Open Access Article
Open Access ArticleAutophagy-deficient Arabidopsis mutant atg5, which shows ultraviolet-B sensitivity, cannot remove ultraviolet-B-induced fragmented mitochondria†
Gönül
Dündar
 a,
Mika
Teranishi
a and
Jun
Hidema
a,
Mika
Teranishi
a and
Jun
Hidema
 *ab
*ab
aGraduate School of Life Sciences, Tohoku University, Sendai 980-8577, Japan. E-mail: j-hidema@ige.tohoku.ac.jp; Fax: +8122 217 5691; Tel: +81 22 217 5690
bDivision for the Establishment of Frontier Sciences of the Organization for Advanced Studies, Tohoku University, Sendai 980-8577, Japan
First published on 25th November 2020
Abstract
Mitochondria damaged by ultraviolet-B radiation (UV-B, 280–315 nm) are removed by mitophagy, a selective autophagic process. Recently, we demonstrated that autophagy-deficient Arabidopsis thaliana mutants exhibit a UV-B-sensitive phenotype like that of cyclobutane pyrimidine dimer (CPD)-specific photolyase (PHR1)-deficient mutants. To explore the relationship between UV-B sensitivity and autophagy in UV-B-damaged plants, we monitored mitochondrial dynamics and autophagy in wild-type Arabidopsis (ecotype Columbia); an autophagy-deficient mutant, atg5; a PHR1-deficient mutant, phr1; an atg5 phr1 double mutant; and AtPHR1-overexpressing (AtPHR1ox) plants following high-dose UV-B exposure (1.5 W m−2 for 1 h). At 10 h after exposure, the number of mitochondria per mesophyll leaf cell was increased and the volumes of individual mitochondria were decreased independently of UV-B-induced CPD accumulation in all genotypes. At 24 h after exposure, the mitochondrial number had recovered or almost recovered to pre-exposure levels in plants with functional autophagy (WT, phr1, and AtPHR1ox), but had increased even further in atg5. This suggested that the high dose of UV-B led to the inactivation and fragmentation of mitochondria, which were removed by mitophagy activated by UV-B. The UV-B-sensitive phenotype of the atg5 phr1 double mutant was more severe than that of atg5 or phr1. In wild-type, phr1, and AtPHR1ox plants, autophagy-related genes were strongly expressed following UV-B exposure independently of UV-B-induced CPD accumulation. Therefore, mitophagy might be one of the important repair mechanisms for UV-B-induced damage. The severe UV-B-sensitive phenotype of atg5 phr1 is likely an additive effect of deficiencies in independent machineries for UV-B protection, autophagy, and CPD photorepair.
Introduction
Plants use sunlight as the energy source for photosynthesis and are inevitably exposed to harmful ultraviolet-B radiation (UV-B, 280–315 nm). Despite UV-B being a minor proportion of the UV radiation that reaches the earth's surface, it causes substantial damage to plant macromolecules such as lipids, proteins, and membranes, and especially DNA.1UV-B induces lesions in nuclear, mitochondrial, and chloroplast genome structures in plants by specifically inducing photoproducts between adjacent pyrimidines on the same DNA strand.2,3 These lesions, cyclobutane pyrimidine dimers (CPDs) and pyrimidine pyrimidone (6-4) photoproducts, can retard growth and development in Arabidopsis.4,5 The CPDs, which make up the largest component (∼75%) of photoproducts,6 are a principal cause of UV-B-induced growth inhibition in plants, and are often used as a direct indicator of UV-B damage in plants.7 Exposure to UV-B also causes oxidative stress via the production of reactive oxygen species (ROS).1 Because electron leakage from respiration and photosynthesis chains promotes the production of strong oxidants, organelles are major sources of damaging by-products.8 The ROS produced in response to UV-B can also directly damage DNA and proteins by inhibiting transcription and replication and inducing mutations or cell death, thereby inhibiting plant growth and development.9–12
Plants have evolved many response mechanisms to adapt to UV-B stress, e.g., repair of CPDs by photoreactivation and nucleotide excision repair.2 CPD-specific photolyases, which absorb UVA (315–400 nm) and blue radiation as energy to monomerize dimers, are widely distributed among species ranging from eubacteria to eukaryotes, but not in placental mammals.13 In plants, photoreactivation by CPD photolyase is the primary mechanism of CPD repair.3,4 We previously demonstrated that CPD photolyase is crucial for determining the resistance of rice to UV-B and that elevated CPD photolyase activity can significantly alleviate UV-B-induced growth inhibition in rice.7,14 In Arabidopsis, deficiency of CPD photolyase (PHR1, also named UV RESISTANCE 2 [UVR2]; AT1G12370) induces UV-B sensitivity,4 indicating that UV-B-induced CPD accumulation is a principal cause of UV-B-induced cell death and growth inhibition in plants. In addition to CPD, other types of DNA damage and ROS also activate damage and stress response signaling pathways that control the expression of genes related to protection against UV-B-induced damage in plants.15–18
Recently, we reported that both artificial UV-B radiation and natural sunlight cause vacuolar transport of whole photo-damaged chloroplasts through an autophagy process known as chlorophagy.19 In that study, we showed that mutants of the autophagy-related genes 2 (ATG2), 5 (ATG5) and 7 (ATG7) exhibit UV-B-sensitive phenotypes like that of PHR1-deficient Arabidopsis thaliana ecotype Landsberg erecta mutants (uvr2).19 These results indicated that autophagy also plays an important role in the plant response to UV-B-induced damage.
