Open Access Article
Open Access ArticleThe contribution of achiral residues in the laspartomycin family of calcium-dependent lipopeptide antibiotics†
Thomas M.
Wood
 ab,
Kristine
Bertheussen
a and
Nathaniel I.
Martin
ab,
Kristine
Bertheussen
a and
Nathaniel I.
Martin
 *a
*a
aBiological Chemistry Group, Institute of Biology Leiden, Leiden University, Sylviusweg 72, 2333 BE Leiden, The Netherlands. E-mail: n.i.martin@biology.leidenuniv.nl
bDepartment of Chemical Biology & Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Universiteitsweg 99, 3584 CG Utrecht, The Netherlands
First published on 12th December 2019
Abstract
The growing threat of antibacterial resistance is a global concern. The so-called calcium-dependent lipopeptide antibiotics (CDAs) have emerged as a promising source of new antibiotic agents that are rich in structural and mechanistic diversity. Over forty unique CDAs have been identified to date and share a number of common features. Recent efforts in our group have provided new mechanistic and structural insights into the laspartomycin family of CDAs. We here describe investigations aimed at probing the role of the three glycine residues found in the laspartomycin peptide macrocycle. In doing so laspartomycin analogues containing the achiral 2-aminoisobutyric acid (AIB) as well as L- or D-alanine in place of glycine were prepared and their antibacterial activities evaluated.
Introduction
The accelerated emergence of multi drug-resistant (MDR) bacteria is now considered a top priority and one of the most urgent global threats to human health.1 According to a 2017 report from the Center for Disease Control and prevention (CDC) more than 2 million Americans acquire antibiotic-resistant infections each year leading to at least 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in the US alone.2 Since the so-called “golden age” of antibiotic discovery spanning the 1940s–1960s, very few new antibiotics operating with novel modes of action have been introduced.3 Notable in this regard is daptomycin, the preeminent calcium-dependent antibiotic (CDA) and the most recent first-in-class antibiotic to have entered the clinic.4,5 Daptomycin's precise mechanism of action is a topic of some debate.6,7 By comparison, the mode of action of the structurally similar CDA laspartomycin C (Fig. 1) is more clearly understood.8,9 Unlike daptomycin, laspartomycin C specifically targets the essential bacterial phospholipid undecaprenyl phosphate (C55–P). C55–P represents a novel target and to date there are no clinically approved antibiotics that operate by binding C55–P. Our group recently reported the first total synthesis of laspartomycin C
000 deaths in the US alone.2 Since the so-called “golden age” of antibiotic discovery spanning the 1940s–1960s, very few new antibiotics operating with novel modes of action have been introduced.3 Notable in this regard is daptomycin, the preeminent calcium-dependent antibiotic (CDA) and the most recent first-in-class antibiotic to have entered the clinic.4,5 Daptomycin's precise mechanism of action is a topic of some debate.6,7 By comparison, the mode of action of the structurally similar CDA laspartomycin C (Fig. 1) is more clearly understood.8,9 Unlike daptomycin, laspartomycin C specifically targets the essential bacterial phospholipid undecaprenyl phosphate (C55–P). C55–P represents a novel target and to date there are no clinically approved antibiotics that operate by binding C55–P. Our group recently reported the first total synthesis of laspartomycin C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 as well as the co-crystal structure of laspartomycin C bound to both calcium and a C10 truncated analogue of C55–P.11
10 as well as the co-crystal structure of laspartomycin C bound to both calcium and a C10 truncated analogue of C55–P.11
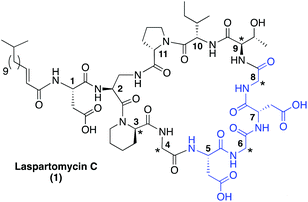 | ||
| Fig. 1 Laspartomycin C. Indicated with a star are the achiral or D-amino acids and highlighted in blue is the conserved Asp-X-Asp-Gly calcium binding motif common to the CDAs. | ||
While the CDAs comprise a diverse family of lipopeptide antibiotics with varying mechanisms of action, they share a number of key structural similarities. These include the positioning of key achiral or D-amino acids as well as the highly conserved Asp-X-Asp-Gly motif.12 This Asp-X-Asp-Gly sequence is essential for the calcium binding that all CDAs utilize in achieving their full antibacterial activity. In the absence of calcium, the activity of the CDAs is significantly reduced or completely lost.13Fig. 1 presents the structure of laspartomycin C and highlights the structural features common to many CDAs.
