Open Access Article
Open Access ArticleHighly selective staining and quantification of intracellular lipid droplets with a compact push–pull fluorophore based on benzothiadiazole†
S. Israel
Suarez
a,
Caroline C.
Warner
 b,
Heather
Brown-Harding
b,
Heather
Brown-Harding
 c,
Andrea M.
Thooft
b,
Brett
VanVeller
c,
Andrea M.
Thooft
b,
Brett
VanVeller
 *b and
John C.
Lukesh
III
*b and
John C.
Lukesh
III
 *a
*a
aDepartment of Chemistry, Wake Forest University, Winston-Salem, NC 27101, USA. E-mail: lukeshjc@wfu.edu
bDepartment of Chemistry, Iowa State University, Ames, IA 50011, USA. E-mail: bvv@iastate.edu
cDepartment of Biology, Wake Forest University, Winston-Salem, NC 27101, USA
First published on 12th December 2019
Abstract
A robust lipophilic dye, based on the structures of the benzothiadiazole heterocycle, was shown to be a potent fluorescent stain for the selective imaging of lipid droplets (LDs) within both live and fixed human cells. Its small molecular framework, large Stokes shift, and vastly improved photostability over that of the current status quo, Nile Red, highlight its tremendous potential as a versatile chemical tool for facilitating LD imaging and research.
Introduction
Lipid droplets (LDs) are highly dynamic cellular organelles involved in the storage and metabolism of neutral lipids. The processing of LDs is tightly controlled, where mis-regulation is closely associated with various metabolic diseases such as obesity and cancer.1–5 Thus, the selective detection and visualization of LDs is critical to the assessment of their relative abundance, size, distribution, and further elucidation of their fundamental roles in human health and disease. To this end, lipophilic dyes have proven to be invaluable chemical tools for advancing LD research by providing a non-destructive method for the imaging and quantification of LDs in real time—a necessary feature for probing LD dynamics within complex cellular environments.6–14Of the reported LD stains, Nile Red (Fig. 1) is most commonly employed as a result of its commercial availability and long history as a reported LD marker.15–17 However, the poor photostability and small Stokes shift of Nile Red results in high levels of noise and background artifacts.18 Consequently, to better assess their primary and secondary roles under physiological conditions, there remains a need to develop new lipophilic fluorophores with enhanced photophysical properties and improved selectivity towards LDs.19
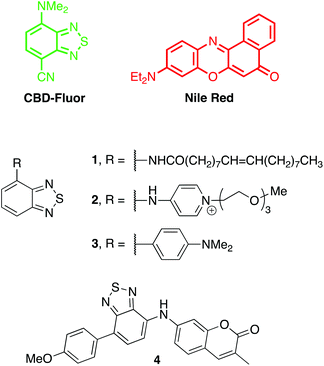 | ||
| Fig. 1 Chemical structures of CBD-Fluor, Nile Red and other probes based on benzothiadiazole that have been used to image lipidic structures. | ||
Chromophores based on the benzothiadiazole scaffold have received increasing attention in recent years for bioimaging applications.20–22 Fluorophores that feature the benzothiadiazole core are often solvochromic and display comparatively large Stokes shifts. Benzothiadiazole derivatives also exhibit differences in emission intensity depending on the local solvent environment. In general, polar environments significantly quench fluorescence, whereas non-polar media ‘turns-on’ emission. These attributes have made benzothiadiazoles attractive for staining lipidic structures23–25 and LDs in particular.13,14
The benzothiadiazole core can be decorated with targeting agents to help localize the fluorophore to the structure of interest.25 For example, the oleamide in 1 was proposed to assist in cellular uptake and the staining of LDs.23 Similarly, the amphiphilic appendage in 2 was proposed to facilitate the targeting of lipid bilayers.24 Notably, however, specific targeting agents are not necessary to image LDs, as evidenced by the prevalence of Nile Red as an LD marker. Indeed, both benzothiadiazole derivatives 3 and 4 have been reported to stain LDs within cells.13,14 Presumably, this selectivity is due to the fluorescence turn-on behaviour of benzothiadiazoles within the hydrophobic core of lipid droplets. In this regard, the large hydrophobic surface area of 3, 4 and even Nile Red leads to low solubility in water and an increase in non-specific staining of other hydrophobic structures that contribute to background. Herein, we report the use of a compact and more polar fluorophore, CBD-Fluor (Fig. 1),26 that leads to more red-shifted wavelengths of emission, higher photostability, higher turn-on fluorescence response, and excellent contrast with background for the selective staining of lipid droplets.