In yeast and mammals, damaged organelles, especially damaged mitochondria, which are the center of energy production and several other important metabolic processes, are removed by a selective autophagic process known as mitophagy.20–22 Mitophagy is linked to several physiological functions and human pathologies, such as neurodegenerative disease.23,24 However, little is known about the relationships between UV-B-induced CPD accumulation and mitochondrial dynamics or between mitochondrial dynamics and autophagy in plants damaged by UV-B. Here, to understand these relationships, we focused on the mitochondrial dynamics in A. thaliana wild-type (WT) (ecotype Columbia); the autophagy-deficient mutant atg5; the PHR1-deficient mutant phr1; the atg5 phr1 double mutant (atg5 phr1); and AtPHR1-overexpressing transgenic plants (AtPHR1ox) exposed to UV-B. Our results indicate that UV-B damages mitochondrial function and causes mitochondrial fragmentation in Arabidopsis, and that these damaged dysfunctional mitochondria are removed by autophagy. Our results also show that, although UV-B induces autophagy, UV-B-induced mitochondrial fragmentation and autophagy occur independently of UV-B-induced CPD accumulation.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia was used as WT. The Arabidopsis T-DNA insertion lines atg5 (SAIL_129_B07)25,26 and phr1 (WiscDsLox368H08) were obtained from the Arabidopsis Biological Resource Center. The atg5 phr1 double mutant (atg5 phr1) was obtained by sexual crossing between atg5 and phr1. Arabidopsis AtPHR1 was transformed into phr1 under the control of the cauliflower mosaic virus 35S promoter (Pro35S) by using pBI121 vector (AtPHR1ox). The following transgenic plants were generated as previously described:27,28 mitochondria-targeted GFP (MT-GFP) driven by Pro35S was introduced into WT, atg5 and phr1 (Mt-WT, Mt-atg5 and Mt-phr1) and chloroplast stroma-targeted GFP (CT-GFP) driven by Pro35S were each introduced into WT, atg5, phr1, atg5 phr1 (Ct-WT, Ct-atg5, Ct-phr1, Ct-atg5 phr1) by gene transformation or sexual crossing. When generating transgenic AtPHR1ox expressing mitochondria- or chloroplast-targeted GFP, the mitochondrial isocitrate dehydrogenase (IDH) promoter (ProIDH) or ribulose bisphosphate carboxylase small chain 2B (RBCS2B) promoter (ProRBCS2B) was used to avoid gene silencing caused by using the same promoter as that driving PHR1 in AtPHR1ox. MT-GFP or CT-GFP was transformed into WT under the control of the ProIDH or ProRBCS2B as previously described.28,29 Then, the plants expressing MT-GFP or CT-GFP were sexually crossed with AtPHR1ox line giving Mt-AtPHR1ox or Ct-AtPHR1ox respectively. Arabidopsis plants were grown in soil in chambers at 23 °C under a 16 h light (6 a.m. to 10 p.m.)/8 h dark (10 p.m. to 6 a.m.) photoperiod under white fluorescent lamps (140 μmol m−2 s−1). Fourteen-day-old seedlings were used for all experiments.UV-B treatments and measurement of plant growth
Fourteen-day-old seedlings were irradiated for 1 h with narrowband UV-B at different intensities (0.5 or 1.5 W m−2) supplied by a xenon light source [MAX-303 (UV-VIS); Asahi Spectra Co., Ltd, Tokyo, Japan] filtered through a 280 nm band pass filter (LX0280; Asahi Spectra Co., Ltd). The spectrum or energy fluence rate was measured with a spectroradiometer (USR-45DA; Ushio Inc., Tokyo, Japan). The UV-B irradiation treatment was started at 10 a.m. (4 h after the start of the photoperiod). After UV-B irradiation, plants were returned to the growth chamber for 10 h, 24 h, or 2 d for further experiments. The above-ground parts of the plants were cut off, and fresh weights (FW) were measured individually. At least 30 plants from three biological repeats were analyzed.Measurement of CPD level
Seedlings were irradiated with 1.5 W m−2 UV-B as described above, and then the above-ground parts of some plants were harvested immediately. The other UV-B-irradiated seedlings were returned to the growth chamber for repairing CPDs under white light. To measure the level of CPD photolyase-independent repair, UV-B-irradiated seedlings were placed in a light-tight box before being returned to the growth chamber. Two days after UV-B exposure, the above-ground parts were harvested. All harvested samples were frozen in liquid nitrogen, then stored at −80 °C until analysis. The following manipulations were carried out under dim red light to minimize uncontrolled photoreactivation. Each seedling was homogenized in liquid nitrogen. Agarose plugs were prepared, and DNA was purified in the gel. Half of the agarose plugs were treated with UV endonuclease as described previously (UV endonuclease specifically cleaves single DNA strands at CPD sites).30 The DNA molecules were separated according to their single-strand molecular lengths in 0.7% alkaline agarose gels by static field electrophoresis and biased sinusoidal field gel electrophoresis (Genofield; ATTO Co., Osaka, Japan).30 The DNA length markers were DNA fragments from Hansenula wingei chromosomes (smallest, 1.05 Mb) (Bio-Rad, Hercules, CA, USA), T4 phage DNA (170 kb), Lambda phage DNA (48.5 kb), and HindIII-digested Lambda phage DNA (23.1, 9.4, 6.6, 4.3, and 2.3 kb). The CPD frequencies were determined using a DNA damage analysis system constructed by the Tohoku Electric Co., (Sendai, Japan) as previously described.30 The frequencies were calculated using a molecular length standard curve, and the quantity of DNA at each migration position was determined from quantitative image data.31 The CPD frequency was measured by subtracting the value of the UV endonuclease-untreated sample from that of the UV endonuclease-treated sample and is expressed as the number of CPD per Mb. Three independent experiments were performed.Laser-scanning confocal microscopy imaging
The laser-scanning confocal microscopy (LSCM) imaging was performed using a C-apochromat LD63× water-immersion objective (numerical aperture = 1.15; LSM800, Carl Zeiss, Oberkochen, Germany). The excitation wavelength for GFP was 488 nm and that for chlorophyll autofluorescence was 640 nm.Detection of dead cells
Dead cells were quantified in transgenic Arabidopsis lines harboring CT-GFP to estimate cell death. Two days after UV-B exposure, plants were observed by LSCM. The percentage of dead cells in a fixed area (containing 50 cells on average) was determined. Four areas of the leaf were analyzed per plant, and three plants for each genotype were analyzed (n = 12). Dead cells were detected as entirely green cells because GFP fluorescence spread throughout the cell after the vacuolar degradation of chloroplast stroma during programmed cell death.32Measurement of mitochondrial number and volume
Transgenic Arabidopsis lines harboring MT-GFP were irradiated with 1.5 W m−2 UV-B. At 10 and 24 h after UV-B exposure, plants were observed by LSCM. Fixed image areas (212- × 212- × 40 μm each) were monitored by changing the focus in LSCM. The Z-stack images from LSCM were converted into three-dimensional pictures and the number and volume of mitochondria were detected using Imaris microscopy analysis software (Bitplane, Zurich, Switzerland). To determine the number of mitochondria, three plants for each timepoint were analyzed [n = 3 (≥40 cells)]. To measure the volume of mitochondria, four cells per plant and four plants for each timepoint were analyzed [n = 4 (≥16 cells)].Detection of active mitochondria
To label active mitochondria, excised leaves of seedlings were infiltrated with 200 nM tetramethylrhodamine, ethyl ester (TMRE) and incubated at room temperature for 20 min,33 then fluorescence images of cells were observed and captured by LSCM. At least three areas (212- × 212- × 40 μm each) were assayed per plant, and four plants for each timepoint were analyzed.Concanamycin A treatment
The transgenic Arabidopsis lines harboring MT-GFP were irradiated with 1.5 W m−2 UV-B. Entire transgenic Arabidopsis plants (including roots) harboring MT-GFP were pulled from the soil. Then, 1 μM Concanamycin A (ConcA) was added to 10 mM MES-NaOH (pH 5.5) solution, and the mixture was infiltrated into the leaves using a 1 ml syringe. After treatment, the seedlings were immersed in the same ConcA solution, with filter paper laid on top to keep them submerged. The seedlings were then returned to the growth chamber for 24 h. Plants in the control were treated in the same way, but with the same amount of DMSO replacing ConcA. The mesophyll cells in the infiltrated area were observed by LSCM.RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA was isolated from rosette leaves using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany), and cDNA was synthesized using random hexamer and oligo dT primers with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Otsu, Japan). An aliquot of the synthesized cDNA derived from 500 ng total RNA was subjected to qRT-PCR analysis using the KAPA SYBR FAST qPCR Kit (KAPA Biosystems, Woburn, MA, USA) and a real-time PCR detection system (CFX96, Bio-Rad). The β-tubulin gene was used as an internal control. Gene-specific primers are listed in ESI Table 1.†Statistical analysis
