosynthetic reactions of higher plants, are unevenly distributed in the thylakoid membrane: PSII, together with its light harvesting complex (LHC)II, is enriched in the appressed grana stacks, while PSI–LHCI resides in the non-appressed stroma thylakoids, which wind around the grana stacks. The two photosystems interact in a third membrane domain, the grana margins, which connect the grana and stroma thylakoids and allow the loosely bound LHCII to serve as an additional antenna for PSI. The light harvesting is balanced by reversible phosphorylation of LHCII proteins. Nevertheless, light energy also damages PSII and the repair process is regulated by reversible phosphorylation of PSII core proteins. Here, we discuss the detailed composition and organization of PSII–LHCII and PSI–LHCI (super)complexes in the thylakoid membrane of angiosperm chloroplasts and address the role of thylakoid protein phosphorylation in dynamics of the entire protein complex network of the photosynthetic membrane. Finally, we scrutinize the phosphorylation-dependent dynamics of the protein complexes in context of thylakoid ultrastructure and present a model on the reorganization of the entire thylakoid network in response to changes in thylakoid protein phosphorylation.
Introduction
The chloroplast enclosed thylakoid system of photosynthetic organisms is a unique membrane network that hosts the energy conversion machinery responsible for producing the chemical energy that fuels life on Earth. The photosynthetic machinery, composed of photosystem (PS) II, cytochrome (Cyt) b6f, PSI and ATP synthase mediates the first phase of photosynthesis, the light reactions. In the linear electron transfer chain, the two photosystems are interconnected by the Cyt b6f complex and the two mobile electron carriers, plastoquinone in the lipid bilayer and plastocyanin in the thylakoid lumen (Fig. 1). The energy of solar irradiance is first captured by PS-bound light harvesting complexes (LHC) and then converted and stabilized as chemical energy by electron and proton transfer reactions mediated by the main thylakoid protein complexes. The light energy is stored in NADPH and ATP molecules, which are used in subsequent energy-demanding reactions such as carbon assimilation.Photosynthetic energy conversion requires highly coordinated interplay between individual thylakoid protein complexes and therefore depends on the molecular architecture of the photosynthetic machinery. While the basic electron and proton transport reactions as well as the involved enzyme complexes have, in principle, remained fundamentally similar in all oxygenic photosynthetic organisms throughout the evolution, the arrangement of the photosynthetic machinery and the organization of the thylakoid membrane varies radically between different organisms. A unique landmark of plant thylakoid membrane is the formation of appressed membrane discs and the uneven lateral distribution of the photosynthetic protein complexes along the membrane plane. This membrane system is capable of adjusting its function upon the most subtle changes in environmental conditions by making use of a multitude of regulatory processes. Reversible protein phosphorylation, i.e. the addition of negatively charged phosphate group (P) from ATP to a sidechain of certain amino acids, is among the most important post-translational modifications of proteins in all eukaryotic cells. In plant thylakoid membrane, the protein phosphorylation regulates the energy conversion reactions and the repair of damaged photosynthetic machinery through remodeling the protein complex arrangement and thylakoid ultrastructure. Here, we provide an overview of the composition and protein phosphorylation-dependent dynamic organization of the photosynthetic machinery in angiosperms.
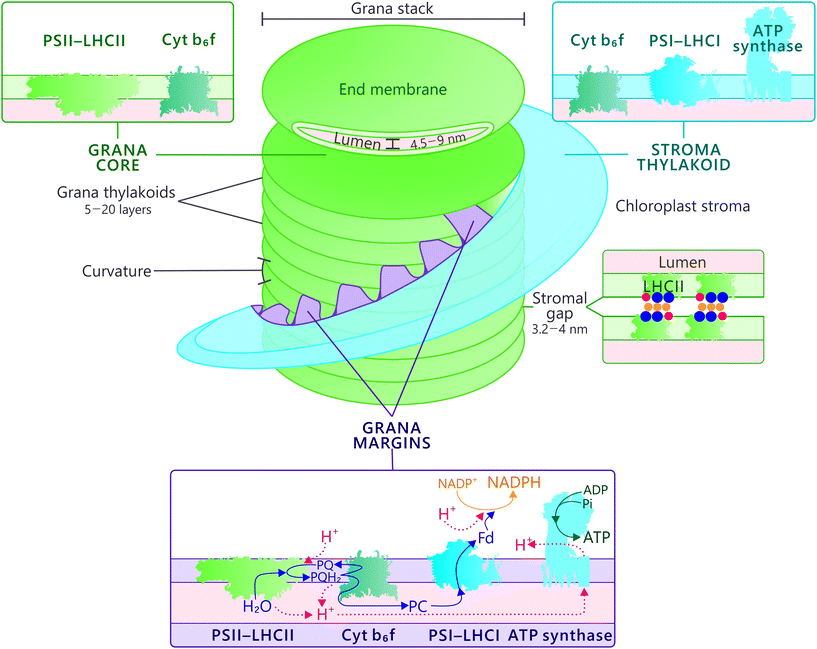
Thylakoid membrane ultrastructure and distribution of thylakoid protein complexes
In plants and algae, the photosynthetic energy transduction occurs in chloroplast-enclosed flattened vesicles, the thylakoids. As illustrated in Fig. 1, the thylakoid network in higher plants is differentiated into two morphologically distinct domains: the cylindrical grana appressions and the interconnecting and non-appressed stroma thylakoids. Each chloroplast contains approximately 40–60 grana stacks1 that are formed usually by 5–20 folded thylakoid layers and have a diameter ranging from 300 to 600 nm, depending on the plant species and the light condition.2,3 The width of the thylakoid enclosed lumen varies between 4.5 nm in darkness up to 9 nm in light, and that of the stromal gap between neighboring grana membranes from 3.2 to 4.0 nm.4 The grana stacks account for the majority of the total membrane area and harbor the majority of the total pigment content,5 but the proportion of grana relative to the stroma thylakoids varies depending on the light condition.6The interface between grana and stroma thylakoids, denoted as grana margins, is a biochemically distinct thylakoid fraction, but the exact composition and location of this domain is under debate. Based on structural analysis, it was proposed that grana margins consist of the highly curved regions of the grana stacks and is protein-free,7–10 whereas another interpretation suggests that grana margin is represented by a larger annular ring at the periphery of the grana stack and is enriched in photosynthetic protein complexes.5,11 A recently characterized thylakoid fraction, obtained by digitonin solubilisation and subsequent centrifugation steps, was reported to be a combination of a highly curved grana margins, straight membranes at the interphase between grana and stroma as well as of the fret-like protrusions (the slits) between grana and stroma membranes.12,13 Subsequent further optimization of thylakoid fractionation yielded a separation of the curved membrane domain from the rest of the margin domain,14 and accordingly, the nomenclature adopted in Fig. 1 includes a distinction between the grana curvature and grana margin domains.
Different structural models exist on the exact three-dimensional configuration of the thylakoid membrane. The overall structure is similar in all models, but the uncertainty arises from different interpretations on how the stroma thylakoids are joined to the grana appressions. Folded fork model postulates that a granum represents a bifurcation of the stroma membranes and is formed of repeated units each containing three grana stacks.15 Shimoni et al.16 proposed that a granum is a bifurcation of a lamellar stroma membrane sheet and the neighboring grana units, each composed of paired layers that are further connected by membrane bridges. Helical fretwork model, originally proposed by Paolillo,17 suggests that the stroma thylakoids wind around the grana stacks as right-handed helices and are interconnected to the grana by several narrow membrane slits,2,17,18 which according to Anderson et al.,6 might represent the grana margins. The membrane slits guarantee the continuum of the lipid and lumenal phases of the thylakoid membrane and allow both functional and structural interaction between the photosynthetic protein complexes6,19 (Fig. 1). Recently, Bussi et al. combined different electron tomography techniques and demonstrated that, in addition to the four to six right handed helices winding around grana, the membrane consists of several left-handed helical junctions that connect adjacent lamellar sheets or helices and consolidate the geometry of the membrane system by minimizing the surface and bending energies.20 Since the helical fretwork model is the most prevalent of the above mentioned models and supported by data obtained with different electron microscopy techniques as well as modelling,2,20–22 we scrutinize the thylakoid protein complex dynamics in the frame of this model.