At present there are more than 40 unique CDAs known and their structural diversity continues to provide great opportunity for discovery.14 All CDAs, except for the newly reported malacidins, contain 10 amino acids in their macrocycle. Using the macrocycle of laspartomycin C as a reference point it can be noted that: (1) The majority of CDAs bear an L-amino acid at position 4 (usually an ornithine, alanine, aspartic acid, or β-methyl-aspartic acid) with the exception being the laspartomycins which include a glycine at position 4; (2) position 6 is nearly always a D-amino acid, again with the exception of the laspartomycins (as well as the related friulimicins and amphomycins) which contain glycine at position 6; and (3) 100% of CDAs contain a glycine at position 8.
The inclusion of glycine at residues 4, 6, and 8 in the laspartomycin C macrocycle is unique among the CDAs. This observation prompted us to investigate the contribution that the absence of chirality at these positions has on antibacterial activity. To this end we investigated the introduction of the achiral 2-aminoisobutyric acid (Aib) at these positions as well as the introduction of either L- or D-alanine residues. We here describe the preparation and evaluation of a series of novel laspartomycin analogues that provide new stereochemical structure–activity insights at positions 4, 6, and 8.
Results and discussion
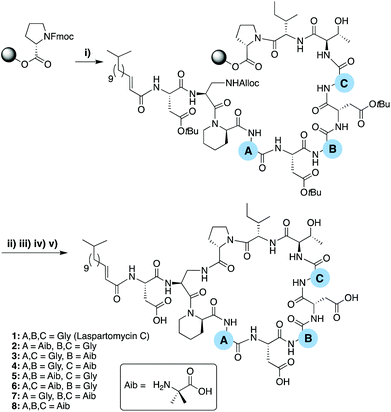
To evaluate the role of the three achiral centers in the laspartomycin C macrocycle we began by preparing a series of analogues containing the achiral amino acid 2-aminoisobutyric acid (Aib) at each position. These laspartomycin C variants were prepared using a robust combined solid- and solution-phase approach that our group as well as Payne and coworkers previously described (Scheme 1).10,15 The linear lipopeptide precursors were first assembled on solid support using the acid sensitive 2-chlorotrityl resin. Starting from C-terminal Pro11 each peptide was assembled using standard Fmoc-SPPS strategies with the notable introduction of an Alloc side chain protected L-diaminopropioinc acid (L-Dap) residue at position 2. Also, for analogues containing Gly at positions 6 and/or 8 (compounds 1–6) DMB-Gly was employed to avoid aspartamide formation. Following N-terminal lipidation, the Alloc side chain protecting of L-Dap2 was removed and the partially protected peptide released from the resin using mildly acidic conditions. The partially protected peptide was then cyclized at high dilution by treatment with BOP/DIPEA resulting in the clean formation of the macrocyclic product. Global deprotection followed by RP-HPLC purification provided the various laspartomycin analogues in yields ranging from 5–13%.To assess the contribution of the achiral positions 4, 6, and 8, analogues 2–8 were prepared wherein one or more of the glycine residues was replaced by the achiral Aib. The rationale for this replacement related to the previously reported ability of the Aib residue to predispose peptides towards their active conformation(s).16 The antimicrobial activity of analogues 2–8 was compared with that of authentic laspartomycin C (Table 1) using a serial dilution assay employing a clinically relevant strain of Methicillin-resistant Staphylococcus aureus (MRSA). Analogues 2 and 3 were found to exhibit calcium-dependent activity, albeit significantly less than laspartomycin C, while analogues 4–8, were essentially devoid of activity. Interesting, analogues 2 and 3, both containing a single Aib at position 4 and 6 respectively, maintain some activity. Conversely, Aib substitution at position 8 as in analog 4 led to complete loss of activity. Notably, analogue 8 containing Aib at all three positions, exhibits modest activity while showing no Ca2+ dependency. These results indicate that modification at positions 4 and 6 can compensate for an otherwise detrimental modification at position 8.
