Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Live cell single molecule tracking and localization microscopy of bioorthogonally labeled plasma membrane proteins
Andres I.
König
ab,
Raya
Sorkin
c,
Ariel
Alon
ab,
Dikla
Nachmias
ab,
Kalyan
Dhara
d,
Guy
Brand
c,
Ofer
Yifrach
a,
Eyal
Arbely
 abd,
Yael
Roichman
ce and
Natalie
Elia
abd,
Yael
Roichman
ce and
Natalie
Elia
 *ab
*ab
aDepartment of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva 84105, Israel. E-mail: elianat@post.bgu.ac.il; Fax: +972-8-6477546; Tel: +972-8-6428735
bNational Institute for Biotechnology in the Negev (NIBN), Ben-Gurion University of the Negev, Beer Sheva 84105, Israel
cRaymond & Beverly Sackler School of Chemistry, Tel Aviv University, Tel Aviv 6997801, Israel
dDepartment of Chemistry, Ben-Gurion University of the Negev, Beer Sheva 84105, Israel
eRaymond & Beverly Sackler School of Physics, and the Center for light-matter interaction, Tel Aviv University, Tel Aviv 6997801, Israel
First published on 3rd September 2020
Abstract
Correction for ‘Live cell single molecule tracking and localization microscopy of bioorthogonally labeled plasma membrane proteins’ by Andres I. König et al., Nanoscale, 2020, 12, 3236–3248, DOI: 10.1039/C9NR08594G.
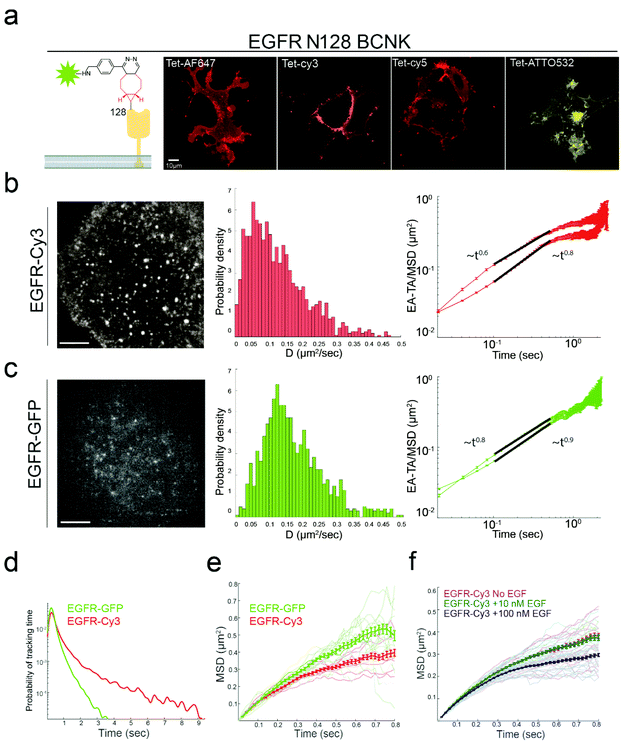
The authors regret that in the original manuscript, position 128 in the EGFR was referred to incorrectly as L128, rather than N128. This error occurred on page 3237 in the following sentence: “To this end, EGFR was mutated to carry a TAG codon at the previously published labeling site, Leu 128, and cloned into a single expression vector that encodes the cognate pair of tRNAcua:tRNA-synthetase17,18 (Fig. S1a†).”
The error also occurred in Fig. 1a, the corrected version of which is displayed below, along with the original, unaltered caption. These errors do not affect any of the experimental results and discussion or conclusions reported in the paper, only the display of the figure and text.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2020 |