The functional relevance and the exact mechanism of the grana stacking are still under debate (comprehensively reviewed by Chow et al.23). Although the Lhcb1 and Lhcb2 proteins – the most abundant constituents of the grana membranes – are not a prerequisite for the grana formation,24 but some level of stacking is observed even without the majority of Lhcb proteins,25 they are generally considered to play an essential role in stabilizing the stacking. The negatively charged stromal loops of Lhcb proteins form salt bridges with cations, most importantly with divalent Mg2+, which are screened between adjacent membrane layers.26,27 Although this electrostatic screening is considered as the most important stabilizing force of the membrane appressions, the interaction between the positively charged N-terminus of one Lhcb protein with the negatively charged stromal loop of another Lhcb protein on the opposite membrane bilayer might also be involved in grana formation.28 Further, CURT1 oligomers located at the curved regions of grana are known to regulate the number and size of the stacks.10
The photosynthetic protein complexes embedded in the thylakoid membrane account for more than 70% of the membrane area and are unevenly distributed in the thylakoid membrane29 (Fig. 1). PSII–LHCII supercomplexes are densely packed in the appressed grana and cover 80% of the membrane area.30 PSI–LHCI and ATP synthase with their 5 nm and 16 nm stromal protrusion are unable to fit to the stromal gap between adjacent grana layers and are enriched in the stroma-exposed thylakoids.31 Cyt b6f is likely located in all thylakoid domains32,33 and its distribution might be regulated according to light conditions.34 The grana margins, which should not be confused with the curvature domain in the edge of the grana membranes,14 are biochemically a fusion of grana and stroma thylakoids, and accommodate both photosystems.5,35–39
Composition and LHCII-mediated supramolecular arrangement of the photosystems
The photosynthetic machinery is composed of large protein complexes, each comprising several nucleus and chloroplast-encoded protein subunits (Table 1 for PSII–LHCII and Table 2 for PSI–LHCI) and prosthetic groups. The first event of photosynthetic light reactions is the absorption of solar irradiation by the pigment-binding light harvesting complex (LHC) antennas. The LHC proteins associate peripherally with both photosystems and are mainly responsible for collecting the solar energy and for transferring the excitation energy to the reaction centers located in the photosystem core complexes. The LHC proteins typically contain three transmembrane α-helices (A, B, C) and two lumenal helices (D, E).| Protein | Gene | AGI identifier | Function |
|---|---|---|---|
| Lhcb1 | Lhcb1.1 | AT1G29920 | Light harvesting in S-, M- and L-LHCII |
| Lhcb1.2 | AT1G29910 | ||
| Lhcb1.3 | AT1G29930 | ||
| Lhcb1.4 | AT2G34430 | ||
| Lhcb1.5 | AT2G34420 | ||
| Lhcb2 | Lhcb2.1 | AT2G05100 | Light harvesting in S- and L-LHCII |
| Lhcb2.2 | AT2G05070 | ||
| Lhcb2.3 | AT3G27690 | ||
| Lhcb2.4 | AT3G27690 | ||
| Lhcb3 | Lhcb3 | AT5G54270 | Light harvesting in M-LHCII |
| CP29 | Lhcb4.1 | AT5G01530 | Light harvesting, M-LHCII binding |
| Lhcb4.2 | AT3G08940 | ||
| Lhcb4.3 | AT2G40100 | ||
| CP26 | Lhcb5 | AT4G10340 | Light harvesting, S-LHCII binding |
| CP24 | Lhcb6 | AT1G15820 | Light harvesting, M-LHCII binding |
| D1 | PsbA | ATCG00020 | Reaction center |
| CP47 | PsbB | ATCG00680 | Light harvesting |
| CP43 | PsbC | ATCG00280 | Light harvesting |
| D2 | PsbD | ATCG00270 | Reaction center |
| Cyt b559 α | PsbE | ATCG00580 | Reaction center |
| Cyt b559 β | PsbF | ATCG00570 | Reaction center |
| PsbH | PsbH | ATCG00710 | Antenna association |
| PsbI | PsbI | ATCG00080 | Reaction center |
| PsbJ | PsbJ | ATCG00550 | Core stabilization |
| PsbK | PsbK | ATCG00070 | Core stabilization |
| PsbL | PsbL | ATCG00560 | PSII dimerization |
| PsbM | PsbM | ATCG00220 | PSII dimerization |
| PsbTc | PsbTc | ATCG00690 | PSII dimerization |
| PsbTn | PsbTn | AT3G21055 | Light acclimation |
| PsbZ | PsbZ | ATCG00300 | Antenna association |
| PsbO | PsbO-1 | AT5G66570 | |
| PsbO-2 | AT3G50820 | Regulation of D1 dephosphorylation and turnover | |
| PsbP | PsbP-1 | AT1G06680 | Stabilization of Mn4CaO5 complex |
| PsbP-2 | AT2G30790 | Stabilization of Mn4CaO5 complex | |
| PsbQ | PsbQ-1 | AT4G21280 | Stabilization of Mn4CaO5 complex |
| PsbQ-2 | AT4G05180 | Stabilization of Mn4CaO5 complex | |
| PsbR | PsbR | AT1G79040 | Stabilization of Mn4CaO5 complex |
| PsbS | PsbS | AT1G44575 | Thermal dissipation |
| PsbW | PsbW | AT2G30570 | Core stabilization, antenna association |
| PsbX | PsbX | AT2G06520 | Core stabilization |
| Protein | Gene | AGI identifier | Function |
|---|---|---|---|
| Lhca1 | Lhca1 | AT3G54890 | Light harvesting |
| Lhca2 | Lhca2 | AT3G61470 | Light harvesting |
| Lhca3 | Lhca3 | AT1G61520 | Light harvesting |
| Lhca4 | Lhca4 | AT3G47470 | Light harvesting |
| Lhca5 | Lhca5 | AT1G45474 | NDH-1 docking |
| Lhca6 | Lhca6 | AT1G19150 | NDH-1 docking |
| PsaA | PsaA | ATCG00350 | Reaction center |
| PsaB | PsaB | ATCG00340 | Reaction center |
| PsaC | PsaC | ATCG01060 | FA and FB binding, ferredoxin docking |
| PsaD | PsaD-1 | AT4G02770 | Ferredoxin docking |
| PsaD-2 | AT1G03130 | ||
| PsaE | PsaE-1 | AT4G28750 | Ferredoxin docking |
| PsaE-2 | AT2G20260 | ||
| PsaF | PsaF | AT1G31330 | Plastocyanin docking |
| PsaG | PsaG | AT1G55670 | LHCI stabilization |
| PsaH | PsaH-1 | AT3G16140 | L-LHCII docking |
| PsaH-2 | AT1G52230 | ||
| PsaI | PsaI | ATCG00510 | Stabilization of LHCII docking |
| PsaJ | PsaJ | ATCG00630 | PsaF stabilization |
| PsaK | PsaK | AT1G30380 | LHCI stabilization |
| PsaL | PsaL | AT4G12800 | L-LHCII docking |
| PsaN | PsaN | AT5G64040 | Plastocyanin docking |
| PsaO | PsaO | AT1G08380 | L-LHCII docking |
LHCII and PSII core subunits
The light harvesting complex II (LHCII), the antenna system of photosystem II, consists of six different Lhcb proteins and binds half of the chlorophyll pigments in the thylakoid membrane.40 The three major Lhcb proteins, Lhcb1, Lhcb2 and Lhcb3, are assembled into trimers that are bound to PSII core via Lhcb4 (CP29), Lhcb5 (CP26) and Lhcb6 (CP24) proteins (Table 1). Each of the major Lhcb monomers bind eight chlorophyll a and six chlorophyll b pigments as well as four carotenoids.41 The three isoforms of the major Lhcb proteins are present in different molar ratios. In Arabidopsis thaliana (Arabidopsis), the Lhcb1:2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio has been estimated to be 7
3 ratio has been estimated to be 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 based on proteomic data,42,43 but the ratio depends on the light condition. Each of the major Lhcb isoforms is present as several subisoforms. In Arabidopsis, five subisoforms are found (encoded by Lhcb1.1–5), and of these, Lhcb1.1–3 are identical, while Lhcb1.4 and Lhcb1.5 show some difference.44 Of the Lhcb2 subisoforms (encoded by Lhcb2.1–4), on the other hand, Lhcb2.1, Lhcb2.2 and Lhcb2.3 are identical and Lhcb2.4 diverges by one amino acid.44 Moreover, instead of a separate gene, Lhcb2.3 appears to represent one of the alleles of Lhcb2.1. From the three Lhcb4 subunits (encoded by Lhcb4.1–3), Lhcb4.3 diverges significantly from Lhcb4.1 and Lhcb4.2 by lacking a fraction of the C-terminal part located in the lumen.44,45 For more information about different Lhcb isoforms and their evolution, see the recent review by Crepin and Caffarri.43
1 based on proteomic data,42,43 but the ratio depends on the light condition. Each of the major Lhcb isoforms is present as several subisoforms. In Arabidopsis, five subisoforms are found (encoded by Lhcb1.1–5), and of these, Lhcb1.1–3 are identical, while Lhcb1.4 and Lhcb1.5 show some difference.44 Of the Lhcb2 subisoforms (encoded by Lhcb2.1–4), on the other hand, Lhcb2.1, Lhcb2.2 and Lhcb2.3 are identical and Lhcb2.4 diverges by one amino acid.44 Moreover, instead of a separate gene, Lhcb2.3 appears to represent one of the alleles of Lhcb2.1. From the three Lhcb4 subunits (encoded by Lhcb4.1–3), Lhcb4.3 diverges significantly from Lhcb4.1 and Lhcb4.2 by lacking a fraction of the C-terminal part located in the lumen.44,45 For more information about different Lhcb isoforms and their evolution, see the recent review by Crepin and Caffarri.43
The PSII core complex functions as a dimer and is surrounded by peripheral LHC antenna system. Thus far, the structure of plant (Pisum sativum) PSII–LHCII supercomplex has been resolved by cryo-electron microscopy at 2.7 Å resolution.46 One PSII core monomer consists of 20–23 protein subunits, depending on the organism, and harbors several pigments, including 35 chlorophylls, two pheophytins and 10 carotenoids as well as several other co-factors such as quinones.41 The PsbA (D1), PsbD (D2) and PsbI proteins as well as the α (PsbE) and β (PsbF) subunits of the Cyt b559 complex form the PSII reaction center complex, which is the minimal unit required for primary photochemistry.47,48 The reaction center is surrounded by two large chlorophyll-binding light harvesting proteins CP47 (PsbB) and CP43 (PsbC) as well as the lumenal oxygen evolving complex. The oxygen evolving complex is composed of PsbO, PsbP, PsbQ and PsbR proteins that stabilize the Mn4CaO5 cluster. PsbO-Q are encoded by two genes, and for PsbO, the two isoforms have been suggested to possess different functions: the major isoform PsbO-1 stabilizes PSII activity, while the minor isoform PsbO-2 has been suggested to regulate the dephosphorylation and turnover of D1.49 In addition, the core complex includes several so-called low molecular mass (<10 kDa) subunits: PsbH-M, PsbTc, PsbTn, PsbW, PsbX and PsbZ.