Statistical analyses were performed with GraphPad Prism version 8.00 (GraphPad Software). Two-way ANOVA was used for the comparison of paired samples followed by Tukey's test or Dunnett's test for the comparison of multiple samples, as indicated in figure legends.Results
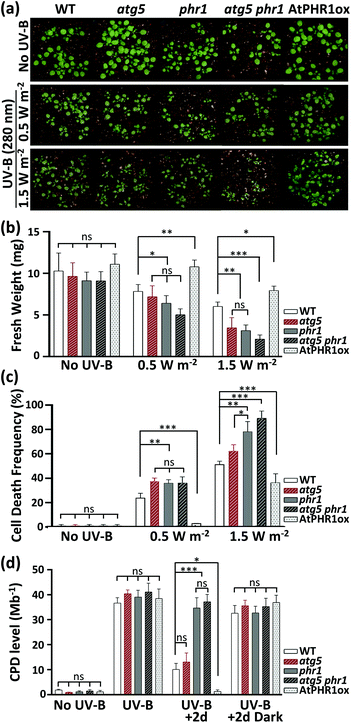
First, to test the relationship between CPD accumulation and autophagy function in the UV-B sensitivity of Arabidopsis, seedlings of WT, atg5, phr1, atg5 phr1, and AtPHR1ox were exposed to different doses of UV-B (1.5 or 0.5 W m−2) for 1 h. Their growth retardation phenotypes (shoot FW) were examined 2 d later (Fig. 1a and b). The atg5 and phr1 mutants were significantly more UV-B-sensitive than WT to the 1.5 W m−2 UV-B treatment, as previously reported.19 The growth retardation of atg5 phr1 in response to UV-B was more severe than that of the UV-B-sensitive single mutants atg5 and phr1. By contrast, the AtPHR1ox line showed significant resistance to UV-B compared with WT. UV-B at 0.5 W m−2 reduced the shoot FW of WT, atg5, phr1, atg5 phr1, and AtPHR1ox plants by 24%, 25%, 27%, 45%, and 11%, respectively; whereas UV-B at 1.5 W m−2 reduced the shoot FW of these lines to a greater extent, by 43%, 62%, 61%, 77%, and 33%, respectively. At 2 d after exposure, the ratio of dead cells to total cells in the plant leaf (cell death frequency) increased with increasing UV-B dose (Fig. 1c and ESI Fig. 1†). At 1.5 W m−2 UV-B irradiation, the cell death frequencies were higher in atg5 and phr1 than in WT, and that in atg5 phr1 was the highest among all the genotypes examined. The cell death frequency in AtPHR1ox was extremely low, significantly lower than in WT. Exposure to UV-B at 0.5 W m−2 caused little cell death in AtPHR1ox.To determine the relationship between CPD accumulation and the UV-B-sensitive phenotype, we measured CPD levels in all the plant genotypes (Fig. 1d). Prior to UV-B exposure, the CPD level in all plants grown under white light alone was less than 1.5 CPD per Mb. Exposure to 1.5 W m−2 UV-B for 1 h induced similar levels of CPDs (approximately 35–40 CPD per Mb) among all the plant genotypes. In plants that were returned to the growth chamber under white light, CPD accumulation decreased dramatically in WT, atg5, and AtPHR1ox, but not in the CPD PHR1-deficient mutants (phr1 or atg5 phr1), compared with the levels immediately after UV-B exposure. The CPD levels in AtPHR1ox decreased to pre-UV-B exposure levels, but those in WT and atg5 remained at ∼12 CPD per Mb. These results strongly indicate that UV-B-induced CPD accumulation induces cell death, thereby explaining the UV-B sensitivity of the PHR1-deficient mutants, phr1 and atg5 phr1. However, atg5 and phr1 showed similar UV-B sensitivity (Fig. 1a and b), even though the CPD level was significantly lower in atg5 than in phr1 (Fig. 1d). Even though the CPD level was similar in atg5 phr1 and phr1 (Fig. 1d), the atg5 phr1 double mutant was more UV-B-sensitive than was the phr1 mutant (Fig. 1a and b). These results indicate that the mechanism behind the UV-B sensitivity of the autophagy-deficient mutant, atg5, might be independent of CPD accumulation.
Next, to investigate the mitochondrial dynamics following UV-B exposure, we expressed a construct with mitochondrial matrix-targeted (MT)-GFP under the control of Pro35S in WT, atg5, and phr1 plants to produce Mt-WT, Mt-atg5, and Mt-phr1 lines, respectively. Before performing these analyses, to confirm the CPD levels in mitochondria in these plants exposed to UV-B radiation, we conducted an organelle-specific assay for detection of CPDs by using T4 endonuclease V digestion and Southern blot analysis (ESI Fig. 2†). We determined the percentage of fragments with enzyme-sensitive sites (ESSs; hereafter, CPD frequency). Exposure to 1.5 W m−2 UV-B for 1 h increased the CPD frequency in mitochondria and nuclei to roughly same level among the plant lines examined here. The CPD frequency in mitochondria and nuclei of Mt-WT or Mt-atg5 lines had decreased by 10 or 24 h of recovery under white light in the growth chamber. The rate of decrease in the CPD frequency was slightly lower in mitochondria than in nuclei in Mt-WT, and a similar trend was observed in Mt-atg5. However, there was no difference in the rate of decrease in the CPD frequency between mitochondria and nuclei in Mt-WT and Mt-atg5. Some CPDs in mitochondria of Mt-WT or Mt-atg5 were not repaired at 24 h after UV-B exposure (approximately 40% of UV-B induced CPD remained in both genotypes). However, the CPD frequency in mitochondria and nuclei of Mt-phr1 was unchanged after 24 h of recovery under white light. These results indicate that CPDs induced by UV-B exposure in mitochondrial DNA are repaired by CPD photolyase, PHR1.
To further investigate the mitochondrial dynamics, the plants were exposed to UV-B (1.5 W m−2 for 1 h) and the mesophyll leaf cells were observed by LSCM. In the non-irradiated leaves, the GFP signals were uniformly detected at the edge of chloroplasts and in the cell center in Mt-WT, Mt-atg5, and Mt-phr1 (Fig. 2a), and a similar number of GFP signals was observed in each line (Fig. 2b). In these plants, the number of GFP-fluorescent dots per cell (hereafter, ‘number of mitochondria’) was significantly increased (Fig. 2a and b) whereas the volumes of individual dots (hereafter, ‘mitochondrial volume’) were significantly decreased at 10 h after UV-B exposure, compared with no UV-B irradiation (Fig. 2c). To determine whether mitochondrial fragmentation caused the phenomena of increased number of mitochondria and decreased mitochondrial volume, we treated the leaves of Mt-WT and Mt-atg5 with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a mitochondrial uncoupler that causes rapid and dramatic fragmentation of mitochondria34 (ESI Fig. 3†). Compared with the control, the CCCP treatment significantly increased the number of mitochondria in Mt-WT and Mt-atg5 (p < 0.01), and to a greater extent in Mt-atg5 than in Mt-WT (ESI Fig. 3b†). Damaged mitochondria fragment and then are degraded by the ATG5-dependent autophagy system. The CCCP treatment also led to decreased mitochondrial volume (ESI Fig. 3c†). Therefore, this finding supports that the increased number of mitochondria and decreased mitochondrial volume following UV-B exposure might signify mitochondrial fragmentation resulting from UV-B-induced damage, which accelerates mitochondrial fission and suppresses mitochondrial fusion.
By contrast, at 24 h after UV-B exposure, the mitochondrial dynamics were significantly different among Mt-WT, Mt-atg5, and Mt-phr1 (Fig. 2). The number of mitochondria in Mt-WT was significantly increased at 10 h after UV-B exposure, but it had almost returned to the pre-exposure level at 24 h after UV-B exposure (Fig. 2b). In the case of mitochondrial volume, a significant decreasing was observed at 10 h after UV-B exposure in Mt-WT, but it tended to return to the pre-exposure level at 24 h after UV-B exposure (Fig. 2c). In Mt-atg5, the number of mitochondria was significantly increased by 10 h and further increased at 24 h after UV-B exposure; the number of mitochondria increased in the center of the cell at 24 h after UV-B exposure (Fig. 2a and b). However, the mitochondrial volume was significantly decreased by 10 h and further decreased by 24 h after UV-B exposure (Fig. 2c). The mitochondrial number and mitochondrial volume showed similar trends in Mt-phr1 and Mt-WT at 10 h after UV-B exposure (Fig. 2b and c). However, at 24 h after UV-B exposure, the number of mitochondria decreased to lower than the pre-exposure level, and the volume increased to higher than the pre-exposure level. At 24 h after UV-B exposure, the GFP-fluorescent dots were aggregated in cells of the Mt-phr1 mutant (Fig. 2a). The observed aggregation was thought to be responsible for the decreased number and the increased volume of mitochondria in Mt-phr1 at 24 h.
