Open Access Article
Open Access ArticleRevisiting cyanobacterial state transitions
Pablo I.
Calzadilla
and
Diana
Kirilovsky
 *
*
Université Paris-Saclay, CNRS, CEA, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif sur Yvette, France. E-mail: diana.kirilovsky@cea.fr
First published on 12th March 2020
Abstract
Photosynthetic organisms are exposed to a fluctuating environment in which light intensity and quality change continuously. Specific illumination of either photosystem (PSI or PSII) creates an energy imbalance, leading to the reduction or oxidation of the intersystem electron transport chain. This redox imbalance could trigger the formation of dangerous reactive oxygen species. Cyanobacteria, like plants and algae, have developed a mechanism to re-balance this preferential excitation of either reaction center, called state transitions. State transitions are triggered by changes in the redox state of the membrane-soluble plastoquinone (PQ) pool. In plants and green algae, these changes in redox potential are sensed by Cytochrome b6f, which interacts with a specific kinase that triggers the movement of the main PSII antenna (the light-harvesting complex II). By contrast, although cyanobacterial state transitions have been studied extensively, there is still no agreement about the molecular mechanism, the PQ redox state sensor and the signaling pathways involved. In this review, we aimed to critically evaluate the results published on cyanobacterial state transitions, and discuss the “new” and “old” models in the subject. The phycobilisome and membrane contributions to this physiological process were addressed and the current hypotheses regarding its signaling transduction pathway were discussed.
1. Introduction
Light is essential for photosynthetic organisms that harvest solar energy and convert it into chemical energy (ATP) and reducing power (NADPH). These molecules are used in the assimilation of CO2 and synthesis of organic carbon molecules, in a process called photosynthesis, in which molecular oxygen is produced as a sub-product. However, light can become dangerous in certain cases. For example, when the light energy reaching the photosystems exceeds the energy consumption by multiple cellular processes, the entire photosynthetic chain is reduced. Under these conditions, secondary reactions are induced, and the accumulation of high energy reactive intermediates is promoted, subsequently leading to the production of singlet oxygen and oxygen radicals (ROS). These molecules cause severe cell damage, especially at the level of the two photosystems: photosystem I (PSI) and photosystem II (PSII). In addition, changes in light quality can imbalance the photosystems’ activity, decreasing their photosynthetic efficiency, and/or altering the redox state of the electron transport chain, which again leads to ROS production.In order to efficiently use the incoming light and to avoid damage, sensing and adapting to environmental changes becomes crucial. Thus, photo-protective mechanisms were evolutionarily developed by photosynthetic organisms. These mechanisms induce a rapid reorganization of the photosynthetic apparatus, changing the interaction between the antennae and reaction centers, and adjusting light absorption and utilization. Particularly in cyanobacteria, two photo-protective mechanisms, the Orange Carotenoid Protein (OCP)-related Non-Photochemical-Quenching (NPQ) (see reviews1–5) and state transitions (see reviews6–8), regulate the transfer of energy arriving at the photochemical reaction centers, sensing the intensity and the quality of light. While OCP-related NPQ simultaneously decreases the energy arriving at both the photosystems under high light conditions,9,10 state transitions decrease (or increase) the activity of only one of the two photosystems under fluctuating light quality.
The state transitions mechanism was first described fifty years ago in green and red algae.11,12 Two states were characterized, one induced by light preferentially absorbed by PSII (State II), and the other triggered by light preferentially absorbed by PSI (State I). Specific illumination of PSI or PSII creates an imbalance in the electron transport chain, rendering the plastoquinone (PQ) pool more oxidized or more reduced. This effect is rebalanced by state transitions. Each state differs in its fluorescence characteristics, i.e., in State II, the PSII to PSI fluorescence ratio is low, while in State I this ratio is higher.
During the last few decades, state transitions have been well characterized in plants and green algae (see review13). In these organisms, the differences in the fluorescence of PSI and PSII are related to a partial migration of the Light-Harvesting Complex II (LHCII), which is the main membrane chlorophyll-carotenoid PSII antenna. This migration is triggered by redox changes in the PQ pool, in a process involving Cytochrome b6f (Cyt b6f) and phosphorylation of LHCII (see reviews13–15). When the PQ pool gets reduced, due to preferential illumination of PSII, a specific kinase (STN7/STT716,17) is activated by the binding of plastoquinol (PQH2) to the Cyt b6f Qo site.18–20 This binding leads to conformational changes in the Cyt b6f, particularly in the Rieske protein,20–23 and in specific amino acids on the peripheral stromal structure of subunit IV which are in contact with the kinase.24 The activated kinase phosphorylates the mobile trimers of LHCII, resulting in their detachment from PSII and attachment to PSI, triggering the transition from State I to State II.25,26 Conversely, when the PQ pool gets oxidized, in darkness or due to preferential absorption of light by PSI, the kinase is deactivated, and a specific phosphatase (TAP38/PPH127,28) dephosphorylates LHCII, which migrates back to PSII, leading to State I transition. It must be noted that in plants and green algae, Cyt b6f is the redox sensor of the PQ pool, and in its absence, state transitions are inhibited.29
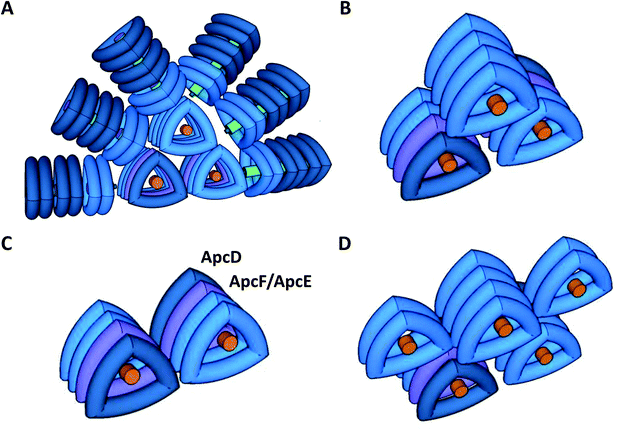
Cyanobacteria do not contain LHCII; instead, the PSII antenna is a huge membrane-extrinsic complex, the phycobilisome (PBS), which is bound to the stromal surface of the thylakoid membranes. The PBS is constituted of water soluble phycobiliproteins (which bind blue, violet and red bilins) and linker proteins. In most cyanobacteria, these proteins are organized as rods radiating from a central core (see reviews30–34) (Fig. 1). Each rod is formed by hexamers of phycocyanin (PC, λmax = 620 nm) and, in some cases, phycoerythrin (PE, λmax = 577 nm) and phycoerythrocyanin (PEC, λmax = 625 nm).31,35 When these last two phycobiliproteins are present, PC is localized in the core proximal hexamers, and PEC and PE form the distal hexamers.31
The core structure varies between the different cyanobacterial strains, and can be formed by two (Synechococcus elongatus PCC 7942, hereafter S. elongatus), three (Synechocystis PCC 6803, hereafter Synechocystis), or five cylinders (Nostoc PCC 7120, Thermosynechococcus elongatus) of allophycocyanin (APC). Each cylinder is made of 4 APC trimers, composed of αAPC-βAPC heterodimers, which absorb at 650 nm and emit at 660 nm. Within each basal cylinder, one αAPC subunit in an external trimer is replaced by an αAPC-like subunit, called ApcD, and one βAPC in an internal trimer is replaced by a βAPC-like subunit, called ApcF. Adjacent to ApcF, another APC dimer is formed by a βAPC subunit and the bilin-linker region of an αAPC-like domain, called ApcE (or LCM) (Fig. 1). ApcD and ApcE emit at 680 nm, and are terminal emitters of the PBS, which transfer the energy absorbed by this antenna to the photosystems.
The PBS architecture favors the funneling of absorbed photons from the peripheral rods to the central core, and then to the photosystems (ref. 36 and 37, see review38). This energy transfer model is based on Förster Resonance Energy Transfer (FRET), where the excitation energy is transmitted from the donor to the acceptor molecule by resonance of excited states.39 The down-hill energy transfer is possible due to the overlapping of the emission spectra of the different chromophores (ref. 36 and 37 see review38). Mutations in the PBS terminal emitters affect the energy transfer to one or both photosystems in different cyanobacterial strains (see discussion in following sections).40–44
The entire scientific community agrees that in cyanobacteria, like in plants and green algae, state transitions are triggered by redox changes in the PQ pool.45,46 However, no agreement can be found in literature about the molecular mechanism, the sensor of the PQ redox state, or the signaling pathways involved. The present review critically evaluates the different propositions regarding the aforementioned knowledge gaps, and describes the most recent working models of cyanobacterial state transitions, based on the results obtained in the last few years.
2. Following photosystem fluorescence changes during state transitions
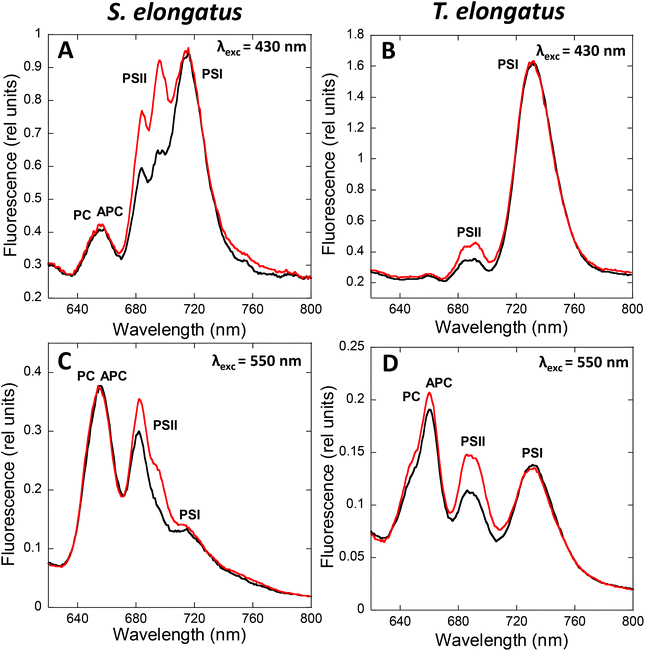
State transitions can be studied by fluorescence measurements at room temperature or at 77 K. At room temperature, changes in only PSII fluorescence can be measured, while at 77 K, both PSII and PSI fluorescence can be resolved. The fluorescence emission spectra of cyanobacterial cells depend on the wavelength of the excitation light (Fig. 2).When cells are excited at 430 nm, which is preferentially absorbed by the Chl antennae, fluorescence emitted by Chl is principally observed. Thus, under these conditions, the changes observed are mainly related to processes occurring in the membrane Chl antennae and reaction centers. When cells are excited by light absorbed mainly by the PBS (550 to 620 nm, depending on the presence or absence of PE), fluorescence emitted by phycobiliproteins, phycobilisome final emitters, and Chl is detected. In this case, the fluorescence changes observed are related to processes occurring in the PBS and the reaction center, and to the energy transfer from PBS to the photosystems. Independently of the excitation light, the peak at 695 nm is related to CP47 and Reaction Center II fluorescence, while the peak at 715–740 nm is related to PSI.47,48 Emissions at 575, 650, and 660 nm, related to PE, PC, and APC, respectively, contribute much more to the 77 K fluorescence spectrum when PBS are excited.49,50 In addition, with PBS excitation, the peak at 685 nm is mainly related to the PBS terminal emitter, with a contribution of CP43 emission.48 In contrast, when Chl is excited, the 685 nm peak is mainly related to CP43 emission.
