Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efflux pump insensitive rhodamine–jasplakinolide conjugates for G- and F-actin imaging in living cells†
Rūta
Gerasimaitė
 a,
Jan
Seikowski
b,
Jens
Schimpfhauser
b,
Georgij
Kostiuk
a,
Jan
Seikowski
b,
Jens
Schimpfhauser
b,
Georgij
Kostiuk
 a,
Tanja
Gilat
c,
Elisa
D'Este
a,
Tanja
Gilat
c,
Elisa
D'Este
 d,
Sebastian
Schnorrenberg‡
d,
Sebastian
Schnorrenberg‡
 c and
Gražvydas
Lukinavičius
c and
Gražvydas
Lukinavičius
 *a
*a
aChromatin Labeling and Imaging Group, Department of Nanobiophotonics, Max Planck Institute for Biophysical Chemistry, Am Faßberg 11, 37077, Göttingen, Germany. E-mail: grazvydas.lukinavicius@mpibpc.mpg.de
bFacility for Synthetic Chemistry, Max Planck Institute for Biophysical Chemistry, Am Faßberg 11, 37077, Göttingen, Germany
cDepartment of Nanobiophotonics, Max Planck Institute for Biophysical Chemistry, Am Faßberg 11, 37077, Göttingen, Germany
dOptical Microscopy Facility, Max Planck Institute for Medical Research, Jahnstraße 29, 69120 Heidelberg, Germany
First published on 26th March 2020
Abstract
The actin cytoskeleton is crucial for endocytosis, intracellular trafficking, cell shape maintenance and a wide range of other cellular functions. Recently introduced cell-permeable fluorescent actin probes, such as SiR-actin, suffer from poor membrane permeability and stain some cell populations inhomogeneously due to the active efflux by the plasma membrane pumps. We analyzed a series of new probes composed of jasplakinolide and modified rhodamine fluorophores and found that rhodamine positional isomerism has a profound effect on probe performance. The probes based on the 6'-carboxy-carbopyronine scaffold are considerably less susceptible to efflux and allow efficient staining without efflux pump inhibitors. They can be used for 2D and 3D fluorescence nanoscopy at high nanomolar concentrations without significant cytotoxicity. We show that jasplakinolide-based fluorescent probes bind not only to actin filaments, but also to G-actin, which enables imaging highly dynamic actin structures. We demonstrate an excellent performance of the new probes in multiple organisms and cell types: human cell lines, frog erythrocytes, fruit fly tissues and primary neurons.
Introduction
Exploiting the full potential of fluorescence microscopy requires tools and methods for specific labelling of targets in living cells.1 Small molecule probes, directed at organelles or proteins, are gaining momentum, as they do not require genetic modification and allow the choice of fluorophore with the desired optical properties.2–5 Such probes are synthesized by coupling a fluorophore to a well-characterized, cell permeable ligand, which binds its target with high affinity and specificity. SiR-actin, consisting of 6′-carboxy silicon rhodamine (6′-SiR) 9 linked to the toxin jasplakinolide (JAS), is a widely used probe for staining actin in living cells.6,7 Due to fluorogenicity, it yields high contrast confocal and stimulated emission depletion (STED) microscopy images. However, SiR-actin is susceptible to the active efflux by the plasma membrane pumps, which results in poor or mosaic staining of some cell lines.6,8,9 Herein we introduce new JAS-based probes for actin that are considerably less sensitive to efflux and thus allow uniform staining of cancer cell lines that possess high multidrug resistance pump activity. In addition, we demonstrate that the fluorescent jasplakinolide derivatives are able to interact with both G- and F-actin.The modular probes for live-cell imaging often exploit rhodamines as reporters.10 These dyes exist in a dynamic equilibrium between fluorescent zwitterionic state and non-fluorescent spirolactone. A more hydrophobic spirolactone facilitates passage through the cell membrane, whereas binding to the target shifts the equilibrium towards the fluorescent form, thus ensuring high contrast images.11 Given this simple mechanism, most of the efforts to improve the performance of modular probes were directed at fine-tuning the spirolactone–zwitterion equilibrium by modifying the dye structure.12,13 However, the optimized probes still required the use of verapamil to inhibit efflux pumps and to achieve efficient staining.12 This suggests that either the improvements were insufficient or factors other than the spirolactone–zwitterion equilibrium might affect probe performance.
Previously, we and others have shown that finding an optimal fluorophore–ligand pair can make all the difference between useless and excellent-performing probes.6,14–16 The changes as subtle as the attachment position on the dye can have a dramatic effect on staining of the target.8,16–18 These examples call for a better understanding of the interplay between the dye and the targeting ligand in determining the probe properties and underscore the importance of combinatorial screening when seeking new probes. Thus, we explored the possibility to identify an improved living cell compatible actin probe.