PsbTc and PsbTn that were previously considered as plastidial and nucleic paralogues of PsbT but are now recognized as functionally separate subunits. The transmembrane protein PsbTc has been suggested to contribute to PSII dimerization, whereas the lumenal extrinsic PsbTn seems to be important for light acclimation.50,51 Noteworthy, PsbN is no longer considered as a structural subunit of PSII.52
PSII–LHCII supercomplexes
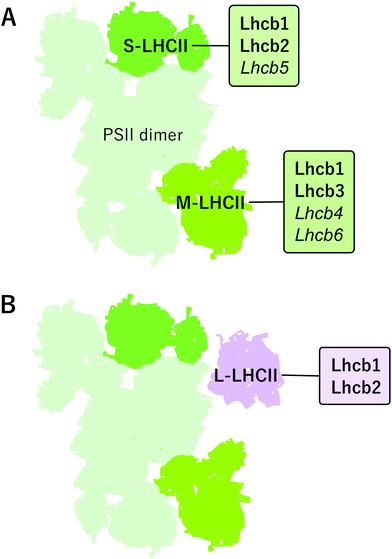
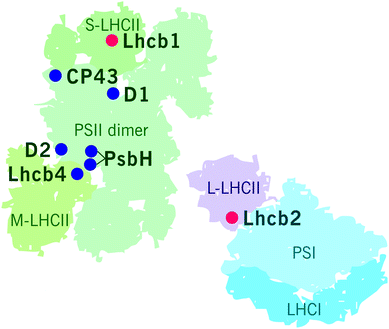
PSII–LHCII supercomplexes are comprised of PSII core dimers and the peripheral LHCII antenna complexes. In the most abundant form of PSII–LHCII supercomplexes, C2S2M2, both core monomers (C) are associated with a strongly bound S-LHCII trimer and a moderately bound M-LHCII trimer, which are associated to the PSII core via minor antenna proteins Lhcb5 and Lhcb4/6, respectively (Fig. 2A). In addition to the S- and M-trimers, a large pool of LHCII trimers, denoted as L-LHCII (loosely-bound), exists in the thylakoid membrane and likely occupies the spaces between individual PSII–LHCII supercomplexes in the grana core8,19,53 and interconnects the two photosystems in grana margins54 (discussed in more detail later) (Fig. 2B).The distinct LHCII trimers of the major Lhcb proteins play different roles in the structure of the photosynthetic apparatus. In the model plant Arabidopsis, the S-LHCII trimer is composed of Lhcb1 and Lhcb2 proteins, whereas the M-trimer consists of Lhcb1 (especially Lhcb1.4) and Lhcb3 proteins.42 Lhcb3 and Lhcb6 are unique to land plants, but intriguingly, are absent from the gymnosperm family Pinaceae.45,55 In land plants, particular structural features in the N-terminus and in the AC- and BC-loops of the Lhcb3 protein facilitate its interaction with Lhcb6 and determine the binding orientation of the M-trimer to the core complex.46 The structure of C2S2M2 supercomplex has been determined with cryo-EM at 5.3 Å resolution from Arabidopsis41 and at 2.7–3.2 Å resolution from Pisum sativum,46 and, based on the location of individual pigments, these structures suggest potential energy transfer routes from LHCII antenna to the core. The composition of L-trimers is not yet well defined, but they probably consist mainly of Lhcb1 (especially Lhcb1.5) and Lhcb2 (Lhcb2.2) proteins.42,56
In certain conditions, the PSII–LHCII supercomplexes have been reported to form semi-crystalline arrays in grana core.3,57,58 Besides such lateral re-arrangement, the PSII–LHCII supercomplexes in the array are also vertically associated with complexes across the stromal gap.46,59 Although the arrays are relatively rarely observed60 and even suggested to represent only artefacts arising from sample preparation,61 they might also serve a function in changing light conditions: switching of the supramolecular organization of PSII–LHCII complexes from disordered to well-arranged semi-crystalline arrays is assumed to be important in regulation of the diffusion efficiency of plastoquinol as well as for photoprotection and protein repair.62 The semi-crystalline PSII–LHCII arrays are the most abundant in low light-acclimated plants. The highly ordered organization of the C2S2M2 supercomplexes in grana probably allows faster energy transfer between the complexes and more fluent lateral diffusion of lipophilic plastoquinol between PSII and Cyt b6f but, concomitantly, restricts the mobility of protein complexes, for instance during the PSII repair cycle (discussed in more detail later).62 Under prolonged high light conditions, on the other hand, the frequency of semi-crystalline arrays decreases, facilitating the lateral mobility of membrane protein complexes and enabling thermal dissipation from LHCII in a process involving PsbS and zeaxanthin.60,63 Such a dissipative mode of PSII–LHCII complexes has been suggested to represent an LHCII aggregate,64,65 a dissociated LHCII trimer66 or a dissociated pentamer LHCII-CP24-CP29 i.e. M-LHCII.67
PSI–LHCI subunits and protein supercomplexes
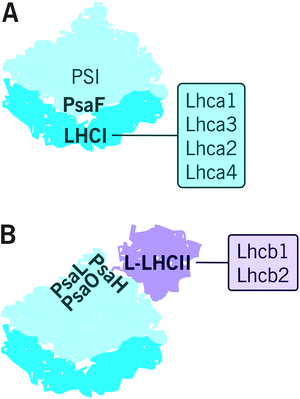
Subunits of PSI–LHCI are presented in Table 2. So far, the structure of plant (Pisum sativum) PSI–LHCI has been resolved with X-ray crystallography at 2.6 Å resolution.68 This structure contains 16 protein subunits and 232 cofactors. PSI reaction center is formed by PsaA/PsaB heterodimer that harbors most of the cofactors involved in the electron transfer reactions i.e. 6 chlorophyll a, 2 phylloquinones and a Fe4S4 cluster FX.69 The reaction center is surrounded by PsaC-PsaL, PsaN and PsaO, subunits of which the PsaH, PsaG and PsaO are unique to plants and algae.70 PsaC binds the rest of the redox active cofactors i.e. Fe4S4 clusters FA and FB and is involved, together with PsaD and PsaE, in docking ferredoxin on the stromal side of PSI. PsaF, stabilized by PsaJ, docks plastocyanin together with PsaN and mediates the binding of LHCI antenna, whereas PsaG and PsaK are involved in stabilization of the LHCI antenna.71As illustrated in Fig. 3A, the PSI core forms a stable supercomplex with the LHCI antenna, which is associated to the PsaF side of the core complex.72 The LHCI comprises of Lhca1–4, which are assembled into dimers Lhca1/Lhca4 and Lhca2/Lhca3.73,74 The fact that the Lhca proteins form dimers while the Lhcb proteins preferentially oligomerize as trimers likely derives from the slightly different orientation of their N-terminus.70 It is important to note that, despite the highly conserved PSI–LHCI complex with four Lhca proteins, some species diverge from this model by dividing the Lhca proteins between the PsaF and the PsaH side75 or by binding additional Lhca proteins on the PsaH76 or PsaF77 side of the PSI core. Interestingly, an additional Lhca1/Lhca4 dimer bound to PsaH side was recently found in Arabidopsis.78 (For a recent review on the supercomplex diversity, see ref. 79 and 80.) PSI–LHCI has also been reported to physically interact with Cyt b6f in Chlamydomonas reinhardtii81 and recently also in Arabidopsis.82 Moreover, up to six PSI–LHCI complexes have been found attached to type I NADH dehydrogenase (NDH-1) complex via LHCI proteins Lhca5 and Lhca6.82–84 Importantly, PSI–LHCI is also able to bind L-LHCII. As depicted in Fig. 3B, PsaL, PsaH and PsaO form the docking site for L-LHCII85 (see the next chapters), and the docking has been suggested to be stabilized by PsaI.86
L-LHCII connecting PSII and PSI
Although both photosystems have their own light harvesting antennas, the majority of the light energy is harvested and transferred to both PSII and PSI reaction centers by mutual L-LHCII system that likely resides in the grana margins.54,87,88 A megacomplex containing both photosystems and the shared L-LHCII antenna has been suggested to exist based on analysis with native acrylamide gel electrophoresis.37,87,89 However, the migration of the complex in the native gel electrophoresis system corresponds a mass of several megadaltons90 and therefore likely represents a larger cluster of proteins (here assigned as PSII–LHCII–PSI–LHCI cluster) rather than a defined complex. Nevertheless, since the mechanical fractionation of the thylakoid membrane revealed both PSII and PSI in the grana margin fraction,35 the evidence has accumulated at accelerating rate to show that the two photosystems are enriched in grana margins and become interconnected via mutual LHCII antenna system.38,39,42,54,85,91,92 We argue that the digitonin-soluble PSII–LHCII–PSI–LHCI cluster originates from such network.88 Nevertheless, such a cluster has not been successfully confirmed by electron microscopy, apparently due to a weak interaction via L-LHCII, and for this reason, more detailed biophysical, biochemical and structural analyses are needed.Protein phosphorylation – flexible organization of protein complexes and dynamics of the entire thylakoid ultrastructure upon changing light conditions ensure fluent photosynthesis
As discussed above, a high-level organization of thylakoid protein complexes is essential for the key reactions of photosynthesis, yet the protein–protein interactions are highly dynamic and dependent on environmental cues, particularly on changes in light conditions. The amount and spectral properties of sunlight entering a plant leaf undergo seasonal and cyclic variations, but the sun flecks, caused by brief movements of the canopy or clouds in the sky, introduce rapid changes in the input of irradiation taking place in timescale of seconds or minutes. These changes come along with substantial impact on the photosynthetic machinery, as an equilibrium between optimal light utilization and avoidance of destructive consequences of excess light energy requires subtle fine-tuning of the entire network of photosynthetic membrane system. The variations in environmental cues ensue structural changes of the extraordinary plastic thylakoid membrane system.6 Alterations in grana stacking, changes in the width of the lumen and stromal gap and changes in the complex interactions between proteins93–95 can take place in timescale of minutes.96 Subsequent systematic research on the interaction, activation and relaxation of different regulatory mechanisms of photosynthetic light reactions, operating efficiently in fluctuating light conditions (e.g.ref. 97–99), have paved the way towards unifying models of the structural and functional dynamics of the thylakoid membrane.Post-translational protein phosphorylation constitutes one of the most important and extensively scrutinized short-term regulatory mechanisms that affects the thylakoid ultrastructure and the enzymatic activity, affinity and stability of the proteins, and contributes to many other aspects in cellular regulation in all eukaryotes. Addition of a phosphate group to serine, threonine (Thr) or tyrosine residue of the client proteins is catalyzed by specific protein kinases, while the removal of the phosphate group is mediated by counteracting protein phosphatases. Almost 200 phosphoproteins have been found in the chloroplast of the model plant Arabidopsis and at least 28 of them are located in the thylakoid membrane.100 Here, the reversible phosphorylation of PSII–LHCII proteins and subsequent changes in protein complex interactions and thylakoid structural dynamics are discussed.