State I is characterized by a high PSII (685 and 695 nm) to PSI (715–720 nm) fluorescence ratio in the 77 K spectra. Conversely, State II is characterized by a low PSII to PSI fluorescence ratio. These differences are visible when the PBS or the Chl are excited (Fig. 2), suggesting that changes in both PBS and Chl-containing-complexes are involved in state transitions.51,52 It has been observed that PSII-related fluorescence is lower in State II compared to that in State I (see Tables 1 and 2). However, only one third of the realized studies showed that PSI-related fluorescence is lower in State I compared to that in State II (see Tables 1 and 2, and discussion about spillover below).
| Article | Species | Changes in PSII fluorescence | Changes in PSI fluorescence | Internal standard used |
|---|---|---|---|---|
| Calzadilla et al. (2019)52 | S. elongatus and Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at 800 nm |
| McConnell et al. (2002)51 | Synechococcus 7002 and Synechocystis PCC 6803 | Increase in State I | No change | No |
| Zlenko et al. (2019)44 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at 650 nm |
| Zhao et al. (2014)94 | Synechocystis PCC 6803 | Increase in State I | Decrease in State I | No – normalization at 707 nm |
| Ogawa et al. (2013)74 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at PSI peak |
| Kaňa et al. (2012)105 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at PSI peak |
| Kondo et al. (2009)107 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at PSI peak |
| Yang et al. 2009155 | Synechocystis PCC 6803 | Increase in State I | Decrease in State I | No – normalization at 709 nm |
| Fujimori et al. (2005)156 | Synechocystis PCC 6803 | No change | Increase in State I | No – normalization at 695 nm |
| El-Bissati et al. (2000)65 | Synechocystis PCC 6803 | Increase in State I | No change | Yes – fluorescein |
| Emlyn-Jones et al. (1999)78 | Synechocystis PCC 6803 | Increase in State I | Slight increase in State I | No – normalization at phycocyanin fluorescence peak |
| Olive et al. (1997)61 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at 665 nm |
| Joshua and Mullineaux, (2005)92 | S. elongatus | Increase in State I | No change | No – normalization at 654 nm |
| Joshua and Mullineaux, (2004)89 | S. elongatus | Increase in State I | No change | No –normalization at 654 nm |
| Ranjbar Choubeh et al. (2018)106 | S. elongatus | Increase in State I | No change | Yes Rhodamine B |
| Aspinwall et al. (2004)99 | S. elongatus | Increase in State I | No change | No |
| Deng et al. (2012)93 | Synechococcus PCC 7002 | Increase in State I | Slight decrease in State I | No |
| Huang et al. (2003)73 | Synechococcus PCC 7002 | Increase in State I | Slight decrease in State I | No – normalization at 640 nm |
| Schluchter et al. (1996)98 | Synechococcus PCC 7002 | Increase in State I | No change | No |
| Bruce et al. (1989)81 | Synechococcus PCC 7002 | Increase in State I | Decrease in State I | No – normalization at 660 nm |
| Li et al. (2006)68 | Spirulina platensis | Increase in State I | Decrease in State I | No – normalization at 710 nm |
| Li et al. (2004)83 | Spirulina platensis | Increase in State I | Decrease in State I | No – normalization at 660 nm |
| Dong and Zhao, (2008)85 | Anabaena PCC 7120 | Increase in State I | Decrease in State I | No |
| Bruce and Biggins, (1985)157 | Anacystis nidulans | Increase in State I | No change | No – normalization at 650 nm |
| Mullineaux and Allen, (1990)46 | Synechococcus PCC 6301 | Increase in State I | No change | No – normalization at 654 nm |
| Olive et al. (1986)110 | Synechocystis PCC 6714 | Increase in State I | Decrease in State I | No |
| Article | Species | Changes in PSII fluorescence | Changes in PSI fluorescence | Internal standard used | Conclusions about spill-over |
|---|---|---|---|---|---|
| Calzadilla et al. (2019)52 | S. elongatus and Synechocystis PCC 6803 | Increase in State I | No change | Yes – Rhodamine B | No involvement |
| Ranjbar Choubeh et al. (2018)106 | S. elongatus and Synechocystis PCC 6803 | Increase in State I | No change | Yes – Rhodamine B | No involvement |
| Aspinwall et al. (2004)99 | S. elongatus | Increase in State I | No change | No | Not addressed |
| Ogawa et al. (2013)74 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at PSI peak | Not addressed |
| Stadnichuk et al. (2009)80 | Synechocystis PCC 6803 | Increase in State I | No change | Yes – Rhodamine B | No involvement |
| Kondo et al. (2009)107 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at PSI peak | Not addressed |
| Olive et al. (1997)61 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at 665 nm | Spillover |
| Emlyn-Jones et al. (1999)78 | Synechocystis PCC 6803 | Increase in State I | No change | No – normalization at PSI peak | Minor role, if any, in cyanobacterial state transitions |
| McConnel et al. (2002)51 | Synechococcus 7002 and Synechocystis PCC 6803 | Increase in State I | No change | No | Spillover |
| Deng et al. (2012)93 | Synechococcus PCC 7002 | No change | Increase in State I | No | Not addressed |
| Mullineaux and Allen, (1990)46 | Synechococcus 6301 | Increase in State I | No change | No – normalization at 654 nm | Not addressed |
| Salehian and Bruce, (1992)158 | Synechococcus PCC 6301 | Increase in State I | Slight increase in State I | No | Spillover |
| Li et al. (2006)68 | Spirulina platensis | Increase in State I | Decrease in State I | No – normalization at 708 nm | Spillover |
| Li et al. (2004)83 | Spirulina platensis | Increase in State I | Decrease in State I | No | Spillover |
| Bruce et al. (1989)81 | Synechococcus PCC 7002 | Increase in State I | Decrease in State I | No – normalization at 660 nm | Spillover |
State transitions kinetics can be studied using modulated fluorometers, which record the fluorescence resulting from excitation by non-actinic pulses (non-induced charge separation),53 and measuring changes in fluorescence yield. The most frequently used fluorometers are PAM101–103, dual-PAM-100, and multicolor PAM. The only fluorometer used in state transition studies, i.e., PAM101–103, uses a detecting light of 650 nm, which is absorbed by both PBS and Chl. Thus, changes in fluorescence yield could be a result of a change in the PBS and/or the antenna-Chl emission, or a change in the excitation energy transfer from PBS to PSII. In multicolor PAM, several detecting pulses with different wavelengths (going from blue to red) can be used. Since fluorescence emission spectra of cyanobacterial cells depend markedly on the excitation wavelength, the contribution of PBS and Chl fluorescence will be different depending on the wavelength of the detecting pulses (see ref. 54 for a detailed explanation). Changes at the PBS level will mainly be observed when orange detecting light (590–630 nm) is used, while Chl emission changes will be more prominent when using blue detecting light. In dual-PAM, red, which is the most frequently used, and blue detecting pulses are also available.
In PAM fluorometers, three levels of PSII fluorescence can be measured (Fig. 3). The minimal fluorescence level, F0, is determined using only the detecting modulated light in dark-adapted cells. No measurable charge separation is induced under these conditions, and all PSII are open. Strong pulses are applied to close all PSII centers and measure the maximal fluorescence level (Fm – in darkness and F′m – in light) (Fig. 3). Under continuous illumination, the steady-state fluorescence level (Fs) is measured, which depends on the relative PSII and PSI activity influencing the PQ pool redox state. The contributions of PBS and Chl fluorescence in F0 and Fm are different.54
When State II adapted cells are illuminated with far-red light (absorbed by PSI) or low intensities of blue light (principally absorbed by Chl), an increase in all levels of fluorescence yield is observed, which is related to State II to State I transition (Fig. 3). The Chl antenna of PSI is almost three times larger than that of PSII, i.e., 100 and 35 Chl molecules, respectively.55,56 Thus, illumination with light principally absorbed by Chl results in a higher activity of PSI compared to that of PSII, and oxidation of the PQ pool occurs.
When State I adapted cells are illuminated with orange or green light (principally absorbed by phycobiliproteins), a decrease in all levels of fluorescence related to State I to State II transition is induced (Fig. 3). Since PBS is the principal antenna of PSII,57 this illumination increases PSII activity, resulting in reduction of the PQ pool. However, PSI is still active under orange and green light, because PBS can transfer energy directly to both photosystems.58,59 Moreover, PSI can also receive energy from PSII (known as spillover), when the excess energy arriving at PSII is transferred to PSI via the Chl antennae.60–66 The percentage of energy arriving at one or the other photosystem depends on the cyanobacterial strain and the environmental conditions.59,67
It is worth mentioning that upon illumination of dark-adapted cells with any kind of light (including orange light), PSI is activated and the PQ pool becomes more oxidized. Consequently, as mentioned previously, an increase in fluorescence yield related to State I transition is observed. However, blue light is more effective than orange light in inducing this transition.46 Thus, both dark to light and orange to blue transitions induce an increase in fluorescence related to State I. Although majority of published results suggest that the mechanisms involved in these transitions are identical, one publication proposed that while changes in light quality induce only PBS movement, dark-light transition involves changes in concentration of PSI trimers and photosystem movements as well.68
In the dark, in contrast to plants and green algae, cyanobacterial cells are in State II.69,70 In cyanobacteria, both photosynthesis and respiration take place in the thylakoid membranes, and PQ, Cyt b6f and plastocyanin (or cytochrome c6) are common to both electron transport chains (see review71). Homologs of mitochondrial Complex I (NDH-1) and Complex II (succinate dehydrogenase, SDH) reduce the PQ pool, and terminal oxidases oxidize it via the respiratory chain.71 Since the PQ pool reduction is generally faster than its oxidation via respiration, the PQ pool is reduced in the dark.69,70 However, due to differences in this redox state balance between cyanobacterial strains, the level of PQ reduction state varies, leading to different levels of State II fluorescence in the dark.52,72 Mutants in the respiratory process also affect state transitions. For instance, NDH-1 and SDH mutants are in State I in the dark,73–76 while terminal oxidase mutants have a more pronounced dark State II.75
The different “levels” of State II in the dark lead to different amplitudes of fluorescence changes during dark to light transitions. In Synechocystis,51,52,61,77–80Synechococcus PCC 700240,51,81 and Spirulina platensis,68,82,83 these changes are smaller than in S. elongatus or T. elongatus (ref. 52 and Fig. 3). Interestingly, both S. elongatus and T. elongatus lack the photo-protective mechanism OCP-related NPQ.84 Thus, studying cyanobacterial state transitions in S. elongatus could lead to a better understanding of the PBS and membrane contributions to the process, which are further discussed in the following sections.
