Open Access Article
Open Access ArticleNitric oxide-dependent biodegradation of graphene oxide reduces inflammation in the gastrointestinal tract†
Guotao
Peng
 a,
Marcelo F.
Montenegro
a,
Marcelo F.
Montenegro
 b,
Chifundo N. M.
Ntola
c,
Sandra
Vranic
c,
Kostas
Kostarelos
b,
Chifundo N. M.
Ntola
c,
Sandra
Vranic
c,
Kostas
Kostarelos
 cd,
Carmen
Vogt
e,
Muhammet S.
Toprak
cd,
Carmen
Vogt
e,
Muhammet S.
Toprak
 e,
Tianbo
Duan
f,
Klaus
Leifer
e,
Tianbo
Duan
f,
Klaus
Leifer
 f,
Lars
Bräutigam
g,
Jon O.
Lundberg
b and
Bengt
Fadeel
f,
Lars
Bräutigam
g,
Jon O.
Lundberg
b and
Bengt
Fadeel
 *a
*a
aInstitute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden. E-mail: bengt.fadeel@ki.se
bDepartment of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden
cNational Graphene Institute, and Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK
dCatalan Institute of Nanoscience and Nanotechnology (ICN2), Barcelona, Spain
eDepartment of Applied Physics, KTH Royal Institute of Technology, Stockholm, Sweden
fDepartment of Engineering Sciences, Uppsala University, Uppsala, Sweden
gComparative Medicine, Karolinska Institutet, Stockholm, Sweden
First published on 10th August 2020
Abstract
Understanding the biological fate of graphene-based materials such as graphene oxide (GO) is crucial to assess adverse effects following intentional or inadvertent exposure. Here we provide first evidence of biodegradation of GO in the gastrointestinal tract using zebrafish as a model. Raman mapping was deployed to assess biodegradation. The degradation was blocked upon knockdown of nos2a encoding the inducible nitric oxide synthase (iNOS) or by pharmacological inhibition of NOS using L-NAME, demonstrating that the process was nitric oxide (NO)-dependent. NO-dependent degradation of GO was further confirmed in vitro by combining a superoxide-generating system, xanthine/xanthine oxidase (X/XO), with an NO donor (PAPA NONOate), or by simultaneously producing superoxide and NO by decomposition of SIN-1. Finally, by using the transgenic strain Tg(mpx:eGFP) to visualize the movement of neutrophils, we could show that inhibition of the degradation of GO resulted in increased neutrophil infiltration into the gastrointestinal tract, indicative of inflammation.
Graphene and its derivatives have been intensively explored for a multitude of biomedical and other applications. This, therefore, mandates a comprehensive assessment of the potential impact of these materials on human health.1 In recent years, attention has been focused on the biological fate of graphene-based materials, including their potential biodegradability. Thus, enzymatic degradation of graphene oxide (GO) by plant and animal peroxidases, including myeloperoxidase (MPO), has been documented.2,3 Purified human MPO was shown to degrade highly dispersed GO, but failed to metabolize aggregated GO.3 Furthermore, we demonstrated in a previous study that primary human neutrophils are capable of degrading GO when activated to undergo degranulation with release of MPO,4 while GO functionalized with a chemotactic peptide triggered neutrophil activation with subsequent degradation of GO.5 Single- and few-layer graphene is also susceptible to enzymatic degradation though the process was found to be less efficient as compared to GO.6
Previous studies on single- and multi-walled carbon nanotubes (CNTs) have revealed the propensity of these materials to undergo peroxidase-mediated biodegradation.7–10 Furthermore, CNTs may also undergo peroxynitrite (ONOO−) dependent biodegradation in macrophages11,12 and chondrocytes.13 Peroxynitrite can be generated by the rapid reaction of superoxide radicals with nitric oxide (NO) according to the following reaction:
| O2˙− + NO˙ → ONOO− |
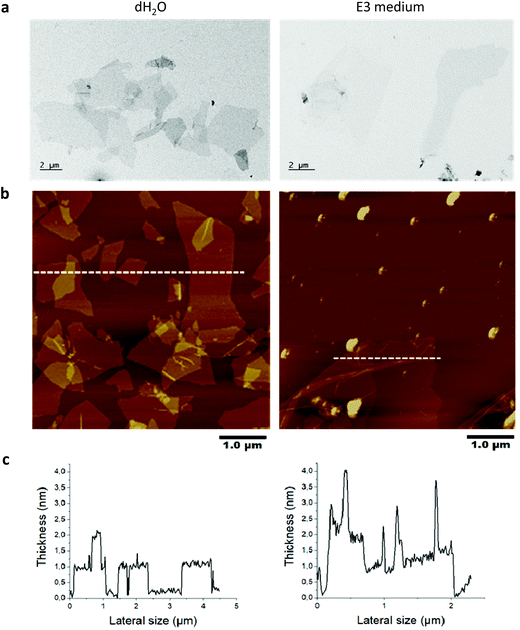
The peroxynitrite anion produced in this manner displays high diffusibility,14 obviating the need for a direct interaction of CNTs with the participating enzymes.11 However, no study has been conducted to investigate the possibility of NO-driven biodegradation of GO. Here we studied GO produced by the Hummers’ method to investigate the in vitro and in vivo degradability. The physicochemical characterization results are summarized in Table S1.† Based on the evaluation of GO by TEM, SEM, optical microscopy and AFM, we found that 90% of the sheets ranged between 0.1 to 15 μm in lateral dimensions (representative TEM images are shown in Fig. 1a). AFM analysis revealed that the thickness of the GO sheets was between 1 to 2 nm, allowing us to conclude that GO was mainly present as single- or few-layer sheets in water and after incubation for 24 h in E3 medium (Fig. 1b and c). Note that the spikes in thickness following incubation in E3 medium are due to wrinkling and folding of GO sheets as a result of incubation in a high salt containing medium.
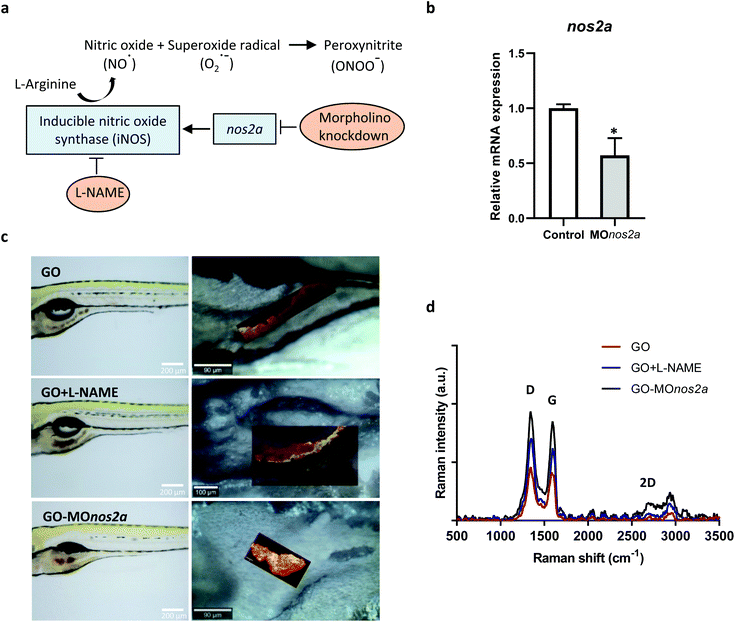
Inhalation is the main route of unintentional exposure to engineered nanomaterials, and the lungs therefore represent the most immediate target organ for their toxic effects. However, extrapulmonary effects of inhaled nanomaterials including effects on the gastrointestinal (GI) tract have been reported; additionally, nanomaterials can be ingested leading to a direct exposure of the GI tract.15 Despite the potential implications on human health, the impact of nanomaterials on the GI tract has been largely overlooked.16 We focused here on oral exposure to GO. It is well known that NO is produced in the GI tract through enzymatic and non-enzymatic reactions,17 and we hypothesized that GO may undergo NO-dependent biodegradation (Fig. 2a). To address this question, we used zebrafish embryos that have emerged in recent years as a robust in vivo model in toxicology and drug discovery.18 The degradation of GO in zebrafish larvae (5 days postfertilization, dpf) was evaluated using confocal Raman mapping. To investigate the mechanism of biodegradation of GO in the GI tract, we employed two approaches: (i) downregulation of the inducible NO synthase (iNOS) by using morpholino oligonucleotides directed against nos2a; and (ii) inhibition of NOS by using the cell-permeable inhibitor, N-omega-nitro-L-arginine methyl ester hydrochloride (L-NAME). The successful downregulation of nos2a was documented by RT-qPCR (Fig. 2b). As demonstrated in Fig. 2c, GO accumulated in the GI tract and could be detected on the basis of its characteristic Raman signature, i.e., the D band (1354 cm−1), G band (1582 cm−1), and 2D band (2690 cm−1).19 The background signal of the fish tissues did not interfere with the detection of GO by Raman (data not shown). Importantly, our interventions (shown schematically in Fig. 2a) resulted in a pronounced change in the intensity of the D and G band when compared to untreated, GO-exposed fish, indicating that GO was biodegraded in the latter case through an NO-dependent mechanism (Fig. 2d). It is worth noting that a previous study showed no degradation of GO when immersed in simulated digestive juices representative of the GI tract, as evaluated by Raman analysis.20 This allows us to rule out a role of gastrointestinal fluids in the degradation of GO.