Experimental methods
Spectra and fluorogenicity of the probes
The absorbance and fluorescence spectra of the probes were recorded under four conditions: (i) in phosphate buffered saline pH 7.4 (PBS), (ii) in PBS with bovine serum albumin (BSA), (iii) in general actin buffer (5 mM Tris-HCl pH 8.0, 0.2 mM CaCl2) with F-actin and (iv) in PBS with 0.1% SDS. The following reagents were mixed to a total volume of 250 μl in a black glass-bottom 96-well plate (MatTek, cat. no. PBK96G-1.5-5-F): respective buffer, 2 μM probe, and 0.5 mg ml−1 actin or BSA. Then, 25 μl of 10× actin polymerization buffer (actin sample) or PBS (all other samples) was added, the solution was mixed by pipetting and the plates were incubated for 1 h at 37 °C (10× actin polymerization buffer contains 500 mM KCl, 20 mM MgCl2, 0.05 mM guanidine carbonate and 10 mM ATP). The spectra were acquired using a Tecan Spark20 M plate reader in the bottom reading mode. The spectra were processed and analyzed using a|e v2.2 (background subtraction and normalization) (FluorTools, http://www.fluortools.com) and Spectragryph v1.2.11 (spectra averaging) (F. Menges “Spectragryph – optical spectroscopy software”, version 1.2.11, 2019, http://www.effemm2.de/spectragryph/). To account for light scattering, the spectra of the solutions containing no probes but an equivalent amount of DMSO were acquired and subtracted from the respective probe spectra. Then the absorbance and fluorescence spectra of a probe were normalized to Amax or Fmax of the SDS sample. The experiment was repeated three times; the normalized spectra were averaged and are presented in Fig. S1.†Maintenance of cells
U-2 OS, human primary dermal fibroblasts and HeLa cells were cultured in high-glucose DMEM (Dulbecco's Modified Eagle's Medium, Life Technologies, cat. no. 31053-028) supplemented with GlutaMAX-1 (Life Technologies, cat. no. 35050-038) and 10% foetal bovine serum (FBS, Life Technologies, cat. no. 10270-106) in a humidified 5% CO2 incubator at 37 °C.The medium for normal neonatal human melanocytes (MatTek Corporation, cat. NHM-CRY-NEO) was prepared using MGM™-4 Melanocyte Growth Medium-4 BulletKit™ (Lonza, cat. no. CC-3249), and the cells were grown in a humidified 5% CO2 incubator at 37 °C.
All cell types were split every 3–4 days or at confluence. Staining was performed in the growth media.
Staining and imaging of living cells
U-2 OS cells were grown in 12-well uncoated glass bottom plates (MatTek) in DMEM supplemented with 10% FBS. For staining, the cells were incubated for 2 h at 37 °C in DMEM containing 250 nM or 1 μM probe and 0.1 μg ml−1 Hoechst in the presence or absence of 10 μM verapamil. When using 6-LIVE 510-JAS, 6-LIVE 515-JAS and 6-530RH-JAS probes, the cells were stained and imaged in FluoroBrite™ DMEM medium (Thermo Fisher Scientific, cat. no. A1896701). After the incubation, the cells were washed once with DMEM and once with Hank's Balanced Salt Solution (HBSS). Fresh DMEM was added and the cells were imaged using a wide-field Lionheart FX Automated Microscope (BioTek) with a 20× objective, using laser autofocus. 16 fields of view in 3 focusing planes, spanning 6 μm in thickness, were acquired per well. Image stitching and focus stacking were performed using in-built Gene 5 software (BioTek). The final images encompassed 1380 × 950 μm field of view and contained at least 220 and up to 2000 cells, which ensured analysis with no bias towards a well-stained sub-population. Each experiment was repeated independently 3 times. Actin staining was quantified with the automated pipeline built in CellProfiler v.3.1.8 (Fig. S2†).19 Statistical analysis was performed using GraphPad Prism v. 6.03. In each experiment, we compared verapamil treated and untreated populations using the Mann–Whitney test with a confidence level of 95%. To estimate the magnitude of differences between conditions, we used the ratio of medians (treated versus untreated). We compared staining in each of the three untreated samples versus each of the three verapamil treated samples, thus yielding 9 ratios for 3 independent experiments. We employed the one-sample t-test to check whether the effect was statistically different from 1, the ratio that would mean no difference.Normal neonatal human melanocytes were cultured in 12-well uncoated glass bottom plates in normal human melanocyte cell growth medium in a humidified 5% CO2 incubator at 37 °C. For staining, the cells were incubated for 1 h at 37 °C in the growth media containing 1 μM 6-580CP-JAS and 100 nM 5-SiR-Hoechst and imaged without washing.
Formaldehyde fixation and determination of KF-actinD