3. PBS contribution to state transitions
PBS contribution in cyanobacterial state transitions has been largely studied due to the role of LHCII in plant and green alga state transitions. First, it was shown that PBS mutations affect this process in different cyanobacterial strains.40,41,51,85 Then, FRAP experiments showing that PBS are capable of moving along the surface of the thylakoid membranes,86–88 and that the same chemicals that inhibit PBS movement inhibit state transitions,68,83,89 seemed to confirm that PBS movement from one photosystem to the other could be involved in state transitions. However, more recent experiments and results showed that the recovery of fluorescence after PBS bleaching is not only due to PBS diffusion but also to internal processes in this macromolecular complex.90 The role of PBS in cyanobacterial state transitions remains a matter of discussion.3.1 PBS mutants impaired in state transitions
Mutations in ApcD, ApcF40,41,51,85 and a particular domain of ApcE44 affect state transitions in several cyanobacterial strains (Table 3). Dong et al. (2009)40 suggested that in the ΔApcD Synechococcus PCC 7002 mutant, the inhibition of state transitions was due to a permanent decrease in energy transfer from PBS to PSI. However, it has recently been shown that a lack of ApcD also decreases the energy transfer from PBS to PSI in S. elongatus, without impairing state transitions.43 Furthermore, in Synechocystis, the lack of ApcD does not decrease energy transfer from PBS to any of the photosystems, but it inhibits state transitions.41,43 In the ApcE-C190S and the PB-loop ApcE Synechocystis mutants, the energy transfer from PBS to the photosystems is significantly affected.43,44 However, state transitions in the former are not impaired,91 while they appear to be decreased in the latter.44| Article | Species | Mutation | State transitions λexc PBS | State transitions λexc Chl | Energy transfer PSII | Energy transfer PSI |
|---|---|---|---|---|---|---|
| Calzadilla et al. (2019)43 | S. elongatus and Synechocystis PCC 6803 | ApcD, ApcF | Inhibited in Synechocystis ΔApcD, impaired in Synechocystis ΔApcF | Yes | Affected in Synechocystis ΔApcF | Affected in S. elongatus ΔApcD and Synechocystis ΔApcF |
| Ashby and Mullineaux, (1999)41 | Synechocystis PCC 6803 | ApcD, ApcF | Inhibited in Synechocystis ΔApcD, impaired in Synechocystis ΔApcF | — | Affected in Synechocystis ΔApcF | Affected in Synechocystis ΔApcF |
| Zlenko et al. (2019)44 | Synechocystis PCC 6803 | PB-loop in ApcE | Impaired | — | Affected | Not affected |
| Deng et al. (2012)93 | Synechococcus PCC 7002 | ApcD | Inhibited | — | — | — |
| Dong et al. (2009)40 | Synechococcus PCC 7002 | ApcD | Inhibited | — | — | Affected |
| McConnel et al. (2002)51 | Synechococcus PCC 7002 | ApcD | Inhibited | Yes | — | — |
| Dong and Zhao (2008)85 | Anabaena PCC 7120 | ApcD | Inhibited | — | — | Affected |
It has been proposed that the impairment of state transitions in the ΔApcD Synechocystis mutant is due to a permanent binding of PBS to PSII, which increases the PSII antenna even in darkness and PSII illumination (when the PQ pool is reduced).41 This hypothesis can explain the relatively high fluorescence observed in the dark-adapted ΔApcD Synechocystis cells. In addition, a lack of state transitions in the ΔApcD mutants was observed in Synechocystis, Synechococcus PCC 7002 and Anabaena PCC 7120,40,41,51,85 but not in S. elongatus43 (Table 3). These results could be due to the different roles of ApcD in the PBS–photosystem interaction in different strains, which could also explain the different roles of ApcD in energy transfer to photosystems.40–43 As mentioned previously, absence of ApcD decreases energy transfer from PBS to PSI in S. elongatus and Synechococcus PCC 7002, but not in Synechocystis, while the ΔApcF mutation affects energy transfer to PSII and PSI only in Synechocystis.40–43
A mutation in the rpaC gene also affects state transitions in Synechocystis, but not in S. elongatus.78,92 This gene, which is strongly conserved among different cyanobacteria, codes for a 9 kDa protein, that has been proposed to be localized on the cytoplasmatic side of the thylakoid membrane.7 The rpaC Synechocystis mutant was suggested to present a stronger PBS–PSII interaction, like the ΔApcD mutant, impairing state transitions.7 In contrast, a large decrease in the RpaC concentration did not affect this process in S. elongatus, which might again be due to different PBS–photosystem interaction mechanisms in different strains.92 A complete segregation of this mutation was not viable in S. elongatus, suggesting an essential role of this gene in this strain.
The different effects of the ApcD and RpaC mutants in S. elongatus and Synechocystis could also be explained by a different PBS contribution to state transitions in these strains. If this is the case, membrane processes could have a more important role in S. elongatus than in Synechocystis. It is worth mentioning that in the ΔRpaC Synechocystis, ΔApcD Synechocystis, Synechococcus 7002 and Anabaena PCC 7120 mutants, only the fluorescence changes related to PBS were inhibited40,41,43,51,78,85,92,93 (Table 3). When Chl were preferentially excited at 77 K, the small fluorescence changes between State I and State II conditions were still observable.43,51,78,92 Thus, although the results related to PBS mutants are in favor of PBS participation in cyanobacterial state transitions, the relevance of this contribution is still not clear, and it might depend on the cyanobacterial strain studied and the prevailing growth conditions.
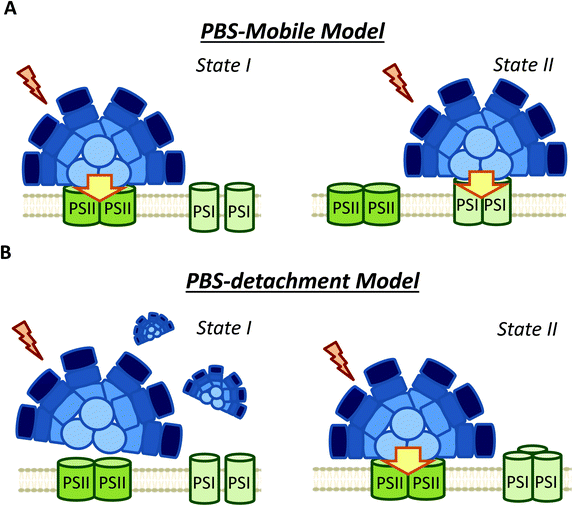
3.2 PBS long distance movement versus PBS detachment and small changes in PBS–photosystems interaction
The possible diffusion of PBS along the thylakoid membrane was first showed by fluorescence recovery after photobleaching (FRAP) experiments in Dactylococcopsis salina and S. elongatus, showing that PBS–photosystem association is a dynamic process which depends on weak interactions with membrane lipids.86,87 Based on FRAP experiments and to the possible association of PBSs with PSI and PSII,57,82,94 the hypothesis of a light-harvesting complex moving and modulating the energy reaching each photosystem, analogous to plant and green algae state transitions, has frequently been proposed (Fig. 4).95 Within this hypothesis, it was suggested that in State II, the low PSII fluorescence is related to a smaller antenna, since most of the PBS are connected to PSI, while in State I, the high PSII fluorescence is related to a bigger PSII antenna, due to the migration of PBS from PSI to PSII.51,58,96,97However, FRAP results were suspected of some artificial artefacts due to the high energy laser used51 and possible effects induced in the PBS. Indeed, although PBS mobility was confirmed by FRAP and the fluorescence loss in photobleaching (FLIP) technique in T. elongatus, reversible fluorescence processes within the PBS were detected.88 This led to the correction of PBS diffusion coefficient to a lower value (1.7 ± 0.4 × 10−10 cm2 s−1, which might differ depending on experimental conditions).88 More recently, it was demonstrated by single molecule spectroscopy that PBS can lose (dark state) and recover their fluorescence spontaneously (blinking).90 Under high light intensities, the “dark state” is formed more often and the PBSs remain longer time in this state.90 Then, in darkness the PBSs recover their fluorescence. Because this blinking also affects FRAP experiments, the movement of PBS could be largely slower than it was supposed in the first studies.
Another argument for the involvement of PBS movement in state transitions was the fact that PBS mobility is inhibited by high osmotic buffers, which also inhibit state transitions in S. elongatus and Spirulina platensis.68,83,89 Incubation of cyanobacterial cells with phosphate (>0.5 M), betaine (1 M) or sucrose (1 M) suppresses the fluorescence changes related to state transitions observed at 77 K when PBS are excited (λexc ≈ 590 nm).68,83,89 These results seemed to support the hypothesis that the movement of PBS was the principal mechanism involved in cyanobacterial state transitions.89 However, it was shown that these hyper-osmotic buffers also impair membrane processes, inhibiting the changes observed in 77 K fluorescence spectra when Chl is excited (λexc ≈ 430).52,83 In addition, these chemical were also shown to inhibit quenching processes in the PBS.90 Thus, it is not possible to conclude from these experiments that PBS long distance movements are involved in state transitions.
It was also shown that PSI oligomerization affects both state transitions98,99 and PBS diffusion.99 In ΔPsaL strains of Synechococcus 7002 and S. elongatus, PSI trimerization is impaired, and State II to State I transition is faster than in the WT.98,99 In the S. elongatus mutant, it was also shown that the PBS diffusion rate was increased.99 Although Aspinwall et al. (2004)99 ascribed the faster kinetics of state transitions to the increase in PBS diffusion rate, the effect of differing distribution of protein complexes in the membrane cannot be discarded.
It has recently been shown that cyanobacterial thylakoid membranes are organized in a mosaic-like structure defined by domains.100 Three types of domains were described, the first domain was enriched mainly by PSI, the second by PSII and PBS, and in the third domain, the abundance of PSII, PSI and PBS was balanced. These microdomains seem to be very stable and not change with light conditions. The fact that macrocomplexes containing PSII–PSI–PBS were isolated,57 most probably from the third type of microdomain, suggests that very small position modification of the PBS (and/or photosystems) could induce changes in energy transfer from PBS to one or other photosystem. Thus, small PBS movement could be involved in state transitions. However, although the heterogeneous organization of cyanobacterial thylakoid membranes have been shown in several works,101–104 a conclusive model especially concerning the stability of the domains is still not available.