Results and discussion
A critical element of design for benzothiadiazole chromophores that has been under utilized in the examples given in Fig. 1 is the application of a push–pull substitution pattern. The cooperative effects of appending an electron-releasing functional group (the push) in conjugation with an electron-withdrawing group (the pull) serves to dramatically red-shift the wavelengths of absorption and emission of chromophores. In the context of benzothiadiazoles, push–pull substitution patterns (such as the one exhibited by CBD-Fluor) can red-shift spectral features while maintaining a small size.26,28–31Spectral characteristics of CBD-Fluor are ideal for LD imaging
The compact and polar structure of CBD-Fluor compared to other polycyclic aromatic dyes has the potential to improve solubility in water in order to diminish non-specific staining and disruption of native interactions. Despite this compact structure, however, CBD-Fluor emits similarly to larger cell-imaging dyes such as Nile Red, BODIPY, and fluorescein (>400 nm, Fig. 2A), but with a larger Stokes shift to limit signal from background. Moreover, CBD-Fluor displays marked photostability under continuous irradiation, in contrast to Nile Red, which rapidly photobleaches (Fig. 2B). Notably, CBD-Fluor is also more photostable than other benzothiadiazole dyes reported to stain LDs selectively.14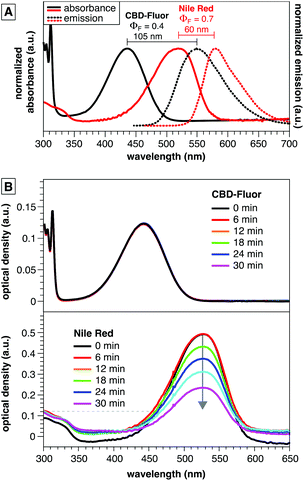 | ||
| Fig. 2 (A) Emission spectra and quantum yield of CBD-Fluor and Nile Red in dioxane (B) photostability of CBD-Fluor26versus Nile Red27 demonstrated by an unchanged absorption spectrum for CBD-Fluor versus photodecomposition of Nile Red. Solutions of each dye were prepared in air saturated THF with identical optical density (0.12 absorbance units) at 455 nm and were continuously irradiated using an LED source with an irradiance of 370 mW cm−2. | ||
CBD-Fluor displays environmentally sensitive emission—high emission intensity in non-polar environments and diminished emission in polar environments (Fig. 3A). CBD-Fluor, however, displayed a stronger bias in this turn-on behavior relative to other LD-specific dyes (Fig. 3B),13,14,23,26 with a limit of detection in dioxane that was less than 200 pM (Fig. S1†). Thus, we hypothesized that CBD-Fluor would be ideal for applications in lipid staining to address current shortcomings in standard LD analysis with Nile Red.