U-2 OS cells were grown as before, briefly washed with 1 ml HBSS and incubated for 10 min with 1 ml of 4% formaldehyde in PBS (pH 7.4). The solution was removed, and 1 ml of 30 mM glycine in PBS (pH 7.4) was added for 5 min. Then the samples were washed 3 × 5 min with 1 ml PBS, incubated for 5 min with 1 ml of 0.1% reduced Triton X-100 in PBS and washed again 3 × 5 min with 1 ml PBS. The samples were blocked by incubating with 1 ml of 1% BSA in PBS for 30 min and stored in PBS at 4 °C. The cells were stained by incubating with 1 ml of 1–1000 nM probe and 0.1 μg ml−1 Hoechst in PBS for 30 min at RT, washed 2 times with 1 ml PBS and imaged on a wide-field Lionheart FX Automated Microscope (BioTek) with a 4× objective, using laser autofocus. Actin staining was measured using the CellProfiler v.3.1.8 (ref. 19) pipeline, as explained in Fig. S2,† and the data were fitted into dose–response eqn (1):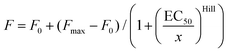 | (1) |
Probe interaction with G-actin
The reactions were performed in general actin buffer, supplemented with 0.2 mM ATP and 0.5 mM DTT. Eleven 2-fold dilutions of actin were prepared, starting from 8 μM, and aliquots of 25 μl were transferred into a black 96-well half-area plate (Greiner Bio-One, cat. no. 675076). Immediately before use, probes were diluted from DMSO stock solution to 20 nM in the same buffer at room temperature, and 25 μl aliquots were mixed with actin dilutions. The samples were incubated at 37 °C for 1 h and the fluorescence was read with a Tecan Spark20 M plate reader in the top-reading mode. Excitation/emission wavelengths (nm) were as follows: 470/530 for LIVE 510 and LIVE 515, 510/560 for 530RH, 570/610 for 580CP, 570/630 for 610CP and 590/650 for SiR; excitation and emission bandwidths were in all the cases 15 nm. Background fluorescence of the sample without actin was subtracted. When assessing the effects of latrunculin A or DNase I, actin dilutions were prepared in the buffer containing 10 μM of these inhibitors, thus making their concentration constant in the whole titration series and equal to 5 μM in the final reaction mixture. The data were fitted into full eqn (2) of a single site binding: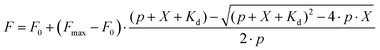 | (2) |
STED nanoscopy of neurons
Primary rat hippocampal neurons were prepared as previously described.6 Cells were labelled in the growth medium at 30 days in vitro for 45 min with 100 nM probe and washed prior to imaging. Axons were identified by labelling with an anti-neurofascin antibody (NeuroMab, 75-172) and an anti-mouse AlexaFluor 488 secondary antibody.6 Imaging was performed in ACSF buffer on an Abberior easy3D STED/RESOLFT QUAD scanning microscope (Abberior Instruments GmbH, Göttingen, Germany) built on a motorized inverted microscope IX83 (Olympus, Tokyo, Japan). The microscope was equipped with pulsed STED lasers at 595 nm and 775 nm, and with 355 nm, 405 nm, 485 nm, 561 nm, and 640 nm excitation lasers. Spectral detection was performed with avalanche photodiodes (APD) in the following spectral windows: 650–800 nm and 505–550 nm. Images were acquired with a 100×/1.40 UPlanSApo oil immersion objective lens (Olympus) and the pixel size was 30 nm.Confocal, 2D and 3D STED imaging
Confocal imaging was performed on a Leica SP8 (Leica Microsystems, Mannheim, Germany) inverted confocal microscope equipped with an HC PL APO CS2 63×/1.40 oil objective. Images were acquired using a 1000 Hz bidirectional scanner, with a voxel size of 80 nm × 80 nm × 1000 nm, a pinhole of 1 AU and line averaging of 3. Hoechst 33342 was excited with a 405 nm laser and detected with a HyD detector in the 430–480 nm range. Fluorescent actin probes were excited and detected using the following parameters: LIVE 515 probe – excited with a 488 nm laser and detected with a Leica HyD detector set within the spectral range of 530–580 nm, 580CP probes – excited with a 561 nm laser and detected in the range of 610–660 nm, 610CP and SiR probes – exited with a 633 nm laser and detected in the range of 650–700 nm.Comparative confocal and STED images were acquired on an Abberior STED 775 QUAD scanning microscope (Abberior Instruments GmbH, Germany) equipped with 561 nm and 640 nm 40 MHz pulsed excitation lasers, a pulsed 775 nm 40 MHz STED laser, and an UPlanSApo 100×/1.40 oil objective. The following detection windows were used: 580CP channel 615/20 nm and 610CP/SiR channel 685/70 nm. In this setup, the pixel size was 30 nm in the xy plane and the pinhole was set to 1 AU for 2D STED images. The laser power was optimized for each sample. 3D STED images were acquired with the pinhole set to 0.8 AU, the voxel size set to 40 × 40 × 40 nm, the 3D STED doughnut set to 90%, single line accumulation and xyz scanning mode. The acquired images were processed using Fiji20 and SVI Huygens deconvolution software.