As an alternative to the long distance PBS movement model, recent publications have associated the high PSII fluorescence in State I to PBS detachment from thylakoid membranes (Fig. 4).79,105,106 Kaňa et al. (2012)105 studied chlorophyll fluorescence induction curves and spectrally resolved fluorescence induction (SRFI) in different cyanobacterial strains, and proposed that transition from State II to State I involves PBS decoupling from PSII. Furthermore, Chukhutsina et al. (2015)79 and Ranjbar Choubeh et al. (2018)106 demonstrated, by measurements of fast time-resolved fluorescence, that the duration of PBS fluorescence was higher in State I than in State II, correlating with a small population of functionally disconnected PBS. However, in Synechocystis, PBS decoupling was estimated to be around 13% of the total PBS pool,79 once again questioning the relevance of this light-harvesting complex in cyanobacterial state transitions.
As mentioned earlier, PBS contribution to state transitions can be observed by 77 K fluorescence emission spectra. All authors have reported an increase in PSII fluorescence (685 and 695 nm) during transition from State II to State I, on preferential excitation of the PBS (λexc ≈ 590) (Table 1). Conversely, for PSI fluorescence (715–740 nm), some authors have reported no changes between State I and State II, while others have observed a decrease in its fluorescence during State I to State II transition, and still others have reported an increase during this transition (Table 1). The variability in the results can be ascribed at least partially to the different cyanobacterial strains studied. However different results were obtained with the same strains. Thus, different growth conditions and lights used to trigger state transitions could influence the obtained results. In addition, the normalization method employed in each case could significantly affect the interpretation of these results. In most cases, the fluorescence emission spectra were normalized within the spectra themselves (to PC, APC, PSII or even PSI fluorescence peaks) (Table 1), hindering the analysis of the fluorescence changes observed. For instance, some authors did not observe changes in PSI fluorescence between State II and State I in Synechocystis, but the PSI peak was used for normalization.74,105,107 In addition, several studies cited in Table 1 normalized their results to the APC peak, which was also found to change during state transitions.108
In only two of the papers mentioned in Table 1,65,106 the fluorescence changes were normalized to an internal standard. When these chemicals, such as fluorescein or rhodamine B,65,106 are used, the fluorescence emission values are normalized to a wavelength outside the spectrum, allowing a better calibration of the fluorescence changes observed. Similar results can be obtained when normalization is done to wavelengths outside the spectrum (such as 800 nm), even in the absence of internal standards.52 In summary, the normalization of 77 K fluorescence emission spectra needs to be correctly addressed in order to facilitate the understanding of the PBS contribution to cyanobacterial state transitions. This subject is further discussed in section 4, where the contribution of membrane fluorescence to this physiological process is analyzed.
As a conclusion, although the analysis of PBS fluorescence changes during state transitions has been largely addressed in different cyanobacterial strains, it is still unclear how PBS participates in this process. As it is depicted in Table 1, the use of different experimental procedures to analyze the PBS-fluorescence related changes, hinders the possibility of finding general mechanisms for cyanobacterial state transitions. Nevertheless, based on the more recent results and our own interpretation of them, we propose that long distance movement of PBS is not involved in state transitions. Moreover, changes in the relative interaction of PBS and photosystems (involving slight PBS position changes) are most probably not involved neither. We suggest that only partial PBS detachment from one or both photosystems could contribute to the increase of PSII fluorescence in State I.
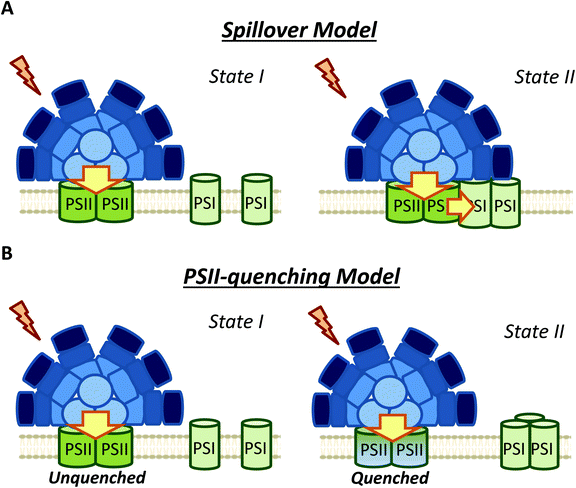
4. Membrane processes contribution to state transitions: spillover versus PSII quenching
Membrane participation in cyanobacterial state transitions was first proposed by Bruce et al. (1985) in the cyanobacteria Anacystis nidulans.109 The involvement of the membrane was also suggested in Synechocystis cells, due to the changes in the thylakoid membrane topology of PSII distribution between the two states.61,110,111 In this cyanobacterium, a row disposition was observed for PSII in State I, and a random distribution (favoring spillover) was observed in State II. Further evidence supporting the involvement of the movement of membrane complexes, or changes in the oligomerization state of these complexes was presented by El-Bissati et al. (2000),65 who demonstrated that this process is affected by alterations in membrane fluidity. However, more recently, Maksimov et al. (2017)112 have suggested that the rate of oxidation of the PQ pool depends on membrane fluidity, and therefore on temperature. Thus, the changes observed by Bissati et al. could have been related to slower kinetics of reduction and oxidation of the PQ pool in less fluid membranes.65 Finally, state transitions were also observed in mutants of Synechocystis and Synechococcus 7002 lacking PBS.81,113It was first proposed that the rearrangement of photosystem positions in the membrane induced changes in spillover (energy transfer from PSII to PSI) during state transitions.61,109 However, although changes in spillover have long been studied (Fig. 5),51,61,77 there are no clear results confirming the participation of this phenomenon in cyanobacterial state transitions. A key evidence supporting the spillover model would be a decrease in PSI fluorescence at 77 K (when Chl is preferentially excited) during State II to State I transition (when photosystems move apart), concomitant with an increase in PSII fluorescence under the same conditions. However, most studies on spillover did not show this result (Table 2). Of the articles listed in Table 2, only one work realized by Bruce et al. (1989) in Synechococcus PCC 7002,81 and two studies on Spirulina platensis,68,83 showed an apparent increase in PSI fluorescence in State II, which can be considered as an evidence for spillover. However, none of these works analyzed the 77 K steady-state fluorescence emission spectra using internal standards (such as fluorescein or rhodamine B). The utilization of these standards allows the study of the absolute variation in the different fluorescence contributions.52,79,80,106 Interestingly, when these internal standards were used, authors lead to the conclusion that due to the constant level of PSI emission between State I and II, spillover is an unlikely mechanism of cyanobacterial state transitions.52,80,106 Moreover, a recent study employing a streak camera to measure fast (ps to ms) fluorescence decays, strongly suggested that at least in S. elongatus (and most probably in Synechocystis) changes in spillover are not involved in state transitions.106 These authors measured fast fluorescence decay kinetics (at 77 K) of cells adapted to State I and II, and subsequently obtained the decay-associated spectra (DAS) after global analysis of the data. When 430 nm excitation wavelengths were used, PSII emission decreased in State II, however, the DAS showed that PSI emission was similar in State I and State II.
Ranjbar Choubeh et al. (2018) recently proposed that the decrease in PSII fluorescence is related to a reversible PSII-quenching in State II (Fig. 5), at least in S. elongatus and Synechocystis cells.106 This quenching has been suggested to take place at the level of PSII, and is not related to the NPQ mechanism described for cyanobacteria, which occurs in the PBS.9 It is worth noting that higher PSII quenching should lead to extra heat emission in State II. However, optoacoustic studies did not show differences between the heat production in States I and II.114,115 As previously discussed, Van Amerongen et al. showed that in both S. elongatus and Synechocystis, a partial disconnection of PBS occurs in State I,79,106 which should increase the conversion of excitation energy into heat. This increase in heat could be adventitiously similar to the heat increase due to PSII quenching in State II, which might explain the absence of heat emission changes during state transitions. Overall, the results of Ranjbar Choubeh et al. (2018) demonstrated that PSII is not quenched by PSI in State II in S. elongatus.106 The quenching mechanism related to PSII remains to be elucidated.
Stadnichuk et al. (2009) also suggested that state transitions do not involve PSI, and could be completely attributed to PSII.80 In their study, fluorescence emission changes at 77 K were observed for PSII, but not for PSI, when Chl were excited in Synechocystis cells. Based on these results, they suggested that spillover is an unlikely mechanism implicated in cyanobacterial state transitions.80 These observations were made in WT, CK (mutant without PC) and PAL (mutant depleted in PBS) strains, leading to the conclusion that thylakoid membranes participate in state transitions, and their contribution is not negligible.
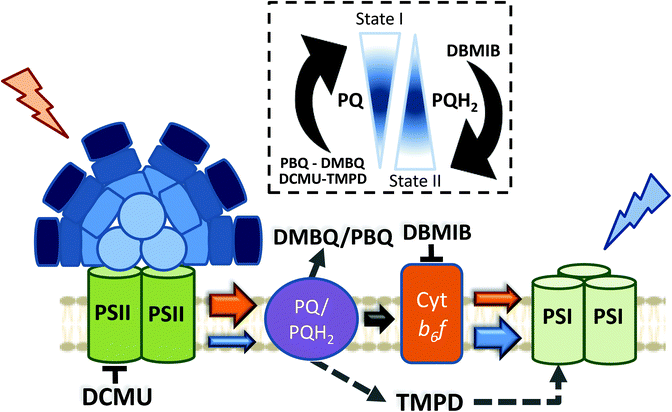
5. The signal transduction pathways and the role of Cyt b6f
Apart from the mechanism involved in cyanobacterial state transitions, the signal transduction pathways leading to State I/II transition remain to be deciphered. It is well established that changes in the redox state of the PQ pool trigger state transitions in cyanobacteria, like in plants and green algae.46,69,73,74,116,117 While a reduced PQ pool induces State II transition, State I transition is induced by its oxidation. As previously discussed, in the dark, cyanobacterial cells are in State II due to PQ pool reduction via respiration.69,70 In addition, chemicals oxidizing or reducing the PQ pool also trigger state transitions46 (Fig. 6). Among these chemicals, some quinones, such as p-benzoquinone (PBQ) or 2,6-dimethoxy-1,4-benzoquinone (DMBQ), induce State I transition through oxidation of the PQ pool in dark-adapted cells.73,117 In contrast, addition of 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) induces State I to State II transition even under blue-light illumination.46,73,117 DBMIB binds close to the [2Fe–2S] cluster of the Cyt b6f and blocks photosynthetic and respiratory electron transport,118,119 inhibiting the re-oxidation of the PQ pool by the complex. Thus, the PQ pool is reduced in its presence.There are two DBMIB binding sites in Cyt b6f, one with high affinity and the other with low affinity.119 The high affinity binding site is located at the periphery of the complex, outside the Qo pocket. At low concentrations (0.5–1 μM), DBMIB binds only to the high affinity site, and in darkness, it does not inhibit electron transport in vivo, as reported in Synechococcus 7002.119 For inhibition of electron transport, the cells must be illuminated to induce the migration of DBMIB from its peripheral position to the low affinity site (close to the [2Fe–2S] cluster).119 However, at higher concentrations (>2 μM), DBMIB fully inhibits Cyt f re-reduction also in darkness.