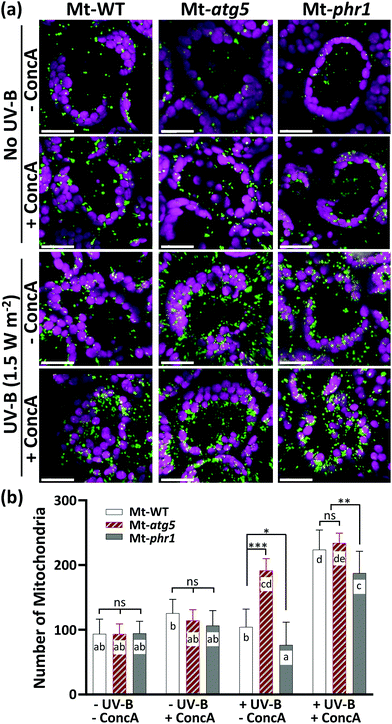
The number of mitochondria had increased by 10 h after UV-B exposure in all genotypes examined, suggesting that UV-B induces mitochondrial fragmentation independently of PHR1-deficiency or autophagy. If this is the case, then the degradation of UV-B-damaged mitochondria in the vacuole by autophagy should occur in Mt-WT and Mt-phr1, but not in the autophagy-deficient mutant Mt-atg5, between 10 and 24 h after UV-B exposure. To test this possibility, we investigated changes in the number of mitochondria in Mt-WT, Mt-atg5, and Mt-phr1 treated with the vacuolar type H+-ATPase-inhibitor ConcA, which inhibits autophagic degradation in vacuoles35,36 (Fig. 3). The ConcA treatment did not affect the number of mitochondria in non-UV-B-irradiated Mt-atg5 and Mt-phr1. Under UV-B irradiation, the number of mitochondria increased only in Mt-atg5 (Fig. 2b). In contrast, ConcA treatment after UV-B irradiation increased the number of mitochondria in Mt-WT and Mt-phr1, and to a lesser degree in Mt-atg5. Thus, without ConcA treatment, UV-B-induced fragmented mitochondria were removed by autophagic degradation in Mt-WT and MT-phr1. These results suggest that a large proportion of mitochondria removal is mediated by atg5-dependent autophagy.
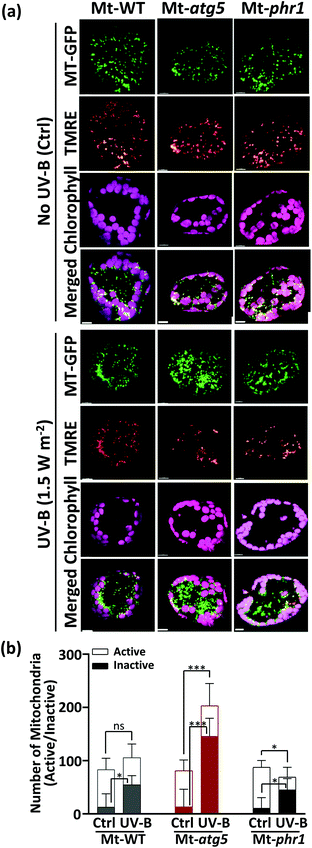
To determine that whether the GFP signals were indicative of functional living mitochondria or not, we stained the leaves of Mt-WT, Mt-atg5, and Mt-phr1 with TMRE at 24 h after UV-B exposure (1.5 W m−2 for 1 h). This dye is a cationic lipophilic compound that accumulates in active mitochondrial membranes. The stained cells were observed by LSCM. Prior to UV-B-radiation, more than 80% of MT-GFP signals were clearly co-localized with TMRE signals in all lines tested (Fig. 4). However, at 24 h after UV-B exposure, there were significantly more GFP-fluorescent dots that were not merged with TMRE-fluorescent dots (hereafter, ‘inactive mitochondria’) than were merged (hereafter, ‘active mitochondria’) in mesophyll leaf cells. The percentages of inactive mitochondria were significantly higher in Mt-atg5 and Mt-phr1 than in Mt-WT (Mt-WT: 50%, Mt-atg5: 71%, Mt-phr1: 69%). Thus, dysfunctional mitochondria remained in the cells of Mt-atg5 and Mt-phr1 mutants at 24 h after UV-B exposure. In Mt-atg5, UV-B irradiation reduced the number of active mitochondria even though the total number of mitochondria increased. Thus, a larger proportion of the increased number of mitochondria in Mt-atg5 was dysfunctional.
Next, we examined whether mitochondrial fragmentation or dysfunction also occurred following UV-B exposure in the AtPHR1ox line, which has a UV-B-resistant phenotype. To answer this question, we generated an AtPHR1ox line (Mt-AtPHR1ox). In this Mt-AtPHR1ox line, ProIDH-GFP was expressed as a mitochondrial matrix marker under the control of the native IDH promoter, instead of Pro35S. to minimize the effects of gene silencing.37 In plants grown in a chamber, the CPD removal rates in nuclei and mitochondria were higher in Mt-AtPHR1ox than in Mt-WT (ESI Fig. 4†). At 10 h following UV-B exposure (1.5 W m−2 for 1 h), the number of mitochondria in Mt-AtPHR1ox was significantly increased compared with the pre-exposure level; and by 24 h the number had decreased although it was still significantly higher than the pre-exposure level (Fig. 5). Thus, the mitochondrial dynamics of the Mt-AtPHR1ox line were similar but not identical to those of Mt-phr1 (Fig. 2b). Compared with their respective non-irradiated controls, Mt-AtPHR1ox contained more mitochondria at 24 h after UV-B exposure (Fig. 5b), while Mt-phr1 contained fewer mitochondria at 24 h after UV-B exposure (Fig. 2b). The number of inactive mitochondria was similar between UV-B-irradiated plants and non-irradiated plants in Mt-AtPHR1ox (ESI Fig. 5†) but was significantly increased in Mt-phr1 at 24 h after UV-B exposure (Fig. 4b). The CPD level at 2 d after UV-B exposure was significantly lower in AtPHR1ox than in phr1 (Fig. 1d), providing further evidence that UV-B-induced CPD accumulation induces cell death. However, the finding that mitochondrial fragmentation occurred in both UV-B-irradiated Mt-phr1 and Mt-AtPHR1ox plants strongly indicates that mitochondrial fragmentation induced by UV-B was caused independently of CPD accumulation. The loss of mitochondrial function due to the UV-B-induced CPD accumulation can be interpreted from the result that many active mitochondria remained in the AtPHR1ox plants after UV-B exposure, in comparison to UV-B-irradiated Mt-WT.