Reversible LHCII antenna phosphorylation
LHCII proteins account for half of the total protein mass of the thylakoid membrane. They are responsible for maximizing the trapping of solar irradiance, delivering equal amount of excitation energy to the two photosystems and thermally dissipating the energy, which exceeds the utilization capacity of the photosynthetic machinery. Several light harvesting proteins are reversibly phosphorylated by specific protein kinases and phosphatases. The phosphorylation of the N-terminal Thr residues of Lhcb1 and Lhcb2 proteins, first described and linked to the regulation of light harvesting already decades ago,101 has an important role in short-term light acclimation. The addition of a phosphate group to the Lhcb proteins is catalyzed by redox-dependent STN7 kinase,102 which is activated by the binding of plastoquinol to the Q0 site of the Cyt b6f complex.103 The STN7 kinase itself contains three or four phosphosites100,104 and the activity and turn-over of the kinase is likely regulated by phosphorylation of the Ser and two of the Thr residues of the STN7 kinase.104 Upon a shift from low to high light, the STN7 kinase becomes inactivated due to changes in stromal redox state.105 In addition to Lhcb1 and Lhcb2, STN7 can phosphorylate to a minor extent also the PSII core proteins D1, D2 and CP43 in specific conditions as well as the monomeric antenna protein Lhcb4 (CP29).102,106,107The removal of the phosphate group from Lhcb proteins is catalyzed by THYLAKOID-ASSOCIATED PHOSPHATASE OF 38 kDa /PROTEIN PHOSPHATASE1 (TAP38/PPH1).108,109 The TAP38/PPH1 phosphatase is constitutively expressed and seems to be redox-independent.108 Nevertheless, the flexible association of the phosphatase with large protein complexes might affect the activity or stability of TAP38/PPH1.110 Unlike the STN7 kinase, the TAP38 phosphatase selectively targets only the Lhcb1 and Lchb2 proteins, not the PSII core proteins, by two specific surface cleft sites on the phosphatase.111
LHCII antenna phosphorylation and reorganization of protein complexes in grana margins allow fluent electron flow between the two photosystems
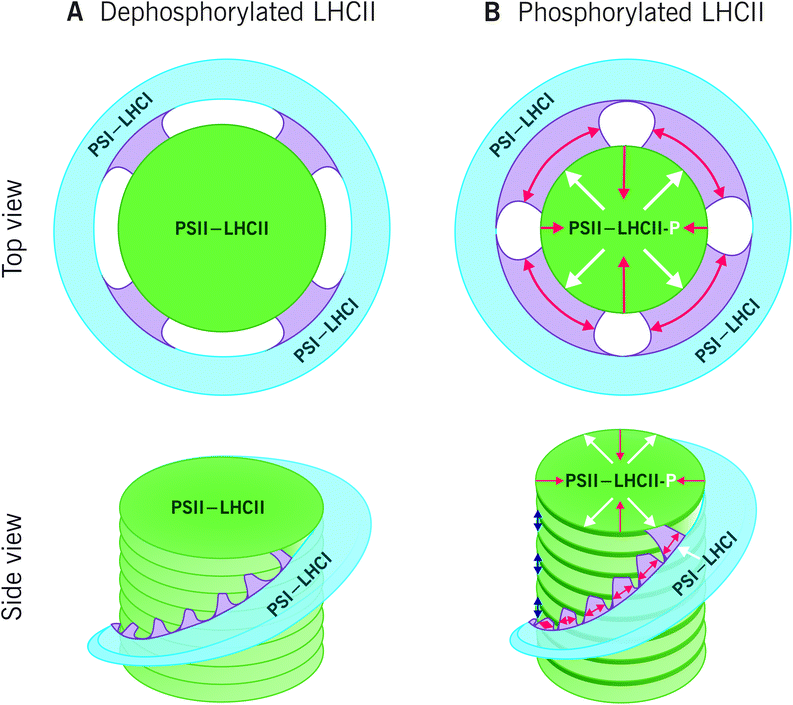
The reversible LHCII phosphorylation is an important regulatory mechanism that ensures balanced excitation energy distribution between PSII and PSI. The two photosystems work in series and must obtain similar amount of excitation energy in order to maintain ETC in balance despite prompt short-term changes in light quantity and quality. The STN7 kinase phosphorylates Lhcb1 and Lhcb2 proteins in different pools of LHCII trimers (S, M and L) (Fig. 4). The phosphorylation of Lhcb2 occurs almost exclusively in the pool of loosely bound L-LHCII trimers.88,90 When phosphorylated, the affinity of L-LHCII trimer to the PSI–LHCI complex increases and it attaches to the LHCII docking site in PSI, thus forming an isolatable LHCII–PSI–LHCI complex.42,90,112–114Remarkably, only one phosphorylated Lhcb2 protein is required for the association of LHCII trimer to PSI.90 Recent cryo-EM structure of the PSI–LHCII complex demonstrated that the phosphorylated Thr-3 of the Lhcb2 protein directly interacts with PsaL, whereas the two preceding arginine residues, Arg-1 and Arg-2, bind to PsaH, PsaL and PsaO residues.85 In addition to the formation of an isolatable PSI–LHCII complex, the accumulation of the aforementioned PSII–LHCII–PSI–LHCI cluster (demonstrated by native-PAGE) is largely dependent on LHCII phosphorylation: upon complete LHCII dephosphorylation, as demonstrated by the stn7 mutant, the amount of the PSII–LHCII–PSI–LHCI cluster is clearly reduced, and the amount of large PSI–LHCI supercomplexes concomitantly increased.87 Conversely, upon low-light-induced LHCII phosphorylation, also the amount of PSII–LHCII–PSI–LHCI cluster increases.87 As depicted in Fig. 4, the phosphorylation of Lhcb1 takes place mainly in PSII–LHCII supercomplexes,88,90 mostly in the S-trimer, since the M-trimer contains large amount of Lhcb1.4 isoform, which does not contain a phosphosite.42 Importantly, the Lhcb1 phosphorylation does not induce the detachment of the LHCII trimer from PSII–LHCII supercomplexes54,115,116 and the phosphorylated Lhcb1 does not function as an interaction surface between LHCII and PSI–LHCI.85 Instead, the Lhcb1 phosphorylation seems to indirectly regulate the excitation energy distribution through remodeling of the grana.113To merge the biochemical data that has been obtained on phosphorylation-dependent dynamics of thylakoid protein complexes and supercomplexes (e.g.ref. 39, 42, 54, 87, 89, 90, 113, 115, 117 and 118) with the dynamics of the entire thylakoid ultrastructure, we suggest a hypothetical model illustrated in Fig. 5. The model assumes that the biochemically distinct grana margin domains are located in the junctional slits between the grana and stroma membranes. Yet, further research is necessary for validation of the structural domains and, in particular, to reach a consensus about the 3D structure of the thylakoid membrane network. As depicted in Fig. 5A, when Lhcb1 and Lhcb2 proteins are dephosphorylated, as generally is the case in the dark, strong lateral heterogeneity between the two photosystems exist: PSII–LHCII complexes are tightly packed in grana, whereas PSI–LHCI is located in the stroma thylakoids. In light (Fig. 5B), the Lhcb1 proteins in the PSII–LHCII supercomplexes and the Lhcb2 proteins in the L-LHCII trimers become phosphorylated, the stromal gap get wider (blue arrows in Fig. 5B)14 and the LHCII phosphorylation induces lateral diffusion of PSII–LHCII complexes and L-LHCII to the margins of grana membranes39 (white arrows in Fig. 5B). Concomitantly, the phosphorylated PSII–LHCII complexes from the grana core and the PSI–LHCI complexes from stroma thylakoids get partially mixed,38 thus increasing the relative area of the margin domain and decreasing that of the grana (red arrows in Fig. 5B). Indeed, the decreasing of the grana core area in response to LHCII phosphorylation was recently demonstrated with 3D structured illumination microscopy.119 The lateral movement of PSII–LHCII complexes towards grana margins, where also PSI–LHCI complexes are present, locally decreases the lateral heterogeneity and allows the contact of the two photosystems, thus directing excitation energy from L-LHCII towards PSI–LHCI in this domain.87,117 In the very core of grana, the Lhcb1 in the PSII–LHCII supercomplexes remains dephosphorylated90 and the complexes might form semicrystalline arrays,57,120 where excitation energy transfers fast from one complex to another and where also the diffusion of plastoquinol is fast in the lipid channels between the arrays thus rapidly reaching the Cyt b6f complex.
Better understanding of the extraordinary complexity of the Lhcb1 and Lhcb2 phosphorylation and their role in modulation of the thylakoid membrane ultrastructure requires further research. Intriguingly, both Lhcb1 and Lhcb2 proteins contain several additional phosphosites that do not seem to be phosphorylated by STN7 or STN8,121 leaving their function to be solved. In addition to phosphorylation, ion fluxes are also known to be involved in the regulation of thylakoid ultrastructure122 but their interaction with phosphorylation of Lhcb proteins is likewise still elusive. Further, recent research has shown that thylakoid protein acetylation is also required for the regulation of excitation energy balancing between the two photosystems,123 introducing a whole new aspect of post-translational protein modification for regulation of photosynthesis. In addition, acetylation of N-terminal regions of PSII–LHCII proteins was recently shown to be extensive, likely indicating an important role of acetylation in grana stacking.124
PSII core protein phosphorylation – disassembly and repair of PSII complex
Light energy drives the photosynthetic reactions, but the energy beyond that utilized by photochemistry can lead to oxidative damage of the photosynthetic apparatus, and consequently result in reduction of the photosynthetic capacity. The strong oxidative chemistry performed by the water splitting PSII makes the enzyme complex particularly vulnerable to oxidative damage in light. In fact, even low light induces damage to the PSII reaction center protein D1 leading to irreversible photoinactivation of the entire reaction centre.125 The degree of light damage is directly proportional to the intensity of light the plants are exposed to,126 but the photoinhibitory phenotype arises only when the rate of the damage exceeds the rate of repair. The restoration of PSII function occurs via a complex repair cycle in which the injured PSII complexes migrate from the grana core to the non-appressed membrane regions, where the damaged D1 protein is degraded by specific proteases and replaced with de novo synthesized protein.127–131 As the damage is targeted to the D1 protein, PSII photoinhibition may be considered as evolutionary strategy to protect the rest of the PSII subunits and even PSI.132The majority of the auxiliary proteins involved in the PSII repair cycle, such as the Deg and FtsH proteases, are located in the non-appressed thylakoids.38,133 The PSII core protein phosphorylation appears to affect PSII repair cycle, although the exact role is poorly understood. As depicted in Fig. 4, the N-terminal Thr residues of D1, D2, CP43 and Thr-4 in PsbH are phosphorylated by the STN8 kinase upon exposure to high light106,134 and, conversely, dephosphorylated by PSII CORE PHOSPHATASE (PBCP) when plants are transferred to low light or darkness.135 Initially, the phosphorylation was considered to mark the D1 protein for degradation, but later investigations of the stn8 and stn7snt8 mutants demonstrated that the mutants are not more susceptible to light damage and made the authors to conclude that the PSII protein phosphorylation is not crucial for PSII repair.106 Although not essential for the repair per se, the PSII core phosphorylation was later shown to facilitate the disassembly of the PSII–LHCII supercomplexes136,137 and to increase the mobility of the PSII complexes95,138 under photoinhibitory conditions. Locations of the PSII core phosphorylation sites at monomer–monomer interface and at the sites of Lhcb association to the core139 are in support of this suggestion. Opposing to this view, Fristedt et al.140 argued that the PSII core phosphorylation instead induces macroscopic rearrangements to the thylakoid membrane and allow the PSII repair cycle by decreasing the membrane cohesion. The different hypotheses on the roles of PSII core protein phosphorylation are not necessarily mutually exclusive.