Given the relationship between the redox state of the PQ pool and state transitions in both cyanobacteria and plants,45,46 and the involvement of Cyt b6f in the signal transduction pathway of plants and algae,29 the participation of this complex in cyanobacterial state transitions has also been hypothesized. Results suggesting the participation of phosphorylation reactions in this signal transduction pathway strengthened this idea.120–125 Hence, Mao et al. (2002) and Huang et al. (2003) proposed Cyt b6f as the redox sensor of the PQ pool in cyanobacterial state transitions.73,117 They based their conclusions on the effect of DBMIB on PBQ or DMBQ poisoned cells, suggesting that DBMIB induces transition to State II by binding to the Cyt b6f complex, even when the PQ pool remains oxidized in the presence of the aforementioned quinones.73,117 However, they did not measure the redox state of the pool under these conditions.
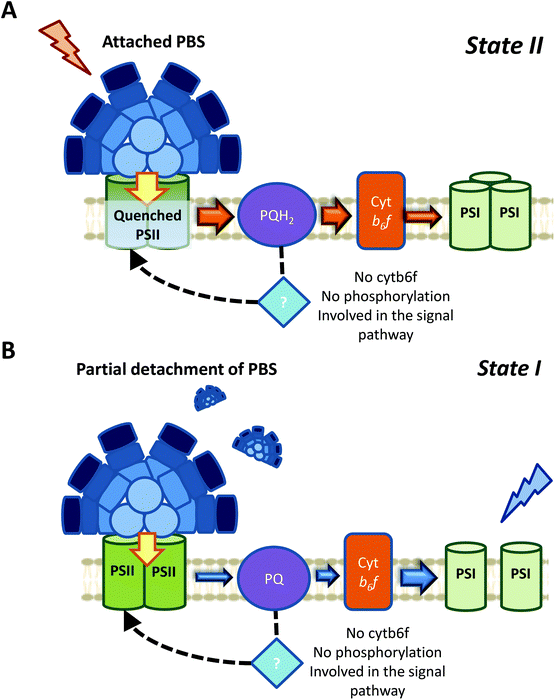
It has recently been shown, using chlorophyll fluorescence induction curves, that the PQ pool is rapidly reduced by addition of DBMIB, even in the presence of DMBQ.52 Moreover, the same concentration of DBMIB has different effects on state transitions under light-induced or DMBQ-induced State I-adapted cells. The concentration of DBMIB used were higher than 2 μM. These experiments strongly suggest that DBMIB induces State II transition due to the reduction of the PQ pool, and not due to its binding to Cyt b6f. In another set of experiments, the same authors showed that it was possible to induce State I transition when illuminating DBMIB-poisoned cells, using N,N′,N′-tetramethyl-p-phenylenediamine (TMPD).52 This chemical was shown to restore photosynthetic electron transport, even when Cyt b6f was inhibited by DBMIB.126 Thus, the oxidation of the PQ pool triggers State I transition, through a signal transduction pathway that does not involve Cyt b6f.
It has also been proposed that the Chl a molecule of the Cyt b6f complex is the signal transmitter in cyanobacterial state transitions.127 In this model, the Chl a senses the changes in the Qo site, and participates in this signal transduction pathway by altering its volume. This hypothesis proposed by Vladkova (2016) was based on the analysis of various Cyt b6f X-ray structures, differing in the volume of Chl a.127 Changes in the Chl a volume, along with changes in the redox states of the PQ pool, would affect the hydrophobic thickness of Cyt b6f, which would induce a hydrophobic mismatch in the membrane. This hydrophobic mismatch is the difference between the hydrophobic thickness of the protein and the hydrophobic thickness of the lipid bilayer, and would be the driving force behind membrane reorganization during the progression of state transitions.127
If the Chl a volume is the signal transmitter in cyanobacterial state transitions, it would be expected that inhibition of Cyt b6f would have an irreversible effect on this process. The volume of Chl a must not change in DBMIB-poisoned cells, however, addition of TMPD was found to induce state transitions.52 This suggests that the involvement of this Chl a molecule is also unlikely. It is worth mentioning that under the conditions tested by Calzadilla et al. (2019) (DBMIB concentration >2 μM),52 the two DBMIB binding sites in Cyt b6f were occupied, fully inhibiting the complex and photosynthetic electron transport.119
The possible participation of phosphorylation reactions in cyanobacterial state transitions has also been studied. First reports showing a correlation between changes in thylakoid phosphorylation patterns, PQ pool redox changes and state transitions came from studies conducted on Synechococcus 6301.120,122,128 A model was proposed, in which protein phosphorylation and PBS migration were requisites for State I/II transition (ref. 121, see review129). However, none of these studies showed a direct connection between the changes in phosphorylation and state transitions. Moreover, the results of Biggins et al. (1984)130 suggest that state transitions in organisms with PBS do not involve reversible protein phosphorylation. This has been discussed in detail by Allen (1992).129
In more recent works, it has been observed that PBS and photosystem proteins can undergo phosphorylation, indicating the participation of these reactions in cyanobacterial state transitions.123–125 However, only Chen et al. (2015) directly addressed this possible relationship.124 These authors showed that four residues of the β subunits of PC (Ser22, Ser49, Thr94 and Ser154) were phosphorylated. Mutation of these amino acids avoiding phosphorylation seemed to affect state transitions. A slower and smaller increase of fluorescence was observed upon illumination of dark-adapted mutant cells. However, these cells were not extensively characterized, for instance, alterations in the energy transfer within the PBS were suggested, which might affect PBS concentration and/or PSI/PSII ratio between strains.124 Moreover, the dark PQ pool redox state was not measured; in addition, the differences between mutants cannot be ignored. Consequently, the observed impairment in state transitions could not conclusively be linked to the lack of PBS phosphorylation.
Ser/Thr kinases have been widely described as key factors in the signal transduction pathways of cyanobacteria (ref. 131 and 132, see review133). Synechocystis presents 12 genes coding for Ser/Thr kinases (Spk),134,135 of which seven are classified in the PKN2 subfamily (spkA to spkG),136 and the rest have been assigned to the ABC1 subfamily (spkH to spkL).137 These kinases have been shown to participate in different cell functions,138 such as cell motility,139 oxidative stress response,140 salt and low temperature stress,141,142 and regulation of carbon143 and nitrogen metabolism.144 In addition, the GroES chaperone protein and ferredoxin were also reported to be targets of some Spk.145,146
As previously mentioned, Chen et al. (2015) suggested the possible participation of Ser/Thr kinases in cyanobacterial state transitions,124 like in plants and green algae.16,17,124 Recently, Calzadilla et al. (2019) addressed this issue by constructing single mutants for all the Ser/Thr kinases and phosphatases identified in Synechocystis.52 None of them were impaired in state transitions. In addition, experiments with kinase (Staurosporine and K252a) and phosphatase inhibitors (NaF and Na3VO4) were also performed. Incubation with these chemicals had no effect on state transitions in Synechocystis and S. elongatus, although they affected other processes in the cells.52 Thus, these results lead to the conclusion that phosphorylation reactions are not essential in cyanobacterial state transitions.
6. Conclusions and perspectives
Despite the large number of studies conducted on cyanobacterial state transitions, the mechanism behind this physiological process has not yet been determined. Four different mechanisms (PBS movement, PBS detachment, spillover and PSII quenching), and combinations of them, were proposed to be related to the changes of fluorescence observed when PSII or PSI is specifically illuminated. Moreover, different models were proposed for the same cyanobacteria strain (Table 4).| Organism | Model proposed | References |
|---|---|---|
| S. elongatus | PBS-mobile | Sarcina et al. (2001)87; Joshua and Mullineaux (2004)89; Aspinwall et al. (2004)99; Joshua and Mullineaux (2005)92 |
| PBS-detachment | Ranjbar Choubeh et al. (2018)106 | |
| PSII-quenching | Ranjbar Choubeh et al. (2018)106 | |
| Synechocystis PCC 6803 | PBS-mobile | Ashby and Mullineaux (1999)41; Joshua and Mullineaux (2005)92 |
| PBS-mobile/spillover | McConnell et al. (2002)51 | |
| PBS-detachment | Chukhutsina et al. (2015)79; Kaňa et al. (2012)105; Ranjbar Choubeh et al. (2018)106 | |
| Spillover | Olive et al. (1997)61; Folea et al. (2008)111; El-Bissati et al. (2000)65; Vernotte et al. (1990)77 | |
| PSII-quenching | Ranjbar Choubeh et al. (2018)106; Stadnichuk et al. (2009)80 | |
| Synechococcus PCC 7002 | PBS-mobile | Dong et al. (2009)40 |
| PBS-mobile/spillover | McConnell et al. (2002)51 | |
| Spillover | Bruce et al. (1989)81 | |
| Spirulina platensis | PBS-mobile/spillover | Li et al. (2006)68; Li et al. (2004)83 |
| Dactylococcopsis salina | PBS-mobile | Mullineaux et al. (1997)86 |
Nevertheless, considering the literature included in the present review, it is possible to make some conclusions about the molecular mechanism and the signal transduction pathways involved. Our proposed model is described in Fig. 7. First, it is clear that cyanobacterial state transitions cannot be reduced to changes in the energy transfer from PBS to the photosystems, as was proposed several years ago. Although it was demonstrated that PBS could move on the thylakoid surface, there is no direct evidence showing that this movement has a physiological role in state transitions. Furthermore, recent results suggest that only a partial detachment of PBS from one or both photosystems is involved in state transitions. However, we cannot totally discard small changes in their position favoring a better energy transfer to one or other photosystem, at least in some cyanobacteria strains. Finally, PBS contribution seems to depend on the cyanobacterial strain and growth conditions.