 | ||
| Fig. 5 Changes in mitochondrial number and volume in Arabidopsis PHR1-overexpressing line (Mt-AtPHR1ox) following UV-B irradiation. (a) Laser-scanning confocal microscopy image of leaf mesophyll cells in WT and Arabidopsis PHR1 (AtPHR1)-overexpressing transgenic seedlings expressing MT–GFP (Mt-WT and Mt-AtPHR1ox) that were non-irradiated (control) or exposed to UV-B (1.5 W m−2 for 1 h). Images were taken at 10 and 24 h after UV-B treatment. Green, mitochondria; red, chlorophyll. Scale bars, 20 μm. (b, c) Number of mitochondria per cell (b) and volume of individual mitochondria (c) before UV-B exposure and 10 h and 24 h after UV-B exposure. Mitochondrial number and volume were determined as described in Fig. 2b and c. Images from three different replicates were used for calculations. Data are means ± SE (n ≥ 40 cells). Asterisks denote significant differences based on two-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001). ns, not significant. | ||
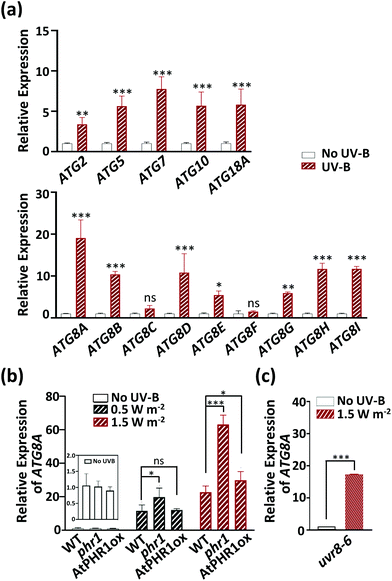
Because UV-B-induced fragmented mitochondria were removed by autophagic degradation in Arabidopsis, we wondered whether UV-B radiation activated the expression of autophagy-related genes. To answer this question, the transcript levels of autophagy-related genes in the ATG2, 5, 7, 10, 18A, and ATG8 families were monitored in UV-B-exposed WT Arabidopsis plants (Fig. 6a). ATG8 family proteins play a role in autophagosome formation and are essential in the execution of autophagy. Among the ATG8 genes, ATG8A is a common autophagy marker.38 Except for the ATG8 gene family members ATG8C and ATG8F, all the autophagy-related genes examined here showed significantly higher transcript levels in UV-B-irradiated than in non-irradiated WT plants, with ATG8A showing the largest change in transcript levels (the level in UV-B-irradiated plants was 19 times that in non-irradiated plants). Next, to understand the relationship between CPD accumulation and induction of the autophagy marker gene ATG8A, we analyzed its transcript levels in WT, phr1, and AtPHR1ox seedlings exposed to UV-B (0.5 W m−2 or 1.5 W m−2 for 1 h). At 2 d after UV-B exposure, ATG8A transcript levels were increased in all genotypes, with the largest increase in phr1 despite it being a PHR1-deficient mutant (Fig. 6b). To confirm the autophagic activity caused by UV-B exposure, we performed a DALGreen-staining assay of root cells of transgenic Arabidopsis plant over-expressing mitochondria-targeted yellow fluorescent protein (MtY-WT) (ESI Fig. 6†). DALGreen is an autophagosome-specific dye that is useful for monitoring late-phase autophagy in HeLa cells.39,40 To test whether this assay can also be used in plant cells, we performed this assay using the root of MtY-WT treated with CCCP. A weak DALGreen fluorescent signal indicative of autophagosomes was detected in the non-treated root cells of MtY-WT. In contrast, by staining for 4 h after CCCP treatment, there were strong DALGreen fluorescent signals in the root cells of MtY-WT. These results would mean that these signals indicate autophagosomes. When the seedlings including roots of MtY-WT were exposed to UV-B radiation (1.5 W m−2 for 1 h), strong DALGreen fluorescent signals were appeared in the root cells of MtY-WT by staining for 4 h after UV-B exposure. These results indicate that UV-B radiation induces autophagy-related genes and activates autophagy, and that this induction or activation by UV-B exposure is independent of CPD accumulation.
Plants have a UV-B-specific photoreceptor, UV RESISTANCE LOCUS 8 (UVR8), which regulates the expression of many genes related to photomorphogenic UV-B responses and UV-B protection.41,42 To test whether UVR8 also regulates autophagy-related genes, we investigated the effect of UV-B irradiation (1.5 W m−2 for 1 h) on the transcript levels of ATG8A in the UVR8-deficient Arabidopsis mutant, uvr8-6 (Fig. 6c).43 The transcript levels of ATG8A at 2 d after UV-B exposure were similar in WT (Fig. 6a) and uvr8-6 (Fig. 6c), indicating that UV-B induces the expression of at least one autophagy-related gene, ATG8A, independently of the UVR8 signaling pathway.
Discussion
High doses of UV-B damage the biological components of plant cells, decreasing plant growth.1 We previously demonstrated that both CPD photorepair7,14 and autophagy19 play important roles in UV-B resistance in plants. Here, to understand the role of autophagy in UV-B protection in plant cells, we focused on the mitochondrial dynamics and UV-B-induced CPD accumulation in WT, atg5, phr1, atg5 phr1, and AtPHR1ox plants. For the mitochondrial dynamics experiments, we analyzed WT, atg5, phr1, and atg5 phr1 lines expressing MT-GFP and constructed an AtPHR1ox line expressing IDH-GFP.Before discussing the relationship between mitochondrial dynamics and CPD accumulation in mitochondria of Arabidopsis, we should discuss the function of PHR1 in mitochondria of Arabidopsis because it is unclear whether AtPHR1 is localized/active in mitochondria in Arabidopsis. Draper and Hays demonstrated the blue light-dependent removal of CPDs from mitochondrial DNA in Arabidopsis leaves.8 By contrast, Kaiser et al. demonstrated that GFP fused to AtPHR1 (AtPHR1::GFP) was localized exclusively in the nuclei, and was not transported into mitochondria in green protoplasts prepared from an Arabidopsis mesophyll cell culture.44 Previously, we identified a mitochondrial targeting signal sequence (MTS) in OsPHR1, which functions in nuclei, mitochondria, and chloroplasts, and found that the MTS of OsPHR1 is highly conserved among class-II CPD photolyases including AtPHR1.45 In this study, a site-specific assay for detecting CPDs showed that light-dependent removal of UV-B-induced CPD in mitochondrial DNA occurred in leaves of WT Arabidopsis, but not in those of the AtPHR1-deficient mutant (phr1) (ESI Fig. 2†). In addition, CPDs in mitochondrial DNA were more effectively removed in the AtPHR1ox line (ESI Fig. 4†). These results strongly indicate that AtPHR1 functions in mitochondria in Arabidopsis.
In all the lines used for mitochondrial dynamics experiments, a UV-B treatment increased the number of mitochondria per cell while decreasing the volumes of individual mitochondria observed as GFP-fluorescent dots in the cells at 10 h after UV-B exposure (Fig. 2 and 5). However, the number of mitochondria in plants with autophagy function recovered fully (Mt-WT and Mt-phr1) or partially (Mt-AtPHR1ox) to pre-UV-B exposure levels at 24 h after UV-B exposure (Fig. 2 and 5). These results indicate that UV-B induces mitochondrial fragmentation in the plant cells. Mitochondrial fragmentation occurs in response to various genotoxic stresses, including UV-B, in a dose-dependent manner in mammals,46–48 and also occurs in response to the accumulation of oxidized mitochondrial components (DNA, proteins, and phospholipids), or instability of mitochondrial DNA.49–51 However, our results do not clarify whether the UV-B-induced mitochondrial fragmentation is caused by accumulation of DNA damage other than CPD on mitochondrial or nuclear DNA, or by oxidized mitochondrial components. In any case, our results suggest that CPD accumulation is not directly involved in UV-B-induced mitochondrial fragmentation in Arabidopsis.
Interestingly, in the autophagy-deficient Mt-atg5 mutant, the number of mitochondria continued to increase between 10 h and 24 h after UV-B exposure (Fig. 2b). By differentiating between inactive and active mitochondria with TMRE staining (Fig. 4), we found that great portion of those increased mitochondria were inactive in Mt-atg plants at 24 h after UV-B exposure compared with pre-exposure levels. These results indicate that UV-B damages mitochondria, and the resultant dysfunctional mitochondria are removed by autophagy in Arabidopsis. Our findings suggest that UV-B-induced damage activates mitophagy in Arabidopsis as previously shown for other organelles that are degraded under different stresses. For example, autophagy can remove photo-damaged chloroplasts (chlorophagy),19 oxidized peroxisomes,52–55 and stressed endoplasmic reticulum.56 A mitophagy deficiency leads to the accumulation of ROS, mutations in mitochondrial DNA, and consequently cell death, or cancer initiation and progression in human cells.57,58 Thus, the UV-B-sensitive phenotype of the atg5 mutant that accumulated dysfunctional mitochondria suggests that mitophagy might be one of the important responses to UV-B induced damage.