Beside the PSII core protein phosphorylation, dynamic thylakoid rearrangements under high light exposure might also arise from changes in the oligomerization state of the CURVATURE THYLAKOID1 proteins CURT1A-D.10,141 The CURT1 proteins have, in fact, been suggested to facilitate the PSII repair cycle by unstacking the edges of the grana stacks.142 This regulation likely involves phosphorylation, since CURT1B was recently shown to be phosphorylated by STN8 and to follow the phosphorylation dynamics of PSII core proteins D1 and D2.14 Moreover, CURT1 proteins might even play a role in the successful connection between LHCII and PSI.14,143 On the contrary, Reduced Induction of Non-Photochemical Quenching (RIQ) proteins RIQ1 and RIQ2 seem to act oppositely to the CURT1 proteins with respect to grana stacking, yet both the RIQ and CURT1 proteins seem to be essential for sufficient thermal dissipation from LHCII upon exposure of plants to high light.144
Concluding remarks
Discussion above focuses on protein phosphorylation-dependent rearrangements of the photosynthetic machinery in the thylakoid membrane of angiosperms. More generally speaking, the photosynthetic light reactions are regulated by a plethora of mechanisms, with distinct evolutionary diversity between cyanobacteria, algae and land plants, and such differences are closely related to the ultrastructure of the thylakoid membrane network.145 Phosphorylation of thylakoid proteins and consequent modifications in thylakoid ultrastructure make a specific example of short-term dynamic regulation of light reactions with concomitant rearrangements in thylakoid architecture, in order to allow rapid acclimation of the photosynthetic apparatus to changing environmental cues. Yet, different land plants, and to a minor extent also different angiosperms, show modifications in phosphorylation target proteins of both the LHCII and PSII core proteins as well as in the number and location of phosphosites in each protein. Thus, we still miss a wealth of information related to the exact roles and mechanisms of thylakoid protein phosphorylation involved in response to environmental cues of a wide range of different plant species. Although the phosphorylation dynamics of LHCII and PSII core proteins are the most extensively scrutinized, and the only phosphoproteins in our model of thylakoid reorganization (Fig. 5), they likely represent only a tip of an iceberg of the factors that regulate the thylakoid ultrastructure and optimize photosynthetic reactions. As discussed above, also the CURT1B protein, which are phosphorylated by the same kinase as the PSII core proteins, have an important role in regulation of thylakoid rearrangements. Moreover, other post-translational protein modifications, such as lysine acetylation, seem to bring a completely new layer of regulatory mechanisms for thylakoid protein phosphorylation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Center of excellence of the Academy of Finland (307335), Jane and Aatos Erkko Foundation and Turku University Foundation. Dr Andrea Trotta is thanked for critically reviewing and commenting the manuscript.References
- L. A. Staehelin, Chloroplast structure: from chlorophyll granules to supra-molecular architecture of thylakoid membranes, Photosynth. Res., 2003, 76, 185–196 CrossRef CAS PubMed.
- L. Mustárdy and G. Garab, Granum revisited. A three-dimensional model – where things fall into place, Trends Plant Sci., 2003, 8, 117–122 CrossRef.
- B. Daum, D. Nicastro, J. Austin, J. R. McIntosh and W. Kühlbrandt, Arrangement of Photosystem II and ATP Synthase in Chloroplast Membranes of Spinach and Pea, Plant Cell, 2010, 22, 1299–1312 CrossRef CAS PubMed.
- H. Kirchhoff, C. Hall, M. Wood, M. Herbstová, O. Tsabari, R. Nevo, D. Charuvi, E. Shimoni and Z. Reich, Dynamic control of protein diffusion within the granal thylakoid lumen, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 20248–20253 CrossRef CAS PubMed.
- P.-Å. Albertsson, A quantitative model of the domain structure of the photosynthetic membrane, Trends Plant Sci., 2001, 6, 349–354 CrossRef CAS PubMed.
- J. M. Anderson, P. Horton, E.-H. Kim and W. S. Chow, Towards elucidation of dynamic structural changes of plant thylakoid architecture, Philos. Trans. R. Soc., B, 2012, 367, 3515–3524 CrossRef CAS PubMed.
- D. J. Murphy, The molecular organisation of the photosynthetic membranes of higher plants, Biochim. Biophys. Acta, Rev. Biomembr., 1986, 864, 33–94 CrossRef CAS.
- J. P. Dekker and E. J. Boekema, Supramolecular organization of thylakoid membrane proteins in green plants, Biochim. Biophys. Acta, Bioenerg., 2005, 1706, 12–39 CrossRef CAS PubMed.
- R. Kouřil, G. T. Oostergetel and E. J. Boekema, Fine structure of granal thylakoid membrane organization using cryo electron tomography, Biochim. Biophys. Acta, Bioenerg., 2011, 1807, 368–374 CrossRef PubMed.
- U. Armbruster, M. Labs, M. Pribil, S. Viola, W. Xu, M. Scharfenberg, A. P. Hertle, U. Rojahn, P. E. Jensen, F. Rappaport, P. Joliot, P. Dörmann, G. Wanner and D. Leister, Arabidopsis CURVATURE THYLAKOID1 Proteins Modify Thylakoid Architecture by Inducing Membrane Curvature, Plant Cell, 2013, 25, 2661–2678 CrossRef CAS PubMed.
- R. Danielsson, P.-Å. Albertsson, F. Mamedov and S. Styring, Quantification of photosystem I and II in different parts of the thylakoid membrane from spinach, Biochim. Biophys. Acta, Bioenerg., 2004, 1608, 53–61 CrossRef CAS PubMed.
- S. Puthiyaveetil, O. Tsabari, T. Lowry, S. Lenhert, R. R. Lewis, Z. Reich and H. Kirchhoff, Compartmentalization of the protein repair machinery in photosynthetic membranes, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15839–15844 CrossRef CAS PubMed.
- H. Koochak, S. Puthiyaveetil, D. L. Mullendore, M. Li and H. Kirchhoff, The structural and functional domains of plant thylakoid membranes, Plant J., 2019, 100, 213 CrossRef PubMed.
- A. Trotta, A. A. Bajwa, I. Mancini, V. Paakkarinen, M. Pribil and E.-M. Aro, The role of phosphorylation dynamics of CURVATURE THYLAKOID 1B in plant thylakoid membranes, Plant Physiol., 2019, 181, 1615–1631 CrossRef CAS PubMed.
- P.-O. Arvidsson and C. Sundby, A model for the topology of the chloroplast thylakoid membrane, Funct. Plant Biol., 1999, 26, 687–694 CrossRef CAS.
- E. Shimoni, O. Rav-Hon, I. Ohad, V. Brumfeld and Z. Reich, Three-Dimensional Organization of Higher-Plant Chloroplast Thylakoid Membranes Revealed by Electron Tomography, Plant Cell, 2005, 17, 2580–2586 CrossRef CAS PubMed.
- D. J. Paolillo, The Three-Dimensional Arrangement of Intergranal Lamellae in Chloroplasts, J. Cell Sci., 1970, 6, 243–253 Search PubMed.
- J. Brangeon and L. Mustardy, Ontogenetic assembly of intra-chloroplastic lamellae viewed in 3-dimension, Biol. Cell, 1979, 36, 71–80 Search PubMed.
- A. V. Ruban and M. P. Johnson, Visualizing the dynamic structure of the plant photosynthetic membrane, Nat. Plants, 2015, 1, 15161 CrossRef CAS PubMed.
- Y. Bussi, E. Shimoni, A. Weiner, R. Kapon, D. Charuvi, R. Nevo, E. Efrati and Z. Reich, Fundamental helical geometry consolidates the plant photosynthetic membrane, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 22366–22375 CrossRef CAS PubMed.
- L. Mustárdy, K. Buttle, G. Steinbach and G. Garab, The Three-Dimensional Network of the Thylakoid Membranes in Plants: Quasihelical Model of the Granum-Stroma Assembly, Plant Cell, 2008, 20, 2552–2557 CrossRef PubMed.
- J. R. Austin and L. A. Staehelin, Three-Dimensional Architecture of Grana and Stroma Thylakoids of Higher Plants as Determined by Electron Tomography, Plant Physiol., 2011, 155, 1601–1611 CrossRef CAS PubMed.
- W. S. Chow, E.-H. Kim, P. Horton and J. M. Anderson, Granal stacking of thylakoid membranes in higher plant chloroplasts: the physicochemical forces at work and the functional consequences that ensue, Photochem. Photobiol. Sci., 2005, 4, 1081 RSC.
- J. Andersson, M. Wentworth, R. Walters, C. Howard, A. Ruban, P. Horton and S. Jansson, Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II – effects on photosynthesis, grana stacking and fitness, Plant J., 2003, 35, 350–361 CrossRef CAS PubMed.
- D. J. Simpson, Freeze-fracture studies on barley plastid membranes. III. Location of the light-harvesting chlorophyll-protein, Carlsberg Res. Commun., 1979, 44, 305 CrossRef CAS PubMed.
- N. Murata, Control of excitation transfer in photosynthesis. II. Magnesium ion-dependent distribution of excitation energy between two pigment systems in spinach chloroplasts, Biochim. Biophys. Acta, Bioenerg., 1969, 189, 171–181 CrossRef CAS.
- J. Barber, An explanation for the relationship between salt-induced thylakoid stacking and the chlorophyll fluorescence changes associated with changes in spillover of energy from photosystem II to photosystem I, FEBS Lett., 1980, 118, 1–10 CrossRef CAS.
- J. Standfuss, A. C. T. van Scheltinga, M. Lamborghini and W. Kühlbrandt, Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution, EMBO J., 2005, 24, 919–928 CrossRef CAS PubMed.
- B. Andersson and J. M. Anderson, Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts, Biochim. Biophys. Acta, Bioenerg., 1980, 593, 427–440 CrossRef CAS.
- H. Kirchhoff, I. Tremmel, W. Haase and U. Kubitscheck, Supramolecular Photosystem II Organization in Grana Thylakoid Membranes: Evidence for a Structured Arrangement, Biochemistry, 2004, 43, 9204–9213 CrossRef CAS PubMed.
- R. Nevo, D. Charuvi, O. Tsabari and Z. Reich, Composition, architecture and dynamics of the photosynthetic apparatus in higher plants, Plant J., 2012, 70, 157–176 CrossRef CAS PubMed.
- J. M. Anderson, The role of chlorophyll-protein complexes in the function and structure of chloroplast thylakoids, Mol. Cell. Biochem., 1982, 46, 161–172 CrossRef CAS PubMed.