| Compound | [Ca2+] | |||
|---|---|---|---|---|
| 0 | 2.5 | 5.0 | 10 | |
| a Minimum inhibitory concentrations reported in μg mL−1 against MRSA USA300 at calcium concentration indicated. | ||||
| 1 (LaspC) | ≥128 | 4 | 4 | 2 |
| 2 | ≥128 | 16 | 16 | 16 |
| 3 | ≥128 | 64 | 32 | 32 |
| 4 | ≥128 | ≥128 | ≥128 | ≥128 |
| 5 | 64 | 64 | 64 | 64 |
| 6 | ≥128 | ≥128 | ≥128 | ≥128 |
| 7 | ≥128 | ≥128 | ≥128 | ≥128 |
| 8 | 64 | 64 | 64 | 64 |
All known CDAs contain a glycine at position 8 which is part of the Asp-X6-Asp-Gly8 calcium binding region. It therefore stands to reason that changes at position 8 significantly impact in the peptide's ability to chelate calcium and bind to its bacterial target. In this regard it is notable that position 6, also part of the CDA calcium binding region, is more amenable to variation. In the case of the laspatomycins, friulimicins, and amphomycins the inclusion of Gly at position 6 sets them apart from the majority of other CDAs which generally contain a D-amino acid at this site. In the case of daptomycin D-alanine is found at position 6 while in the A54145 class it is D-lysine and in the confusingly named “CDA-class” it is D-phenylglycine.14 With this in mind we prepared four additional analogues examining the effect in introducing L- or D-alanine at positions 4 and 6 (Fig. 2).
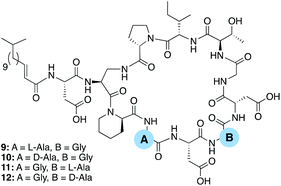 | ||
| Fig. 2 Laspartomycin C analogues prepared to investigate the effect of introducing L- or D-alanine at positions 4 and 6. | ||
The antimicrobial activity of analogues 9–12 was compared with that of authentic laspartomycin C (Table 2). Using the same activity assay employing MRSA USA300 as an indicator strain, all four analogues were found to be more active than their Aib containing counterparts with analogues 10 and 12 showing activity on par with that of laspartomycin itself. Interesting, the introduction of L- or D-alanine at position 4 yields compounds of similar activity while at position 6 the D-Ala variant was 16-fold more active than the L-analogue. These findings further demonstrate the importance of a D-stereocenter at position 6 of the CDA calcium-binding motif.
| Compound | [Ca2+] | |||
|---|---|---|---|---|
| 0 | 2.5 | 5.0 | 10 | |
| a Minimum inhibitory concentrations reported in μg mL−1 against MRSA USA300 at calcium concentration indicated. | ||||
| 1 (LaspC) | ≥128 | 8 | 4 | 2 |
| 9 | ≥128 | 16 | 8 | 4 |
| 10 | ≥128 | 16 | 8 | 2 |
| 11 | ≥128 | 64 | 32 | 16 |
| 12 | 64 | 16 | 8 | 1 |
Conclusion
In summary, we have generated a focused library of laspartomycin C analogues designed to explore the contribution of the achiral 4, 6, and 8 positions on antibacterial activity. Using a combined solid- and solution-phase approach, analogues were first prepared wherein positions 4, 6 and 8 were modified to contain the achiral amino acid Aib. These analogues showed a significant reduction in activity compared to the parent compound. Incorporation of Aib at position 8 led to complete loss of activity while the same modification at positions 4 or 6 led to analogues that retained some activity. A second series of analogues incorporating either D or L-alanine at positions 4 and 6 were also prepared. The introduction of both D- and L-Ala at position 4 led to compounds that showed activity similar to laspatpomcyin C. However, at position 6 the D-Ala variant is 16-times more active than the L-Ala and even showed slightly better activity than laspartomycin C.While the various members of the CDA family contain diverse amino acids at position 4 they are always either glycine or a L-amino acid. Our findings indicate that the incorporation of D-Ala at position 4 does not negatively affect activity. Position 6 plays a key role in the calcium-binding motif (Asp-X6-Asp-Gly) and is always either a glycine of a D-amino acid. Our findings demonstrate that positions 4 and 6 of Laspartomycin C are amenable to substitution provided the correct stereochemical constraints are respected. To this end ongoing studies in our laboratory are aimed at establishing whether the antibacterial activity of the laspartomycins can be enhanced by structural variation at these positions.