In the case of the PHR1-deficient mutant, Mt-phr1, which cannot repair CPDs by photoreactivation, the number of mitochondria was even lower at 24 h after UV-B exposure than before UV-B exposure (Fig. 2b). Although the number of inactive mitochondria per cell was increased in both Mt-WT and Mt-phr1 at 24 h after UV-B exposure, there were significantly fewer active mitochondria per cell in Mt-phr1 than in Mt-WT (Fig. 4). Thus, at 24 h after UV-B exposure, the ratio of active mitochondria to total mitochondria in cells was lower in Mt-phr1 than in Mt-WT (Fig. 4). By contrast, at 24 h after UV-B exposure, many active mitochondria still remained in the UV-B-irradiated Mt-AtPHR1ox line (ESI Fig. 5†). These results indicate that CPD accumulation caused a significant loss of mitochondrial functionality, and that some of the damaged mitochondria were removed by autophagy in the Mt-phr1 mutant. As shown in Fig. 2 and 3, some other dysfunctional mitochondria, which appeared to aggregate, remained in cells of the MT-phr1 mutant, despite it retaining autophagy function. Autophagy might not be able to remove such aggregated dysfunctional mitochondria. Therefore, accumulation of dysfunctional mitochondria, which cannot be removed by autophagy, might contribute to the UV-B-sensitive phenotype of phr1 plants.
All the tested autophagy-related genes in the ATG2, 5, 7, 10, 18A and ATG8 gene families, except for ATG8C and ATG8F, were strongly expressed in WT plants following UV-B exposure (Fig. 6a). Activation of ATG8A expression occurred even in the absence of the UVR8-dependent signaling pathway (Fig. 6c). UVR8 is a UV-B-specific photoreceptor that responds to UV-B at low fluence rates (<1 μmol m−2 s−1 UV-B). In this experiment, we measured the transcript levels of ATG8A in the uvr8-6 mutant in response to 1.5 W m−2 UV-B. This intensity is more than enough to activate the UVR8 photoreceptor, and is sufficient to induce ROS and DNA damage. Therefore, expression of ATG8A in response to 1.5 W m−2 UV-B was not dependent on UVR8, but rather on ROS and/or DNA damage. The expression of autophagy-related genes was up-regulated by UV-B exposure, suggesting that mitophagy is activated in response to UV-B damage in Arabidopsis, as we observed previously for chlorophagy.19 Numerous studies on human cells have reported that induction of autophagy, mainly mitophagy, by UV-B is associated with ROS generation and DNA damage,23,59 although the mechanistic details remain unclear. In yeast, animals, and plants, high doses of UV-B cause molecular damage within cells directly and/or indirectly via the production of ROS and DNA damage, and can induce the expression of genes involved in damage and stress responses via signaling pathways. The ROS can cause genomic instability through direct damage to DNA (e.g., formation of 7,8-dihydro-8-oxo-guanine (8-oxo-G) residues).60 The accumulation of DNA damage such as 8-oxo-G in mitochondrial DNA was shown to trigger autophagy in human cells.59 In addition, in human cells, the transcription factors p53 and E2F1, which regulate the expression of numerous DNA damage response genes involved in cell cycle progression and apoptosis, can activate autophagy after DNA damage, including the formation of UV-B-induced CPDs.61–64 Autophagy is positively regulated by the recognition of UV-B-induced CPDs or pyrimidine pyrimidone (6-4) photoproducts in mouse embryonic fibroblasts (MEF) cells.65 However, our results suggest that UV-B-induced mitochondrial fragmentation and autophagy occur independently of CPD accumulation, and therefore, may be regulated by factors other than CPDs. Taken together, our results indicate that the severe UV-B-sensitive phenotype of the atg5 phr1 double mutant compared with the atg5 and phr1 mutants is caused by the additive effect of deficiency in two independent machineries for UV-B protection mechanism, i.e., autophagy and CPD photorepair, in Arabidopsis.
Conclusions
Plants use sunlight as the energy source for photosynthesis and are inevitably exposed to harmful UV-B. As sessile organisms, plants are at risk of UV-B-induced damage, which reduces growth and productivity. We have previously shown that chlorophagy is activated and chloroplasts damaged by sunlight are removed to vacuoles in Arabidopsis plants grown under natural sunlight.19 The results of the current study show that irradiation with a high dose of UV-B leads to the inactivation and fragmentation of mitochondria, which are removed by mitophagy in Arabidopsis. Our results also show that the functions of CPD photorepair and autophagy play important and independent roles in resistance to UV-B. Considerable progress has been made in autophagy studies on yeast and mammals, in which a selective mitophagy process has been shown to be mediated by phosphatase, a tensin homolog (PTEN)-induced putative kinase (PINK1), and an E3 ubiquitin ligase known as Parkin.66–68 Homologs of these two proteins have been found in most of the animals tested, including flies, worms, and mice, but not in bacteria or plants.69 Therefore, plants likely have a different mitophagy mechanism to that in yeast and animals; this mechanism requires further exploration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank all members of our laboratory team, especially Drs. Atsushi Higashitani, Shusei Sato, and Masanori Izumi, for valuable discussions. We thank Dr. Kohki Yoshimoto for providing seeds of the atg5 mutant, and Dr. Maureen Hanson for providing chloroplast-targeted GFP (Pro35S:CT-GFP) and MT-GFP mitochondria-targeted GFP (Pro35S:MT-GFP) transgenic Arabidopsis seeds. This research was supported by Grants-in-Aid for Scientific Research (KAKENHI) (No. 19K12317 to M. T., and No. 15H05945, 15K21745, 17H01827, and 20H04330 to J. H.).References
- M. Tevini, W. Iwanzik and A. H. Teramura, Effects of UV-B radiation on plants during mild water stress II. Effects on growth, protein and flavonoid content, Z. Pflanzenphysiol., 1983, 110, 459–467 CrossRef CAS.
- A. B. Britt, DNA damage and repair in plants, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1996, 47, 75–100 CrossRef CAS.
- M. Takahashi, M. Teranishi, H. Ishida, J. Kawasaki, A. Takeuchi, T. Yamaya, M. Watanabe, A. Makino and J. Hidema, Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA, Plant J., 2011, 66, 433–442 CrossRef CAS.
- L. G. Landry, A. E. Stapleton, J. Lim, P. Hoffman, J. B. Hays, V. Walbot and R. L. Last, An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 328–332 CrossRef CAS.
- A. B. Britt, J. J. Chen, D. Wykoff and D. Mitchell, A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone(6-4) dimers, Science, 1993, 261, 1571–1574 CrossRef CAS.
- D. L. Mitchell and R. S. Nairn, The biology of the (6-4) photoproduct, Photochem. Photobiol., 1989, 49, 805–819 CrossRef CAS.
- J. Hidema, T. Taguchi, T. Ono, M. Teranishi, K. Yamamoto and T. Kumagai, Increase in CPD photolyase activity functions effectively to prevent growth inhibition caused by UVB radiation, Plant J., 2007, 50, 70–79 CrossRef CAS.
- C. K. Draper and J. B. Hays, Replication of chloroplast, mithochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves, Plant J., 2000, 23, 255–265 CrossRef CAS.
