Open Access Article
Open Access ArticleAdaptable DNA interactions regulate surface triggered self assembly†
Roberta
Lanfranco
a,
Pritam Kumar
Jana
b,
Gilles
Bruylants
 c,
Pietro
Cicuta
c,
Pietro
Cicuta
 a,
Bortolo Matteo
Mognetti
a,
Bortolo Matteo
Mognetti
 b and
Lorenzo
Di Michele
b and
Lorenzo
Di Michele
 *ad
*ad
aCavendish Laboratory, University of Cambridge, JJ Thomson Avenue, Cambridge, CB3 0HE, UK
bInterdisciplinary Center for Nonlinear Phenomena and Complex Systems, Université libre de Bruxelles (ULB), Campus Plaine, CP 231, Blvd. du Triomphe, B-1050 Brussels, Belgium
cEngineering of Molecular NanoSystems, Université libre de Bruxelles (ULB), CP165/64, 50 av. F.D. Roosevelt, 1050 Brussels, Belgium
dDepartment of Chemistry, Imperial College London, Molecular Sciences Research Hub, 80 Wood Lane, London W12 0BZ, UK. E-mail: l.di-michele@imperial.ac.uk
First published on 25th August 2020
Abstract
DNA-mediated multivalent interactions between colloidal particles have been extensively applied for their ability to program bulk phase behaviour and dynamic processes. Exploiting the competition between different types of DNA–DNA bonds, here we experimentally demonstrate the selective triggering of colloidal self-assembly in the presence of a functionalised surface, which induces changes in particle–particle interactions. Besides its relevance to the manufacturing of layered materials with controlled thickness, the intrinsic signal-amplification features of the proposed interaction scheme make it valuable for biosensing applications.
Inspired by biology, self-assembly has been extensively investigated for its relevance to the manufacturing of advanced materials. Much of the effort to date has focused on engineering the interactions between nanoscale and colloidal building blocks, with the goal of controlling their bulk phase behaviour and ultimately tailoring the morphological, mechanical, and dynamic features of the resulting materials.1–6
Very few of the biological examples of self-assembly, however, can rightfully be regarded as taking place “in the bulk”. Indeed, biological macromolecules often operate in heterogeneous environments or close to functional interfaces, which can trigger self-assembly and regulate the properties and size of the aggregates. For example, microtubule organisation centres such as centrosomes and basal bodies are known to control the self-assembly of microtubules and shape them into morphologically and functionally distinct architectures.7,8 While centrosomes sculpt the spindle apparatus, crucial for cell division, basal bodies regulate the emergence of eukaryotic cilia and flagella.7,8 Many more instances of interface-mediated self-assembly can be identified in biology, including the ubiquitous complexation of cell-membrane receptors9 and the self-assembly of viral capsids templated by their nucleic acid cargo.10
Inspired by the recent numerical study of Jana and Mognetti,11 here we propose an experimental colloidal system in which self-assembly of finite-size aggregates only occurs in the presence of a functional interface, while in its absence the particles exist in a stable colloidal gas phase. The sought outcome is obtained thanks to the open-ended programmability of DNA-mediated multivalent interactions,12 and the arsenal of design strategies that ourselves and other have developed to engineer their equilibrium and kinetic features.13–19
To demonstrate experimentlly this new mechanism, we choose silica colloidal particles with diameter of ∼1 μm as building blocks. The microspheres are coated with a Supported Lipid Bilayer (SLB), to which DNA constructs (linkers) mediating particle–particle and particle–surface interactions are anchored, as demonstrated in Fig. 1.19–21 DNA linkers feature a 36 base-pair (bp), rigid, double-stranded (ds)DNA: the “spacer”. One end of the spacer is decorated by two hydrophobic cholesterol moieties (“anchors”), which cause the linkers to irreversibly insert in the bilayers. Fluorescence-Recovery After Photobleaching (FRAP) measurements, shown in Fig. S1a,† demonstrate the fluidity of the SLBs. FRAP also confirms that the linkers themselves are capable of undergoing lateral diffusion – a key characteristic for the designed response of the system (Fig. S1b and c†).19 At the other end of the spacer, linkers feature a single-stranded (ss)DNA “sticky end”, the base sequence of which dictates linker-linker interactions.