In contrast, membrane processes seem to be an intrinsic part of cyanobacterial state transitions. Chl fluorescence changes can be ascribed to the proposed PSII-quenching, and/or changes in spillover involvement. The different membrane topologies observed in State I and State II, favoring or hindering direct energy transfer from PSII to PSI, supported changes in spillover during state transitions. However, recent studies employing a streak camera to measure fast (ps to ms) fluorescence decays, strongly suggested that changes in spillover are not involved in state transitions.106
Thus, many questions remain to be answered: How changes in membrane topology participate in cyanobacterial state transitions? Do these membrane changes also affect PBS energy transfer to photosystems? What is the mechanism involved in PSII-specific quenching? Regarding this last question, it has recently been shown that the thermal phase of the O–J–I–P curve can be ascribed to a light-induced conformational change in PSII, triggered by saturating multiple turnover flashes (ref. 147–149, see review150). This increase in the fluorescence yield of Chl a is temperature dependent,147 and its kinetics depends on the length, rather than the intensity, of the flashes.148 This increase was further found to be related with changes in the interaction between the PSII reaction center and the antenna proteins CP43 and CP47.149,151 Although it has been established that this phenomenon is not related with state transitions, because it takes place at the ms scale and can relax and be regenerated without changes in redox state of the PQ pool,147 similar PSII conformational changes can be associated with the PSII-specific quenching suggested by Ranjbar Choubeh et al. (2018).106 If the relevance of this quenching is confirmed, the definition of cyanobacterial state transitions as a re-balance of the excitation energy reaching the photosystems needs to be reconsidered.
The different contributions of PBS and membrane processes in cyanobacterial state transitions seem to be dependent on the cyanobacterial strain. Hence, comparing the same processes in different strains would lead to more conclusive results regarding state transitions. Along these lines, the utilization of S. elongatus and T. elongatus as model organisms can facilitate this work, due to the larger fluorescence differences observed between State I and II, compared to the other organisms used in the past (such as Synechocystis or Synechococcus 7002). It is also worth mentioning that state transitions could be dependent on growth conditions of the cultured strains. If this is the case, analyzing the fluorescence changes of cells acclimated to different conditions would allow a better understanding of the relative contributions of PBS and membrane processes to state transitions.
In case of the signal transduction pathways, showing that Cyt b6f and phosphorylation reactions are not involved in cyanobacterial state transitions was a step forward. However, how changes in the redox state of the PQ pool could be transmitted to the PBS and/or photosystems remains to be discovered. If we consider that one of the main fluorescence changes between State I and II might come from the membrane contribution, and might involve a PSII-specific quenching, one tempting hypothesis would be that PSII itself is able to sense the redox state of the PQ pool. Accumulation of QA− cannot be this redox sensor, because while its accumulation due to 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) addition induces State I, its accumulation due to the reduction of the PQ pool induces State II.
It was suggested that a Qc channel between Cyt b559 and PsbJ could exist.152 This channel, which is connected to the QB site, has been suggested to have a quinone-binding function, and can participate in PSII regulation. Thus, a possible hypothesis would be to consider the Qc hydrophobic tunnel as the redox sensor of the PQ pool. However, the existence of this Qc site is still unclear, since its presence was not confirmed in the most recent high resolution PSII structure model.56 Notwithstanding, it was shown that mutations in Cyt b559, PsbJ, and near the putative Qc site, can alter the kinetics and amplitude of state transitions in Synechocystis cells.153 These results present an interesting starting point for further studies, although the participation of novel factors or known proteins in this signal transduction pathway cannot be discarded. New approaches, such as the utilization of mutant library screenings,154 need to be developed to shed light on the mechanism connecting the redox state of the PQ pool and the fluorescence changes observed in cyanobacterial state transitions.
Funding
This work was supported by grants from the Agence Nationale de la Recherche (ANR projects RECYFUEL (ANR-16-CE05-0026)) and by the European Union's Horizon 2020 Research and Innovation program under grant agreement no. 675006 (SE2B). P. C. salary is financed by RECYFUEL (ANR project). The research is also supported by the Centre National de la Recherche Scientifique (CNRS) and the Commissariat à l'Energie Atomique (CEA).Conflicts of interest
There are no conflicts to declare.References
- N. Sluchanko, Y. Slonimskiy and E. Maksimov, Biochemistry, 2017, 82, 1592–1614 CAS.
- D. Kirilovsky and C. A. Kerfeld, Nat. Plants, 2016, 2, 16180 CrossRef CAS PubMed.
- D. Kirilovsky, Photosynth. Res., 2007, 93, 7 CrossRef CAS PubMed.
- D. Kirilovsky and C. A. Kerfeld, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 158–166 CrossRef CAS PubMed.
- C. A. Kerfeld, M. R. Melnicki, M. Sutter and M. A. Dominguez-Martin, New Phytol., 2017, 215, 937–951 CrossRef CAS PubMed.
- J. Van Thor, C. Mullineaux, H. Matthijs and K. Hellingwerf, Bot. Acta, 1998, 111, 430–443 CrossRef CAS.
- C. W. Mullineaux and D. Emlyn-Jones, J. Exp. Bot., 2004, 56, 389–393 CrossRef PubMed.
- D. Kirilovsky, Photosynth. Res., 2015, 126, 3–17 CrossRef CAS PubMed.
- A. Wilson, G. Ajlani, J.-M. Verbavatz, I. Vass, C. A. Kerfeld and D. Kirilovsky, Plant Cell, 2006, 18, 992–1007 CrossRef CAS PubMed.
- M. G. Rakhimberdieva, I. V. Elanskaya, W. F. Vermaas and N. V. Karapetyan, Biochim. Biophys. Acta, Bioenerg., 2010, 1797, 241–249 CrossRef CAS PubMed.
- N. Murata, Biochim. Biophys. Acta, Bioenerg., 1969, 172, 242–251 CrossRef CAS.
- C. Bonaventura and J. Myers, Biochim. Biophys. Acta, Bioenerg., 1969, 189, 366–383 CrossRef CAS.
- J. Minagawa, Biochim. Biophys. Acta, Bioenerg., 2011, 1807, 897–905 CrossRef CAS PubMed.
- S. Lemeille and J.-D. Rochaix, Photosynth. Res., 2010, 106, 33–46 CrossRef CAS PubMed.
- P. Pesaresi, M. Pribil, T. Wunder and D. Leister, Biochim. Biophys. Acta, Bioenerg., 2011, 1807, 887–896 CrossRef CAS PubMed.
- N. Depège, S. Bellafiore and J.-D. Rochaix, Science, 2003, 299, 1572–1575 CrossRef PubMed.
- S. Bellafiore, F. Barneche, G. Peltier and J.-D. Rochaix, Nature, 2005, 433, 892 CrossRef CAS PubMed.
- A. V. Vener, P. J. van Kan, A. Gal, B. Andersson and I. Ohad, J. Biol. Chem., 1995, 270, 25225–25232 CrossRef CAS PubMed.
- A. V. Vener, P. J. Van Kan, P. R. Rich, I. Ohad and B. Andersson, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 1585–1590 CrossRef CAS PubMed.
- F. Zito, G. Finazzi, R. Delosme, W. Nitschke, D. Picot and F. A. Wollman, EMBO J., 1999, 18, 2961–2969 CrossRef CAS PubMed.
- Z. Zhang, L. Huang, V. M. Shulmeister, Y.-I. Chi, K. K. Kim, L.-W. Hung, A. R. Crofts, E. A. Berry and S.-H. Kim, Nature, 1998, 392, 677 CrossRef CAS PubMed.
- C. Breyton, J. Biol. Chem., 2000, 275, 13195–13201 CrossRef CAS PubMed.
- G. Finazzi, F. Zito, R. P. Barbagallo and F.-A. Wollman, J. Biol. Chem., 2001, 276, 9770–9774 CrossRef CAS PubMed.
- L. Dumas, F. Zito, S. Blangy, P. Auroy, X. Johnson, G. Peltier and J. Alric, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 12063–12068 CrossRef CAS PubMed.
- U. K. Larsson, B. Jergil and B. Andersson, Eur. J. Biochem., 1983, 136, 25–29 CrossRef CAS PubMed.
- D. Kyle, L. Staehelin and C. Arntzen, Arch. Biochem. Biophys., 1983, 222, 527–541 CrossRef CAS PubMed.
- M. Pribil, P. Pesaresi, A. Hertle, R. Barbato and D. Leister, PLoS Biol., 2010, 8, e1000288 CrossRef PubMed.
- A. Shapiguzov, B. Ingelsson, I. Samol, C. Andres, F. Kessler, J.-D. Rochaix, A. V. Vener and M. Goldschmidt-Clermont, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4782–4787 CrossRef CAS PubMed.
- F.-A. Wollman and C. Lemaire, Biochim. Biophys. Acta, Bioenerg., 1988, 933, 85–94 CrossRef CAS.
- A. N. Glazer, Biochim. Biophys. Acta, 1984, 768, 29–51 CrossRef CAS.
- R. MacColl, J. Struct. Biol., 1998, 124, 311–334 CrossRef CAS PubMed.
- N. Adir, Photosynth. Protein Complexes, 2008, 243–274 CAS.
- N. Adir, S. Bar-Zvi and D. Harris, Biochim. Biophys. Acta, Bioenerg., 2019 DOI:10.1016/j.bbabio.2019.07.002.
- M. Watanabe and M. Ikeuchi, Photosynth. Res., 2013, 116, 265–276 CrossRef CAS PubMed.
- D. A. Bryant, Microbiology, 1982, 128, 835–844 CAS.
- A. R. Holzwarth, Physiol. Plant., 1991, 83, 518–528 CrossRef CAS.
- I. Eisenberg, F. Caycedo-Soler, D. Harris, S. Yochelis, S. F. Huelga, M. B. Plenio, N. Adir, N. Keren and Y. Paltiel, J. Phys. Chem. B, 2017, 121, 1240–1247 CrossRef CAS PubMed.
- T. Mirkovic, E. E. Ostroumov, J. M. Anna, R. Van Grondelle and G. D. Scholes, Chem. Rev., 2016, 117, 249–293 CrossRef PubMed.
- M. Şener, J. Strümpfer, J. Hsin, D. Chandler, S. Scheuring, C. N. Hunter and K. Schulten, ChemPhysChem, 2011, 12, 518–531 CrossRef PubMed.
- C. Dong, A. Tang, J. Zhao, C. W. Mullineaux, G. Shen and D. A. Bryant, Biochim. Biophys. Acta, Bioenerg., 2009, 1787, 1122–1128 CrossRef CAS PubMed.
- M. K. Ashby and C. W. Mullineaux, Photosynth. Res., 1999, 61, 169–179 CrossRef CAS.
- J. Zhao, J. Zhou and D. Bryant, Photosynth. Res, 1992, 34, 83 Search PubMed.
- P. I. Calzadilla, F. Muzzopappa, P. Sétif and D. Kirilovsky, Biochim. Biophys. Acta, Bioenerg., 2019, 1860, 488–498 CrossRef CAS PubMed.
- D. V. Zlenko, I. V. Elanskaya, E. P. Lukashev, Y. V. Bolychevtseva, N. E. Suzina, E. S. Pojidaeva, I. A. Kononova, A. V. Loktyushkin and I. N. Stadnichuk, Biochim. Biophys. Acta, Bioenerg., 2019, 1860, 155–166 CrossRef CAS PubMed.