- K. Asada, The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1999, 50, 601–639 CrossRef CAS.
- S. A. H. Mackerness, B. R. Jordan and B. Thomas, Reactive oxygen species in the regulation of photosynthetic genes by ultraviolet-B radiation (UV-B: 280-320 nm) in green and etiolated buds of pea (Pisum sativum L.), J. Photochem. Photobiol., B, 1999, 48, 180–188 CrossRef CAS.
- S. Karpinski, C. Escobar, B. Karpinska, G. Creissen and P. M. Mullineaux, Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress, Plant Cell, 1997, 9, 627–640 CAS.
- T. Fujibe, H. Saji, K. Arakawa, N. Yabe, Y. Takeuchi and K. T. Yamamoto, A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-b irradiation, Plant Physiol., 2004, 134, 275–285 CrossRef CAS.
- A. Sancar, L. A. Lindsey-Boltz, K. Ünsal-Kaçmaz and S. Linn, Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints, Annu. Rev. Biochem., 2004, 73, 39–85 CrossRef CAS.
- J. Hidema, M. Teranishi, Y. Iwamatsu, T. Hirouchi, T. Ueda, T. Sato, B. Burr, B. M. Sutherland, K. Yamamoto and T. Kumagai, Spontaneously occurring mutations in the cyclobutane pyrimidine dimer photolyase gene cause different sensitivities to ultraviolet-B in rice, Plant J., 2005, 43, 57–67 CrossRef CAS.
- M. Brosché and Å. Strid, Molecular events following perception of ultraviolet-B radiation by plants, Physiol. Plant., 2003, 117, 1–10 CrossRef.
- S. A. H. Mackerness, C. F. John, B. Jordan and B. Thomas, Early signaling components in ultraviolet-B responses: Distinct roles for different reactive oxygen species and nitric oxide, FEBS Lett., 2001, 489, 237–242 CrossRef CAS.
- H. Frohnmeyer and D. Staiger, Ultraviolet-B radiation-mediated responses in plants. balancing damage and protection, Plant Physiol., 2003, 133, 1420–1428 CrossRef CAS.
- N. Li, M. Teranish, H. Yamaguchi, T. Matsushita, M. K. Watahiki, T. Tsuge, S. S. Li and J. Hidema, UV-B-induced CPD photolyase gene expression is regulated by UVR8-dependent and-independent pathways in Arabidopsis, Plant Cell Physiol., 2015, 56, 2014–2023 CrossRef CAS.
- M. Izumi, H. Ishida, S. Nakamura and J. Hidema, Entire photodamaged chloroplasts are transported to the central vacuole by autophagy, Plant Cell, 2017, 29, 377–394 CrossRef CAS.
- D. R. Green, L. Galluzzi and G. Kroemer, Mitochondria and the autophagy-inflammation-cell death axis in organismal aging, Science, 2011, 333, 1109–1112 CrossRef CAS.
- R. J. Youle and D. P. Narendra, Mechanisms of mitophagy, Nat. Rev. Mol. Cell Biol., 2011, 12, 9–14 CrossRef CAS.
- T. Kanki, K. Furukawa and S. Yamashita, Mitophagy in yeast: Molecular mechanisms and physiological role, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853, 2756–2765 CrossRef CAS.
- A. T. Vessoni, E. C. Filippi-Chiela, C. F. M. Menck and G. Lenz, Autophagy and genomic integrity, Cell Death Differ., 2013, 20, 1444–1454 CrossRef CAS.
- D. B. Zorov, I. Vorobjev, V. Popkov, A. V. Babenko, L. D. Zorova, I. B. Pevzner, D. N. Silachev, S. D. Zorov, N. V. Andrianova and E. Y. Plotnikov, Lessons from the discovery of mitochondrial fragmentation (fission): A review and update, Cells, 2019, 8, 175 CrossRef CAS.
- A. R. Thompson, J. H. Doelling, A. Suttangkakul and R. D. Vierstra, Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways, Plant Physiol., 2005, 138, 2097–2110 CrossRef CAS.
- D. Hofius, T. Schultz-Larsen, J. Joensen, D. I. Tsitsigiannis, N. H. T. Petersen, O. Mattsson, L. B. Jørgensen, J. D. G. Jones, J. Mundy and M. Petersen, Autophagic components contribute to hypersensitive cell death in Arabidopsis, Cell, 2009, 137, 773–783 CrossRef CAS.
- R. H. Köhler, W. R. Zipfel, W. W. Webb and M. R. Hanson, The green fluorescent protein as a marker to visualize plant mitochondria in vivo, Plant J., 1997, 11, 613–621 CrossRef.
- Y. Ono, S. Wada, M. Izumi, A. Makino and H. Ishida, Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana, Plant, Cell Environ., 2013, 36, 1147–1159 CrossRef CAS.
- T. Hirota, M. Izumi, S. Wada, A. Makino and H. Ishida, Vacuolar protein degradation via autophagy provides substractes to amino acid catabolic pathways as an adaptive response to sugar starvation in Arabidopsis thaliana, Plant Cell Physiol., 2018, 59, 1363–1376 CAS.
- J. Hidema and T. Kumagai, UVB-induced cyclobutyl pyrimidine dimer and photorepair with progress of growth and leaf age in rice, J. Photochem. Photobiol., B, 1998, 43, 121–127 CrossRef CAS.
- S. E. Freeman, H. Hacham, R. W. Gange, D. J. Maytum, J. C. Sutherland and B. M. Sutherland, Wavelength dependence of pyrimidine dimer formation in DNA of human skin irradiated in situ with ultraviolet light, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 5605–5609 CrossRef CAS.
- K. E. Swearingen, W. P. Loomis, B. Kehimkar, B. T. Cookson and N. J. Dovichi, Quantification of green fluorescent protein in cellular supernatant by capillary electrophoresis with laser-induced fluorescence detection for measurement of cell death, Talanta, 2010, 81, 948–953 CrossRef CAS.
- K. Okatsu, T. Oka, M. Iguchi, K. Imamura, H. Kosako, N. Tani, M. Kimura, E. Go, F. Koyano, M. Funayama, K. Shiba-Fukushima, S. Sato, H. Shimizu, Y. Fukunaga, H. Taniguchi, M. Komatsu, N. Hattori, K. Mihara, K. Tanaka and N. Matsuda, PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria, Nat. Commun., 2012, 3, 1016 CrossRef.
- S. R. Yoshii, C. Kishi, N. Ishihara and N. Mizushima, Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane, J. Biol. Chem., 2011, 286, 19630–19640 CrossRef CAS.
- M. Izumi and S. Nakamura, Vacuolar digestion of entire damaged chloroplasts in Arabidopsis thaliana is accomplished by chlorophagy, Autophagy, 2017, 13, 1239–1240 CrossRef CAS.
- H. Ishida, K. Yoshimoto, M. Izumi, D. Reisen, Y. Yano, A. Makino, Y. Ohsumi, M. R. Hanson and T. Mae, Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process, Plant Physiol., 2008, 148, 142–155 CrossRef CAS.
- G. Zhong, Q. Zhu, Y. Li, Y. Liu and H. Wang, One for all: a novel robust system for co-expression of multiple chimeric fluorescent fusion proteins in plants, Front. Plant Sci., 2017, 8, 1–8 Search PubMed.
- V. V. Ryabovol and F. V. Minibayeva, Molecular mechanisms of autophagy in plants: Role of ATG8 proteins in formation and functioning of autophagosomes, Biochemistry, 2016, 81, 348–363 CAS.