Airyscan microscopy
Freshly extracted frog erythrocytes were diluted 1000× in RBC buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.1% glycerol) supplemented with 250 nM 6-610CP-JAS; 200 μl aliquots were seeded into a 10-well plate (Greiner Bio-One culture slides, PS, 75/25 mm (Art. Nr.: 543079)) and incubated for 2 h at room temperature. Imaging was performed on a Zeiss LSM880 system equipped with an oil immersion Plan-Apochromat 63×/1.40 Oil Corr M27 objective (Carl Zeiss) at room temperature. Stained erythrocytes were imaged in the Z-stack mode according to Nyquist parameters (x × y × z = 55 × 55 × 150 nm). A 633 nm diode laser was used for excitation, and emission was detected using a BP 570 − 620 + LP 645 filter onto an Airyscan fast detector. The signal was averaged from 2 frame scans. Live-cell imaging was performed on the same system equipped with an incubator set to 37 °C and 5% CO2. The images were reconstructed using Zen software (Carl Zeiss) and processed using Fiji.20Human fibroblasts were stained in a 10-well plate (Greiner Bio-One culture slides, PS, 75/25 mm (Art. Nr.: 543079) with 200 μl of DMEM supplemented with 10% FBS and 250 nM 6-610CP-JAS for 2 h at 37 °C and 5% CO2. Imaging was performed on a Zeiss LSM880 system equipped with an oil immersion Plan-Apochromat 63×/1.40 Oil Corr M27 objective (Carl Zeiss) at 37 °C and 5% CO2. Stained primary human fibroblasts were imaged in the time-lapse mode for 15 min every 15 s in one Z-plane (x × y = 55 × 55 nm). A 633 nm diode laser was used for excitation, and emission was detected using a BP 570 − 620 + LP 645 filter onto an Airyscan fast detector. The signal was averaged from 4 frame scans. The images were processed using Fiji.20
Maintenance and preparation of Drosophila melanogaster larvae
Wild type Oregon-R Drosophila melanogaster was raised on standard cornmeal–yeast–agar medium at 25 °C and used for all experiments. For staining of living Drosophila melanogaster tissues, wandering third instar larvae were dissected in 1× PBS pH 7.4 (Phosphate Buffered Saline) (Life Technologies, Carlsbad, California, USA) and the inverted front half of the larva was incubated with probes of 1 μM concentration in 1× PBS for 1 h at room temperature. After a single washing step with 1× PBS, isolated tissues were mounted in Schneider's Drosophila Medium (Thermo Fisher Scientific) under a coverslip and sealed with nontoxic duplicating silicone (picodent, Wipperfuerth, Germany) to prevent evaporation of the medium during imaging.Results and discussion
Design and synthesis of actin probes
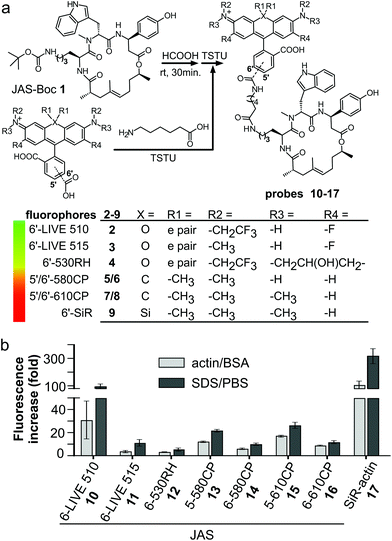
We prepared a jasplakinolide Nω-Boc-lysine analogue7,21 (JAS-Boc 1) and conjugated it to a series of STED nanoscopy compatible fluorophores – 6′-530RH,22 5′-580CP,8 5′-610CP,8 and 6′-610CP23 – and compared them to SiR-actin (Fig. 1a). In order to obtain a better insight into the structure–performance relationship, we also analyzed recently published 6-LIVE 510-JAS, 6-LIVE 515-JAS24 and 6-580CP-JAS25 probes.Chromogenicity and fluorogenicity of the probes
All probes were chromogenic and fluorogenic, i.e. showed absorbance and fluorescence increase upon SDS detergent addition and actin binding (Fig. 1b and Fig. S1a–h†). The probe's optical properties in actin-bound state versus free state determine the image contrast and possibility of no-wash imaging. SDS dissolves the aggregates, shifts the equilibrium to the fluorescent zwitterionic state and thus allows the measurement of the maximum fluorescence and absorbance values in an aqueous environment.11 BSA was used to estimate the fluorescence increase due to unspecific interactions, which was negligible in all the cases (Fig. S1†). Fluorogenicity in the presence of actin or SDS followed the same trend: SiR-actin and 6-LIVE 510-JAS were the most fluorogenic, while 6-530RH-JAS and 6-LIVE 515-JAS were the least fluorogenic (Fig. 1b). In agreement with the previous studies, high fluorogenicity was determined by low fluorescence in the unbound state, rather than fluorescence enhancement after interaction with the target (Fig. S1†).Most probes achieved ∼80% of the maximal absorbance and fluorescence, when bound to actin (Fig. S1i†). Noteworthily, 6-LIVE 510-JAS and 6-LIVE 515-JAS gained ∼80% in absorbance, indicating that the binding had switched the equilibrium towards the zwitterionic state, but the fluorescence increase reached only 30–50% of that in SDS. This suggests fluorescence quenching due to the interaction with actin or aromatic residues of jasplakinolide.26
Staining actin in living cells
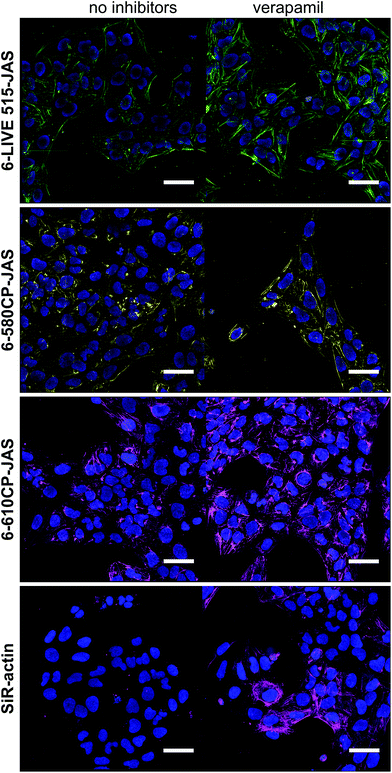