- P.-Å. Albertsson, E. Andreasson, P. Svensson and S.-G. Yu, Localization of cytochrome f in the thylakoid membrane: evidence for multiple domains, Biochim. Biophys. Acta, Bioenerg., 1991, 1098, 90–94 CrossRef CAS.
- O. Vallon, L. Bulte, P. Dainese, J. Olive, R. Bassi and F. A. Wollman, Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 8262–8266 CrossRef CAS PubMed.
- L. Wollenberger, H. Stefansson, S.-G. Yu and P.-Å. Albertsson, Isolation and characterization of vesicles originating from the chloroplast grana margins, Biochim. Biophys. Acta, Bioenerg., 1994, 1184, 93–102 CrossRef CAS.
- R. Danielsson and P.-Å. Albertsson, Fragmentation and separation analysis of the photosynthetic membrane from spinach, Biochim. Biophys. Acta, Bioenerg., 2009, 1787, 25–36 CrossRef CAS PubMed.
- S. Järvi, M. Suorsa, V. Paakkarinen and E.-M. Aro, Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes, Biochem. J., 2011, 439, 207–214 CrossRef PubMed.
- M. Suorsa, M. Rantala, R. Danielsson, S. Järvi, V. Paakkarinen, W. P. Schröder, S. Styring, F. Mamedov and E.-M. Aro, Dark-adapted spinach thylakoid protein heterogeneity offers insights into the photosystem II repair cycle, Biochim. Biophys. Acta, Bioenerg., 2014, 1837, 1463–1471 CrossRef CAS PubMed.
- S. Rantala and M. Tikkanen, Phosphorylation-induced lateral rearrangements of thylakoid protein complexes upon light acclimation, Plant Direct, 2018, 2, 1–12 CrossRef PubMed.
- Z. Liu, H. Yan, K. Wang, T. Kuang, J. Zhang, L. Gui, X. An and W. Chang, Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution, Nature, 2004, 428, 287–292 CrossRef CAS PubMed.
- L. S. van Bezouwen, S. Caffarri, R. S. Kale, R. Kouřil, A.-M. W. H. Thunnissen, G. T. Oostergetel and E. J. Boekema, Subunit and chlorophyll organization of the plant photosystem II supercomplex, Nat. Plants, 2017, 3, 17080 CrossRef CAS PubMed.
- P. Galka, S. Santabarbara, T. T. H. Khuong, H. Degand, P. Morsomme, R. C. Jennings, E. J. Boekema and S. Caffarri, Functional Analyses of the Plant Photosystem I–Light-Harvesting Complex II Supercomplex Reveal That Light-Harvesting Complex II Loosely Bound to Photosystem II Is a Very Efficient Antenna for Photosystem I in State II, Plant Cell, 2012, 24, 2963–2978 CrossRef CAS PubMed.
- A. Crepin and S. Caffarri, Functions and Evolution of Lhcb Isoforms Composing LHCII, the Major Light Harvesting Complex of Photosystem II of Green Eukaryotic Organisms, Curr. Protein Pept. Sci., 2018, 19, 699–713 CrossRef CAS PubMed.
- S. Jansson, A guide to the Lhc genes and their relatives in Arabidopsis, Trends Plant Sci., 1999, 4, 236–240 CrossRef PubMed.
- S. Grebe, A. Trotta, A. A. Bajwa, M. Suorsa, P. J. Gollan, S. Jansson, M. Tikkanen and E.-M. Aro, The unique photosynthetic apparatus of Pinaceae: analysis of photosynthetic complexes in Picea abies, J. Exp. Bot., 2019, 70, 3211–3225 CrossRef CAS PubMed.
- X. Su, J. Ma, X. Wei, P. Cao, D. Zhu, W. Chang, Z. Liu, X. Zhang and M. Li, Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex, Science, 2017, 357, 815–820 CrossRef CAS PubMed.
- O. Nanba and K. Satoh, Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 109–112 CrossRef CAS PubMed.
- A. N. Webber, L. Packman, D. J. Chapman, J. Barber and J. C. Gray, A fifth chloroplast-encoded polypeptide is present in the photosystem II reaction centre complex, FEBS Lett., 1989, 242, 259–262 CrossRef CAS.
- B. Lundin, M. Hansson, B. Schoefs, A. V. Vener and C. Spetea, The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein, Plant J. Cell Mol. Biol., 2007, 49, 528–539 CrossRef CAS PubMed.
- X. Wei, X. Su, P. Cao, X. Liu, W. Chang, M. Li, X. Zhang and Z. Liu, Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution, Nature, 2016, 534, 69–74 CrossRef CAS PubMed.
- Y.-E. Chen, S. Yuan, L. Lezhneva, J. Meurer, S. Schwenkert, F. Mamedov and W. P. Schröder, The Low Molecular Mass Photosystem II Protein PsbTn Is Important for Light Acclimation, Plant Physiol., 2019, 179, 1739–1753 CrossRef CAS PubMed.
- M. Plöchinger, S. Schwenkert, L. von Sydow, W. P. Schröder and J. Meurer, Functional Update of the Auxiliary Proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in Maintenance and Assembly of PSII, Front. Plant Sci., 2016, 7, 423 Search PubMed.
- K. Sznee, J. P. Dekker, R. T. Dame, H. van Roon, G. J. L. Wuite and R. N. Frese, Jumping Mode Atomic Force Microscopy on Grana Membranes from Spinach, J. Biol. Chem., 2011, 286, 39164–39171 CrossRef CAS PubMed.
- M. Grieco, M. Suorsa, A. Jajoo, M. Tikkanen and E.-M. Aro, Light-harvesting II antenna trimers connect energetically the entire photosynthetic machinery—including both photosystems II and I, Biochim. Biophys. Acta, Bioenerg., 2015, 1847, 607–619 CrossRef CAS PubMed.
- R. Kouřil, L. Nosek, J. Bartoš, E. J. Boekema and P. Ilík, Evolutionary loss of light-harvesting proteins Lhcb6 and Lhcb3 in major land plant groups – break-up of current dogma, New Phytol., 2016, 210, 808–814 CrossRef PubMed.
- M. M. Koskela, A. Brünje, A. Ivanauskaite, L. S. Lopez, D. Schneider, R. A. DeTar, H.-H. Kunz, I. Finkemeier and P. Mulo, Comparative analysis of thylakoid protein complexes in state transition mutants nsi and stn7: focus on PSI and LHCII, Photosynth. Res., 2020 DOI:10.1007/s11120-020-00711-4.
- H. Kirchhoff, W. Haase, S. Wegner, R. Danielsson, R. Ackermann and P.-A. Albertsson, Low-Light-Induced Formation of Semicrystalline Photosystem II Arrays in Higher Plant Chloroplasts, Biochemistry, 2007, 46, 11169–11176 CrossRef CAS PubMed.
- R. Kouřil, J. P. Dekker and E. J. Boekema, Supramolecular organization of photosystem II in green plants, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 2–12 CrossRef PubMed.
- P. Albanese, J. Nield, J. A. M. Tabares, A. Chiodoni, M. Manfredi, F. Gosetti, E. Marengo, G. Saracco, J. Barber and C. Pagliano, Isolation of novel PSII-LHCII megacomplexes from pea plants characterized by a combination of proteomics and electron microscopy, Photosynth. Res., 2016, 130, 19–31 CrossRef CAS PubMed.
- T. K. Goral, M. P. Johnson, C. D. P. Duffy, A. P. R. Brain, A. V. Ruban and C. W. Mullineaux, Light-harvesting antenna composition controls the macrostructure and dynamics of thylakoid membranes in Arabidopsis, Plant J., 2012, 69, 289–301 CrossRef CAS PubMed.
- N. M. Tsvetkova, A. P. R. Brain and P. J. Quinn, Structural characteristics of thylakoid membranes of Arabidopsis mutants deficient in lipid fatty acid desaturation, Biochim. Biophys. Acta, Biomembr., 1994, 1192, 263–271 CrossRef CAS.
- S. Tietz, S. Puthiyaveetil, H. M. Enlow, R. Yarbrough, M. Wood, D. A. Semchonok, T. Lowry, Z. Li, P. Jahns, E. J. Boekema, S. Lenhert, K. K. Niyogi and H. Kirchhoff, Functional Implications of Photosystem II Crystal Formation in Photosynthetic Membranes, J. Biol. Chem., 2015, 290, 14091–14106 CrossRef CAS PubMed.
- S. Kereïche, A. Z. Kiss, R. Kouřil, E. J. Boekema and P. Horton, The PsbS protein controls the macro-organisation of photosystem II complexes in the grana membranes of higher plant chloroplasts, FEBS Lett., 2010, 584, 759–764 CrossRef PubMed.
- P. Horton, A. V. Ruban, D. Rees, A. A. Pascal, G. Noctor and A. J. Young, Control of the light-harvesting function of chloroplast membranes by aggregation of the LHCII chlorophyll-protein complex, FEBS Lett., 1991, 292, 1–4 CrossRef CAS PubMed.
- P. Horton, M. Wentworth and A. Ruban, Control of the light harvesting function of chloroplast membranes: The LHCII-aggregation model for non-photochemical quenching, FEBS Lett., 2005, 579, 4201–4206 CrossRef CAS PubMed.
- A. R. Holzwarth, Y. Miloslavina, M. Nilkens and P. Jahns, Identification of two quenching sites active in the regulation of photosynthetic light-harvesting studied by time-resolved fluorescence, Chem. Phys. Lett., 2009, 483, 262–267 CrossRef CAS.
- N. Betterle, M. Ballottari, S. Zorzan, S. de Bianchi, S. Cazzaniga, L. Dall'Osto, T. Morosinotto and R. Bassi, Light-induced Dissociation of an Antenna Hetero-oligomer Is Needed for Non-photochemical Quenching Induction, J. Biol. Chem., 2009, 284, 15255–15266 CrossRef CAS PubMed.
- Y. Mazor, A. Borovikova, I. Caspy and N. Nelson, Structure of the plant photosystem I supercomplex at 2.6 Å resolution, Nat. Plants, 2017, 3, 17014 CrossRef CAS PubMed.
- S. Caffarri, T. Tibiletti, R. Jennings and S. Santabarbara, A comparison between plant photosystem I and photosystem II architecture and functioning, Curr. Protein Pept. Sci., 2014, 15, 296–331 CrossRef CAS PubMed.
- I. Caspy and N. Nelson, Structure of the plant photosystem I, Biochem. Soc. Trans., 2018, 46, 285–294 CrossRef CAS PubMed.