- J. F. Allen, J. Bennett, K. E. Steinback and C. J. Arntzen, Nature, 1981, 291, 25 CrossRef CAS.
- C. W. Mullineaux and J. F. Allen, Photosynth. Res., 1990, 23, 297–311 CrossRef CAS PubMed.
- R. Van Dorssen, J. Breton, J. Plijter, K. Satoh, H. Van Gorkom and J. Amesz, Biochim. Biophys. Acta, Bioenerg., 1987, 893, 267–274 CrossRef CAS.
- D. Siefermann-Harms, J. Chromatogr. A, 1988, 448, 411–416 CrossRef CAS.
- A. N. Glazer and L. Stryer, Biophys. J., 1983, 43, 383–386 CrossRef CAS.
- M. Wu, P. M. Goodwin, W. P. Ambrose and R. A. Keller, J. Phys. Chem., 1996, 100, 17406–17409 CrossRef CAS.
- M. D. McConnell, R. Koop, S. Vasil'ev and D. Bruce, Plant Physiol., 2002, 130, 1201–1212 CrossRef CAS PubMed.
- P. I. Calzadilla, J. Zhan, P. Sétif, C. Lemaire, D. Solymosi, N. Battchikova, Q. Wang and D. Kirilovsky, Plant Cell, 2019, 31, 911–931 CrossRef CAS PubMed.
- U. Schreiber, W. Bilger and C. Neubauer, in Ecophysiology of photosynthesis, Springer, 1995, pp. 49–70 Search PubMed.
- W. Remelli and S. Santabarbara, Biochim. Biophys. Acta, Bioenerg., 2018, 1859, 1207–1222 CrossRef CAS PubMed.
- P. Jordan, P. Fromme, H. T. Witt, O. Klukas, W. Saenger and N. Krauß, Nature, 2001, 411, 909 CrossRef CAS PubMed.
- Y. Umena, K. Kawakami, J.-R. Shen and N. Kamiya, Nature, 2011, 473, 55 CrossRef CAS PubMed.
- H. Liu, H. Zhang, D. M. Niedzwiedzki, M. Prado, G. He, M. L. Gross and R. E. Blankenship, Science, 2013, 342, 1104–1107 CrossRef CAS PubMed.
- C. W. Mullineaux, Biochim. Biophys. Acta, Bioenerg., 1992, 1100, 285–292 CrossRef CAS.
- M. G. Rakhimberdieva, V. A. Boichenko, N. V. Karapetyan and I. N. Stadnichuk, Biochemistry, 2001, 40, 15780–15788 CrossRef CAS PubMed.
- A. Ley and W. Butler, Biochim. Biophys. Acta, Bioenerg., 1980, 592, 349–363 CrossRef CAS.
- J. Olive, G. Ajlani, C. Astier, M. Recouvreur and C. Vernotte, Biochim. Biophys. Acta, Bioenerg., 1997, 1319, 275–282 CrossRef CAS.
- J. Biggins and D. Bruce, Photosynth. Res., 1989, 20, 1–34 CrossRef CAS PubMed.
- J. Biggins, N. A. Tanguay and H. A. Frank, FEBS Lett., 1989, 250, 271–274 CrossRef CAS PubMed.
- C. Vernotte, M. Picaud, D. Kirilovsky, J. Olive, G. Ajlani and C. Astier, Photosynth. Res., 1992, 32, 45–57 CrossRef CAS PubMed.
- K. El Bissati, E. Delphin, N. Murata, A.-L. Etienne and D. Kirilovsky, Biochim. Biophys. Acta, Bioenerg., 2000, 1457, 229–242 CrossRef CAS.
- S. Federman, S. Malkin and A. Scherz, Photosynth. Res., 2000, 64, 199 CrossRef CAS PubMed.
- L. Tian, I. H. van Stokkum, R. B. Koehorst, A. Jongerius, D. Kirilovsky and H. van Amerongen, J. Am. Chem. Soc., 2011, 133, 18304–18311 CrossRef CAS PubMed.
- H. Li, D. Li, S. Yang, J. Xie and J. Zhao, Biochim. Biophys. Acta, Bioenerg., 2006, 1757, 1512–1519 CrossRef CAS PubMed.
- C. W. Mullineaux and J. F. Allen, FEBS Lett., 1986, 205, 155–160 CrossRef CAS.
- M. Aoki and S. Katoh, Biochim. Biophys. Acta, Bioenerg., 1982, 682, 307–314 CrossRef CAS.
- C. W. Mullineaux, Biochim. Biophys. Acta, Bioenerg., 2014, 1837, 503–511 CrossRef CAS PubMed.
- M. Misumi, H. Katoh, T. Tomo and K. Sonoike, Plant Cell Physiol., 2015, 57, 1510–1517 Search PubMed.
- C. Huang, X. Yuan, J. Zhao and D. A. Bryant, Biochim. Biophys. Acta, Bioenerg., 2003, 1607, 121–130 CrossRef CAS PubMed.
- T. Ogawa, T. Harada, H. Ozaki and K. Sonoike, Plant Cell Physiol., 2013, 54, 1164–1171 CrossRef CAS PubMed.
- Y. V. Bolychevtseva, F. Kuzminov, I. Elanskaya, M. Y. Gorbunov and N. Karapetyan, Biochemistry, 2015, 80, 50–60 CAS.
- T. Ogawa and K. Sonoike, J. Photochem. Photobiol., B, 2015, 144, 61–67 CrossRef CAS PubMed.
- C. Vernotte, C. Astier and J. Olive, Photosynth. Res., 1990, 26, 203–212 CrossRef CAS PubMed.
- D. Emlyn-Jones, M. K. Ashby and C. W. Mullineaux, Mol. Microbiol., 1999, 33, 1050–1058 CrossRef CAS PubMed.
- V. Chukhutsina, L. Bersanini, E.-M. Aro and H. Van Amerongen, Sci. Rep., 2015, 5, 14193 CrossRef CAS PubMed.
- I. N. Stadnichuk, E. P. Lukashev and I. V. Elanskaya, Photosynth. Res., 2009, 99, 227–241 CrossRef CAS PubMed.
- D. Bruce, S. Brimble and D. A. Bryant, Biochim. Biophys. Acta, Bioenerg., 1989, 974, 66–73 CrossRef CAS.
- D. Li, J. Xie, Y. Zhao and J. Zhao, Biochim. Biophys. Acta, Bioenerg., 2003, 1557, 35–40 CrossRef CAS.
- D. Li, J. Xie, J. Zhao, A. Xia, D. Li and Y. Gong, Biochim. Biophys. Acta, Bioenerg., 2004, 1608, 114–121 CrossRef CAS PubMed.
- C. Boulay, L. Abasova, C. Six, I. Vass and D. Kirilovsky, Biochim. Biophys. Acta, Bioenerg., 2008, 1777, 1344–1354 CrossRef CAS PubMed.
- C. Dong and J. Zhao, Chin. Sci. Bull., 2008, 53, 3422–3424 CrossRef CAS.
- C. W. Mullineaux, M. J. Tobin and G. R. Jones, Nature, 1997, 390, 421 CrossRef CAS.
- M. Sarcina, M. J. Tobin and C. W. Mullineaux, J. Biol. Chem., 2001, 276, 46830–46834 CrossRef CAS PubMed.
- S. Yang, Z. Su, H. Li, J. Feng, J. Xie, A. Xia, Y. Gong and J. Zhao, Biochim. Biophys. Acta, Bioenerg., 2007, 1767, 15–21 CrossRef CAS PubMed.
- S. Joshua and C. W. Mullineaux, Plant Physiol., 2004, 135, 2112–2119 CrossRef CAS PubMed.
- M. Gwizdala, R. Berera, D. Kirilovsky, R. Van Grondelle and T. P. Krüger, J. Am. Chem. Soc., 2016, 138, 11616–11622 CrossRef CAS PubMed.
- D. Jallet, M. Gwizdala and D. Kirilovsky, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 1418–1427 CrossRef CAS PubMed.
- S. Joshua and C. W. Mullineaux, Biochim. Biophys. Acta, Bioenerg., 2005, 1709, 58–68 CrossRef CAS PubMed.
- G. Deng, F. Liu, X. Liu and J. Zhao, FEBS Lett., 2012, 586, 2342–2345 CrossRef CAS PubMed.
- J. Zhao, L. Chen, F. Gao, Q. Wang, Z. Qiu and W. Ma, Acta Biochim. Biophys. Sin., 2014, 46, 911–916 CrossRef CAS PubMed.
- C. W. Mullineaux, Photosynth. Res., 2008, 95, 175 CrossRef CAS PubMed.
- C. W. Mullineaux, Biochim. Biophys. Acta, Bioenerg., 1994, 1184, 71–77 CrossRef CAS.
- D. Bald, J. Kruip and M. Rögner, Photosynth. Res., 1996, 49, 103–118 CrossRef CAS PubMed.
- W. M. Schluchter, G. Shen, J. Zhao and D. A. Bryant, Photochem. Photobiol., 1996, 64, 53–66 CrossRef CAS PubMed.
- C. L. Aspinwall, M. Sarcina and C. W. Mullineaux, Photosynth. Res., 2004, 79, 179 CrossRef CAS PubMed.
- A. Strašková, G. Steinbach, G. Konert, E. Kotabová, J. Komenda, M. Tichý and R. Kaňa, Biochim. Biophys. Acta, Bioenerg., 2019 DOI:10.1016/j.bbabio.2019.07.008.
- W. F. Vermaas, J. A. Timlin, H. D. Jones, M. B. Sinclair, L. T. Nieman, S. W. Hamad, D. K. Melgaard and D. M. Haaland, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4050–4055 CrossRef CAS PubMed.
- D. M. Sherman, T. A. Troyan and L. A. Sherman, Plant Physiol., 1994, 106, 251–262 CrossRef CAS PubMed.
- S. Casella, F. Huang, D. Mason, G.-Y. Zhao, G. N. Johnson, C. W. Mullineaux and L.-N. Liu, Mol. Plant, 2017, 10, 1434–1448 CrossRef CAS PubMed.
- G. Steinbach, F. Schubert and R. Kaňa, J. Photochem. Photobiol., B, 2015, 152, 395–399 CrossRef CAS PubMed.
- R. Kaňa, E. Kotabová, O. Komárek, B. Šedivá, G. C. Papageorgiou and O. Prášil, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 1237–1247 CrossRef.
- R. R. Choubeh, E. Wientjes, P. C. Struik, D. Kirilovsky and H. van Amerongen, Biochim. Biophys. Acta, Bioenerg., 2018, 1859, 1059–1066 CrossRef.
- K. Kondo, C. W. Mullineaux and M. Ikeuchi, Photosynth. Res., 2009, 99, 217–225 CrossRef CAS PubMed.
