Open Access Article
Open Access ArticleRetracted Article: Convenient synthesis of pyrimidine 2′-deoxyribonucleoside monophosphates with important epigenetic marks at the 5-position†
Song
Zheng
a,
Ai
Tran
a,
Alyson M.
Curry
b,
Dawanna S.
White
b and
Yana
Cen
 *bc
*bc
aDepartment of Pharmaceutical Sciences, Albany College of Pharmacy and Health Sciences, Colchester, VT 05446, USA
bDepartment of Medicinal Chemistry, Virginia Commonwealth University, Richmond, VA 23219, USA. E-mail: ceny2@vcu.edu; Tel: +1 804-828-7405
cInstitute for Structural Biology, Drug Discovery and Development, Virginia Commonwealth University, Richmond, VA 23219, USA
First published on 23rd June 2020
Abstract
Methyl groups of thymine and 5-methylcytosine (5mC) bases in DNA undergo endogenous oxidation damage. Additionally, 5mC residues can be enzymatically deaminated or oxidized through either genetic alterations or the newly identified epigenetic reprogramming pathway. Several methods have been developed to measure the formation of modified DNA nucleobases including 32P-postlabeling. However, the postlabeling method is often limited by the absence of authentic chemical standards. The synthesis of monophosphate standards of nucleotide oxidation products is complicated by the presence of additional functional groups on the modified bases that require complex protection and deprotection strategies. Due to the emerging interest in the pyrimidine oxidation products, the corresponding protected 3′-phosphoramidites needed for solid-phase oligonucleotide synthesis have been reported, and several are commercially available. We report here an efficient synthesis of 3′-monophosphates from 3′-phosphoramidites and the subsequent enzymatic conversion of 3′-monophosphates to the corresponding 5′-monophosphates using commercially available enzymes.
Introduction
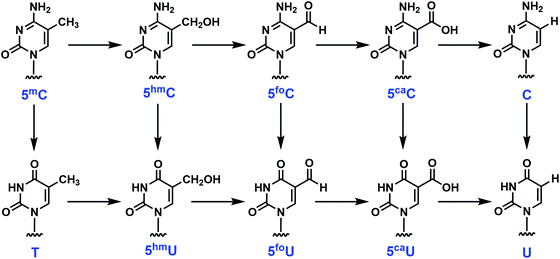
The importance of DNA oxidation and hydrolysis in the development of human diseases is increasingly recognized.1–3 Recently a biochemical pathway has been identified in which 5-methylcytosine (5mC), an important component of the epigenetic landscape in higher organisms, can be enzymatically oxidized to the corresponding 5-hydroxymethyl (5hmC), formyl (5foC) and carboxyl (5caC) analogs, mediated by ten eleven translocation (TET) enzymes (Scheme 1).4–9 This epigenetic reprogramming pathway is essential to normal development and differentiation, yet, defective in most human cancer cells.4–11 The spontaneous hydrolysis of 5mC to T at methylated CpG islands has long been considered the mechanism underlying the single most frequent type of single-base mutation found in human cancer cells.12–14 This pathway might also include enzymatic deamination to the corresponding uracil analogs followed by glycosylase removal and DNA repair (Scheme 1).8,9 In addition to the active demethylation pathway, an alternative proposal for restoration of cytosine (C) involves the direct C–C bond cleavage at the 5-position to remove the substitution.15 The potential coexistence of the enzymatic 5mC modification pathway and established endogenous DNA damage and repair pathways creates significant analytical challenges.Multiple methods have been developed to quantify the abovementioned epigenetic modifications including chromatographic separation by HPLC, GC and TLC and identification and quantification by mass spectrometry, UV detection or 32P-labeling. The 32P-labeling approach has proven to be very sensitive for measuring a multitude of DNA damage products;16–19 however, this approach is often limited by the absence of authentic standards. While MS methods have emerged as extremely powerful and sensitive approaches to the measurement and unequivocal identification of DNA damage products, 32P-labeling strategies provide an approach for following sequential conversions of specific nucleotides through multiple chemical or enzymatic pathways in complicated matrices including intact cells and cell extracts and at specific sequence positions within the oligonucleotides.
In the case of the modified pyrimidines examined here, chemical synthesis of the 2′-deoxynucleosides can be challenging, and conversion to the corresponding monophosphates is complicated by the additional functional groups present on the modified bases that must be selectively protected prior to chemical phosphorylation.20–23 Due to the growing interest in the modified pyrimidines, several groups have reported the synthesis of phosphoramidite derivatives needed for the solid-phase assembly of oligonucleotides containing these modifications (Table 1), and some of these modified phosphoramidites are now commercially available. The investment made in developing methods for the protection and deprotection of the 3′-phosphoramidites could be leveraged to provide a convenient source for both the corresponding 3′- and 5′-monophosphates. Here we report a novel and versatile synthesis of 3′-dNMPs using nucleoside phosphoramidites as building blocks. With the availability of fully characterized dNMPs, not only the composition of synthetic oligonucleotides can be determined, but also the 32P-labeling strategy can be applied to this group of biologically important pyrimidine analogs.
| 3′-dNMPs | Elemental composition [M − H]− | Exact mass, m/z [M − H]− | Measured mass, m/z [M − H]− | λ max (nm) | Extinction coefficient (103 M−1 cm−1) | Retention timea (min) |
|---|---|---|---|---|---|---|
| a The retention times were obtained on an Aquasil C18 column using an ammonium acetate (20 mM, pH 5.5) – methanol solvent system. | ||||||
| 3′-cadUMP | C10H12N2O10P− | 351.0230 | 351.0236 | 271 | 12.3 ± 0.5 | 3.25 |
| 3′-cadCMP | C10H13N3O9P− | 350.0389 | 350.0390 | 280 | 10.6 ± 0.5 | 3.52 |
| 3′-dCMP | C9H13N3O7P− | 306.0491 | 306.0490 | 269 | 9.7 ± 0.7 | 5.78 |
| 3′-hmdCMP | C10H15N3O8P− | 336.0597 | 336.0586 | 274 | 12.5 ± 0.7 | 6.22 |
| 3′-dUMP | C9H12N2O8P− | 307.0331 | 307.0345 | 261 | 13.7 ± 0.8 | 6.71 |
| 3′-hmdUMP | C10H14N2O9P− | 337.0437 | 337.0444 | 263 | 11.9 ± 0.5 | 6.97 |
| 3′-mdCMP | C10H15N3O7P− | 320.0648 | 320.0643 | 277 | 10.2 ± 0.3 | 10.17 |
| 3′-fodUMP | C10H12N2O9P− | 335.0280 | 335.0289 | 275 | 17.2 ± 0.3 | 10.88 |
| 3′-dTMP | C10H14N2O8P− | 321.0488 | 321.0493 | 267 | 10.3 ± 0.6 | 11.47 |
| 3′-fodCMP | C10H13N3O8P− | 334.0440 | 334.0424 | 283 | 14.1 ± 0.3 | 13.27 |
Results and discussion
Synthesis of 3′-dNMPs
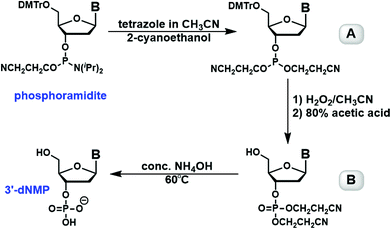
The formation of deoxynucleoside-3′-phosphate triester with a free 5′-hydroxyl group was further confirmed by NMR analysis. 1H NMR showed complete disappearance of signals corresponding to trityl group protons, and 31P signals shifted from around 148 ppm (doublet) in phosphoramidites to −3.5 ppm (singlet) in deoxynucleoside-3′-phosphate triesters.
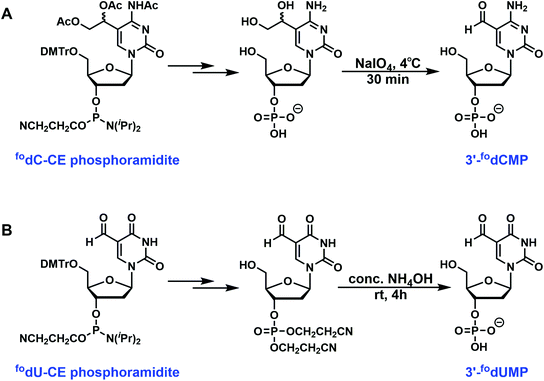
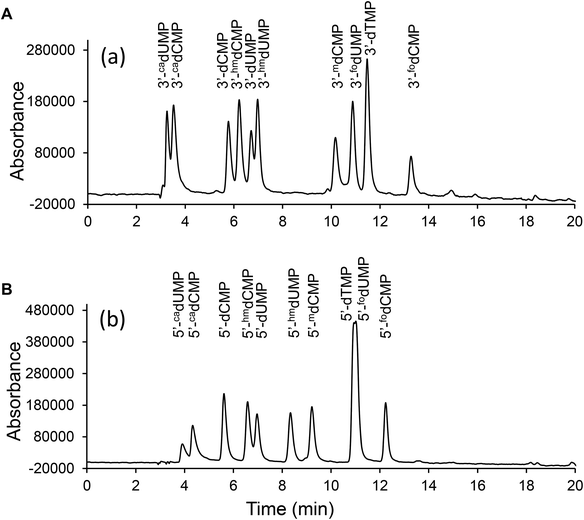
The aforementioned synthetic protocols of 3′-dNMPs have several beneficial features. First of all, phosphoramidites are either commercially available or easily accessible, which enables a straightforward construction of 3′-dNMPs. Secondly, already-existing protecting groups in phosphoramidites prevent undesired side reactions. Reactions were easily monitored by TLC, and the 3′-dNMPs were obtained in 70–90% yields with minimal purification (most synthesis only requires one chromatographic isolation). The method is, therefore, suitable for large scale synthesis for a broad class of compounds. Corresponding MS (Table 1), UV (Table 1), HPLC (Table 1, Fig. 3 and S1†) and NMR (ESI†) data are provided for each of the ten 3′-monophosphates prepared here.
Enzymatic conversion of 3′-dNMP to 5′-dNMP
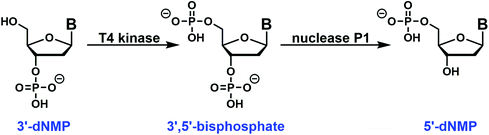
The easy access to the 3′-dNMPs made the facile synthesis of 5′-dNMPs possible. This enzymatic conversion was inspired by phosphoglycerate mutase, a glycolytic enzyme.24 It catalyzes the conversion of 3-phosphoglycerate to 2-phosphoglycerate through a 2,3-bisphosphoglycerate intermediate.24,25 In our two-enzyme coupled synthesis, the 3′-dNMP was first incubated with ATP and a T4 kinase lacking the 3′-phosphatase activity to generate the corresponding 3′,5′-bisphosphate (Fig. 4).26,27 Subsequently, the 3′,5′-bisphosphate was dephosphorylated by nuclease P1 to afford the 5′-dNMP.28 Both T4 kinase and nuclease P1 demonstrated excellent functional group tolerance as 5′-dNMPs carrying various substituents were generated in decent to good yields by this robust sequence (Table 2). Similar to the 3′-dNMPs, the 5′-dNMPs can be readily resolved by HPLC (Fig. 3).| 5′-dNMPs | Yielda (%) | Retention timeb (min) |
|---|---|---|
| a Yield for two steps starting from the 3′-dNMP. b The retention times were obtained on an Aquasil C18 column using an ammonium acetate (20 mM, pH 5.5) – methanol solvent system. | ||
| 5′-cadUMP | 80.2 | 3.91 |
| 5′-cadCMP | 79.2 | 4.33 |
| 5′-dCMP | 53.2 | 5.61 |
| 5′-hmdCMP | 85.7 | 6.58 |
| 5′-dUMP | 54.0 | 6.97 |
| 5′-hmdUMP | 81.5 | 8.34 |
| 5′-mdCMP | 56.2 | 9.22 |
| 5′-dTMP | 83.3 | 10.94 |
| 5′-fodUMP | 59.3 | 11.01 |
| 5′-fodCMP | 31.5 | 12.24 |
Determination of synthetic oligonucleotide composition using dNMP standards
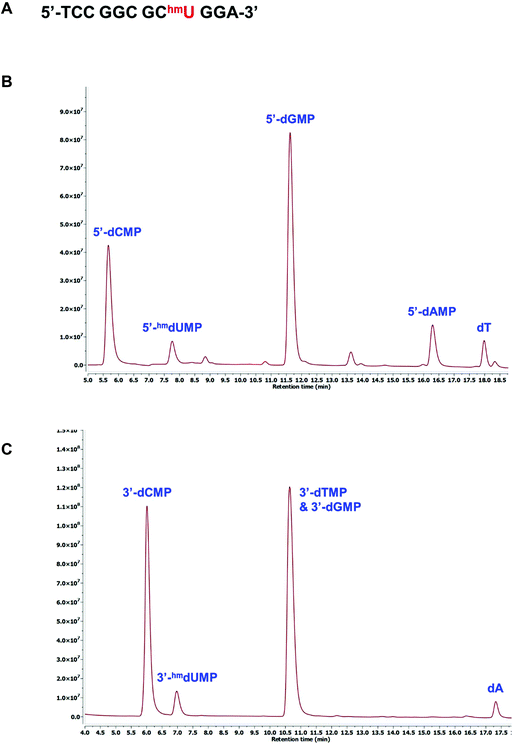
The presence of modified pyrimidines in synthetic oligonucleotides can be confirmed using either the 3′- or 5′-dNMP standards. A 12mer oligonucleotide containing an internal 5hmU residue was generated by standard solid-phase DNA synthesis (Fig. 5A). This oligonucleotide was incubated with snake venom phosphodiesterase I, an exonuclease capable of successively hydrolyzing 5′-dNMPs from the 3′-terminus of a DNA sequence.29,30 The digested sample was then injected onto HPLC for further analysis. Comparison with the 5′-dNMP standards indicated that in addition to G, T, C and A, 5hmU was also present in the sequence (Fig. 5B). The composition of the oligonucleotide can also be determined by treating the same 12mer with bovine spleen phosphodiesterase II, an exonuclease that can remove 3′-dNMPs sequentially from the 5′-terminus.31 HPLC analysis suggested the formation of 3′-hmdUMP along with other nucleoside monophosphates (Fig. 5C).Probing conversions of nucleobase using dNMP standards
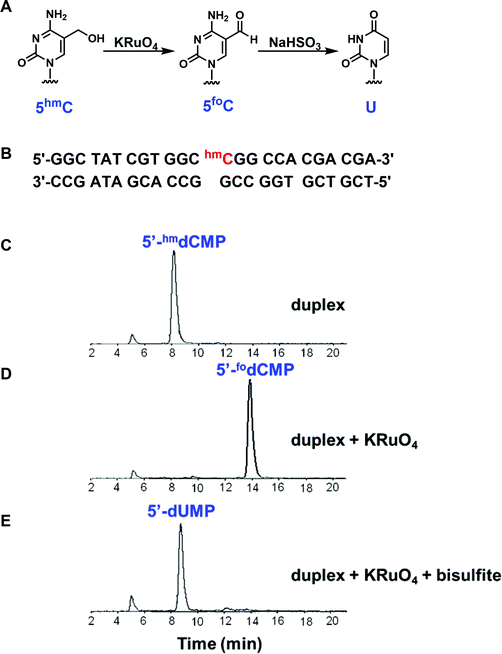
The availability of the dNMP standards should facilitate experiments probing both chemical and enzymatic conversions of target residues in oligonucleotides. We demonstrate this by examining the chemical reactivity of 5hmC in a duplex oligonucleotide. Bisulfite sequencing has become extremely powerful in distinguishing cytosine from 5mC residues and thus tracking methylation patterns in genomic DNA.32–34 However, 5mC and 5hmC cannot be distinguished by this method.35 It has been reported that 5hmC can be oxidized to 5foC by KRuO4.36–38 Subsequently, bisulfite treatment can convert 5foC to uracil (U).39 This two-step sequence could serve as an alternative approach in detecting 5hmC level (Fig. 6A). In order to evaluate this conversion sequence directly, a 12mer oligonucleotide containing a 5hmC residue at the 5′-position was 32P-labeled, annealed to the unlabeled 24mer complementary strand and then ligated to an unlabeled 12mer oligonucleotide to form a duplex (Fig. 6B). A fraction of the duplex was digested with DNase I and snake venom phosphodiesterase I. The resulting 5′-dNMPs were then resolved by HPLC with an in-line radio-detector. The 32P label was found exclusively in the 5′-hmdCMP (Fig. 6C). A second fraction of the duplex was treated with KRuO4 and digested as described above. The formation of 5foC was readily detected (Fig. 6D). The complete conversion of 5hmC to U was observed when the duplex was incubated with KRuO4 followed by bisulfite treatment and enzymatic digestion. In this case, the radioactivity was detected in 5′-dUMP exclusively (Fig. 6E). While analytical methods have been used to confirm the conversion of 5hmC to U,36 ours is the first study to directly follow the transformations in on oligonucleotide with 32P-labeling. The selective 32P-labeling of a target base, followed by enzymatic digestion and HPLC analysis, provides a method to monitor either enzymatic or chemical conversions, even with crude cellular extracts.Conclusions
Phosphoramidites prepared for the synthesis of oligonucleotides containing biologically important modified bases can be conveniently converted to the corresponding 3′-dNMPs. 5′-dNMPs are then obtained through enzyme-mediated phosphorylation and dephosphorylation. Authentic 3′- and 5′-dNMP standards, such as those described here, are essential for the validation of highly sensitive analytical methods using 32P-labeling. We have demonstrated this approach by following the conversion of a selectively-labeled 5hmC residue in an oligonucleotide to 5foC, and then on to U, using Rh oxidation and bisulfite conversion methods currently employed to sequence 5mC and 5hmC residues in DNA. This approach can be extended to study future base modification strategies needed to follow additional base conversions in DNA, including those involved with epigenetic reprogramming.Materials and methods
Materials
Reagent grade solvents including dichloromethane, methanol, and ethyl acetate were purchased from Fisher Scientific (Pittsburgh, PA) and used without further purification. Anhydrous grade solvents including tetrahydrofuran, pyridine, trimethylamine, and acetonitrile were purchased from Sigma-Aldrich (St Louis, MO) and used as received unless otherwise noted. Di-tert-butylsilyl bis(trifluoromethanesulfonate), dimethylformamide, 5-iodo-2′-deoxyuridine, 4,4′-dimethoxytrityl chloride, tris(dibenzylideneacetone)dipalladium(0), triphenylphosphine, toluene, imidazole, tributyltin hydride, HF-pyridine, diisopropylethylamine, 2-cyanoethanol, 2-cyanoethyl N,N,N′,N′-tetraisopropylphosphordiamidite, and carbon monoxide were purchased from Sigma-Aldrich (St Louis, MO). Commercially available phosphoramidites and sublimed 1H-tetrazole in anhydrous acetonitrile were purchased from Glen Research (Sterling, VA). Thin layer chromatography (TLC) was performed on precoated silica gel 60 F254, 5 cm × 20 cm, 250 μM thick plates purchased from EMD (Gibbstown, NJ). Snake venom phosphodiesterase I and bovine spleen phosphodiesterase II were purchased from Worthington Biochemical. T4 polynucleotide kinase and T4 DNA ligase were purchased from New England Biolabs. Nuclease P1 was purchased from US Biologicals.1H, 13C, and 31P NMR spectra were obtained using a Bruker Ultrashield 300 MHz spectrometer (Manning Park, MA). HPLC analysis was performed with a ThermoFinnigan (Waltham, MA) Surveyor HPLC system with a photodiode array and a single quad mass spectrometer detector. For the analysis of 32P-labeled monophosphates, a similar HPLC system was interfaced with a Berthold Technologies FlowStar LB513 radioactivity detector (Wildbad, Germany). Oligonucleotides were synthesized with a Millipore Expedite nucleic acids synthesis system (Billerica, MA). High-resolution mass spectra were obtained with either a Waters Micromass Q-tof Ultima (Milford, MA) or a Thermo Scientific Q-Exactive Hybrid Quadrupole Orbitrap (Waltham, MA).
General procedures for deoxynucleoside-3′-monophosphate synthesis
Special deprotection strategies
Enzymatic conversion of a 3′-dNMP to the corresponding 5′-dNMP
Each 3′-dNMP was enzymatically converted to the corresponding 5′-dNMP using a series of two enzymatic reactions. In the first step, the 3′-dNMP was converted to the 3′,5′-bisphosphate. In this reaction, 400 nmol of the 3′-dNMP was combined with 20 μL 1 M Tris-HCl, pH 7.0, 20 μL 0.1 M MgCl2, 20 μL 50 mM DTT, 40 μL 20 mM ATP, and 20 μL T4 kinase (10 U μL−1) lacking 3′-phosphatase activity in a total volume of 200 μL as described previously.26,27 The reaction mixture was incubated at 37 °C, and progress was followed by HPLC analysis of a 5 μL aliquot of the reaction. In some cases, the 3′-dNMP co-eluted with ATP. However, the ATP can be reduced by incubation with 1 μL of 1 U μL−1 apyrase for 10 min prior to HPLC analysis.The 3′,5′-bisphosphate generated in the above reaction was purified by HPLC as described below and converted to the 5′-dNMP with nuclease P1 as described previously.28 HPLC fractions containing the bisphosphate were combined and dried. The bisphosphate was incubated in water (50 μL), buffer (25 μL 0.25 M NaOAc, pH 5.0, 15 μL 2 mM ZnCl2) and nuclease P1 (10 μL 5 μg μL−1) at 37 °C. The reaction was monitored by HPLC, and the final product 5′-dNMP isolated by HPLC and characterized by UV and mass spectrometry as described below.
Determination of extinction coefficient
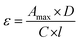 | (1) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by start-up funding from VCU (to Y. C.) and from NSF (CHE-1846785 to Y. C.).References
- E. Mullaart, P. H. Lohman, F. Berends and J. Vijg, Mutat. Res., 1990, 237, 189–210 CAS.
- B. N. Ames, L. S. Gold and W. C. Willett, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 5258–5265 CrossRef CAS PubMed.
- P. Lonkar and P. C. Dedon, Int. J. Cancer, 2011, 128, 1999–2009 CrossRef CAS PubMed.
- S. Kriaucionis and N. Heintz, Science, 2009, 324, 929–930 CrossRef CAS PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Science, 2009, 324, 930–935 CrossRef CAS PubMed.
- S. Ito, A. C. D'Alessio, O. V. Taranova, K. Hong, L. C. Sowers and Y. Zhang, Nature, 2010, 466, 1129–1133 CrossRef CAS PubMed.
- M. Munzel, D. Globisch and T. Carell, Angew. Chem., Int. Ed., 2011, 50, 6460–6468 CrossRef PubMed.
- Y. Fu and C. He, Curr. Opin. Chem. Biol., 2012, 16, 516–524 CrossRef CAS PubMed.
- L. Shen and Y. Zhang, Curr. Opin. Cell Biol., 2013, 25, 289–296 CrossRef CAS PubMed.
- C. Liu, L. Liu, X. Chen, J. Shen, J. Shan, Y. Xu, Z. Yang, L. Wu, F. Xia, P. Bie, Y. Cui, X. W. Bian and C. Qian, PLoS One, 2013, 8, e62828 CrossRef CAS PubMed.
- H. Yang, Y. Liu, F. Bai, J. Y. Zhang, S. H. Ma, J. Liu, Z. D. Xu, H. G. Zhu, Z. Q. Ling, D. Ye, K. L. Guan and Y. Xiong, Oncogene, 2013, 32, 663–669 CrossRef CAS PubMed.
- P. A. Jones, W. M. Rideout 3rd, J. C. Shen, C. H. Spruck and Y. C. Tsai, Bioessays, 1992, 14, 33–36 CrossRef CAS PubMed.
- S. Tornaletti and G. P. Pfeifer, Oncogene, 1995, 10, 1493–1499 CAS.
- V. Rusmintratip and L. C. Sowers, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 14183–14187 CrossRef CAS PubMed.
- A. Schon, E. Kaminska, F. Schelter, E. Ponkkonen, E. Korytiakova, S. Schiffers and T. Carell, Angew. Chem., 2020, 59, 5591–5594 CrossRef PubMed.
- N. J. Gorelick, Mutat. Res., 1993, 288, 5–18 CrossRef CAS PubMed.
- M. V. Reddy and K. Randerath, Carcinogenesis, 1986, 7, 1543–1551 CrossRef CAS PubMed.
- M. Zeisig, T. Hofer, J. Cadet and L. Moller, Carcinogenesis, 1999, 20, 1241–1245 CrossRef CAS PubMed.
- D. H. Phillips and V. M. Arlt, Nat. Protoc., 2007, 2, 2772–2781 CrossRef CAS PubMed.
- D. K. Rogstad, J. Heo, N. Vaidehi, W. A. Goddard 3rd, A. Burdzy and L. C. Sowers, Biochemistry, 2004, 43, 5688–5697 CrossRef CAS PubMed.
- K. Sato, W. Hirose and A. Matsuda, Curr. Protoc. Nucleic Acid Chem., 2008, 1.21.1–1.21.19 Search PubMed , chapter 1, unit 1 21.
- Q. Dai and C. He, Org. Lett., 2011, 13, 3446–3449 CrossRef CAS PubMed.
- A. S. Schroder, J. Steinbacher, B. Steigenberger, F. A. Gnerlich, S. Schiesser, T. Pfaffeneder and T. Carell, Angew. Chem., Int. Ed., 2014, 53, 315–318 CrossRef PubMed.
- L. A. Fothergill-Gilmore and H. C. Watson, Adv. Enzymol. Relat. Areas Mol. Biol., 1989, 62, 227–313 CAS.
- H. I. Fraser, M. Kvaratskhelia and M. F. White, FEBS Lett., 1999, 455, 344–348 CrossRef CAS PubMed.
- J. R. Lillehaug and K. Kleppe, Biochemistry, 1975, 14, 1221–1225 CrossRef CAS PubMed.
- E. Hilario, Mol. Biotechnol., 2004, 28, 77–80 CrossRef CAS PubMed.
- D. H. Phillips, A. Hewer and V. M. Arlt, Methods Mol. Biol., 2005, 291, 3–12 CAS.
- M. Laskowski, in The Enzymes, ed. P. D. Boyer, Academic Press, 1971, vol. 4, pp. 313–328 Search PubMed.
- N. W. Ho and P. T. Gilham, Biochim. Biophys. Acta, 1973, 308, 53–58 CrossRef CAS.
- A. Bernardi and G. Bernardi, in The Enzymes, ed. P. D. Boyer, Academic Press, 1971, vol. 4, pp. 329–336 Search PubMed.
- M. Frommer, L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy and C. L. Paul, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 1827–1831 CrossRef CAS PubMed.
- A. Chatterjee, E. J. Rodger, I. M. Morison, M. R. Eccles and P. A. Stockwell, Methods Mol. Biol., 2017, 1537, 249–277 CrossRef CAS PubMed.
- F. Leti, L. Llaci, I. Malenica and J. K. DiStefano, Methods Mol. Biol., 2018, 1706, 233–254 CrossRef CAS PubMed.
- Y. Huang, W. A. Pastor, Y. Shen, M. Tahiliani, D. R. Liu and A. Rao, PLoS One, 2010, 5, e8888 CrossRef PubMed.
- M. J. Booth, M. R. Branco, G. Ficz, D. Oxley, F. Krueger, W. Reik and S. Balasubramanian, Science, 2012, 336, 934–937 CrossRef CAS PubMed.
- W. Mao, J. Hu, T. Hong, X. Xing, S. Wang, X. Chen and X. Zhou, Org. Biomol. Chem., 2013, 11, 3568–3572 RSC.
- P. Schuler and A. K. Miller, Angew. Chem., Int. Ed., 2012, 51, 10704–10707 CrossRef PubMed.
- L. Jin, W. Wang, D. Hu and J. Lu, Phys. Chem. Chem. Phys., 2014, 16, 3573–3585 RSC.
- O. Shimelis, X. Zhou, G. Li and R. W. Giese, J. Chromatogr. A, 2004, 1053, 143–149 CrossRef CAS PubMed.
- M. J. Gait, Oligonucleotide synthesis : a practical approach, IRL Press, Oxford Oxfordshire, Washington, DC, 1984 Search PubMed.
- J. Fujimoto, L. Tran and L. C. Sowers, Chem. Res. Toxicol., 1997, 10, 1254–1258 Search PubMed.
- V. Guerniou, D. Gasparutto, S. Sauvaigo, A. Favier and J. Cadet, Nucleosides, Nucleotides Nucleic Acids, 2003, 22, 1073–1075 CrossRef CAS PubMed.
- Z. Cui, J. A. Theruvathu, A. Farrel, A. Burdzy and L. C. Sowers, Anal. Biochem., 2008, 379, 196–207 CrossRef CAS PubMed.
- R. P. Darst, C. E. Pardo, L. Ai, K. D. Brown and M. P. Kladde, Curr. Protoc. Mol. Biol., 2010, 7.9.1–7.9.17 CrossRef PubMed , chapter 7, unit 7 9 1–17.
- Y. Li and T. O. Tollefsbol, Methods Mol. Biol., 2011, 791, 11–21 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ob00884b |
| This journal is © The Royal Society of Chemistry 2020 |