U-2 OS cells display drug efflux activity; thus their uniform and efficient staining with rhodamine probes requires verapamil.8 We used these “difficult” cells to reveal the performance differences among the new probes. We quantified the actin staining in the absence and in the presence of verapamil (Fig. S2†). The ratio of the measured values close to 1 indicates that the active efflux does not limit actin staining. Indeed, the probes behaved very differently (Fig. 2a–c). As previously reported, SiR-actin stained the cells brightly in the presence of verapamil,6 but only a sub-population of the cells was stained without verapamil. 6-LIVE 510-JAS, 5-580CP-JAS and 5-610CP-JAS performed similarly. 6-530RH-JAS stained cells weakly, required a higher concentration and produced numerous fluorescent aggregates. Surprisingly, 6-LIVE 515-JAS, 6-580CP-JAS and 6-610CP-JAS stained the cell population uniformly and verapamil addition only moderately increased the staining intensity (Fig. 2a–c). Thus, jasplakinolide-based actin probes represent yet another example where the isomerism of fluorophores is influential in probe performance. Interestingly, we found no correlation between high fluorogenicity and good performance in the present series of probes. In fact, our best performing probes, 6-580CP-JAS, 6-610CP-JAS and 6-LIVE 515-JAS, are among the least fluorogenic (Fig. 1b). There was little difference in performance of 5′- and 6′-isomers of 580CP-JAS and 610CP-JAS in the presence of verapamil (Fig. 2d). Thus improved staining by the 6′-isomers is likely caused by their impaired efflux.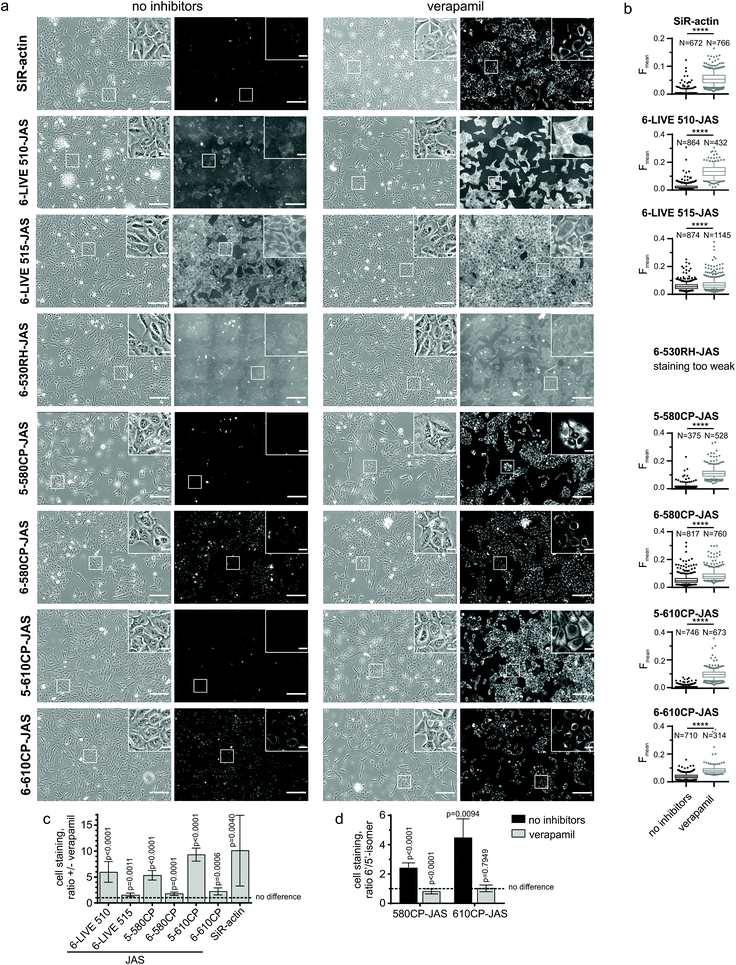 | ||
| Fig. 2 Requirement of verapamil for efficient staining of actin in U-2 OS cells. (a) Living cells were incubated with 250 nM probes for 2 h at 37 °C in the presence or absence of 10 μM verapamil, washed and imaged on an automated wide-field microscope. Phase contrast and the fluorescence channels are shown; the imaging parameters are listed in Table S2.† Scale bar – 200 μm, in the insets – 25 μm. (b) Quantification of images (a) using a CellProfiler pipeline described in Fig. S2†. The data are depicted as 5–95 percentile box plots with outliers; N – number of analysed cells. In all the cases, staining enhancement by verapamil was statistically significant, as determined by the Mann–Whitney test with a confidence level of 95%. ****, two-tailed p < 0.0001. (c) Enhancement of staining by verapamil. (d) A difference of staining between 5′- and 6′-isomers. (c and d) Data are presented as mean ± SD, N = 9 comparisons. The one-sample t-test was used to assess whether the values are statistically different from 1, and two-tailed p values are reported. | ||
We confirmed our observations by confocal imaging of living U-2 OS cells (Fig. 3). Here also, 6-LIVE 515-JAS, 6-580CP-JAS and 6-610CP-JAS, but not SiR-actin, stained cells efficiently without verapamil.
Interaction with F-actin
An alternative cause of varied performance might be impaired probes’ interaction with actin. To assess binding affinities, we stained formaldehyde-fixed and permeabilized U-2 OS cells with 1–1000 nM probes and quantified the resulting fluorescence in the cytoplasm. The data fitted well into a dose–response equation with the Hill coefficient equal to 1, thus the derived ECstaining50 is equivalent to apparent KF-actinD (Fig. S3† and Table 1). Interestingly, KF-actinD of all probes was very similar and fell in the range of 15–60 nM, which is very close to the reported KF-actinD = 15 nM of jasplakinolide.27| Probe | λ ex/λem (nm) | Brightnessa (M−1 cm−1) ×103 | K F-actinD (nM) | Toxicityb (μM) |
|---|---|---|---|---|
| a Brightness was calculated using quantum yields determined relative to the sample with SDS and the published ε of free fluorophores (Table S2†). b Minimal concentration at which progression through the cell cycle is impaired in HeLa cells after 24 h of incubation. | ||||
| JAS-Boc 1 | — | — | — | 0.025 |
| 6-LIVE 510-JAS 10 | 500/525 | 24 ± 1.3 | 14 ± 7 | 2 |
| 6-LIVE 515-JAS 11 | 517/544 | 19.4 ± 0.5 | 16 ± 6 | 2 |
| 6-530RH-JAS 12 | 537/560 | 44 ± 1.9 | 55 ± 25 | >1.4 |
| 5-580CP-JAS 13 | 588/612 | 50 ± 2 | 19 ± 4 | 0.5 |
| 6-580CP-JAS 14 | 590/614 | 50.1 ± 0.9 | 21 ± 9 | 1 |
| 5-610CP-JAS 15 | 616/639 | 54.6 ± 1.8 | 30 ± 11 | >1 |
| 6-610CP-JAS 16 | 615/639 | 55.8 ± 0.2 | 27 ± 9 | 0.25 |
| SiR-actin 17 | 652/672 | 41.9 ± 2.5 | 19 ± 7 | >1 |
This indicates that attachment of the fluorophore does not change significantly jasplakinolide's affinity to actin. Thus differences in probe performance cannot be explained by differences of interaction with actin, but they arise from differences in the cell entry and/or retention.