- W. Zhao, J. Xie and J. Zhao, Chin. Sci. Bull., 2014, 59, 4712–4719 CrossRef CAS.
- D. Bruce, J. Biggins, T. Steiner and M. Thewalt, Biochim. Biophys. Acta, Bioenerg., 1985, 806, 237–246 CrossRef CAS.
- J. Olive, O. Vallon, F.-A. Wollman, M. Recouvreur and P. Bennoun, Biochim. Biophys. Acta, Bioenerg., 1986, 851, 239–248 CrossRef CAS.
- I. M. Folea, P. Zhang, E.-M. Aro and E. J. Boekema, FEBS Lett., 2008, 582, 1749–1754 CrossRef CAS PubMed.
- E. G. Maksimov, K. S. Mironov, M. S. Trofimova, N. L. Nechaeva, D. A. Todorenko, K. E. Klementiev, G. V. Tsoraev, E. V. Tyutyaev, A. A. Zorina and P. V. Feduraev, Photosynth. Res., 2017, 133, 215–223 CAS.
- K. El Bissati and D. Kirilovsky, Plant Physiol., 2001, 125, 1988–2000 CrossRef CAS PubMed.
- C. W. Mullineaux, S. Griebenow and S. E. Braslavsky, Biochim. Biophys. Acta, 1991, 1060, 315–318 CrossRef CAS.
- D. Bruce and O. Salehian, Biochim. Biophys. Acta, Bioenerg., 1992, 1100, 242–250 CrossRef CAS.
- W. P. Williams and P. J. Dominy, Biochim. Biophys. Acta, Bioenerg., 1990, 1015, 121–130 CrossRef CAS.
- H.-B. Mao, G.-F. Li, X. Ruan, Q.-Y. Wu, Y.-D. Gong, X.-F. Zhang and N.-M. Zhao, FEBS Lett., 2002, 519, 82–86 CrossRef CAS PubMed.
- A. G. Roberts, M. K. Bowman and D. M. Kramer, Biochemistry, 2004, 43, 7707–7716 CrossRef CAS PubMed.
- J. Yan, G. Kurisu and W. A. Cramer, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 69–74 CrossRef CAS PubMed.
- J. F. Allen, C. E. Sanders and N. G. Holmes, FEBS Lett., 1985, 193, 271–275 CrossRef CAS.
- J. F. Allen and N. G. Holmes, FEBS Lett., 1986, 202, 175–181 CrossRef CAS.
- M. A. Harrison, N. F. Tsinoremas and J. F. Allen, FEBS Lett., 1991, 282, 295–299 CrossRef CAS PubMed.
- M.-k. Yang, Z.-x. Qiao, W.-y. Zhang, Q. Xiong, J. Zhang, T. Li, F. Ge and J.-d. Zhao, J. Proteome Res., 2013, 12, 1909–1923 CrossRef CAS PubMed.
- Z. Chen, J. Zhan, Y. Chen, M. Yang, C. He, F. Ge and Q. Wang, Plant Cell Physiol., 2015, 56, 1997–2013 CrossRef CAS PubMed.
- P. Spaet, B. Maček and K. Forchhammer, Fron. Microbiol., 2015, 6, 248 Search PubMed.
- M. Nanba and S. Katoh, Biochim. Biophys. Acta, Bioenerg., 1985, 809, 74–80 CrossRef CAS.
- R. Vladkova, J. Biomol. Struct. Dyn., 2016, 34, 824–854 CrossRef CAS PubMed.
- C. E. Sanders, A. Melis and J. F. Allen, Biochim. Biophys. Acta, Bioenerg., 1989, 976, 168–172 CrossRef CAS.
- J. F. Allen, Biochim. Biophys. Acta, Bioenerg., 1992, 1098, 275–335 CrossRef CAS.
- J. Biggins, C. L. Campbell and D. Bruce, Biochim. Biophys. Acta, Bioenerg., 1984, 767, 138–144 CrossRef CAS.
- A. Krupa and N. Srinivasan, BMC Genomics, 2005, 6, 129 CrossRef CAS PubMed.
- J. Perez, A. Castaneda-Garcia, H. Jenke-Kodama, R. Müller and J. Munoz-Dorado, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15950–15955 CrossRef CAS PubMed.
- N. H. Mann, Microbiology, 1994, 140, 3207–3215 CrossRef CAS PubMed.
- T. Kaneko, S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura and S. Sasamoto, DNA Res., 1996, 3, 109–136 CrossRef CAS PubMed.
- C.-C. Zhang, L. Gonzalez and V. Phalip, Nucleic Acids Res., 1998, 26, 3619–3625 CrossRef CAS PubMed.
- A. Kamei, T. Yuasa, X. Geng and M. Ikeuchi, DNA Res., 2002, 9, 71–78 CrossRef CAS PubMed.
- C. J. Leonard, L. Aravind and E. V. Koonin, Genome Res., 1998, 8, 1038–1047 CrossRef CAS PubMed.
- A. Zorina, Russ. J. Plant Physiol., 2013, 60, 589–596 CrossRef CAS.
- A. Kamei, T. Yuasa, K. Orikawa, X. X. Geng and M. Ikeuchi, J. Bacteriol., 2001, 183, 1505–1510 CrossRef CAS PubMed.
- A. Mata-Cabana, M. García-Domínguez, F. J. Florencio and M. Lindahl, Antioxid. Redox Signaling, 2012, 17, 521–533 CrossRef CAS PubMed.
- C. Liang, X. Zhang, X. Chi, X. Guan, Y. Li, S. Qin and H. bo Shao, PLoS One, 2011, 6, e18718 CrossRef CAS PubMed.
- A. Zorina, V. Bedbenov, G. Novikova and V. Panichkin, Mol. Biol., 2014, 48, 390–398 CrossRef CAS.
- S. Laurent, J. Jang, A. Janicki, C.-C. Zhang and S. Bedu, Microbiology, 2008, 154, 2161–2167 CrossRef CAS PubMed.
- A. Galkin, L. Mikheeva and S. Shestakov, Mikrobiologiia, 2003, 72, 64–69 CAS.
- A. Zorina, N. Stepanchenko, G. V. Novikova, M. Sinetova, V. B. Panichkin, I. E. Moshkov, V. V. Zinchenko, S. V. Shestakov, I. Suzuki and N. Murata, DNA Res., 2011, 18, 137–151 CrossRef CAS PubMed.
- M. Angeleri, A. Zorina, E. M. Aro and N. Battchikova, FEBS Lett., 2018, 592, 411–421 CrossRef CAS PubMed.
- G. Schansker, S. Z. Tóth, L. Kovács, A. R. Holzwarth and G. Garab, Biochim. Biophys. Acta, Bioenerg., 2011, 1807, 1032–1043 CrossRef CAS PubMed.
- M. Magyar, G. Sipka, L. Kovács, B. Ughy, Q. Zhu, G. Han, V. Špunda, P. H. Lambrev, J.-R. Shen and G. Garab, Sci. Rep., 2018, 8, 2755 CrossRef PubMed.
- G. Sipka, P. Müller, K. Brettel, M. Magyar, L. Kovács, Q. Zhu, Y. Xiao, G. Han, P. H. Lambrev and J. R. Shen, Physiol. Plant., 2019, 166, 22–32 CrossRef CAS PubMed.
- G. Schansker, S. Z. Tóth, A. R. Holzwarth and G. Garab, Photosynth. Res., 2014, 120, 43–58 CrossRef CAS PubMed.
- Y. Shibata, S. Nishi, K. Kawakami, J.-R. Shen and T. Renger, J. Am. Chem. Soc., 2013, 135, 6903–6914 CrossRef CAS PubMed.
- A. Guskov, J. Kern, A. Gabdulkhakov, M. Broser, A. Zouni and W. Saenger, Nat. Struct. Mol. Biol., 2009, 16, 334 CrossRef CAS PubMed.
- J.-Y. Huang, Y.-F. Chiu, J. M. Ortega, H.-T. Wang, T.-S. Tseng, S.-C. Ke, M. Roncel and H.-A. Chu, Biochemistry, 2016, 55, 2214–2226 CrossRef CAS PubMed.
- B. E. Rubin, K. M. Wetmore, M. N. Price, S. Diamond, R. K. Shultzaberger, L. C. Lowe, G. Curtin, A. P. Arkin, A. Deutschbauer and S. S. Golden, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E6634–E6643 CrossRef CAS PubMed.
- S. Yang, R. Zhang, C. Hu, J. Xie and J. Zhao, Photosynth. Res., 2009, 99, 99–106 CrossRef CAS PubMed.
- T. Fujimori, Y. Hihara and K. Sonoike, J. Biol. Chem., 2005, 280, 22191–22197 CrossRef CAS PubMed.
- D. Bruce and J. Biggins, Biochim. Biophys. Acta, Bioenerg., 1985, 810, 295–301 CrossRef CAS.
- O. Salehian and D. Bruce, J. Lumin., 1992, 51, 91–98 CrossRef CAS.
| This journal is © The Royal Society of Chemistry and Owner Societies 2020 |



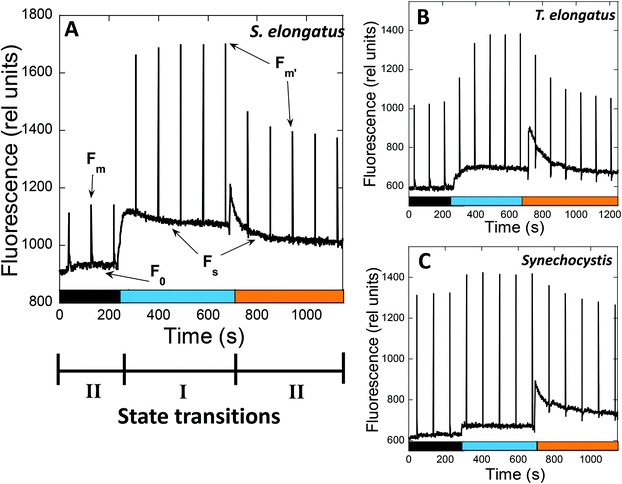
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μmol photons per m2 per s) were applied each 90 s. The PAM used in the measurements has a 650 nm detecting light, which is absorbed by PBS and Chl. Consequently, the changes in fluorescence yield observed under different conditions can be due to changes in the emission of PBS and/or Chl-antenna, or changes in excitation energy transfer between PBS and PSII. Three levels of fluorescence can be observed:
000 μmol photons per m2 per s) were applied each 90 s. The PAM used in the measurements has a 650 nm detecting light, which is absorbed by PBS and Chl. Consequently, the changes in fluorescence yield observed under different conditions can be due to changes in the emission of PBS and/or Chl-antenna, or changes in excitation energy transfer between PBS and PSII. Three levels of fluorescence can be observed: