Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Simultaneous introduction of multiple biomacromolecules into plant cells using a cell-penetrating peptide nanocarrier†
Chonprakun
Thagun
a,
Yoko
Motoda
a,
Takanori
Kigawa
 b,
Yutaka
Kodama
*ac and
Keiji
Numata
b,
Yutaka
Kodama
*ac and
Keiji
Numata
 *ad
*ad
aBiomacromolecules Research Team, RIKEN Center for Sustainable Resource Science, 2-1 Hirosawa, Wako-shi, Saitama 351-0198, Japan. E-mail: keiji.numata@riken.jp; Fax: +81-48-462-4664; Tel: +81-48-467-9525
bLaboratory for Cellular Structural Biology, RIKEN Center for Biosystems Dynamics Research, Yokohama 230-0045, Japan
cCenter for Bioscience Research and Education, Utsunomiya University, Tochigi 321-8505, Japan
dDepartment of Material Chemistry, Graduate School of Engineering, Kyoto University, Kyoto-Daigaku-Katsura, Nishikyo-ku, Kyoto 615-8510, Japan
First published on 8th September 2020
Abstract
Plant cells contain groups of biomolecules that participate together in a particular biological process. Exogenous codelivery of multiple biomolecules is an essential step for elucidation of the biological significance of these molecules and enables various biotechnological applications in plants. However, the currently existing biomolecule delivery methods face difficulties in delivering multiple components into plant cells, mediating transgene expression, and maintaining the stability of the numerous components and lead to delays in biomolecular function. Cell-penetrating peptides (CPPs) have demonstrated remarkable abilities to introduce diverse biomolecules into various plant species. Here, we employed the engineered CPP KH9-BP100 as a carrier to deliver multiple biomolecules into plant cells and performed a bimolecular fluorescence complementation assay to assess the simultaneous introduction of multiple biomolecules. We demonstrate that multiple biomolecule/CPP cargos can be simultaneously internalized by a particular plant cell, albeit with different efficiencies. We present a cutting-edge technique for codelivery of multiple biomolecules into plant cells that can be used for elucidation of functional correlations and for metabolic engineering.
Introduction
Plant cells create functional proteins that synergistically interact with each other to function in particular metabolic processes and in cellular mechanism. How these interacting biomolecules participate in specific biological reactions is intriguing, because protein interactions are highly complex and dynamic. The bimolecular fluorescence complementation (BiFC) assay is one powerful tool for visualizing interactions between two proteins during a biological process.1–4 In protein–protein interaction studies using BiFC, two proteins thought to potentially interact with each other are fused to two nonfluorescent fragments of a fluorescent protein.4,5 Expression cassettes of the two fusion proteins can be transiently introduced into plant cells through conventional plant transformation.6 Protein–protein interactions can be subsequently elucidated by observing the fluorescence signals from the restructured fluorescent proteins under a fluorescence microscope a few days after transformation.3,7 However, many issues prevent extensive use of this technique, such as an inability to deliver multiple exogenous gene expression cassettes into plant cells, unequal expression of multiple transgenes, delays in interacting protein function, and differences in the stability of the recombinant proteins prior to execution of their related functions.2,4,6Rational fine-tuning of multiple transgene expression approaches in plants is critical but is one of the drawbacks of such biomolecule delivery methods. This fine-tuning is necessary because the regulation of gene expression is strictly controlled at both the transcription and translation steps.8 It is unclear whether different gene expression cassettes produce similar levels of mRNA molecules in the same plant tissue.9 Individual mRNA molecules can correspond to numerous proteins that interact each other and might not behave similarly in a single plant cell.10–12 Furthermore, multiple proteins participating in the same event may have distinct vulnerabilities to protein degradation over the duration of an experiment.13 However, exogenous expression of multiple genes through conventional biomolecule introduction methods constrains the measurable functional parameters to only the average biomolecule synthesis rate. To precisely evaluate the dynamic functions of interacting proteins, advanced delivery systems for simultaneous introduction of multiple biomolecules into plant cells must be developed.
Cell-penetrating peptides (CPPs) are short chains of amino acid residues that have the ability to traverse cellular lipid membrane bilayers.14,15 CPPs are typically positively charged or amphipathic polypeptides that are derived from viral proteins as well as peptide toxins.16 These short functional peptides can efficiently deliver biomacromolecules such as double-stranded DNA (dsDNA) and double-stranded RNA (dsRNA) and high-molecular weight proteins into animal cells as well as several types of plant cells.14–20 The constituent amino acid residues of CPPs interact with various biomolecules to form biomolecule/CPP complexes.20–22 Combination of CPPs with cationic domains, e.g., lysine-rich, arginine-rich, and poly(lysine/histidine) domains, has been reported to improve the efficiency of biomolecule transport into different plant species.17,19 Among the CPPs commonly used for biomolecule delivery into plant cells, BP100, a CPP originally used as a plant antimicrobial peptide, and its cationic domain-fused derivatives have exhibited an exceptional ability to deliver biomolecules into intact plants.17–19,23 The efficiency of functionalized CPPs in transporting multiple components into plant cells remains to be explored.
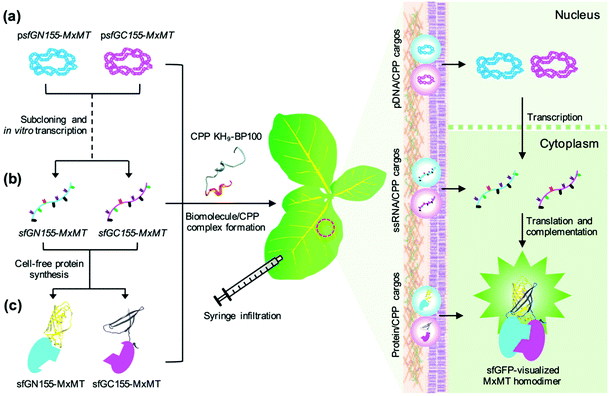
To contribute to advances in the biomolecule delivery techniques for plants, we utilized N-terminal poly(lysine/histidine) domain-fused BP100 (KH9-BP100) as a CPP carrier to transport multiple biomolecules into plant cells. We used a superfolder GFP (sfGFP)-based BiFC assay with a cytosolic homodimer of the Coffee arabica 7-methylxanthine methyltransferase 1 (MxMT) protein24–26 as a model to examine the effects of simultaneous introduction of two molecules with functional complementarity into plant cells. The biomolecules paired for protein–protein interactions, namely, plasmid DNA (pDNA) molecules harbouring transgene expression cassettes, single-stranded RNA (ssRNA) molecules, and fusion proteins, formed biomolecule/CPP complexes with KH9-BP100 (Fig. 1). These biomolecule/CPP complexes were cointroduced into intact plant leaves, and the formation of MxMT homodimers was notably observed in the cytosol by sfGFP-based BiFC.26,27 The efficiency of KH9-BP100 in delivering multiple biocomponents into plant cells was evaluated and compared among pDNA, ssRNA and proteins. Our CPP-mediated multiple-biomolecule delivery system provides a versatile platform with which to elucidate the functions of numerous components that control particular plant metabolic processes and cellular mechanisms.
Results
Multiple pDNA transfer into plant cells via KH9-BP100 peptide carriers
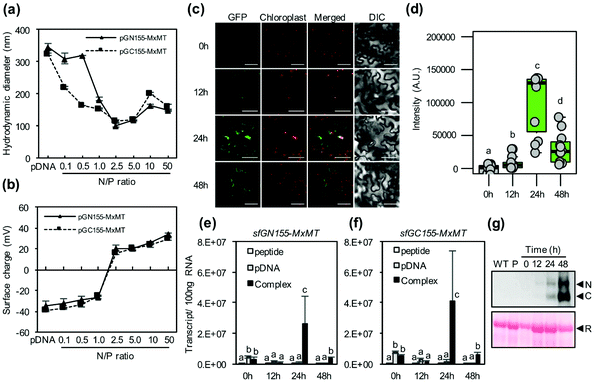
Nucleic acids such as DNA and RNA form nucleic acid/CPP complexes with cationic CPPs by electrostatic interaction.18,19 To produce cargo molecules, we first introduced two different pDNA molecules (psfGN155-MxMT and psfGC155-MxMT) encoding the nonfluorescent N-terminal and C-terminal sfGFP fragment-fused MxMT proteins26,27 (sfGN155-MxMT and sfGC155-MxMT, respectively) into plant cells using KH9-BP100 (Fig. S1a†). Different amounts of KH9-BP100 were added into solutions containing a consistent amount of pDNA at various N/P ratios (the molar ratios of the positively-charged NH3+ groups of basic side chains of lysine and histidine in the KH9-BP100 to the PO4− groups of the pDNA).28 Sub-micron-sized, negative-to-positive surface charge-transitioned pDNA/CPP complexes were formed with increasing N/P ratios (Fig. 2a, b, Fig. S1b, c and Tables S1, S2†). Positively surface-charged pDNA/CPP complexes with particle sizes of approximately 100–150 nm efficiently transfected various plant cell types.15,19 We rationally selected globular-shaped, ∼120 nm pDNA/CPP complexes that formed at an N/P ratio = 2.5 (Fig. 2a, b and Fig. S1d, e†) to monitor the successful delivery of the two different pDNA molecules into plant cells. We then infiltrated a mixture of the psfGN155-MxMT/KH9-BP100 and psfGC155-MxMT/KH9-BP100 complexes into tobacco (Nicotiana benthamiana) leaves. Via confocal laser scanning microscopy (CLSM) imaging analysis, the formation of sfGN155-MxMT/sfGC155-MxMT homodimers was gradually detected as complemented sfGFP fluorescence in the cytosol of leaf cells at different times post cotransfection (Fig. 2c, d and Fig. S2†). The sfGFP intensities in both epidermal and spongy mesophyll cells peaked at 24 hours before declining significantly by 48 hours post transfection (Fig. 2c, d and Fig. S2, S3a†). sfGN155-MxMT and sfGC155-MxMT mRNA accumulated to high levels in the cotransfected leaves by 24 hours post transfection (Fig. 2e and f). Immunoblotting confirmed the high levels of accumulation of both sfGN155-MxMT and sfGC155-MxMT proteins in the cotransfected leaves at 24–48 hours post transfection with pDNA/CPP complexes (Fig. 2g and Fig. S3b†). These results indicate that two pDNA molecules harbouring different gene expression cassettes delivered by a CPP can be coordinately transported into a plant cell and function as templates for the expression of two corresponding proteins that participate in protein–protein interactions.Delivery of multiple ssRNA molecules into plant cells
Although RNA molecules are less stable biochemically than DNA, double-stranded interfering RNA molecules delivered to plant cells by CPPs can efficiently suppress the expression of target genes in plants.18 We therefore examined whether two different RNA molecules could be coordinately transferred into plant cells with a CPP delivery platform (Fig. S4a†). ssRNA molecules of both sfGN155-MxMT and sfGC155-MxMT were synthesized by in vitro transcription (Fig. S4b and S5a, b†). Adding different amounts of KH9-BP100 to solutions containing a constant amount of individual ssRNA molecules changed the physicochemical properties of the ssRNA/CPP complexes at different N/P ratios (Fig. S5c–f and Tables S3, S4†). Taking into consideration the hydrodynamic diameters (Fig. S5c†), surface charges (Fig. S5d†), RNA release characteristics (Fig. S5e and f†) and sphere-shaped appearances of the ssRNA/CPP complexes, a positively charged ssRNA/CPP complex that formed at an N/P ratio of 2.5 (Fig. 3a and b) was chosen for cotransfection into plant cells. Because incorporating dsRNA molecules with peptides can enhance the ability of dsRNA to resist degradation by extra- and intracellular RNases,18 we subsequently monitored the degradation of ssRNA molecules in plant tissues by quantitative reverse transcriptase-PCR (qRT-PCR). Notably, the ssRNA abundances of sfGN155-MxMT and sfGC155-MxMT in the leaves cotransfected with the two ssRNA/CPP complexes were significantly higher than the ssRNA levels in free ssRNA-infiltrated samples (Fig. 3c). These ssRNA molecules accumulated at high levels for up to 4 hours before gradually declining in abundance to ∼30% at 24 hours post transfection (Fig. 3c). Based on the qRT-PCR analysis, the half-lives of the ssRNA molecules in plant leaves cotransfected with ssRNA/CPP complexes were 2.8 ± 1.6 hours for sfGN155-MxMT and 3.2 ± 2.7 hours for sfGC155-MxMT. These results imply that KH9-BP100 protects ssRNA molecules from the RNA degradation process in plant cells.To test the function of KH9-BP100 in transporting multiple ssRNA molecules into plant cells, we coinfiltrated a mixture of sfGN155-MxMT/KH9-BP100 and sfGC155-MxMT/KH9-BP100 complexes into N. benthamiana leaves. Successful internalization of these two ssRNA/CPP complexes by the same cells was confirmed via observation of complemented sfGFP fluorescence of an sfGN155-MxMT/sfGC155-MxMT homodimer in the plant cells. In CLSM analysis, significant increases in fluorescence intensity and a uniform distribution of the complemented sfGFP-visualized MxMT homodimers in the cytosol were observed at 12 and 24 hours after infiltration (Fig. 3d, e and Fig. S6†). Immunoblotting revealed that the accumulation of the sfGN155-MxMT and sfGC155-MxMT proteins in transfected plant cells gradually increased over the incubation periods and reached maximum levels at 24 hours post transfection (Fig. 3f). The CLSM imaging and immunoblotting results suggest that cationic KH9-BP100 can effectively deliver multiple ssRNA molecules for synchronous expression of multiple functional proteins in the same plant cell.
Translocation of multiple proteins into the cytoplasm by KH9-BP100
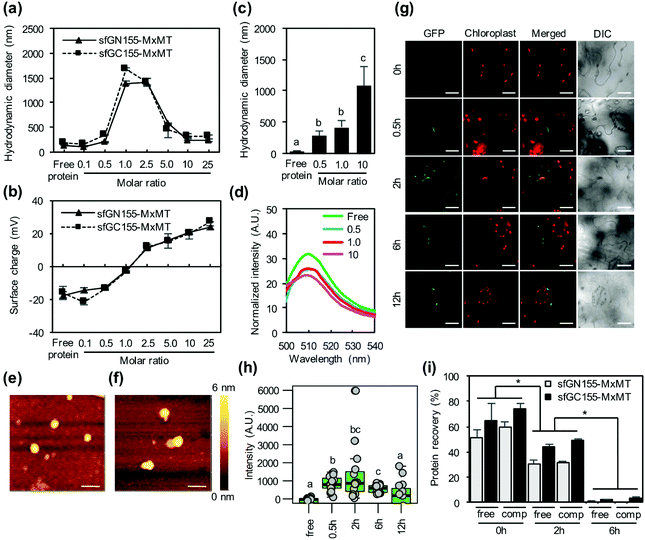
Some functional proteins can be translocated across plant cell boundaries by their CPPs. Negatively charged proteins such as BSA can form complexes with C-terminal-fused cationic (BP100)2-K8 and BP100-KH9 peptides, and BSA/CPP complexes can be delivered into plant cell cytoplasm.17 We analyzed the ability of KH9-BP100 to form protein/CPP complexes with two nonfluorescent N-terminal and C-terminal sfGFP fragment-fused MxMT proteins (sfGN155-MxMT and sfGC155-MxMT, respectively) for translocation into plant cells (Fig. S7a†). Recombinant sfGN155-MxMT and sfGC155-MxMT proteins with predicted molecular weights of 60.5 kDa and 52.4 kDa, respectively, were synthesized by cell-free protein synthesis (Fig. S7b†). Both proteins were calculated to show net charges of −15.2 mV in solution at pH 7.0. Equal amounts of both proteins were added to solutions containing various amounts of KH9-BP100 to form protein/CPP complexes at different molar ratios. Increasing the amounts of KH9-BP100 in the solutions increased the hydrodynamic diameters of both the sfGN155-MxMT/KH9-BP100 and sfGC155-MxMT/KH9-BP100 complexes (molar ratios of 0.1 and 0.5; Fig. 4a and Tables S5, S6†). Unexpectedly, both protein/CPP complexes that formed at molar ratios of 1.0 and 2.5 exhibited hydrodynamic diameters larger than 1 μm (Fig. 4a and Tables S5, S6†). However, the addition of KH9-BP100 to the protein solutions at molar ratios of 5.0 to 25 reduced the hydrodynamic diameters of the protein/CPP complexes to approximately 300 nm (Fig. 4a and Tables S5, S6†). The surface charges of the protein/KH9-BP100 complexes formed at various molar ratios increased progressively with increasing amounts of KH9-BP100 (Fig. 4b and Tables S5, S6†). Native-PAGE analysis revealed that protein mobility gradually decreased with increasing molar ratios (Fig. S8†). Based on the hydrodynamic diameters, surface charges and release of proteins from the protein/CPP complexes in the electrophoretic field, the sfGN155-MxMT/KH9-BP100 and sfGC155-MxMT/KH9-BP100 complexes that formed at molar ratios of 0.5, 1.0, and 10 were chosen for in vitro and in vivo protein–protein interaction assays by sfGFP-based BiFC with the MxMT homodimer.We subsequently investigated the in vitro interaction of two nonfluorescent sfGFP fragment-fused MxMT proteins (sfGN155-MxMT and sfGC155-MxMT) after they formed complexes with KH9-BP100 at molar ratios of 0.5, 1.0, and 10 by dynamic light scattering (DLS) analysis and fluorospectrometry. The hydrodynamic diameters of protein/KH9-BP100 complexes that formed at molar ratios of 0.5, 1.0 and 10 in the in vitro interaction assay buffer showed significant increases at 24 hours post incubation (Fig. 4c). The fluorospectrometric analysis showed that the in vitro BiFC reaction between sfGN155-MxMT and sfGC155-MxMT was reduced by complexation with KH9-BP100 at these molar ratios (Fig. 4d). We further investigated whether the incorporation of KH9-BP100 into sfGN155-MxMT and sfGC155-MxMT could inhibit the spontaneous dimerization of sfGN155-MxMT and sfGC155-MxMT in plant cells. Mixtures of sfGN155-MxMT/KH9-BP100 and sfGC155-MxMT/KH9-BP100 complexes at molar ratios of 0.5, 1.0, and 10 were infiltrated into the tobacco leaves. Complemented sfGFP fluorescence in the cytoplasm was observed in all samples transfected with a mixture of the two protein/KH9-BP100 complexes at 2 hours post infiltration (Fig. S9†). However, sfGFP signals were not detectable in the apoplasts of plant cells transfected with a solution containing free proteins (Fig. S9†). On the basis of the results regarding the inhibition of the in vitro BiFC reaction by KH9-BP100 and the ability to deliver multiple proteins into plant cells, the protein/KH9-BP100 complex that formed at a molar ratio of 10 was chosen for atomic force microscopy (AFM) imaging. The morphologies of the sfGN155-MxMT/KH9-BP100 and sfGC155-MxMT/KH9-BP100 complexes that formed at a molar ratio of 10 were globular in shape with an average size of ∼200 nm (Fig. 4e and f).
The efficiency of KH9-BP100 in transporting multiple proteins into plant cells was evaluated by time-course CLSM imaging. Cellular distribution of complemented sfGFP-visualized MxMT homodimers in the cytoplasm could be observed at 30 minutes post infiltration (Fig. 4g, h and Fig. S10†). The complemented sfGFP signals reached maximum intensity at 2 hours post infiltration and subsequently declined with time (Fig. 4g, h and Fig. S10†). Immunoblotting of the total proteins recovered from leaves transfected with the two protein/CPP complexes showed that up to 50% of the initial amounts of the sfGN155-MxMT and sfGC155-MxMT proteins remained in the plant leaves at 2 hours post transfection (Fig. 4i and Fig. S11†). However, the amounts of these proteins decreased drastically to only 3% of the initial amounts by 6 hours post infiltration (Fig. 4i and Fig. S11†). These results imply that significant degradation of the sfGN155-MxMT and sfGC155-MxMT proteins occurred in plant tissues after transfection with the protein/KH9-BP100 complexes.
KH9-BP100-mediated codelivery of multiple biomolecules into plant cells is an endomembrane-dependent event
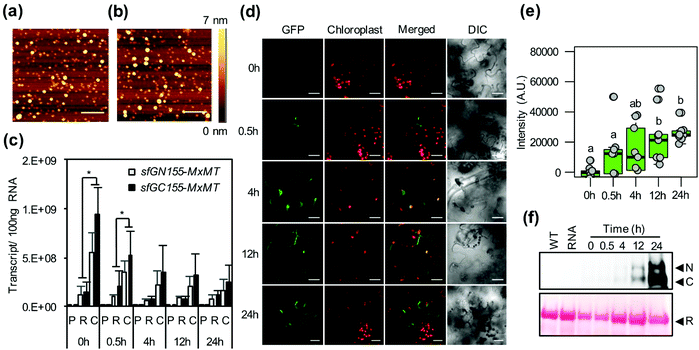
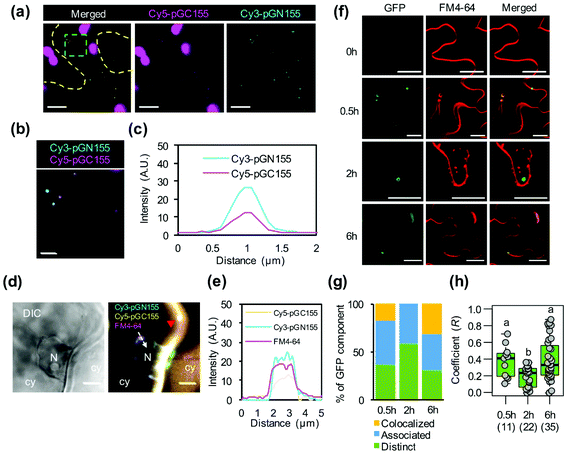
The positively charged helical CPP BP100 induces the formation of endocytic vesicles for transport of biomolecules through plant plasma membranes.23 We thus investigated the cellular uptake of multiple biomolecule/CPP cargos into plant cells. We observed the plant cell internalization of pDNA/KH9-BP100 complexes as representative biomolecules by detecting vesicle formation and colocalization with the plasma membrane-staining fluorescent dye FM4-64.29 Elevated numbers of large FM4-64-stained vesicles (∼1 μm) were found on the plasma membranes of plant cells at 2 hours post transfection (Fig. S12†). This result shows that the pDNA/KH9-BP100 complexes induced the formation of large membrane vesicles in the transfected plant cells.In our experiments, two different pDNA/CPP complexes were codelivered into plant cells. We performed fluorescence microscopy and colocalization analysis of fluorescent dye-labeled psfGN155-MxMT/KH9-BP100 and psfGC155-MxMT/KH9-BP100 complexes in plant cells at 2 hours post transfection. Fluorescent cyanine-3 (Cy3)-labeled psfGN155-MxMT/KH9-BP100 complexes were found to colocalize with fluorescent cyanine-5 (Cy5)-labeled psfGC155-MxMT/KH9-BP100 complexes inside plant cells (Fig. 5a–c). We also observed colocalization of both Cy3-psfGN155-MxMT/KH9-BP100 and Cy5-psfGC155-MxMT/KH9-BP100 complexes with intracellular vesicles stained by FM4-64 (∼1 μm) on the plasma membranes of plant cells at 2 hours post transfection (Fig. 5d and Fig. S13†). Moreover, we surprisingly detected colocalization of the fluorescent dye-labeled pDNA/KH9-BP100 complexes with the intracellular vesicles and observed trafficking to the nuclear membrane via the cellular network (Fig. 5d, e, Fig. S13 and Movie S1†). These results suggest that multiple nucleic acid cargos can be simultaneously internalized by plant cells via large endocytic vesicles and translocated together to the nucleus via intracellular vesicle trafficking.
To investigate how multiple protein/CPP complexes are transported into plant cells, we coinfiltrated intact leaves with a solution containing sfGN155-MxMT/KH9-BP100, sfGC155-MxMT/KH9-BP100, and FM4-64. In a control experiment without KH9-BP100, free proteins could neither be internalized by plant cells nor form complemented sfGFP-visualized MxMT homodimers in the intercellular spaces and apoplasts (Fig. S14a†). We further detected the colocalization of complemented sfGN155-MxMT/sfGC155-MxMT homodimers with FM4-64-stained vesicles in the cytoplasm (Fig. 5f and Fig. S14b†), and the results indicated that the two protein/CPP complexes colocalized with vesicles when the colocalization coefficient (R) was higher than 0.75, associated with vesicles when 0.25 ≤ R ≤ 0.75, or were distinctly released when R was lower than 0.25 (Fig. S14b†). At 30 minutes post transfection, 63% of the complemented sfGFP-visualized MxMT homodimers were colocalized or associated with the membrane-bound vesicles with a median R-value of 0.41 (Fig. 5g and h). Fifty-nine percent of these complemented sfGFP-visualized MxMT homodimers then dissociated from the vesicles and were distinctly located in the cytoplasm at 2 hours post transfection (median R = 0.23) (Fig. 5f–h). Furthermore, 31% of the complemented sfGFP-visualized MxMT homodimers colocalized with large endomembrane vesicles (∼5 μm) bound to the plasma membrane at 6 hours post transfection (Fig. 5f–h). These results imply that the two nonfluorescent sfGFP fragment-fused MxMT proteins delivered by KH9-BP100 were internalized by plant cells via membrane vesicle formation and movement. Additionally, the complemented sfGFP-visualized MxMT homodimers were later released into the cytoplasm (Fig. 5f, 2 hours post transfection).
Discussion
Introduction of multiple biomolecules across plant cell boundaries is a challenging step in investigations regarding the integrative functions of these biomolecules in particular cellular processes.8,30 For example, reprogramming of a significant biosynthesis pathway of plant metabolites requires efficient cotransfection of numerous independent vectors into plant cells.31–33 The translocation efficiencies for miscellaneous biomolecules depend mainly on the availability of the biomolecules after their introduction into and trafficking and accretion within target cells.34 Multiple pDNA molecules that each harbor an individual gene expression cassette can be mechanically and biologically transformed into the nuclei of plant cells.30 In addition, electroporation and PEG-mediated protoplast transfection have been used for direct delivery of short RNA molecules.35,36 Recent advances in the delivery of Cas9/gRNA ribonucleoprotein complexes into protoplasts have also been accomplished via PEG-mediated transfection.37 The applicability of and effort associated with these techniques are decidedly dependent on several factors. These include tedious material preparation and handling steps, the formulation of biomolecule/carrier complexes and the availability of expensive instruments. Our data suggest that multiple pDNA molecules, ssRNA molecules and proteins can be concurrently introduced into plant cells of intact plant tissues using CPP carriers, although with different efficiencies. These biomolecule/CPP cargos can penetrate plant cell barriers without external force or cumbersome plant material preparation, thus overcoming the limitations of conventional plant cell transfection techniques that require mechanical or chemical treatment. The CPP-mediated method also requires fewer biomolecules than gold particle-based microprojectile bombardment and polymeric PEG-mediated protoplast transformation methods.30,35–37Generally, integration of CPPs into biomacromolecules can fortify the molecules against cellular degradation processes or suppress their biological functions.38,39 However, our results suggest that incorporating CPPs into negatively charged pDNA and ssRNA molecules greatly enhances the stability of these polynucleotides without disrupting their biological functions. The functional KH9-BP100 peptide was rationally designed for CPP-based gene delivery.19 Constituent poly(lysine/histidine) (KH9) residues of KH9-BP100 are responsible for the condensation of the pDNA or ssRNA molecule, thus enabling the BP100 region to be freely exposed on the outer surface of the complex.18,19,40,41 This arrangement protects the polynucleotide from plant nucleases while the complex is transported into plant cells and finally to the nucleus. Cationic KH9-BP100 also interacts with the sfGN155-MxMT and sfGC155-MxMT proteins via electrostatic interactions.17 However, our in vitro interaction study results suggested that integration of CPP into the proteins significantly inhibited physical interaction between sfGN155-MxMT and sfGC155-MxMT. Moreover, KH9-BP100 did not contribute a protective effect against protein degradation in plant cells. Unexpectedly, this disorderly protein/peptide complexation could lead to contact of the proteins with proteases, priming the proteins for undesirable degradation before and after entry into plant cells.
The ability of CPPs to enter plant cells depends on the biological and physicochemical properties of both the CPPs and their cargos.20,21,42 The CPP BP100 and its cationic derivatives, KH9-BP100 and BP100-KH9, can destabilize the plasma membrane as well as induce the formation of membrane vesicles.17–19,23 Our results regarding the internalization of multiple biomolecule/KH9-BP100 complexes into plant cells are in accordance with these findings. Consistent with a previous report,23 our study revealed that the numbers of large membrane vesicles increased during the transfection of plant cells with biomolecule/CPP complexes. We also found that the biomolecule/CPP complexes relocated rapidly from the plasma membrane to the cytoplasm. Moreover, two different biomolecule/CPP complexes colocalized inside conspicuous large membrane vesicles. These vesicles subsequently trafficked the biomolecules and released them into the nucleus or cytosol. The two expression vectors were stable and coexisted in the plant cells transfected with pDNA molecules (Fig. S15†). However, the transcript levels of sfGN155-MxMT and sfGC155-MxMT and the complemented sfGFP-visualized MxMT homodimers were significantly decreased in pDNA/CPP complex-infiltrated leaves. Moreover, sfGN155-MxMT and sfGC155-MxMT RNA was rapidly degraded within 3 hours after transfection of the plant leaves with the ssRNA/CPP complexes, indicating that these RNA molecules were relatively unstable. In addition, the complemented sfGFP-visualized MxMT homodimers only showed significant interactions with the protein/CPP complexes within 2–6 hours post infiltration. The reductions in sfGFP fluorescence in the plant cells transfected with the biomolecule/CPP complexes could have been a consequence of the consecutive degradation of mRNA and the recombinant MxMT proteins in plant tissues. Since the trafficking of biomolecule/CPP complexes to the cytoplasm and nucleus occurs via intracellular vesicle movement, the biomolecular cargos could also have been translocated to vacuoles for nonspecific biomolecule degradation.43 It is also possible that the RNA molecules and proteins distributed in the cytoplasm were degraded by cytosolic RNases and proteases upon release from the CPP-based carriers.16
The two MxMT proteins form a homodimeric complex that localizes in the cytosolic fraction to catalyze the second methylation steps in caffeine biosynthesis from xanthosine.24,26,44 Notably, mRNA levels and protein accumulation are not definitively related to the transcriptional and translational activity of the corresponding genes.45,46 Small oscillations in protein accumulation can occur due to variations in the stability of mRNA and protein molecules that are tightly regulated posttranscriptionally and posttranslationally.35,36,45–48 Our findings showed that the transcript (or synthetic RNA) and protein abundances of the two nonfluorescent sfGFP fragment-fused MxMT components in plant leaves transfected with pDNA and ssRNA/CPP cargos were comparable. Additionally, the sfGFP-visualized MxMT homodimers were homogenously distributed in the cytoplasm. However, in plant cells transfected with ssRNA/CPP cargos, sfGFP fluorescence could be detected within 30 minutes post transfection, much faster than in the pDNA/CPP complex-transfected cells. Our results also showed that the sfGFP-visualized MxMT homodimers matured earlier in the endomembrane vesicles after entering the cells via multiple protein/CPP cargos than after entering via pDNA and ssRNA cargos. However, the sfGFP fluorescence intensity and protein accumulation in cells transfected using multiple protein/CPP cargos were much lower than those in cells transfected using multiple pDNA and ssRNA cargos. Moreover, the protein levels in protein/CPP complex-transfected plant cells drastically decreased within 6 hours after transfection, suggesting that protein degradation occurred. Sub-micron-sized (200 to 1000 nm) positively charged protein/CPP complexes can enter cells by membrane-associated macropinocytosis or heparan sulfate-dependent endocytosis.16,17,22,23,49 There is a strong possibility that two BiFC proteins can dissociate from their respective CPP-based carriers and interact with each other inside the endomembrane vesicles. BiFC complexes with MxMT homodimers may then be intermittently released from the vesicles into the cytoplasm, resulting in an uneven distribution of sfGFP signals within the plant cells. These sfGFP-visualized MxMT homodimers could also be transported to the trans-Golgi networks and early endosomes to be recycled back to the plasma membrane or to the vacuoles for complete degradation, as drastic reductions in recombinant MxMT protein levels have been observed after transfection.22,43,50
Biosynthesis of plant metabolites requires multistep enzymatic reactions. The involved enzymes are strictly controlled not only at the expression level but also at the protein level.51 Therefore, comprehensive engineering of targeted metabolic processes can be accomplished by introducing numerous gene expression regulators and corresponding enzymes as well as their activation components into plant cells.52 Multiple genetic modifications at key reaction steps are possible via classic gene transfer techniques.30 However, alteration of multiple genes in a single transgenic line can occasionally cause transgene silencing and unbalanced protein activity as well as cryptic phenotypic defects.30,53 Uncontrolled expression ratios and protein levels in transgenic plants can strongly influence the bifurcated reactions of target metabolic pathways.8,30,53 However, nanocarriers are useful tools for temporary engineering of plant metabolic pathways.54 Among the various types of nanocarriers, CPPs have the ability to deliver diverse biomolecules into plant cells.16–19 Our data highlight the ability of CPPs to enable integrative transport of multiple biocomponents for plant metabolic engineering. A prominent example is the use of CPPs to transfer RNA molecules and protein components into plant cells for genome editing using ribonucleoprotein particles such as CRISPR/Cas complexes, transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases.55,56 Short noncoding RNAs and protein-degrading enzyme inhibitors can also be codelivered into plant cells to transiently inhibit the activity of proteases linked to the rate-limiting steps of a target metabolic pathway.57,58 In addition, biomolecules can be precisely targeted to plastids, mitochondria, peroxisomes and vacuoles using a combination of functionalized CPPs and organelle-targeting peptides (OTPs).59–61 These CPP/OTP-based carriers facilitate effective reprogramming of organellar metabolic pathways in crop species. However, major drawbacks of such peptide-based biomolecule delivery systems are the production costs of the peptides and biomolecules. Further improvements in hybrid technology for peptide synthesis and purification as well as recombinant biomolecule production processes may overcome the manufacturing limitations of peptide-based multiple-biomolecule delivery systems.
Conclusions
Our results show that pairs of molecules that act as interaction partners, i.e., pDNA, ssRNA, and protein pairs, can be codelivered into the same plant cell using cationic CPP-based carriers. The CPP-based multiple-biomolecule delivery system is compatible with different plant species and various biomolecules of interest but does not require specialized equipment or complicated plant material preparation. Furthermore, the CPP can be combined with selected OTPs, i.e., peptides that designate biomolecule translocation to specific organelles,60,61 for targeted delivery of multiple components to various plant organelles. Our peptide-mediated multiple-biomolecule delivery platform provides a robust tool for comprehensive engineering of plant metabolic processes and quality trait improvement.Experimental
Cationic KH9-BP100 peptide
The cationic CPP KH9-BP100 (NH2–KHKHKHKHKHKHKHKHKHKKLFKKILKYL–COO−) was synthesized as previously described.19 The peptide was prepared in a 1.0 mg ml−1 stock solution with water.Plant cultivation
Seeds of N. benthamiana were germinated on soil (Promix, Rivière-du-Loup, Canada) supplemented with vermiculite (ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 25 °C under 12/12-hour light/dark periods with 100 μmol photons per m2 per s1 in a plant growth chamber for 7 days. The seedlings were then transferred to individual pots and cultured at 22 °C under 8/16-hour light/dark periods with 100 μmol photons per m2 per s1 in a plant growth chamber. Fully expanded leaves from 4–5-week-old plants were used for transfection.
1) at 25 °C under 12/12-hour light/dark periods with 100 μmol photons per m2 per s1 in a plant growth chamber for 7 days. The seedlings were then transferred to individual pots and cultured at 22 °C under 8/16-hour light/dark periods with 100 μmol photons per m2 per s1 in a plant growth chamber. Fully expanded leaves from 4–5-week-old plants were used for transfection.
Preparation of pDNA, ssRNA, and protein molecules
pDNA molecules were purified from liquid cultures of Escherichia coli DH5α-harbouring psfGN155-MxMT or psfGC155-MxMT27 using a Qiagen Plasmid Giga Kit (Qiagen, Hilden, Germany). The concentration of pDNA was adjusted to 1.0 mg ml−1 with water and then kept at −20 °C.ssRNA molecules of sfGN155-MxMT and sfGC155-MxMT were synthesized by in vitro transcription.18 The coding sequences of the sfGN155-MxMT and sfGC155-MxMT genes were amplified from psfGN155-MxMT and psfGC155-MxMT plasmids, respectively, with the primers shown in Table S7.† The gene fragments were subcloned into pET-30b(+) vectors at XbaI/BamHI restriction sites. The ssRNA molecules were synthesized from a pET-30b(+) vector harbouring a codon sequence of the sfGN155-MxMT or sfGC155-MxMT gene with a T7 RiboMAX Express System (Promega, Madison, WI, USA) and purified with a QIAquick Nucleotide Removal Kit (Qiagen). The purified ssRNA molecules were analyzed by 20% polyacrylamide gel electrophoresis to determine the integrity of the ssRNA (Fig. S4b†). The ssRNA samples were kept in small aliquots at −80 °C (1.0 mg ml−1 stock solution).
The nonfluorescent sfGFP fragment-fused MxMT proteins sfGN155-MxMT and sfGC155-MxMT were synthesized with a dialysis-mode cell-free protein synthesis system.17 Briefly, pDNA for in vitro transcription was added to a solution containing the necessary substrates, buffer solutions and enzymes for in vitro transcription and translation. His-tagged proteins were purified from total protein in a cell-free synthesis reaction with TALON® Cell Thru HisTrap resin using the manufacturer's protocol (GE Healthcare, Little Chalfont, UK). The polyhistidine affinity tags were removed by in-column TEV protease digestion and purification (Fig. S5a†). The resulting proteins were dialyzed through a membrane with a 15 kDa molecular weight cutoff (Pierce Biotechnology, Rockford, IL, USA) in dialysis buffer at 30 °C for 16 hours. The protein solution recovered from dialysis was analyzed via 4–15% SDS-PAGE (Bio-Rad Laboratories, Hercules, CA, USA) (Fig. S5b†). The proteins were diluted to a 1.0 mg m−1 concentration and stored at 4 °C.
Formation of biomolecule/KH9-BP100 complexes
Two different pDNA/KH9-BP100 complexes were formed at different N/P ratios (0, 0.1, 0.5, 1.0, 2.5, 5.0, 10 and 50) by adding different amounts of KH9-BP100 peptide solution (1.0 mg ml−1 stock solution) into 100 μl of water containing 10 μg of pDNA. The solutions were mixed thoroughly and incubated at ambient temperature for 30 minutes without shaking. After incubation, the solutions were diluted with 700 μl of water. The particle size of the complex was determined by DLS with a Zeta Nanosizer using a 633 nm He–Ne laser at 25 °C with a backscatter detection angle of 173°. The surface charge of the complex in solution was measured by a Zeta potentiometer. The averaged data from three replicates were obtained using Zetasizer software ver. 6.20 (Malvern Instruments, Ltd, Worcestershire, UK). Release of pDNA molecules from the complexes was analyzed by resolving 20 μl of complex solution via 1.0% (w/v) agarose gel retardation assays in 1× TAE buffer at 100 V for 30 minutes.Complexes of ssRNA/KH9-BP100 were prepared by mixing 1.0 μg of ssRNA in 100 μl of RNase-free water containing different amounts of KH9-BP100 peptide (N/P ratios = 0, 0.1, 0.5, 1.0, 2.5, 5.0, 10 and 50). After mixing, each complex solution was incubated at ambient temperature without shaking for 30 minutes. The solution was diluted with 700 μl of RNase-free water before being subjected to measurement of complex size and surface charge as previously described. Release of ssRNA molecules from the complexes was analyzed by resolving 20 μl of complex solution via 2.0% (w/v) agarose gel retardation assays in 0.5× TBE buffer at 80 V for 60 minutes.
To form nonfluorescent sfGFP fragment-fused MxMT protein/KH9-BP100 complexes at various molar ratios (0, 0.1, 0.5, 1.0, 2.5, 5.0, 10 and 25), different volumes of 26.3 μM KH9-BP100 were added to 100 μl of water containing 82.5 μM sfGN155-MxMT protein or 95.5 μM sfGC155-MxMT protein. The complex solutions were incubated at ambient temperature for 30 minutes before dilution with 700 μl of water. The complex size and surface charge were determined with a Zetasizer. The electrostatic interactions of the proteins with the KH9-BP100 peptide were analyzed by protein gel-shift assays on 4–16% Invitrogen NativePAGE Bis-Tris Gels (Life Technologies, Carlsbad, CA, USA).
N. benthamiana leaf infiltration
After complex formation, two biomolecule/KH9-BP100 complexes formed separately were combined in 1.5 ml microcentrifuge tubes at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and gently mixed by pipetting. One hundred microliters of the mixture was infiltrated into the abaxial side of an N. benthamiana leaf with a 1 ml needleless syringe. The transfected plants were incubated at 22 °C under 12/12-hour light/dark periods prior to the expression analyses. Only leaves with complete liquid dispersion through a specific area were used in the subsequent experiments.
1 and gently mixed by pipetting. One hundred microliters of the mixture was infiltrated into the abaxial side of an N. benthamiana leaf with a 1 ml needleless syringe. The transfected plants were incubated at 22 °C under 12/12-hour light/dark periods prior to the expression analyses. Only leaves with complete liquid dispersion through a specific area were used in the subsequent experiments.
CLSM observation and image analysis
Approximately 0.5 × 0.5 cm2 sections of leaves that had been infiltrated with complex solution were cut from the region around the syringe-attached area (indicated by leaf tissue necrosis). The leaf segments were washed twice and deaerated in water prior to observation with a Zeiss LSM 880 confocal microscope (Carl Zeiss, Oberkochen, Germany) with 488 nm/490–535 nm excitation/emission wavelengths for the detection of sfGFP. A 640–700 nm emission wavelength was used to detect chlorophyll autofluorescence in plant cells. Plant cell images with high fluorescence intensity and low background fluorescence were chosen for image analysis with Fiji ImageJ.62 The means of the relative green fluorescence intensity were measured from at least 9 different regions of interest (ROIs) taken from three independent experiments only in the cytoplasm (the nucleus was excluded). The average background signal measured from the 5 ROIs surrounding the cell was subtracted from all fluorescence signals.Transcript analysis
qRT-PCR was used to determine the transcript abundance in pDNA- or ssRNA/KH9-BP100 complex-transfected leaves. Total RNA was extracted from transfected plant leaves with an RNeasy Plant Mini Kit (Qiagen) and treated with DNase I (Qiagen) to eliminate contaminating genomic DNA. Complementary DNA (cDNA) molecules were synthesized from 0.5 μg of purified RNA with a QuantiTect Reverse Transcription Kit (Qiagen). Known concentrations of ssRNA molecules from in vitro transcription were converted to cDNA, which was used as the template in qRT-PCR. qRT-PCR was performed using SYBR™ Green RealTime Master Mix Plus (Toyobo, Osaka, Japan) with the primer sequences provided in Table S7.† A linear regression equation for each mRNA molecule was generated by plotting the threshold cycle of the amplification value (ΔCt) against the amount of transcript (Fig. S16†). The transcript amount per 100 ng of total RNA was determined by fitting the ΔCt value to the linear equation.The RNA decay in the ssRNA/CPP complex-treated plant leaves was determined from qRT-PCR analysis of the transcript abundances. The data points at 0, 0.5 and 4 hours post transfection were used to determine the first-order decay kinetics.63 The transcript change over time was initially log-transformed, and the RNA decay rate constant (k) was determined through linear regression. The half-life (T1/2) was calculated from the RNA decay rate constant as described in eqn (1).