Cytotoxicity of the probes
While SiR-actin is not toxic over a wide concentration range,6 the parent compound, jasplakinolide, is a powerful toxin.28 We assessed the cytotoxicity of the new probes by measuring cell cycle perturbations in HeLa cells after 24 h of incubation (Fig. S4†): JAS-Boc showed a toxicity threshold at 25 nM and for all new probes it was at ∼1 μM (Table 1). The better staining probes were slightly more toxic, which might reflect their reduced efflux. However, even the most toxic probes (250 nM) were ∼10-fold less potent compared to the parent compound. This indicates that cell entry remains a limiting step in staining actin. Rhodamines are known substrates for P-glycoprotein 1 – one of the major players in ATP-dependent efflux.29 Limited passive permeability and active efflux can compromise staining and toxicity by reducing the intracellular probe concentration.30 This is well illustrated by the extreme dependency of SiR-actin staining on verapamil.Interaction with G-actin
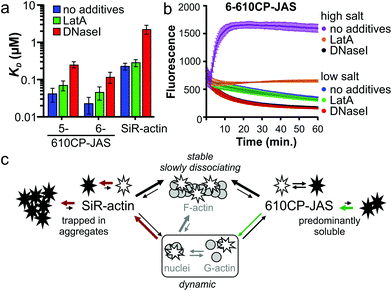
Jasplakinolide binds and stabilizes F-actin filaments.27 The fluorogenic nature of the probes allowed us to investigate their interaction with G-actin. We saw a clear increase in fluorescence while titrating 5-610CP-JAS, 6-610CP-JAS and SiR-actin with G-actin in a low salt buffer that does not support polymerization (Fig. S5†). The apparent KD values for 5/6-610CP-JAS were surprisingly close to KF-actinD (Fig. 4a). To ensure that in our assay actin remains unpolymerized, we repeated titrations in the presence of polymerization inhibitors – the toxin latrunculin A (LatA)31 and DNase I which bind to the sites non-overlapping with the jasplakinolide binding site.27,32,33 LatA had no effect on KD, but in the presence of DNase I, the KD of all three probes increased ∼10-fold (Fig. 4a).To resolve this discrepancy, we tested for possible actin polymerization by monitoring the fluorescence of pyrene-labelled G-actin under the conditions of our titration (Fig. 4b). As the rate of nucleation depends on the G-actin concentration,34 we performed this experiment at the highest G-actin concentration (4 μM) used. The addition of a high salt buffer to the samples induced rapid actin polymerization which was completely blocked by DNase I and largely inhibited by LatA. Slow fluorescence decrease was observed in a low salt buffer used for the titrations, which approached a plateau after 1 h of incubation. Addition of LatA had no effect, while DNase I accelerated this process. Because of the low concentration (10 nM), the probes had no effect on these processes. Taken together, these data indicate that a fraction of actin in a low salt buffer exists in a multimeric state that is completely depolymerized by DNase I.35 The presence of highly polymerized filaments is unlikely, because the actin stock was centrifuged immediately before the experiment, but G-actin can exist under rapid equilibrium with nucleation centres and/or short filaments.
Therefore we assume that KD determined in the presence of DNase I represents the true affinity of the probes to G-actin (KG-actinD).
Comparison of KG-actinD (Fig. 4a) and KF-actinD (Table 1) reveals that 610CP-JAS has ∼10-fold preference, while SiR-actin has ∼100-fold preference for F-actin. We explain the difference by a higher aggregation propensity of SiR-actin.8,14 Assuming that the interaction of jasplakinolide probes with G-actin (and very short filaments) is more dynamic and has a higher koff rate, it would be more sensitive to aggregation than a more stable interaction with F-actin. This effect is stronger for SiR-actin than for 610CP-JAS probes, resulting in a higher preference of SiR-actin to F-actin in vitro (Fig. 4c). However, in living cells the concentrations of both G- and F-actin are in the high micromolar range,36,37 indicating that both forms can be stained by both probes. Indeed, the analysis of fluorescence signal profiles in the Airyscan images has found a similar stress fibre/cytoplasm fluorescence ratio in human fibroblasts stained with SiR-actin and those stained with the new probes (Fig. S6†). This is further supported by the Airyscan video imaging showing highly dynamic diffuse cytoplasm staining in addition to the bright F-actin fibres (Video S1†).