Much larger silica spheres, with diameter of ∼10 μm, play the role of the trigger surface (“substrate”). Substrate spheres are also coated with a fluid SLB and DNA linkers (Fig. 1). Complete information of sample preparation methods and materials are provided in ESI, section S1.†
As shown in Fig. 1b, four types of linkers are present in our system, labelled A1, A2, B and C, hosting different sticky ends. Particles feature linkers A1, A2 and B, while substrates are only functionalised with C (Fig. 1a). In addition to linkers, particles are also decorated with inert constructs I, which feature a longer dsDNA spacer (68 bp) and no sticky ends, and are used to regulate steric repulsion16 (Fig. 1b).
Sticky ends are composed of multiple four-nucleotide (nt) domains, labelled α, β, γ, δ, with their complementary counterparts marked by an asterisk *. The sticky ends of linkers B have domain sequence αβγδ, while those of A1 and A2 feature β*γ* and α*β*, respectively. Consequently, A1B and A2B bonds can form both between linkers anchored to the same particle (loops) and across different particles (bridges). When an A1B (A2B) bond is formed, domain α (γ) on B remains accessible and acts as a toehold for sticky end A2 (A1).22 Consequently A1B and A2B bonds can readily swap thanks to toehold-mediated exchange, as depicted in Fig. 1c, and previously demonstrated for functionalised liposomes16 and DNA hydrogels.23 The sequences of the sticky ends, along with that of all the DNA oligonucleotides employed in this work are shown in the ESI, Table S1.†
While the formation of inter-particle bridges induce particle–particle aggregation, a prominence of intra-particle loops tends to stabilise a colloidal gas phase.24 Free-energy minimisation and combinatorial entropy demand the coexistence of inter- and intra-particle bonds.11,17,24–26 The equilibrium between the two bond-type populations can be systematically tuned by changing the total number of linkers and inert constructs per particle. Owing to combinatorial considerations, related to the number of ways in which a given type of bond (e.g. intra or inter-particle) can be formed, bridges become more favourable when the number of linkers (A1, A2, and B) is higher.19 Instead, inert constructs suppress bridge formation by increasing particle–particle steric repulsion.
At first, we seek to identify experimental conditions under which a colloidal gas phase is stable in the bulk. We will then prove that, under these conditions, the presence of the substrates can trigger the self-assembly of finite-size particle aggregates on their surface.
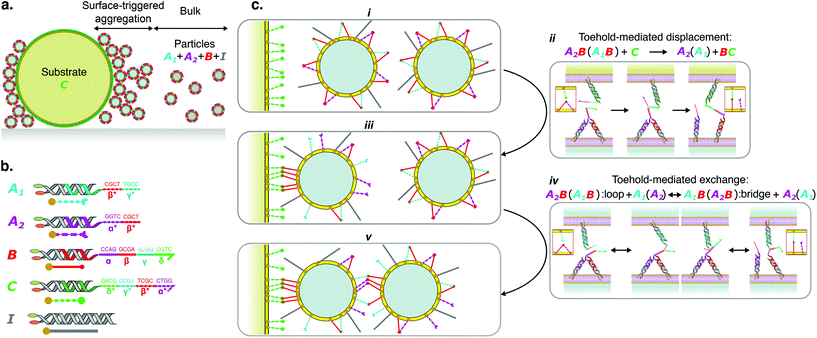
Thanks to a comprehensive theoretical framework recently summarised in ref. 19, we can predict particle–particle interaction potentials and their bulk phase behaviour, as detailed in ESI section S2,† and thus guide experiment design. We consider the experimentally-relevant scenario in which particles are functionalised with 1.6 × 105 constructs, including sticky linkers and inert constructs. The concentration ratios of the three linker species are selected such that [A1] = [A2] = 0.5 × [B]. We define the relative concentration of linkers over the total concentration of particle-tethered constructs as f = [L]/([L] + [I]), with [L] = [A1] + [A2] + [B].
The diagram in Fig. 2a maps the bulk phase behaviour of the particles as a function of f and the temperature T. A stable colloidal gas phase is predicted at room temperature (T = 25 °C) for f < 0.15 ± 0.04, while aggregates emerge at greater fractions of linkers. The errorbar in the phase boundary derives from different assumptions in particle concentration and their coordination within the aggregates (see ESI section S2†). At sufficiently low f, the particle–particle steric repulsion induced by the inert constructs suppresses the formation of inter-particle bridges. Since [B] = [A1] + [A2], in this regime and at sufficiently low temperature, all the available linkers are engaged in loops.
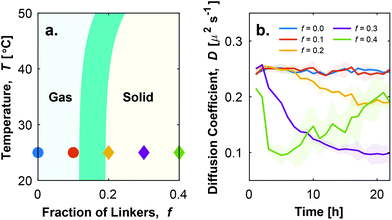 | ||
| Fig. 2 Tuning the proportion of linkers enables the stabilisation of a colloidal gas phase. a. Theoretical phase diagram showing the appearance of a stable colloidal gas phase at sufficiently low fraction of linkers f. Full details on the derivation of the phase diagram are provided in ESI, section S2.† The finite width of the gas–solid phase boundary accounts for the changes in its location following different assumptions in the concentration of the particles and their coordination in the aggregates. Symbols represent conditions tested experimentally and found to display a stable gas phase (○) or emergence of aggregates (◇), as determined from diffusivity data and visual inspection (see ESI Fig. S2†). b. Time evolution of the effective diffusion coefficient D of initially isolated particles, as determined with DDM.27,28 A drop in D indicates particle aggregation. Shadowed regions mark the errorbars of the curves calculated as discussed in the ESI, section S1.2.1.† | ||
To experimentally confirm the predicted bulk phase behaviour of the particles, we prepare samples with f = 0, 0.1, 0.2, 0.3 and 0.4, and study their aggregation (or lack thereof) via bright field light microscopy. Following a strategy developed in ref. 16, particles are initially forced in a state in which only loops can be formed by performing a rapid temperature quench (see ESI section S1.1.3†). Since no bridges are present, in this state samples exhibit a gas phase, which may be either stable or metastable depending on f and T. The samples are then observed over time at room temperature (T ≃ 25 °C). In conditions where bridge formation is thermodynamically feasible, namely at sufficiently high f, aggregation is expected to occur. Note that, as discussed above, loop breakup and bridge formation is kinetically aided by the built-in toehold-mediated-exchange capabilities of the system.16,22 To quantitatively assess the occurrence of particle–particle aggregation we carry out Differential Dynamic Microscopy (DDM) measurements, which allow us to determine the time-evolution of the (apparent) diffusion coefficient D of individual particles and possible aggregates27,28 (see ESI section S1.2.1†). Data are summarised, for different values of f, in Fig. 2b (see also ESI Fig. S2a†). Consistently with theoretical predictions in Fig. 2a, D displays a decreasing trend with time for f ≥ 0.2, following the emergence of aggregates. While for f = 0.2, D exhibits a slight decrease, indicative of small and sparse aggregates (ESI Fig. S2b iii†), the trend becomes more prominent for f = 0.3 where large scale aggregation is observed (ESI Fig. S2b iv†). Samples with f = 0.4 display a sharp drop in diffusion coefficient from the beginning of the observation, followed by a slight increase at later times. The latter trend follows from the formation of very large aggregates (ESI Fig. S2b v†); these are largely immobile and contribute little to the DDM signal, which is in turn dominated by the few smaller aggregates that remain diffusive. The complex fluctuation dynamics of branched colloidal aggregates may also play a role in the upturn of D (Fig. S2a†).29 For f = 0.1, D remains constant over a 22-hour observation window, indicating the lack of particle aggregation (ESI Fig. S2b ii†) and the stability of the colloidal gas phase.