| T(1/2) = ln(2)/k | (1) |
In vitro protein–protein interaction assays
Ten-microgram samples of the nonfluorescent sfGFP fragment-fused sfGN155-MxMT and sfGC155-MxMT proteins were used to form protein/KH9-BP100 complexes at molar ratios of 0.5, 1.0 and 10. Equal amounts of the two proteins without KH9-BP100 were included in the experiment as controls. After complex formation, 25 μl of each protein/CPP complex was mixed together with 50 μl of 20 mM Tris-HCl, pH 7.5 + 0.2% Tween-20. The interaction reaction was incubated at 4 °C for 24 hours without shaking. Each solution was diluted with 800 μl of water and subjected to fluorescence measurement with an FP8500 spectrofluorometer (JASCO, Tokyo, Japan) with a 488 nm excitation wavelength and a scanning emission wavelength of 500 to 580 nm (5 nm bandwidth). The particle size of the interacting complex was determined by DLS measurement with the parameters mentioned above.Internalization of biomolecule/CPP complexes by plant cells
Internalization of biomolecule/KH9-BP100 complexes by plant cells was studied via assessment of colocalization of fluorescent dye-stained pDNA or fluorescent sfGFP with the plasma membrane-staining fluorescent dye FM4-64.29 pDNA molecules were labeled with Cy3 or Cy5 fluorescent dye using a Label IT® Nucleic Acid Labeling Kit (Mirus Bio, Madison, WI, USA). Solutions containing fluorescent dye-labeled pDNA/KH9-BP100 complexes or nonfluorescent sfGFP fragment-fused MxMT protein/KH9-BP100 complexes were introduced to the abaxial sides of fully expanded leaves of N. benthamiana by syringe infiltration. After incubation, the plant leaves were infiltrated with 10 μM FM4-64 solution and incubated for 10 minutes. The colocalization of the fluorescent dye-labeled pDNA/CPP complexes or sfGFP-visualized complemented MxMT homodimers with FM4-64 was observed on 0.5 × 0.5 cm2 sections of leaf samples by CLSM with excitation/emission wavelengths of 555/560–580 nm for Cy3, 633/640–650 nm for Cy5, 488/490–535 nm for sfGFP, and 488/620–700 nm for FM4-64 detection.Immunoblotting
For immunoblot analysis of pDNA- and ssRNA/CPP cargo-transfected plant leaves, total soluble proteins were extracted from at least 5 leaves with protein extraction buffer (100 mM HEPES, pH 7.0, 1 mM EDTA, pH 8.0, 1% (w/v) SDS, 200 mM DTT, 1% (v/v) Halt™ protease inhibitor cocktail (Pierce Biotechnology), and 10% (v/v) glycerol). The soluble proteins were concentrated through a filter membrane with a 10 kDa molecular weight cutoff (Pierce Biotechnology), and the retained proteins were used for immunoblotting. For protein/CPP complex-transfected leaves, total soluble proteins were extracted from one leaf with the protein extraction solution described above. Twenty micrograms of total protein was resolved on 7.5% Mini PROTEAN® TGX™ Precast gels (Bio-Rad Laboratories, Hercules, CA, USA), and the separated proteins were blotted onto Hybond-P PVDF membranes (GE Healthcare, Buckinghamshire, UK). The sfGN155-MxMT and sfGC155-MxMT proteins were detected with a primary antibody solution containing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution of a rabbit anti-GFP polyclonal antibody (NB600-308) (Novus Biologicals, Littleton, CO, USA) and then with a secondary antibody solution containing a 1
5000 dilution of a rabbit anti-GFP polyclonal antibody (NB600-308) (Novus Biologicals, Littleton, CO, USA) and then with a secondary antibody solution containing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgC polyclonal antibody (ab6721) (Abcam, Tokyo, Japan). The signal of HRP activity on the membrane was detected after applying 1 ml of SuperSignal™ West Pico PLUS chemiluminescent substrate (Thermo Scientific, Waltham, MA, USA) onto the membrane using a LAS3000 imaging system (FujiFilm, Tokyo, Japan). The band intensities of sfGFP fusion proteins were quantified using Fiji ImageJ.62 Linear regressions of the purified sfGN155-MxMT- and sfGC155-MxMT proteins were performed as shown in Fig. S10b.† The abundance of the sfGFP fusion protein in each sample was calculated using the linear regression equation generated for each standard protein.
000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgC polyclonal antibody (ab6721) (Abcam, Tokyo, Japan). The signal of HRP activity on the membrane was detected after applying 1 ml of SuperSignal™ West Pico PLUS chemiluminescent substrate (Thermo Scientific, Waltham, MA, USA) onto the membrane using a LAS3000 imaging system (FujiFilm, Tokyo, Japan). The band intensities of sfGFP fusion proteins were quantified using Fiji ImageJ.62 Linear regressions of the purified sfGN155-MxMT- and sfGC155-MxMT proteins were performed as shown in Fig. S10b.† The abundance of the sfGFP fusion protein in each sample was calculated using the linear regression equation generated for each standard protein.
Statistical analysis
Multiple comparisons among groups of samples in the same experiment were performed using analysis of variance (ANOVA) with Tukey's HSD test at p = 0.05. Significant differences in means between two samples were determined by Student's t-test with StatPlus:mac statistical software (AnalystSoft, Walnut, CA, USA).Live subject statement
This study was performed in strict accordance with the RIKEN guidelines for the genetically modified plants and was approved by the Wako safety center of RIKEN (Saitama, Japan).Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST ERATO) Grant Number JPMJER1602.References
- M. Walter, C. Chaban, K. Schutze, O. Batistic, K. Weckermann, C. Nake, D. Blazevic, C. Grefen, K. Schumacher, C. Oecking, K. Harter and J. Kudla, Plant J., 2004, 40, 428–438 CrossRef CAS.
- M. Morell, A. Espargaro, F. X. Aviles and S. Ventura, Nat. Protoc., 2008, 3, 22–33 CrossRef CAS.
- J. Kudla and R. Bock, Plant Cell, 2016, 28, 1002–1008 CrossRef CAS.
- Y. Kodama and C. D. Hu, BioTechniques, 2012, 53, 285–298 CrossRef CAS.
- T. K. Kerppola, Nat. Protoc., 2006, 1, 1278–1286 CrossRef.
- K. W. Berendzen, M. Bohmer, N. Wallmeroth, S. Peter, M. Vesic, Y. Zhou, F. K. Tiesler, F. Schleifenbaum and K. Harter, Plant Methods, 2012, 8, 25 CrossRef CAS.
- N. Ohad, K. Shichrur and S. Yalovsky, Plant Physiol., 2007, 145, 1090–1099 CrossRef CAS.
- M. A. Jackson, P. R. Sternes, S. R. Mudge, M. W. Graham and R. G. Birch, Plant Biotechnol. J., 2014, 12, 925–933 CrossRef CAS.
- N. Esmaeili, X. Yang, Y. Cai, L. Sun, X. Zhu, G. Shen, P. Payton, W. Fang and H. Zhang, Sci. Rep., 2019, 9, 7642 CrossRef.
- R. Zoschke, K. P. Watkins and A. Barkan, Plant Cell, 2013, 25, 2265–2275 CrossRef CAS.
- G. W. Li, D. Burkhardt, C. Gross and J. S. Weissman, Cell, 2014, 157, 624–635 CrossRef CAS.
- T. Fujita, Y. Kurihara and S. Iwasaki, Plant Cell Physiol., 2019, 60, 1917–1926 CrossRef CAS.
- K. Goslin, L. Eschen-Lippold, C. Naumann, E. Linster, M. Sorel, M. Klecker, R. de Marchi, A. Kind, M. Wirtz, J. Lee, N. Dissmeyer and E. Graciet, Plant Physiol., 2019, 180, 2272–2289 CrossRef CAS.
- A. Bolhassani, B. S. Jafarzade and G. Mardani, Peptides, 2017, 87, 50–63 CrossRef CAS.
- M. Ahmed, Biomater. Sci., 2017, 5, 2188–2211 RSC.
- M. Chang, Y. W. Huang, R. S. Aronstam and H. J. Lee, Curr. Pharm. Biotechnol., 2014, 15, 267–275 CAS.
- K. K. Ng, Y. Motoda, S. Watanabe, A. Sofiman Othman, T. Kigawa, Y. Kodama and K. Numata, PLoS One, 2016, 11, e0154081 CrossRef.