Imaging actin in living cells and tissues
Our new probes are compatible with various nanoscopy setups and can be used to stain a variety of samples with high specificity for actin (Fig. 5). 6-580CP-JAS can be combined with SiR-based probes for two colour STED imaging in cell cultures and in living tissues, where the small probe size ensures efficient volume penetration. Efficient staining by 6-610CP-JAS allows imaging actin in erythrocytes that show high haemoglobin absorption. Also, it was successfully used for 3D isotropic STED imaging of actin in human fibroblasts (Video S2†). To help in choosing a probe that is best suited for a particular application, we provide a summary of each probe's advantages and limitations in Table 2.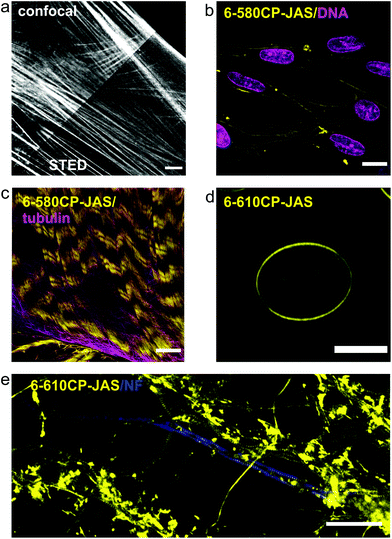 | ||
| Fig. 5 Confocal and nanoscopy images of living cells and tissues stained with the new actin probes. (a) Comparison of confocal and STED images of human fibroblasts stained with 6-610CP-JAS. (b) Human melanocytes co-stained with 6-580CP-JAS and 5-SiR-Hoechst.8 (c) Live-cell imaging of body wall muscle of dissected D. melanogaster larva co-stained with 6-580CP-JAS and 6-SiR-CTX.14 (d) Maximum intensity projection of a frog erythrocyte stained with 6-610CP-JAS. (e) Rat primary neuron culture co-stained with 6-610CP-JAS and neurofascin (AlexaFluor 488). Scale bars – 10 μm (a–d), 5 μm (e). | ||
| Probe | No wash | STED | Sensitivity to efflux | Toxicity | Remarks |
|---|---|---|---|---|---|
| 6-LIVE 515-JAS 11 | Not possible | Yes | Low | Low | Dim |
| 6-580CP-JAS 14 | Suboptimal | Yes | Low | Low | Compatible with SiR probes |
| 6-610CP-JAS 16 | Suboptimal | Yes | Low | Medium | Brighter than SiR-actin |
| SiR-actin 17 | Good | Yes | High | Low | High contrast |
Conclusions
In summary, we demonstrate that subtle differences in the probe structure might affect its interaction with efflux pumps, which can dramatically affect probe performance in living cells. 6-LIVE 515-JAS, 6-580CP-JAS and 6-610CP-JAS represent a valuable addition to the actin imaging toolbox as they can be used for staining cells with high efflux pump activity without inhibitors. Notably, all probes are able to bind both G- and F-actin which should be taken into account while interpreting in vitro and in vivo observations. The membrane permeability remains a limiting factor and work directed at its improvement is currently under way.Conflicts of interest
G. L. has a patent on SiR derivatives.Acknowledgements
The authors acknowledge funding of the Max Planck Society. G. L. is grateful to the Max Planck Society for a Nobel Laureate Fellowship. G. K. acknowledges the Max Planck Institute for Biophysical Chemistry for the Manfred Eigen Fellowship. The authors thank Dr Vladimir Belov and Dr Jonas Bucevičius for the discussions and critical reading of the manuscript. Open Access funding provided by the Max Planck Society.Notes and references
- S. J. Sahl, S. W. Hell and S. Jakobs, Nat. Rev. Mol. Cell Biol., 2017, 18, 685–701 CrossRef CAS PubMed.
- L. Wang, M. S. Frei, A. Salim and K. Johnsson, J. Am. Chem. Soc., 2019, 141, 2770–2781 CrossRef CAS PubMed.
- L. D. Lavis, Annu. Rev. Biochem., 2017, 86, 825–843 CrossRef CAS PubMed.
- J. Bucevičius, G. Lukinavičius and R. Gerasimaitė, Chemosensors, 2018, 6, 18 CrossRef.
- G. Kostiuk, J. Bucevičius, R. Gerasimaitė and G. Lukinavičius, J. Phys. D: Appl. Phys., 2019, 52, 504003 CrossRef CAS.
- G. Lukinavičius, L. Reymond, E. D'Este, A. Masharina, F. Gottfert, H. Ta, A. Guther, M. Fournier, S. Rizzo, H. Waldmann, C. Blaukopf, C. Sommer, D. W. Gerlich, H. D. Arndt, S. W. Hell and K. Johnsson, Nat. Methods, 2014, 11, 731–733 CrossRef PubMed.
- L. G. Milroy, S. Rizzo, A. Calderon, B. Ellinger, S. Erdmann, J. Mondry, P. Verveer, P. Bastiaens, H. Waldmann, L. Dehmelt and H. D. Arndt, J. Am. Chem. Soc., 2012, 134, 8480–8486 CrossRef CAS PubMed.
- J. Bucevičius, J. Keller-Findeisen, T. Gilat, S. W. Hell and G. Lukinavičius, Chem. Sci., 2019, 10, 1962–1970 RSC.
- G. Lukinavičius, L. Reymond, K. Umezawa, O. Sallin, E. D'Este, F. Gottfert, H. Ta, S. W. Hell, Y. Urano and K. Johnsson, J. Am. Chem. Soc., 2016, 138, 9365–9368 CrossRef PubMed.
- L. Xue, I. A. Karpenko, J. Hiblot and K. Johnsson, Nat. Chem. Biol., 2015, 11, 917–923 CrossRef CAS PubMed.
- G. Lukinavičius, K. Umezawa, N. Olivier, A. Honigmann, G. Yang, T. Plass, V. Mueller, L. Reymond, I. R. Correa Jr., Z. G. Luo, C. Schultz, E. A. Lemke, P. Heppenstall, C. Eggeling, S. Manley and K. Johnsson, Nat. Chem., 2013, 5, 132–139 CrossRef PubMed.
- L. Wang, M. Tran, E. D’Este, J. Roberti, B. Koch, L. Xue and K. Johnsson, Nat. Chem., 2020, 12, 165–172 CrossRef CAS PubMed.