- P. E. Jensen, R. Bassi, E. J. Boekema, J. P. Dekker, S. Jansson, D. Leister, C. Robinson and H. V. Scheller, Structure, function and regulation of plant photosystem I, Biochim. Biophys. Acta, Bioenerg., 2007, 1767, 335–352 CrossRef CAS PubMed.
- E. J. Boekema, P. E. Jensen, E. Schlodder, J. F. L. van Breemen, H. van Roon, H. V. Scheller and J. P. Dekker, Green Plant Photosystem I Binds Light-Harvesting Complex I on One Side of the Complex †, Biochemistry, 2001, 40, 1029–1036 CrossRef CAS PubMed.
- M. Ballottari, C. Govoni, S. Caffarri and T. Morosinotto, Stoichiometry of LHCI antenna polypeptides and characterization of gap and linker pigments in higher plants Photosystem I, Eur. J. Biochem., 2004, 271, 4659–4665 CrossRef CAS PubMed.
- E. Wientjes and R. Croce, The light-harvesting complexes of higher-plant Photosystem I: Lhca1/4 and Lhca2/3 form two red-emitting heterodimers, Biochem. J., 2011, 433, 477–485 CrossRef CAS PubMed.
- A. Alboresi, C. Le Quiniou, S. K. N. Yadav, M. Scholz, A. Meneghesso, C. Gerotto, D. Simionato, M. Hippler, E. J. Boekema, R. Croce and T. Morosinotto, Conservation of core complex subunits shaped the structure and function of photosystem I in the secondary endosymbiont alga Nannochloropsis gaditana, New Phytol., 2017, 213, 714–726 CrossRef CAS PubMed.
- S. Kereïche, R. Kouřil, G. T. Oostergetel, F. Fusetti, E. J. Boekema, A. B. Doust, C. D. van der Weij-de Wit and J. P. Dekker, Association of chlorophyll a/c2 complexes to photosystem I and photosystem II in the cryptophyte Rhodomonas CS24, Biochim. Biophys. Acta, Bioenerg., 2008, 1777, 1122–1128 CrossRef PubMed.
- B. Drop, M. Webber-Birungi, F. Fusetti, R. Kouřil, K. E. Redding, E. J. Boekema and R. Croce, Photosystem I of Chlamydomonas reinhardtii Contains Nine Light-harvesting Complexes (Lhca) Located on One Side of the Core, J. Biol. Chem., 2011, 286, 44878–44887 CrossRef CAS PubMed.
- A. Crepin, Z. Kučerová, A. Kosta, E. Durand and S. Caffarri, Isolation and characterization of a large photosystem I–light-harvesting complex II supercomplex with an additional Lhca1–a4 dimer in Arabidopsis, Plant J., 2019 DOI:10.1111/tpj.14634.
- E. Boekema and A. Dmitry, in Light Harvesting in Photosynthesis, 2018 Search PubMed.
- X. Pan, P. Cao, X. Su, Z. Liu and M. Li, Structural analysis and comparison of light-harvesting complexes I and II, Biochim. Biophys. Acta, Bioenerg., 2019, 148038 Search PubMed.
- M. Iwai, M. Yokono, N. Inada and J. Minagawa, Live-cell imaging of photosystem II antenna dissociation during state transitions, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2337–2342 CrossRef CAS PubMed.
- K. N. S. Yadav, D. A. Semchonok, L. Nosek, R. Kouřil, G. Fucile, E. J. Boekema and L. A. Eichacker, Supercomplexes of plant photosystem I with cytochrome b6f, light-harvesting complex II and NDH, Biochim. Biophys. Acta, Bioenerg., 2017, 1858, 12–20 CrossRef CAS PubMed.
- L. Peng, Y. Fukao, M. Fujiwara, T. Takami and T. Shikanai, Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis, Plant Cell, 2009, 21, 3623–3640 CrossRef CAS PubMed.
- R. Kouřil, O. Strouhal, L. Nosek, R. Lenobel, I. Chamrád, E. J. Boekema, M. Šebela and P. Ilík, Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex, Plant J., 2014, 77, 568–576 CrossRef PubMed.
- X. Pan, J. Ma, X. Su, P. Cao, W. Chang, Z. Liu, X. Zhang and M. Li, Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II, Science, 2018, 360, 1109–1113 CrossRef CAS PubMed.
- M. Plöchinger, S. Torabi, M. Rantala, M. Tikkanen, M. Suorsa, P.-E. Jensen, E. M. Aro and J. Meurer, The Low Molecular Weight Protein PsaI Stabilizes the Light-Harvesting Complex II Docking Site of Photosystem I, Plant Physiol., 2016, 172, 450–463 CrossRef PubMed.
- M. Suorsa, M. Rantala, F. Mamedov, M. Lespinasse, A. Trotta, M. Grieco, E. Vuorio, M. Tikkanen, S. Järvi and E. Aro, Light acclimation involves dynamic re-organization of the pigment–protein megacomplexes in non-appressed thylakoid domains, Plant J., 2015, 84, 360–373 CrossRef CAS PubMed.
- M. Rantala, M. Tikkanen and E. Aro, Proteomic characterization of hierarchical megacomplex formation in Arabidopsis thylakoid membrane, Plant J., 2017, 92, 951–962 CrossRef CAS PubMed.
- M. Yokono, A. Takabayashi, S. Akimoto and A. Tanaka, A megacomplex composed of both photosystem reaction centres in higher plants, Nat. Commun., 2015, 6, 6675 CrossRef CAS PubMed.
- A. Crepin and S. Caffarri, The specific localizations of phosphorylated Lhcb1 and Lhcb2 isoforms reveal the role of Lhcb2 in the formation of the PSI-LHCII supercomplex in Arabidopsis during state transitions, Biochim. Biophys. Acta, Bioenerg., 2015, 1847, 1539–1548 CrossRef CAS PubMed.
- P. Svensson and P.-Å. Albertsson, Preparation of highly enriched photosystem II membrane vesicles by a non-detergent method, Photosynth. Res., 1989, 20, 249–259 CAS.
- S. L. Benson, P. Maheswaran, M. A. Ware, C. N. Hunter, P. Horton, S. Jansson, A. V. Ruban and M. P. Johnson, An intact light harvesting complex I antenna system is required for complete state transitions in Arabidopsis, Nat. Plants, 2015, 1, 15176 CrossRef CAS PubMed.
- J. M. Anderson, Photoregulation of the Composition, Function, and Structure of Thylakoid Membranes, Annu. Rev. Plant Physiol., 1986, 37, 93–136 CrossRef CAS.
- M. Khatoon, K. Inagawa, P. Pospíšil, A. Yamashita, M. Yoshioka, B. Lundin, J. Horie, N. Morita, A. Jajoo, Y. Yamamoto and Y. Yamamoto, Quality Control of Photosystem II thylakoid unstacking is necessary to avoid further damage to the D1 protein and to facilitate D1 degradation under light stress in spinach thylakoids, J. Biol. Chem., 2009, 284, 25343–25352 CrossRef CAS PubMed.
- M. Herbstová, S. Tietz, C. Kinzel, M. V. Turkina and H. Kirchhoff, Architectural switch in plant photosynthetic membranes induced by light stress, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20130–20135 CrossRef PubMed.
- P. R. Rozak, R. M. Seiser, W. F. Wacholtz and R. R. Wise, Rapid, reversible alterations in spinach thylakoid appression upon changes in light intensity, Plant, Cell Environ., 2002, 25, 421–429 CrossRef.
- M. Tikkanen, M. Grieco, S. Kangasjärvi and E.-M. Aro, Thylakoid Protein Phosphorylation in Higher Plant Chloroplasts Optimizes Electron Transfer under Fluctuating Light, Plant Physiol., 2010, 152, 723–735 CrossRef CAS PubMed.
- M. Suorsa, S. Järvi, M. Grieco, M. Nurmi, M. Pietrzykowska, M. Rantala, S. Kangasjärvi, V. Paakkarinen, M. Tikkanen, S. Jansson and E.-M. Aro, PROTON GRADIENT REGULATION5 Is Essential for Proper Acclimation of Arabidopsis Photosystem I to Naturally and Artificially Fluctuating Light Conditions, Plant Cell, 2012, 24, 2934–2948 CrossRef CAS PubMed.
- M. Tikkanen, M. Grieco, M. Nurmi, M. Rantala, M. Suorsa and E.-M. Aro, Regulation of the photosynthetic apparatus under fluctuating growth light, Philos. Trans. R. Soc., B, 2012, 367, 3486–3493 CrossRef CAS PubMed.
- S. Reiland, G. Messerli, K. Baerenfaller, B. Gerrits, A. Endler, J. Grossmann, W. Gruissem and S. Baginsky, Large-Scale Arabidopsis Phosphoproteome Profiling Reveals Novel Chloroplast Kinase Substrates and Phosphorylation Networks, Plant Physiol., 2009, 150, 889–903 CrossRef CAS PubMed.
- J. Bennett, Phosphorylation of chloroplast membrane polypeptides, Nature, 1977, 269, 344–346 CrossRef CAS.
- S. Bellafiore, F. Barneche, G. Peltier and J.-D. Rochaix, State transitions and light adaptation require chloroplast thylakoid protein kinase STN7, Nature, 2005, 433, 892 CrossRef CAS PubMed.
- A. V. Vener, P. J. M. van Kan, P. R. Rich, I. Ohad and B. Andersson, Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: Thylakoid protein kinase deactivation by a single-turnover flash, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 1585–1590 CrossRef CAS PubMed.
- A. Trotta, M. Suorsa, M. Rantala, B. Lundin and E.-M. Aro, Serine and threonine residues of plant STN7 kinase are differentially phosphorylated upon changing light conditions and specifically influence the activity and stability of the kinase, Plant J., 2016, 87, 484–494 CrossRef CAS PubMed.
- E. Rintamäki, P. Martinsuo, S. Pursiheimo and E.-M. Aro, Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 11644–11649 CrossRef PubMed.
- V. Bonardi, P. Pesaresi, T. Becker, E. Schleiff, R. Wagner, T. Pfannschmidt, P. Jahns and D. Leister, Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases, Nature, 2005, 437, 1179–1182 CrossRef CAS PubMed.
- M. Tikkanen, M. Piippo, M. Suorsa, S. Sirpiö, M. Mulo, J. Vainonen, A. Vener, Y. Allahverdiyeva and E.-M. Aro, State transitions revisited—a buffering system for dynamic low light acclimation of Arabidopsis, Plant Mol. Biol., 2006, 62, 779 CrossRef PubMed.
- M. Pribil, P. Pesaresi, A. Hertle, R. Barbato and D. Leister, Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow, PLoS Biol., 2010, 8, e1000288 CrossRef PubMed.