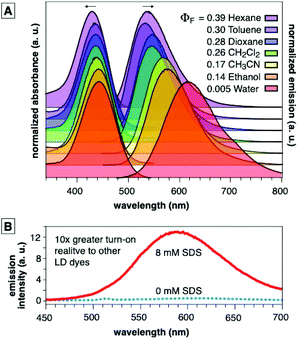 | ||
| Fig. 3 (A) Absorbance and emission spectra for CBD-Fluor showing the solvent polarity and quantum yield effects in various solvents. See ESI† for details. (B) CBD-Fluor (10 μM) before and after addition of SDS (CMC = 8 mM). The turn-on in emission intensity of CBD-Fluor is larger than for previously reported benzothiadiazole dyes.14,23 | ||
Cellular imaging and microscopy
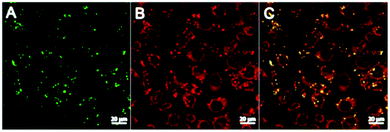
To initially assess the effectiveness of CBD-Fluor as an intracellular stain, both live and fixed HeLa (human cervical cancer) cells were incubated with CBD-Fluor and imaged using a confocal fluorescence microscope. The resulting images clearly depicted punctate staining, indicating that CBD-Fluor was indeed marking subcellular organelles (Fig. 4). To confirm that the spherical structures were lipid droplets, a separate experiment was performed in which live HeLa cells were co-incubated with both CBD-Fluor and Nile Red—the well-established LD stain.15 Although both dyes were excited at 488 nm, given their distinct emission profiles, we were able to use linear unmixing (see ESI† for details) to assign a stained structure to either CBD-Fluor, Nile Red, or both (Fig. 4). Manders's co-localization coefficient (MCC) was then used to determine the co-occurrence of each dye.32 We found that for spherical structures stained by CBD-Fluor, Nile Red would also be present 98% of the time, thus not only confirming co-localization of the two dyes but unequivocally establishing that the stained spherical structures were indeed lipid droplets. A positive correlation between CBD-Fluor and Nile Red was further established by determining Pearson's correlation coefficient (PCC) which was found to be 0.5. Although this value indicates a relatively strong correlation between the two dyes, it is likely reduced by the low signal-to-background of Nile Red.To ensure that CBD-Fluor and Nile Red were staining spherical structures within the cell and not on the cell's surface, an orthogonal analysis was performed (see ESI†). A total of 11 slices at 10 μm were analyzed along the Z-axis. This analysis established that CBD-Fluor was staining spherical structures within the cell. Co-localization studies with Nile Red further confirmed that these spherical structures were in fact lipid droplets.
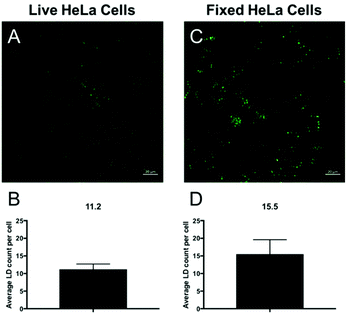
Given their complex structural makeup, one of the few reliable methods for identifying all intracellular LDs is by staining their hydrophobic core with a lipophilic dye. Therefore, to better evaluate CBD-Fluor's LD staining capabilities, we set out to evaluate how easily we could quantify the total number of lipid droplets within both fixed and live cells using CBD-Fluor as a selective fluorescent marker. Given the performance features of CBD-Fluor: (i) improved water solubility—especially compared to other lipophilic dyes (i.e. Nile Red), (ii) impressive photostability, and (iii) remarkable turn-on response within a lipophilic environment, we reasoned that these attributes would reduce background artifacts and provide an enhanced signal-to-noise ratio (S/N). Critically, better S/N performance facilitates the use of standard automated image analysis techniques to quantify intracellular lipids (see ESI†). Using this technique in live HeLa cells, we were able to stain an average of over 11 LDs per cell with CBD-Fluor (Fig. 5A and B). Moreover, similar results were obtained within fixed HeLa cells as an average of nearly 16 LDs per cell were clearly marked with CBD-Fluor (Fig. 5C and D).