- J. B. Grimm, A. K. Muthusamy, Y. Liang, T. A. Brown, W. C. Lemon, R. Patel, R. Lu, J. J. Macklin, P. J. Keller, N. Ji and L. D. Lavis, Nat. Methods, 2017, 14, 987–994 CrossRef CAS PubMed.
- G. Lukinavičius, G. Y. Mitronova, S. Schnorrenberg, A. N. Butkevich, H. Barthel, V. N. Belov and S. W. Hell, Chem. Sci., 2018, 9, 3324–3334 RSC.
- I. R. Correa Jr., B. Baker, A. Zhang, L. Sun, C. R. Provost, G. Lukinavičius, L. Reymond, K. Johnsson and M. Q. Xu, Curr. Pharm. Des., 2013, 19, 5414–5420 CrossRef PubMed.
- F. Stagge, G. Y. Mitronova, V. N. Belov, C. A. Wurm and S. Jakobs, PLoS One, 2013, 8, e78745 CrossRef CAS PubMed.
- C. Deo, S. H. Sheu, J. Seo, D. E. Clapham and L. D. Lavis, J. Am. Chem. Soc., 2019, 141, 13734–13738 CrossRef CAS PubMed.
- P. E. Deal, R. U. Kulkarni, S. H. Al-Abdullatif and E. W. Miller, J. Am. Chem. Soc., 2016, 138, 9085–9088 CrossRef CAS PubMed.
- C. McQuin, A. Goodman, V. Chernyshev, L. Kamentsky, B. A. Cimini, K. W. Karhohs, M. Doan, L. Ding, S. M. Rafelski, D. Thirstrup, W. Wiegraebe, S. Singh, T. Becker, J. C. Caicedo and A. E. Carpenter, PLoS Biol., 2018, 16, e2005970 CrossRef PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- R. Tannert, L. G. Milroy, B. Ellinger, T. S. Hu, H. D. Arndt and H. Waldmann, J. Am. Chem. Soc., 2010, 132, 3063–3077 CrossRef CAS PubMed.
- A. N. Butkevich, V. N. Belov, K. Kolmakov, V. V. Sokolov, H. Shojaei, S. C. Sidenstein, D. Kamin, J. Matthias, R. Vlijm, J. Engelhardt and S. W. Hell, Chem. Eur. J., 2017, 23, 12114–12119 CrossRef CAS PubMed.
- A. N. Butkevich, G. Y. Mitronova, S. C. Sidenstein, J. L. Klocke, D. Kamin, D. N. Meineke, E. D'Este, P. T. Kraemer, J. G. Danzl, V. N. Belov and S. W. Hell, Angew. Chem., Int. Ed., 2016, 55, 3290–3294 CrossRef CAS PubMed.
- F. Grimm, S. Nizamov and V. N. Belov, ChemBioChem, 2019, 20, 2248–2254 CrossRef CAS PubMed.
- A. N. Butkevich, G. Lukinavičius, E. D'Este and S. W. Hell, J. Am. Chem. Soc., 2017, 139, 12378–12381 CrossRef CAS PubMed.
- N. Marme, J. P. Knemeyer, M. Sauer and J. Wolfrum, Bioconjugate Chem., 2003, 14, 1133–1139 CrossRef CAS PubMed.
- M. R. Bubb, A. M. Senderowicz, E. A. Sausville, K. L. Duncan and E. D. Korn, J. Biol. Chem., 1994, 269, 14869–14871 CAS.
- S. J. Robinson, B. I. Morinaka, T. Amagata, K. Tenney, W. M. Bray, N. C. Gassner, R. S. Lokey and P. Crews, J. Med. Chem., 2010, 53, 1651–1661 CrossRef CAS PubMed.
- C. Ludescher, J. Thaler, D. Drach, J. Drach, M. Spitaler, C. Gattringer, H. Huber and J. Hofmann, Br. J. Haematol., 1992, 82, 161–168 CrossRef CAS PubMed.
- G. D. Eytan, R. Regev, G. Oren, C. D. Hurwitz and Y. G. Assaraf, Eur. J. Biochem., 1997, 248, 104–112 CrossRef CAS PubMed.
- M. Coue, S. L. Brenner, I. Spector and E. D. Korn, FEBS Lett., 1987, 213, 316–318 CrossRef CAS PubMed.
- W. M. Morton, K. R. Ayscough and P. J. McLaughlin, Nat. Cell Biol., 2000, 2, 376–378 CrossRef CAS PubMed.
- W. Kabsch, H. G. Mannherz, D. Suck, E. F. Pai and K. C. Holmes, Nature, 1990, 347, 37–44 CrossRef CAS PubMed.
- L. S. Tobacman and E. D. Korn, J. Biol. Chem., 1983, 258, 3207–3214 CAS.
- S. E. Hitchcock, L. Carisson and U. Lindberg, Cell, 1976, 7, 531–542 CrossRef CAS PubMed.
- T. D. Pollard, L. Blanchoin and R. D. Mullins, Annu. Rev. Biophys. Biomol. Struct., 2000, 29, 545–576 CrossRef CAS PubMed.
- S. A. Koestler, K. Rottner, F. Lai, J. Block, M. Vinzenz and J. V. Small, PLoS One, 2009, 4, e4810 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ob00369g |
| ‡ Present address: Advanced Light Microscopy Core Facility, EMBL Heidelberg, Meyerhofstraße 1, 69117 Heidelberg, Germany. |
| This journal is © The Royal Society of Chemistry 2020 |