- H. Iwashita, S. Torii, N. Nagahoro, M. Ishiyama, K. Shioji, K. Sasamoto, S. Shimizu and K. Okuma, Live cell imaging of mitochondria autophagy with a novel fluorescent small molecule, ACS Chem. Biol., 2017, 12, 2546–2551 CrossRef CAS.
- H. Iwashita, H. Tajima-Sakurai, N. Nagahoro, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu and Y. Ueno, Small fluorescent molecules for monitoring autophagic flux, FEBS Lett., 2018, 592, 559–567 CrossRef CAS.
- L. Rizzini, J. J. Favory, C. Cloix, D. Faggionato, A. O'Hara, E. Kaiserli, R. Baumeister, E. Schäfer, F. Nagy, G. I. Jenkins and R. Ulm, Perception of UV-B by the Arabidopsis UVR8 protein, Science, 2011, 332, 103–106 CrossRef CAS.
- J. M. Christie, A. S. Arvai, K. J. Baxter, M. Heilmann, A. J. Pratt, A. O'Hara, S. M. Kelly, M. Hothorn, B. O. Smith, K. Hitomi, G. I. Jenkins and E. D. Getzoff, Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges, Science, 2012, 335, 1492–1496 CrossRef CAS.
- J. J. Favory, A. Stec, H. Gruber, L. Rizzini, A. Oravecz, M. Funk, A. Albert, C. Cloix, G. I. Jenkins, E. J. Oakeley, H. K. Seidlitz, F. Nagy and R. Ulm, Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis, EMBO J., 2009, 28, 591–601 CrossRef CAS.
- G. Kaiser, O. Kleiner, C. Beisswenger and A. Batschauer, Increased DNA repair in Arabidopsis plants overexpressing CPD photolyase, Planta, 2009, 230, 505–515 CrossRef CAS.
- S. Takahashi, M. Teranishi, M. Izumi, M. Takahashi, F. Takahashi and J. Hidema, Transport of rice cyclobutane pyrimidine dimer photolyase into mitochondria relies on a targeting sequence located in its C-terminal internal region, Plant J., 2014, 79, 951–963 CrossRef CAS.
- E. Y. Plotnikov, A. K. Vasileva, A. A. Arkhangelskaya, I. B. Pevzner, V. P. Skulachev and D. B. Zorov, Interrelations of mitochondrial fragmentation and cell death under ischemia/reoxygenation and UV-irradiation: Protective effects of SkQ1, lithium ions and insulin, FEBS Lett., 2008, 582, 3117–3124 CrossRef CAS.
- M. A. Birch-Machin and H. Swalwell, How mitochondria record the effects of UV exposure and oxidative stress using human skin as a model tissue, Mutagenesis, 2010, 25, 101–107 CrossRef CAS.
- R. Jugé, J. Breugnot, C. Da Silva, S. Bordes, B. Closs and A. Aouacheria, Quantification and characterization of UVB-induced mitochondrial fragmentation in normal primary human keratinocytes, Sci. Rep., 2016, 6, 1–9 CrossRef.
- G. Kroemer and J. C. Reed, Mitochondrial control of cell death, Nat. Med., 2000, 6, 513–519 CrossRef CAS.
- P. K. Kamat, A. Kalani, P. Kyles, S. C. Tyagi and N. Tyagi, Autophagy of mitochondria: a promising therapeutic target for neurodegenerative disease, Cell Biochem. Biophys., 2014, 70, 707–719 CrossRef CAS.
- D.-F. Dai, Y. Chiao, D. J. Marcinek, H. H. Szeto and P. S. Rabinovitch, Mitochondrial oxidative stress in aging and healthspan, Longev. Healthspan, 2014, 3, 6 CrossRef.
- L. M. Farmer, M. A. Rinaldi, P. G. Young, C. H. Danan, S. E. Burkhart and B. Bartel, Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation, Plant Cell, 2013, 25, 4085–4100 CrossRef CAS.
- J. Kim, H. Lee, H. N. Lee, S. H. Kim, K. D. Shin and T. Chung, Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth, Plant Cell, 2013, 25, 4956–4966 CrossRef CAS.
- M. Shibata, K. Oikawa, K. Yoshimoto, M. Kondo, S. Mano, K. Yamada, M. Hayashi, W. Sakamoto, Y. Ohsumi and M. Nishimura, Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis, Plant Cell, 2013, 25, 4967–4983 CrossRef CAS.
- K. Yoshimoto, M. Shibata, M. Kondo, K. Oikawa, M. Sato, K. Toyooka, K. Shirasu, M. Nishimura and Y. Ohsumi, Organ-specific quality control of plant peroxisomes is mediated by autophagy, J. Cell Sci., 2014, 127, 1161–1168 CrossRef CAS.
- Y. Liu, J. S. Burgos, Y. Deng, R. Srivastava, S. H. Howell and D. C. Bassham, Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis, Plant Cell, 2012, 24, 4635–4651 CrossRef CAS.
- H. F. Aqbi, M. Wallace, S. Sappal, K. K. Payne and M. H. Manjili, IFN-γ orchestrates tumor elimination, tumor dormancy, tumor escape, and progression, J. Leukocyte Biol., 2018, 103, 1219–1223 CrossRef CAS.
- C. W. Yun and S. H. Lee, The roles of autophagy in cancer, Int. J. Mol. Sci., 2018, 19, 3466 CrossRef.
- A. Sample and Y. Y. He, Autophagy in UV damage response, Photochem. Photobiol., 2017, 93, 943–955 CrossRef CAS.
- B. van Loon, E. Markkanen and U. Hübscher, Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine, DNA Repair, 2010, 9, 604–616 CrossRef CAS.
- L. F. Z. Batista, B. Kaina, R. Meneghini and C. F. M. Menck, How DNA lesions are turned into powerful killing structures: Insights from UV-induced apoptosis, Mutat Res., 2009, 681, 197–208 CAS.
- K. H. Vousden and C. Prives, Blinded by the Light: The growing complexity of p53, Cell, 2009, 137, 413–431 CrossRef CAS.
- E. Zaika, J. Wei, D. Yin, C. Andl, U. Moll, W. El-Rifai and A. I. Zaika, p73 protein regulates DNA damage repair, FASEB J., 2011, 25, 4406–4414 CrossRef CAS.
- Z. Guo, S. Kozlov, M. F. Lavin, M. D. Person and T. T. Paull, ATM activation by oxidative stress, Science, 2010, 330, 517–521 CrossRef CAS.
- L. Qiang, B. Zhao, P. Shah, A. Sample, S. Yang and Y. Y. He, Autophagy positively regulates DNA damage recognition by nucleotide excision repair, Autophagy, 2016, 12, 357–368 CrossRef CAS.
- N. Matsuda, S. Sato, K. Shiba, K. Okatsu, K. Saisho, C. A. Gautier, Y. S. Sou, S. Saiki, S. Kawajiri and F. Sato, PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy, J. Cell Biol., 2010, 189, 211–221 CrossRef CAS.
- D. Narendra, J. E. Walker and R. Youle, Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism, Cold Spring Harbor Perspect. Biol., 2012, 4, 1338 Search PubMed.
- C. Vives-Bauza, C. Zhou, Y. Huang, M. Cui, R. L. A. de Vries, J. Kim, J. May, M. A. Tocilescu, W. Liu, H. S. Ko, J. Magrané, D. J. Moore, V. L. Dawson, R. Grailhe, T. M. Dawson, C. Li, K. Tieu and S. Przedborski, PINK1-dependent recruitment of Parkin to mitochondria in mitophagy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 378–383 CrossRef CAS.
- M. Broda, A. H. Millar and O. Van Aken, Mitophagy: A mechanism for plant growth and survival, Trends Plant Sci., 2018, 23, 434–450 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9pp00479c |
| This journal is © The Royal Society of Chemistry and Owner Societies 2020 |