Experimental
Solid phase peptide synthesis
Chlorotrityl resin (5.0 g, 1.60 mmol g−1) was loaded with Fmoc-Pro-OH. Resin loading was determined to be 0.41–0.62 mmol g−1. Linear peptide encompassing Pro11 to Asp1 were assembled manually via standard Fmoc solid-phase peptide synthesis (SPPS) (resin bound AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fmoc-AA
Fmoc-AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BOP
BOP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DiPEA, 1
DiPEA, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 molar eq.) on a 0.1 mmol scale. DMF was used as solvent and Fmoc deprotections were carried out with piperidine
8 molar eq.) on a 0.1 mmol scale. DMF was used as solvent and Fmoc deprotections were carried out with piperidine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF (1
DMF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 v
4 v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v). Amino acid side chains were protected as follows: tBu for Asp, Alloc for DAP, and DMB for Gly6 and Gly8. D-allo-Thr was introduced without side chain protection. Following coupling and Fmoc deprotection of Asp1, N-terminal acylation was achieved by coupling (E)-13-methyltetradec-2-enoic acid using the same coupling conditions used for SPPS. The resin-bound, Alloc protected intermediate was next washed with CH2Cl2 and treated with Pd(PPh3)4 (30 mg, 0.03 mmol) and PhSiH3 (0.30 mL, 3.0 mmol) in CH2Cl2 (ca. 7 mL) under argon for 1 hour. The resin was subsequently washed with CH2Cl2 (5 × 10 mL), followed by a solution of diethyldithiocarbamic acid trihydrate sodium salt (5 mg mL−1 in DMF, 5 × 10 mL), and DMF (5 × 10 mL). The resin was treated with (CF3)2CHOH
v). Amino acid side chains were protected as follows: tBu for Asp, Alloc for DAP, and DMB for Gly6 and Gly8. D-allo-Thr was introduced without side chain protection. Following coupling and Fmoc deprotection of Asp1, N-terminal acylation was achieved by coupling (E)-13-methyltetradec-2-enoic acid using the same coupling conditions used for SPPS. The resin-bound, Alloc protected intermediate was next washed with CH2Cl2 and treated with Pd(PPh3)4 (30 mg, 0.03 mmol) and PhSiH3 (0.30 mL, 3.0 mmol) in CH2Cl2 (ca. 7 mL) under argon for 1 hour. The resin was subsequently washed with CH2Cl2 (5 × 10 mL), followed by a solution of diethyldithiocarbamic acid trihydrate sodium salt (5 mg mL−1 in DMF, 5 × 10 mL), and DMF (5 × 10 mL). The resin was treated with (CF3)2CHOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2 (1
CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 10 mL) for 1 hour and rinsed with additional (CF3)2CHOH
4, 10 mL) for 1 hour and rinsed with additional (CF3)2CHOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2 and CH2Cl2. The combined washings were then evaporated to yield the linear protected peptide with free C- and N-termini. The residue was dissolved in CH2Cl2 (150 mL) and treated with BOP (0.22 g, 0.5 mmol) and DiPEA (0.17 mL, 1.0 mmol) and the solution was stirred overnight after which TLC indicated complete cyclization. The reaction mixture was concentrated and directly treated with TFA
CH2Cl2 and CH2Cl2. The combined washings were then evaporated to yield the linear protected peptide with free C- and N-termini. The residue was dissolved in CH2Cl2 (150 mL) and treated with BOP (0.22 g, 0.5 mmol) and DiPEA (0.17 mL, 1.0 mmol) and the solution was stirred overnight after which TLC indicated complete cyclization. The reaction mixture was concentrated and directly treated with TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (95
H2O (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5, 10 mL) for 90 minutes. The reaction mixture was added to MTBE
2.5, 10 mL) for 90 minutes. The reaction mixture was added to MTBE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hexanes (1
hexanes (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the resulting precipitate washed once more with MTBE
1) and the resulting precipitate washed once more with MTBE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hexanes (1
hexanes (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The crude cyclic peptide was lyophilized from tBuOH
1). The crude cyclic peptide was lyophilized from tBuOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1
H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and purified with reverse phase HPLC. Pure fractions were pooled and lyophilized to yield the desired cyclic lipopeptide products in >95% purity as white powders, typically in 10–20 mg quantities (4.2–9.3% yield based on resin loading).
1) and purified with reverse phase HPLC. Pure fractions were pooled and lyophilized to yield the desired cyclic lipopeptide products in >95% purity as white powders, typically in 10–20 mg quantities (4.2–9.3% yield based on resin loading).