- K. Numata, M. Ohtani, T. Yoshizumi, T. Demura and Y. Kodama, Plant Biotechnol. J., 2014, 12, 1027–1034 CrossRef CAS.
- M. Lakshmanan, Y. Kodama, T. Yoshizumi, K. Sudesh and K. Numata, Biomacromolecules, 2013, 14, 10–16 CrossRef CAS.
- A. Chugh and F. Eudes, FEBS J., 2008, 275, 2403–2414 CrossRef CAS.
- N. Unnamalai, B. G. Kang and W. S. Lee, FEBS Lett., 2004, 566, 307–310 CrossRef CAS.
- M. Chang, J. C. Chou, C. P. Chen, B. R. Liu and H. J. Lee, New Phytol., 2007, 174, 46–56 CrossRef CAS.
- K. Eggenberger, C. Mink, P. Wadhwani, A. S. Ulrich and P. Nick, ChemBioChem, 2011, 12, 132–137 CrossRef CAS.
- M. Ogawa, Y. Herai, N. Koizumi, T. Kusano and H. Sano, J. Biol. Chem., 2001, 276, 8213–8218 CrossRef CAS.
- K. Mizuno, M. Kato, F. Irino, N. Yoneyama, T. Fujimura and H. Ashihara, FEBS Lett., 2003, 547, 56–60 CrossRef CAS.
- Y. Kodama, T. Shinya and H. Sano, Biochimie, 2008, 90, 547–551 CrossRef CAS.
- Y. Fujii and Y. Kodama, Plant Biotechnol., 2015, 32, 81–87 CrossRef CAS.
- D. J. Gary, J. Min, Y. Kim, K. Park and Y. Y. Won, Macromol. Biosci., 2013, 13, 1059–1071 CrossRef CAS.
- S. Bolte, C. Talbot, Y. Boutte, O. Catrice, N. D. Read and B. Satiat-Jeunemaitre, J. Microsc., 2004, 214, 159–173 CrossRef CAS.
- M. Dafny-Yelin and T. Tzfira, Plant Physiol., 2007, 145, 1118–1128 CrossRef CAS.
- T. Nakatsuka, Y. Abe, Y. Kakizaki, S. Yamamura and M. Nishihara, Plant Cell Rep., 2007, 26, 1951–1959 CrossRef CAS.
- T. Moses, J. Pollier, J. M. Thevelein and A. Goossens, New Phytol., 2013, 200, 27–43 CrossRef CAS.
- P. D. Sonawane, J. Pollier, S. Panda, J. Szymanski, H. Massalha, M. Yona, T. Unger, S. Malitsky, P. Arendt, L. Pauwels, E. Almekias-Siegl, I. Rogachev, S. Meir, P. D. Cardenas, A. Masri, M. Petrikov, H. Schaller, A. A. Schaffer, A. Kamble, A. P. Giri, A. Goossens and A. Aharoni, Nat. Plants, 2016, 3, 16205 CrossRef.
- Y. Cao, H. Chen, R. Qiu, M. Hanna, E. Ma, M. Hjort, A. Zhang, R. S. Lewis, J. C. Wu and N. A. Melosh, Sci. Adv., 2018, 4, eaat8131 CrossRef CAS.
- Y. Qi, X. Zhong, A. Itaya and B. Ding, Nucleic Acids Res., 2004, 32, e179 CrossRef.
- R. Vanitharani, P. Chellappan and C. M. Fauquet, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9632–9636 CrossRef CAS.
- J. W. Woo, J. Kim, S. I. Kwon, C. Corvalan, S. W. Cho, H. Kim, S. G. Kim, S. T. Kim, S. Choe and J. S. Kim, Nat. Biotechnol., 2015, 33, 1162–1164 CrossRef CAS.
- I. Remy, G. Ghaddar and S. W. Michnick, Nat. Protoc., 2007, 2, 2302–2306 CrossRef CAS.
- A. Galarneau, M. Primeau, L. E. Trudeau and S. W. Michnick, Nat. Biotechnol., 2002, 20, 619–622 CrossRef CAS.
- J. A. Chuah, A. Matsugami, F. Hayashi and K. Numata, Biomacromolecules, 2016, 17, 3547–3557 CrossRef CAS.
- Q. R. Chen, L. Zhang, S. A. Stass and A. J. Mixson, Gene Ther., 2000, 7, 1698–1705 CrossRef CAS.
- A. Chugh and F. Eudes, Biochim. Biophys. Acta, 2007, 1768, 419–426 CrossRef CAS.
- C. Eisenach, R. Francisco and E. Martinoia, Curr. Biol., 2015, 25, R136–R137 CrossRef CAS.
- H. Uefuji, S. Ogita, Y. Yamaguchi, N. Koizumi and H. Sano, Plant Physiol., 2003, 132, 372–380 CrossRef CAS.
- D. Drapier, J. Girard-Bascou and F. A. Wollman, Plant Cell, 1992, 4, 283–295 CrossRef CAS.
- J. D. Rochaix, Plant Physiol., 2001, 125, 142–144 CrossRef CAS.
- S. L. Harmer, J. B. Hogenesch, M. Straume, H. S. Chang, B. Han, T. Zhu, X. Wang, J. A. Kreps and S. A. Kay, Science, 2000, 290, 2110–2113 CrossRef CAS.
- S. Eberhard, D. Drapier and F. A. Wollman, Plant J., 2002, 31, 149–160 CrossRef CAS.
- M. Chang, J. C. Chou and H. J. Lee, Plant Cell Physiol., 2005, 46, 482–488 CrossRef CAS.
- M. R. Rosquete, D. J. Davis and G. Drakakaki, Plant Physiol., 2018, 176, 187–198 CrossRef CAS.
- L. J. Sweetlove and A. R. Fernie, New Phytol., 2005, 168, 9–24 CrossRef CAS.
- T. Tohge, F. Scossa and A. R. Fernie, Plant Physiol., 2015, 169, 1499–1511 CAS.
- C. Halpin, Plant Biotechnol. J., 2005, 3, 141–155 CrossRef CAS.
- F. J. Cunningham, N. S. Goh, G. S. Demirer, J. L. Matos and M. P. Landry, Trends Biotechnol., 2018, 36, 882–897 CrossRef CAS.
- A. Malzahn, L. Lowder and Y. Qi, Cell Biosci., 2017, 7, 21 CrossRef.
- A. Bilichak, L. Sastry-Dent, S. Sriram, M. Simpson, P. Samuel, S. Webb, F. Jiang and F. Eudes, Plant Biotechnol. J., 2020, 18, 1307–1316 CrossRef CAS.
- M. Sabzehzari and M. R. Naghavi, Gene, 2019, 682, 13–24 CrossRef CAS.
- R. Hamza, M. Perez-Hedo, A. Urbaneja, J. L. Rambla, A. Granell, K. Gaddour, J. P. Beltran and L. A. Canas, BMC Plant Biol., 2018, 18, 24 CrossRef.
- T. Yoshizumi, K. Oikawa, J. A. Chuah, Y. Kodama and K. Numata, Biomacromolecules, 2018, 19, 1582–1591 CrossRef CAS.
- J. A. Chuah, T. Yoshizumi, Y. Kodama and K. Numata, Sci. Rep., 2015, 5, 7751 CrossRef CAS.
- C. Thagun, J. A. Chuah and K. Numata, Adv. Sci., 2019, 6, 1902064 CrossRef CAS.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS.
- C. Y. Chen, N. Ezzeddine and A. B. Shyu, Methods Enzymol., 2008, 448, 335–357 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr04718j |
| This journal is © The Royal Society of Chemistry 2020 |