Having identified the sought conditions under which particles do not aggregate in bulk, we introduce substrate spheres functionalised with C linkers. The sticky ends of C feature domain sequence δ*γ*β*α*, and are thus fully complementary to B sticky ends. As a result, BC bridges between a particle and a substrate are expected to be substantially more stable than either A1B or A2B bonds. We can thus expect BC bridge formation and adhesion of particles to substrates even for samples with stable bulk gas phase (f = 0.1), as sketched in Fig. 1c i–iii. Note that the breakup of A1B or A2B loops initially present on the particle in favour of BC bridges is made kinetically accessible by toehold-mediated strand displacement22 (Fig. 1c ii). Once formed, BC bridges are effectively irreversible under experimental conditions, given the total length of the complementary domains and that they do not offer any toehold to A1 or A2.
For every BC bridge that forms, either an A1 or an A2 linker initially engaged in a loop becomes free. The lateral mobility of all linkers implies that loop breakup is not limited to the small area of contact between the substrate and the particles. Instead, a large number of bridges can be formed, following the recruitment of B and C linkers in the contact area (Fig. 1c iii).15,30 At the same time, the newly unbound A1 and A2 linkers are free to spread on the surface of the particle, now adhering to the substrate (Fig. 1c iii). As a result, particles adhering to the substrates feature a significantly higher number of unbound A1 and A2 linkers compared to those in bulk. Straightforward combinatorial reasoning prescribes that such an excess of free linkers should facilitate the formation of bridges between adhering particles and those left in the bulk, triggering the deposition of a second particle, as depicted in Fig. 1c iii–v.
Particles in this “second layer” will also display an excess of free A1 and A2, following from the formation of A1B and A2B bridges with the particle initially adhering to the substrate, which could trigger the adhesion of a third particle. This amplification process can lead to the surface-triggered growth of aggregates, as per our objective.
In other words, the presence of the substrate leads to a dynamic adaptation of particle–particle interactions that propagate also to particles which are not in direct contact with the functional interface. Numerical calculations performed by Jana and Mognetti on a closely related system11 demonstrate how the number of linkers freed up following bridge formation, and thus the free energy gain of particle–particle adhesion, decreases with the number of layers, to a point where it is no longer sufficient to stabilise further adhesion of the bulk particles. Hence, the growth of aggregates is expected to be self-limiting, and the size of the aggregates dependent on system parameters such as the relative strength of BC binding, the surface density of linker C and the bulk concentration of particles.11
Experimental results on surface-triggered aggregate growth are summarised in Fig. 3. Confocal and bright-field micrographs demonstrate the presence and morphology of the aggregates, or their absence. Three-dimensional confocal stacks are further analysed to quantify, for each substrate sphere, the average number of particles in each “layer” of adhesion, as detailed in ESI section S1.2.2† and summarised by the histograms in Fig. 3 (bottom).
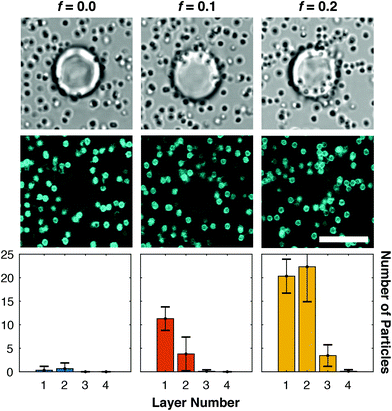 | ||
| Fig. 3 Fraction of linkers and inert constructs regulates the growth of surface-triggered aggregates. Bright-field images (top) and confocal cross-sections (middle) of a typical substrate sphere and surrounding particles for relevant values of the fraction of linkers f. Particle adhesion and substrate-driven aggregation are observed for f = 0.1, although no aggregate formation is detected in bulk for this condition (Fig. 2b). Bottom: Histograms quantifying the average numbers of particles per substrate bead for each “adhesion layer”, with the particles directly adhering to the substrate classified as belonging to layer 1. See ESI section S.1.2.2† for the definition of the layers and the particle-classification procedure. Scale bars: 10 μm. | ||
As expected, for f = 0 no aggregation on the substrate particles is observed, besides a minimal degree of adhesion due to non-specific interactions. For f = 0.1 we observe a significant degree of particle adhesion, and importantly the presence of particles in the second layer of adhesion, confirming the sought effect of surface-triggered assembly of finite-size structures. Samples with f = 0.2, which display only marginal bulk aggregation (ESI Fig. S2b iii†), form instead large particle assemblies on the substrates with a third and even a fourth adhesion layer being observed. Since these large aggregates ultimately originate from a small number of C linker molecules, which produce a larger number of bridges between particles in subsequent layers, our system offers a mechanism for amplifying and visualising molecular signals.