MIC determinations
Minimum inhibitory concentrations (MICs) were determined by broth microdilution according to CLSI guidelines.17 Blood agar plates were inoculated with glycerol stocks of MRSA USA300 and S. simulans 22 (see ESI†) followed by incubation for 16 hours at 37 °C and 30 °C respectively. Cation adjusted Mueller-Hinton broth (MHB) containing 10 mg L−1 Mg2+ was inoculated with individual colonies of S. aureus and Methicillin-resistant S. aureus (MRSA USA300), and incubated for 16 hours at 220 RPM. The peptides were dissolved in MHB (10 mg L−1 Mg2+) and serially diluted on polypropylene microtiter plates with a volume of 50 μL per well. Inoculated MHB (2 × 105 CFU mL−1) containing 10 mg L−1 Mg2+ and varying concentrations of Ca2+ (0–10 mM) was added to reach a total volume of 100 μL per well. The microtiter plates were sealed with an adhesive membrane and after 16 hours of incubation at 37 °C or 30 °C and 220 RPM the wells were visually inspected for bacterial growth. All reported MIC values result from two or more measurements.Abbreviations
| Alloc | Allyloxycarbonyl |
| BOP | (Benzotriazol-1-yloxy)tris-(dimethylamino)phosphonium hexafluorophosphate |
| DIPEA | N,N-Diisopropylethylamine |
| DMB | 2,4-Dimethoxybenzyl |
| Fmoc-SPPS | Fluorenylmethyloxycarbonyl solid phase peptide synthesis |
| RP-HPLC | Reverse phase high performance liquid chromotography |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support provided by the Netherlands Organization for Scientific Research (PhD grant to TMW) and The European Research Council (ERC consolidator grant to NIM, grant agreement no. 725523).Notes and references
- J. O'Neill, Antimicrobial resistance: tackling a crisis for the health and wealth of nations, 2014. https://amr-review.org Search PubMed.
- CDC, Antibiotic use in the United States, 2017: Progress and Opportunities, US Department of Health and Human Services, CDC, Atlanta, GA, 2017 Search PubMed.
- M. N. Gwynn, A. Portnoy, S. F. Rittenhouse and D. J. Payne, Ann. N. Y. Acad. Sci., 2010, 1213, 5–19 CrossRef PubMed.
- A. E. Muller and I. C. Gyssens, Kucers' Use Antibiot. A Clin. Rev. Antibacterial, Antifung. Antiparasit. Antivir. Drugs, Seventh Ed., 2017, 2, 866–907 Search PubMed.
- R. Sauermann, M. Rothenburger, W. Graninger and C. Joukhadar, Pharmacology, 2008, 81, 79–91 CrossRef CAS PubMed.
- R. H. Baltz, Curr. Opin. Chem. Biol., 2009, 13, 144–151 CrossRef CAS PubMed.
- G. Seydlová, A. Sokol, P. Lišková, I. Konopásek and R. Fišer, Antimicrob. Agents Chemother., 2019, 63, 1–14 Search PubMed.
- H. Naganawa, M. Hamada, K. Maeda, Y. Okami, T. Takeuchi and H. Umezawa, J. Antibiot. (Tokyo), 1968, XXI, 55–62 CrossRef PubMed.
- D. B. Borders, R. A. Leese, H. Jarolmen, N. D. Francis, A. A. Fantini, T. Falla, J. C. Fiddes and A. Aumelas, J. Nat. Prod., 2007, 70, 443–446 CrossRef CAS PubMed.
- L. H. Kleijn, S. F. Oppedijk, P. 't Hart, R. M. van Harten, L. A. Martin-Visscher, J. Kemmink, E. Breukink and N. I. Martin, J. Med. Chem., 2016, 59, 3569–3574 CrossRef CAS PubMed.
- L. H. J. Kleijn, H. C. Vlieg, T. M. Wood, J. Sastre Toraño, B. J. C. Janssen and N. I. Martin, Angew. Chem., Int. Ed., 2017, 56, 16546–16549 Search PubMed.
- R. H. Baltz, V. Miao, S. K. Wrigley and V. Miao, Nat. Prod. Rep., 2005, 717–741 RSC.
- M. Strieker and M. A. Marahiel, ChemBioChem, 2009, 10, 607–616 CrossRef CAS PubMed.
- T. M. Wood and N. I. Martin, MedChemComm, 2019, 10, 634–646 RSC.
- L. Corcilius, N. T. Elias, J. L. Ochoa, R. G. Linington and R. J. Payne, J. Org. Chem., 2017, 82, 12778–12785 CrossRef CAS PubMed.
- J. Michael Conlon, R. Al-Kharrge, E. Ahmed, H. Raza, S. Galadari and E. Condamine, Peptides, 2007, 28, 2075–2080 CrossRef PubMed.
- Clinical Laboratory Standard Institute, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Ninth Edition Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement, 2012, vol. 32.
Footnote |
| † Electronic supplementary information (ESI) available: NMR, HPLC and IC50 curves. See DOI: 10.1039/c9ob02534k |
| This journal is © The Royal Society of Chemistry 2020 |