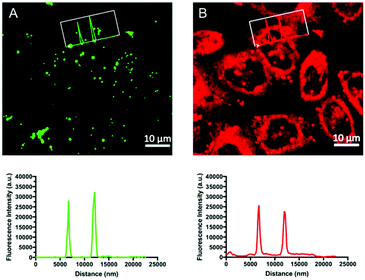
To better visualize the extraordinary turn-on response of CBD-Fluor, a 2D histogram was generated that depicts the relative fluorescence intensity of CBD-Fluor across the entire diameter of a cell (Fig. 6A). Indeed, CBD-Fluor displayed stark turn-on response with a greater than 300-fold increase in relative fluorescence intensity when trapped within the hydrophobic core of a cellular LD. Conversely, with Nile Red (Fig. 6B), a less than 30-fold increase in turn-on fluorescence was observed relative to background. These results further demonstrate CBD-Fluor's potential as a reliable and more selective LD marker within a complex cellular environment.
Finally, and to further demonstrate the overall utility of CBD-Fluor as an intracellular stain, we also showed that it could be used to effectively visualize LDs in the presence of other cellular dyes. Upon co-incubation of CBD-Fluor (green emission) and Hoechst, a common nuclear stain (blue emission), the resulting images were obtained (Fig. 7). Overlay of these signals validated the ease with which CBD-Fluor can be used to image LDs alongside other cellular markers allowing one to track different subcellular organelles simultaneously.
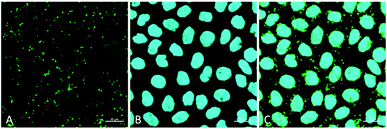 | ||
| Fig. 7 Cellular staining with (A) CBD-Fluor (ex: 488 nm, em: 539 nm and (B) Hoechst (ex: 405 nm, em: 453 nm). (C) Overlay of respective images. Scale bar is set to 20 μm. | ||
Conclusions
CBD-Fluor, a robust and versatile push–pull fluorophore was shown to be a highly effective stain for the imaging and quantification of LDs within both fixed and live human cells. Given its impressive S/N, high biocompatibility, and vastly improved photostability over that of Nile Red, render CBD-Fluor a highly attractive chemical reporter for facilitating both LD imaging and research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
JCL is thankful for financial support from Wake Forest University (Start-up funds). BV is grateful for support of this work by the National Science Foundation under Grant No. 1848261.Notes and references
- Y. Guo, K. R. Cordes, R. V. Farese and T. C. Walther, J. Cell Sci., 2009, 122, 749–752 CrossRef CAS.
- B. Brügger, Annu. Rev. Biochem., 2014, 83, 79–98 CrossRef PubMed.
- P. Shyu, X. F. A. Wong, K. Crasta and G. Thibault, Biosci. Rep., 2018, 38, BSR20180764 CrossRef PubMed.
- M. H. den Brok, T. K. Raaijmakers, E. Collado-Camps and G. J. Adema, Trends Immunol., 2018, 39, 380–392 CrossRef CAS PubMed.
- J. A. Olzmann and P. Carvalho, Nat. Rev. Mol. Cell Biol., 2019, 20, 137–155 CrossRef CAS.
- L. D. Lavis and R. T. Raines, ACS Chem. Biol., 2008, 3, 142–155 CrossRef CAS.
- A. B. Neef and C. Schultz, Angew. Chem., Int. Ed., 2009, 48, 1498–1500 CrossRef CAS.
- J. Spandl, D. J. White, J. Peychl and C. Thiele, Traffic, 2009, 10, 1579–1584 CrossRef CAS.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS PubMed.
- E. Kim, S. Lee and S. B. Park, Chem. Commun., 2012, 48, 2331 RSC.
- H.-J. Yang, C.-L. Hsu, J.-Y. Yang and W. Y. Yang, PLoS One, 2012, 7, e32693 CrossRef CAS PubMed.
- A. Goel, A. Sharma, M. Kathuria, A. Bhattacharjee, A. Verma, P. R. Mishra, A. Nazir and K. Mitra, Org. Lett., 2014, 16, 756–759 CrossRef CAS PubMed.