Similar to bulk particle aggregation (Fig. 2b and Fig. S3, ESI†), surface-triggered clustering develops over the course of a few hours, likely limited by the rate of bond-swapping16 and the diffusivity of the relatively large colloidal particles.
As a control, DDM is also performed on samples containing substrate beads. Since large beads are effectively immobile over the timescales of the DDM observations, and so are the particles adhering to them, we expect this technique to only be sensitive to diffusive particles and aggregates in the bulk. Data in Fig. S3† show trends very similar to those measured for particle-only samples (Fig. 2b), demonstrating that substrate spheres do not affect the bulk behaviour of the system in unwanted ways (e.g. by releasing linkers in solution), but only act locally regulating the growth of the aggregates.
In summary, with this communication we demonstrate the rational design and experimental implementation of a colloidal system in which self-assembly is controlled by a functional interface. The latter triggers the formation of aggregates which are otherwise unstable in the bulk phase, while also limiting their final size. The degree of control of our system derives from the thermodynamic and kinetic programmability of DNA–DNA base-pairing, and our deep understanding of multivalent DNA-mediated interactions.3,12,19
Our proof-of-concept system mimics key features of the spatially regulated assemblies observed in biology, and we argue that similar approaches could be implemented in more complex biomimicry, including engineering the collective behaviour of synthetic cellular systems.31
Furthermore, the assembly of large colloidal aggregates, as triggered by a relatively small number of nucleic acid molecules (C linkers) could be used as an amplification and detection system for nucleic-acid biomarkers, such as disease related microRNA, ciruclating DNA or viral genetic material.32–35 Such a response could be obtained by re-designing A1, A2 and B sticky ends to respond to a new target sequence different from C, then immobilising the (possibly present) target on substrate beads, and checking for the occurrence of substrate-triggered particle aggregation as an amplified readout. Immobilisation could be achieved by pre-functionalising substrate spheres with cholesterolised linkers partially complementary to the target strands. The timescales of aggregate formation could be accelerated for faster readout, e.g. by using smaller, faster diffusing particles, or by slightly increasing the length of the toehold domains to improve the rate of bond-swapping.22
Conflicts of interest
There are no conflicts to declare.Acknowledgements
RL, BMM, PC, LDM, and GB acknowledge support from the Wiener-Anspach Foundation. PKJ and BMM acknowledge support from Fonds de la Recherche Scientifique de Belgique (F.R.S.-FNRS) under grant no. MIS F.4534.17 and by an ARC (ULB) grant of the Fédération Wallonie-Bruxelles. LDM acknowledges support from a Royal Society University Research Fellowship (UF160152) and from the European Research Council (ERC) under the Horizon 2020 Research and Innovation Programme (ERC-STG No. 851667 NANOCELL).References
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2017, 3, 17068 CrossRef.
- G. A. Ozin, K. Hou, B. V. Lotsch, L. Cademartiri, D. P. Puzzo, F. Scotognella, A. Ghadimi and J. Thomson, Mater. Today, 2009, 12, 12–23 CrossRef CAS.
- W. B. Rogers, W. M. Shih and V. N. Manoharan, Nat. Rev. Mater., 2016, 1, 16008 CrossRef CAS.
- J. D. Halverson and A. V. Tkachenko, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2013, 87, 062310 CrossRef.
- Y. Wang, Y. Wang, D. R. Breed, V. N. Manoharan, L. Feng, A. D. Hollingsworth, M. Weck and D. J. Pine, Nature, 2012, 491, 51–55 CrossRef CAS.
- Y. Xiong, S. Yang, Y. Tian, A. Michelson, S. Xiang, H. Xin and O. Gang, ACS Nano, 2020, 14(6), 6823–6833 CrossRef CAS.
- P. T. Conduit, A. Wainman and J. W. Raff, Nat. Rev. Mol. Cell Biol., 2015, 16, 611–624 CrossRef CAS.