- A. Shapiguzov, B. Ingelsson, I. Samol, C. Andres, F. Kessler, J.-D. Rochaix, A. V. Vener and M. Goldschmidt-Clermont, The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4782–4787 CrossRef CAS PubMed.
- M. Rantala, N. Lehtimäki, E.-M. Aro and M. Suorsa, Downregulation of TAP38/PPH1 enables LHCII hyperphosphorylation in Arabidopsis mutant lacking LHCII docking site in PSI, FEBS Lett., 2016, 590, 787–794 CrossRef CAS PubMed.
- X. Wei, J. Guo, M. Li and Z. Liu, Structural Mechanism Underlying the Specific Recognition between the Arabidopsis State-Transition Phosphatase TAP38/PPH1 and Phosphorylated Light-Harvesting Complex Protein Lhcb1, Plant Cell, 2015, 27, 1113–1127 CrossRef CAS PubMed.
- C. Leoni, M. Pietrzykowska, A. Z. Kiss, M. Suorsa, L. R. Ceci, E.-M. Aro and S. Jansson, Very rapid phosphorylation kinetics suggest a unique role for Lhcb2 during state transitions in Arabidopsis, Plant J., 2013, 76, 236–246 CAS.
- M. Pietrzykowska, M. Suorsa, D. A. Semchonok, M. Tikkanen, E. J. Boekema, E.-M. Aro and S. Jansson, The Light-Harvesting Chlorophyll a/b Binding Proteins Lhcb1 and Lhcb2 Play Complementary Roles during State Transitions in Arabidopsis, Plant Cell, 2014, 26, 3646–3660 CrossRef CAS PubMed.
- P. Longoni, D. Douchi, F. Cariti, G. Fucile and M. Goldschmidt-Clermont, Phosphorylation of the Light-Harvesting Complex II Isoform Lhcb2 Is Central to State Transitions, Plant Physiol., 2015, 169, 2874–2883 CAS.
- E. Wientjes, H. van Amerongen and R. Croce, LHCII is an antenna of both photosystems after long-term acclimation, Biochim. Biophys. Acta, Bioenerg., 2013, 1827, 420–426 CrossRef CAS PubMed.
- M. Giovanardi, L. Pantaleoni, L. Ferroni, C. Pagliano, P. Albanese, C. Baldisserotto and S. Pancaldi, In pea stipules a functional photosynthetic electron flow occurs despite a reduced dynamicity of LHCII association with photosystems, Biochim. Biophys. Acta, Bioenerg., 2018, 1859, 1025–1038 CrossRef CAS PubMed.
- N. R. Mekala, M. Suorsa, M. Rantala, E.-M. Aro and M. Tikkanen, Plants actively avoid state transitions upon changes in light intensity: Role of Light-Harvesting Complex II protein dephosphorylation in high light, Plant Physiol., 2015, 168, 721–734 CrossRef CAS PubMed.
- P. Pesaresi, A. Hertle, M. Pribil, T. Kleine, R. Wagner, H. Strissel, A. Ihnatowicz, V. Bonardi, M. Scharfenberg, A. Schneider, T. Pfannschmidt and D. Leister, Arabidopsis STN7 Kinase provides a link between short- and long-term photosynthetic acclimation, Plant Cell, 2009, 21, 2402–2423 CrossRef CAS PubMed.
- W. H. Wood, S. F. H. Barnett, S. Flannery, C. N. Hunter and M. P. Johnson, Dynamic thylakoid stacking is regulated by LHCII phosphorylation but not its interaction with photosystem I, Plant Physiol., 2019, 180, 2152–2166 CrossRef CAS PubMed.
- R. Kouřil, E. Wientjes, J. B. Bultema, R. Croce and E. J. Boekema, High-light vs. low-light: Effect of light acclimation on photosystem II composition and organization in Arabidopsis thaliana, Biochim. Biophys. Acta, Bioenerg., 2013, 1827, 411–419 CrossRef PubMed.
- B. Ingelsson and A. V. Vener, Phosphoproteomics of Arabidopsis chloroplasts reveals involvement of the STN7 kinase in phosphorylation of nucleoid protein pTAC16, FEBS Lett., 2012, 586, 1265–1271 CrossRef CAS PubMed.
- C. Spetea, A. Herdean, G. Allorent, L. Carraretto, G. Finazzi and I. Szabo, An update on the regulation of photosynthesis by thylakoid ion channels and transporters in Arabidopsis, Physiol. Plant., 2017, 161, 16–27 CrossRef CAS PubMed.
- M. M. Koskela, A. Brünje, A. Ivanauskaite, M. Grabsztunowicz, I. Lassowskat, U. Neumann, T. V. Dinh, J. Sindlinger, D. Schwarzer, M. Wirtz, E. Tyystjärvi, I. Finkemeier and P. Mulo, Chloroplast acetyltransferase NSI is required for state transitions in Arabidopsis thaliana, Plant Cell, 2018, 30, 1695–1709 CrossRef CAS PubMed.
- P. Albanese, S. Tamara, G. Saracco, R. A. Scheltema and C. Pagliano, How paired PSII–LHCII supercomplexes mediate the stacking of plant thylakoid membranes unveiled by structural mass-spectrometry, Nat. Commun., 2020, 11, 1–14 CrossRef PubMed.
- B. Demmig-Adams and W. Adams, Photoprotection and other responses of plants to high light stress, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1992, 43, 599–626 CrossRef CAS.
- E. Tyystjärvi and E. M. Aro, The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 2213–2218 CrossRef PubMed.
- E.-M. Aro, I. Virgin and B. Andersson, Photoinhibition of Photosystem II. Inactivation, protein damage and turnover, Biochim. Biophys. Acta, Bioenerg., 1993, 1143, 113–134 CrossRef CAS.
- M. A. Gururani, J. Venkatesh and L. S. P. Tran, Regulation of photosynthesis during abiotic stress-induced photoinhibition, Mol. Plant, 2015, 8, 1304–1320 CrossRef CAS PubMed.
- S. Järvi, M. Suorsa and E.-M. Aro, Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis, Biochim. Biophys. Acta, Bioenerg., 2015, 1847, 900–909 CrossRef PubMed.
- Y. Yamamoto, Quality Control of Photosystem II: The mechanisms for avoidance and tolerance of light and heat stresses are closely linked to membrane fluidity of the thylakoids, Front. Plant Sci., 2016, 7, 1136 Search PubMed.
- E. Baena-González, R. Barbato and E.-M. Aro, Role of phosphorylation in the repair cycle and oligomeric structure of photosystem II, Planta, 1999, 208, 196–204 CrossRef.
- M. Tikkanen, N. R. Mekala and E.-M. Aro, Photosystem II photoinhibition-repair cycle protects Photosystem I from irreversible damage, Biochim. Biophys. Acta, Bioenerg., 2014, 1837, 210–215 CrossRef CAS PubMed.
- K. Nishimura, Y. Kato and W. Sakamoto, Essentials of Proteolytic Machineries in Chloroplasts, Mol. Plant, 2017, 10, 4–19 CrossRef CAS PubMed.
- J. P. Vainonen, M. Hansson and A. V. Vener, STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of Photosystem II core proteins, J. Biol. Chem., 2005, 280, 33679–33686 CrossRef CAS PubMed.
- I. Samol, A. Shapiguzov, B. Ingelsson, G. Fucile, M. Crèvecoeur, A. V. Vener, J.-D. Rochaix and M. Goldschmidt-Clermont, Identification of a Photosystem II phosphatase involved in light acclimation in Arabidopsis, Plant Cell, 2012, 24, 2596–2609 CrossRef CAS PubMed.
- M. Tikkanen, M. Nurmi, S. Kangasjärvi and E.-M. Aro, Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light, Biochim. Biophys. Acta, Bioenerg., 2008, 1777, 1432–1437 CrossRef CAS PubMed.
- R. Fristedt, A. Willig, P. Granath, M. Crèvecoeur, J.-D. Rochaix and A. V. Vener, Phosphorylation of Photosystem II Controls Functional Macroscopic Folding of Photosynthetic Membranes in Arabidopsis, Plant Cell, 2009, 21, 3950–3964 CrossRef CAS PubMed.
- T. K. Goral, M. P. Johnson, A. P. R. Brain, H. Kirchhoff, A. V. Ruban and C. W. Mullineaux, Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation, Plant J., 2010, 62, 948–959 CAS.
- S. Puthiyaveetil and H. Kirchhoff, A phosphorylation map of the photosystem II supercomplex C2S2M2, Front. Plant Sci., 2013, 4, 459 Search PubMed.
- R. Fristedt, P. Granath and A. V. Vener, A Protein Phosphorylation Threshold for Functional Stacking of Plant Photosynthetic Membranes, PLoS One, 2010, 5, e10963 CrossRef PubMed.
- M. Pribil, O. Sandoval-Ibáñez, W. Xu, A. Sharma, M. Labs, Q. Liu, C. Galgenmüller, T. Schneider, M. Wessels, S. Matsubara, S. Jansson, G. Wanner and D. Leister, Fine-Tuning of Photosynthesis Requires CURVATURE THYLAKOID1-Mediated Thylakoid Plasticity, Plant Physiol., 2018, 176, 2351–2364 CrossRef CAS PubMed.
- Y. Yamamoto, H. Hori, S. Kai, T. Ishikawa, A. Ohnishi, N. Tsumura and N. Morita, Quality control of Photosystem II: reversible and irreversible protein aggregation decides the fate of Photosystem II under excessive illumination, Front. Plant Sci., 2013, 4, 433 Search PubMed.
- Q.-B. Yu, G. Li, G. Wang, J.-C. Sun, P.-C. Wang, C. Wang, H.-L. Mi, W.-M. Ma, J. Cui, Y.-L. Cui, K. Chong, Y.-X. Li, Y.-H. Li, Z. Zhao, T.-L. Shi and Z.-N. Yang, Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana, Cell Res., 2008, 18, 1007–1019 CrossRef CAS PubMed.
- R. Yokoyama, H. Yamamoto, M. Kondo, S. Takeda, K. Ifuku, Y. Fukao, Y. Kamei, M. Nishimura and T. Shikanai, Grana-Localized Proteins, RIQ1 and RIQ2, Affect the Organization of Light-Harvesting Complex II and Grana Stacking in Arabidopsis, Plant Cell, 2016, 28, 2261–2275 CrossRef CAS PubMed.
- S. Järvi, M. Rantala and E.-M. Aro, in Photosynthesis and Bioenergetics, World Scientific, 2017, pp. 243–263 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry and Owner Societies 2020 |