- H. Appelqvist, K. Stranius, K. Börjesson, K. Peter, R. Nilsson and C. Dyrager, Bioconjugate Chem., 2017, 28, 1363–1370 CrossRef CAS PubMed.
- A. A. R. Mota, J. R. Correa, L. P. de Andrade, J. A. F. Assumpção, G. A. de Souza Cintra, L. H. Freitas-Junior, W. A. da Silva, H. C. B. de Oliveira and B. A. D. Neto, ACS Omega, 2018, 3, 3874–3881 CrossRef CAS PubMed.
- P. Greenspan, J. Cell Biol., 1985, 100, 965–973 CrossRef CAS.
- I. A. Boldyrev, X. Zhai, M. M. Momsen, H. L. Brockman, R. E. Brown and J. G. Molotkovsky, J. Lipid Res., 2007, 48, 1518–1532 CrossRef CAS.
- J. Rumin, H. Bonnefond, B. Saint-Jean, C. Rouxel, A. Sciandra, O. Bernard, J.-P. Cadoret and G. Bougaran, Biotechnol. Biofuels, 2015, 8, 42 CrossRef PubMed.
- T. Govender, L. Ramanna, I. Rawat and F. Bux, Bioresour. Technol., 2012, 114, 507–511 CrossRef CAS PubMed.
- T. Fam, A. Klymchenko and M. Collot, Materials, 2018, 11, 1768 CrossRef PubMed.
- A. S. Klymchenko, Acc. Chem. Res., 2017, 50, 366–375 CrossRef CAS PubMed.
- B. A. D. Neto, P. H. P. R. Carvalho and J. R. Correa, Acc. Chem. Res., 2015, 48, 1560–1569 CrossRef CAS PubMed.
- A. P. Demchenko, G. Duportail, S. Oncul, A. S. Klymchenko and Y. Mély, in Methods in Membrane Lipids, ed. D. M. Owen, Springer New York, New York, NY, 2015, vol. 1232, pp. 19–43 Search PubMed.
- A. A. R. Mota, P. H. P. R. Carvalho, B. C. Guido, H. C. B. de Oliveira, T. A. Soares, J. R. Corrêa and B. A. D. Neto, Chem. Sci., 2014, 5, 3995 RSC.
- P. H. P. R. Carvalho, J. R. Correa, K. L. R. Paiva, D. F. S. Machado, J. D. Scholten and B. A. D. Neto, Beilstein J. Org. Chem., 2019, 15, 2644–2654 CrossRef CAS PubMed.
- A. S. Klymchenko and R. Kreder, Chem. Biol., 2014, 21, 97–113 CrossRef CAS PubMed.
- A. M. Thooft, K. Cassaidy and B. VanVeller, J. Org. Chem., 2017, 82, 8842–8847 CrossRef CAS PubMed.
- D. L. Sackett and J. Wolff, Anal. Biochem., 1987, 167, 228–234 CrossRef CAS PubMed.
- S. R. Norris, C. C. Warner, B. J. Lampkin, P. Bouc and B. VanVeller, Org. Lett., 2019, 21, 3817–3821 CrossRef CAS PubMed.
- S. Uchiyama, T. Santa, T. Fukushima, H. Homma and K. Imai, J. Chem. Soc., Perkin Trans. 2, 1998, 2165–2174 RSC.
- S. Uchiyama, K. Takehira, S. Kohtani, K. Imai, R. Nakagaki, S. Tobita and T. Santa, Org. Biomol. Chem., 2003, 1, 1067–1072 RSC.
- S. Uchiyama, K. Kimura, C. Gota, K. Okabe, K. Kawamoto, N. Inada, T. Yoshihara and S. Tobita, Chem. – Eur. J., 2012, 18, 9552–9563 CrossRef CAS PubMed.
- K. W. Dunn, M. M. Kamocka and J. H. McDonald, Am. J. Physiol.: Cell Physiol., 2011, 300, C723–C742 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ob02486g |
| This journal is © The Royal Society of Chemistry 2020 |