- C. G. Pearson and M. Winey, Traffic, 2009, 10, 461–471 CrossRef CAS.
- H. Wu, Cell, 2013, 153, 287–292 CrossRef CAS.
- J. D. Perlmutter and M. F. Hagan, Annu. Rev. Phys. Chem., 2015, 66, 217–239 CrossRef CAS.
- P. K. Jana and B. M. Mognetti, Nanoscale, 2019, 11, 5450–5459 RSC.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347, 1260901 CrossRef.
- S. Angioletti-Uberti, B. M. Mognetti and D. Frenkel, Nat. Mater., 2012, 11, 518–522 CrossRef CAS.
- W. B. Rogers and V. N. Manoharan, Science, 2015, 347, 639–642 CrossRef CAS.
- L. Parolini, B. M. Mognetti, J. Kotar, E. Eiser, P. Cicuta and L. Di Michele, Nat. Commun., 2015, 6, 5948 CrossRef CAS.
- L. Parolini, J. Kotar, L. Di Michele and B. M. Mognetti, ACS Nano, 2016, 10, 2392–2398 CrossRef CAS.
- J. D. Halverson and A. V. Tkachenko, J. Chem. Phys., 2016, 144, 094903 CrossRef.
- Y. Zhang, A. McMullen, L.-L. Pontani, X. He, R. Sha, N. C. Seeman, J. Brujic and P. M. Chaikin, Nat. Commun., 2017, 8, 21 CrossRef.
- B. M. Mognetti, P. Cicuta and L. Di Michele, Rep. Prog. Phys., 2019, 82, 116601 CrossRef CAS.
- A.-L. Troutier and C. Ladavière, Adv. Colloid Interface Sci., 2007, 133, 1–21 CrossRef CAS.
- S. A. J. van der Meulen and M. E. Leunissen, J. Am. Chem. Soc., 2013, 135, 15129–15134 CrossRef CAS.
- D. Y. Zhang and E. Winfree, J. Am. Chem. Soc., 2009, 131, 17303–17314 CrossRef CAS.
- F. Bomboi, D. Caprara, J. Fernandez-Castanon and F. Sciortino, Nanoscale, 2019, 9691–9697 RSC.
- S. J. Bachmann, J. Kotar, L. Parolini, A. Šarić, P. Cicuta, L. Di Michele and B. M. Mognetti, Soft Matter, 2016, 12, 7804–7817 RSC.
- L. Di Michele, S. J. Bachmann, L. Parolini and B. M. Mognetti, J. Chem. Phys., 2016, 144, 161104 CrossRef.
- F. Sciortino, Y. Zhang, O. Gang and S. K. Kumar, ACS Nano, 2020, 5, 5628–5635 CrossRef.
- R. Cerbino and V. Trappe, Phys. Rev. Lett., 2008, 100, 188102 CrossRef.
- R. Cerbino and P. Cicuta, J. Chem. Phys., 2017, 147, 110901 CrossRef.
- J. H. Cho, R. Cerbino and I. Bischofberger, Phys. Rev. Lett., 2020, 124, 088005 CrossRef CAS.
- R. Lanfranco, P. K. Jana, L. Tunesi, P. Cicuta, B. M. Mognetti, L. Di Michele and G. Bruylants, Langmuir, 2019, 35, 2002–2012 CrossRef CAS.
- B. C. Buddingh’ and J. C. M. van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef.
- M. van der Vaart and P. J. Pretorius, Clin. Chem., 2007, 53, 2215–2215 CrossRef CAS.
- W. Li, D. Raoult and P.-E. Fournier, FEMS Microbiol. Rev., 2009, 33, 892–916 CrossRef CAS.
- I. M. Mackay, K. E. Arden and A. Nitsche, Nucleic Acids Res., 2002, 30, 1292–1305 CrossRef CAS.
- J. O'Brien, H. Hayder, Y. Zayed and C. Peng, Front. Endocrinol., 2018, 9, 402 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods, theoretical methods, DNA sequences, supplementary figures. See DOI: 10.1039/D0NR04461J |
| This journal is © The Royal Society of Chemistry 2020 |