To support the role of NO in the biodegradation process, we measured NO production in vivo by using the fluorescent probe, DAF-FM-DA. Bacterial lipopolysaccharide (LPS) was used to trigger NO production (Fig. 3a). NO production was suppressed by L-NAME (Fig. 3b). Exposure to GO also led to an increase in NO (Fig. 3b). Importantly, knockdown of nos2a with morpholino oligos resulted in a decline in LPS- and GO-induced NO production (Fig. 3b; representative images of zebrafish embryos are shown in panel a). We also detected peroxynitrite generation in vivo by using DAX-J2 PON Green, a fluorescent probe with high selectivity for peroxynitrite and negligible reactivity with other relevant species including NO. Peroxynitrite production was observed in vivo in response to GO and to a lesser extent with LPS and the signal was suppressed by L-NAME (Fig. S1†).
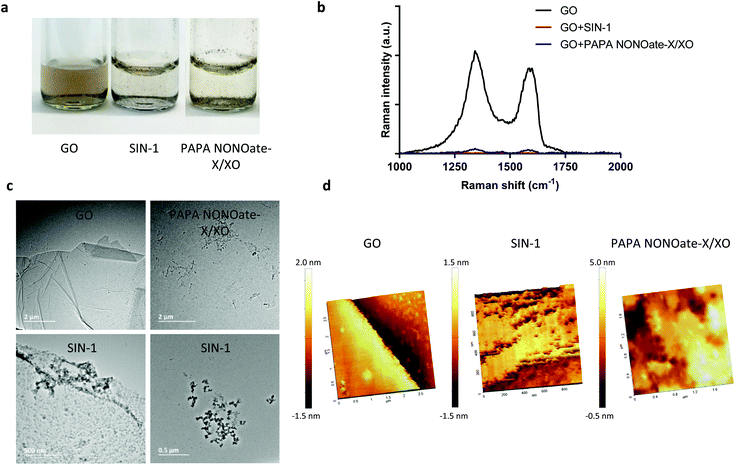
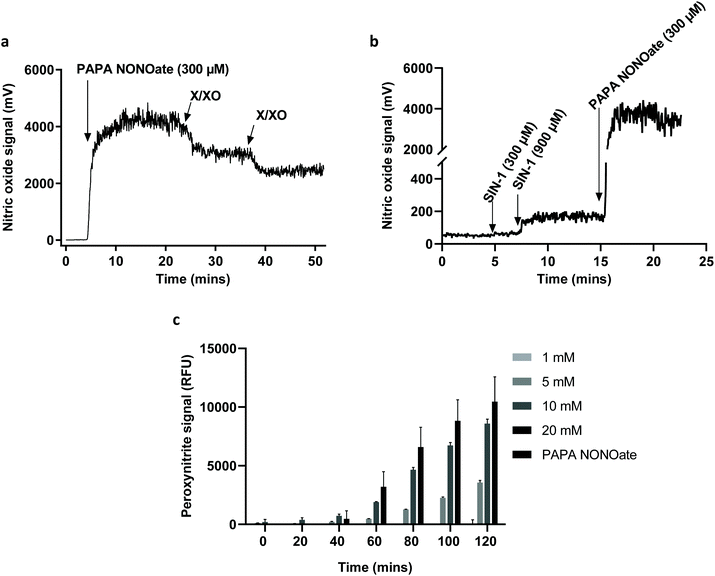
To further confirm the degradation of GO and provide more insight regarding the NO-dependent reaction, we performed in vitro experiments by using two peroxynitrite-generating systems, i.e., simultaneous production of superoxide radicals and NO by decomposition of 3-morpholinosydnonimine hydrochloride (SIN-1), or the combination of a superoxide-generating system, xanthine/xanthine oxidase (X/XO), with an NO donor (PAPA NONOate).11 As shown in Fig. 4a, GO degradation after 5 days of continuous incubation in the presence of SIN-1 or PAPA NONOate-X/XO was visually detectable as the suspensions turned translucent in both cases. To more objectively document the degradation, we performed Raman spectroscopy. The characteristic D and G bands of GO were remarkably diminished in both systems suggesting a near-complete loss of structure of GO (Fig. 4b). The topography and structure of GO incubated with the two peroxynitrite-generating systems was further characterized by TEM (Fig. 4c) and AFM (Fig. 4d). Specifically, after PAPA NONOate-X/XO treatment, the typical morphology of the GO sheets disappeared, and only small fragments could be observed. In addition, SIN-1 treatment led to numerous perforations of the GO sheets, as shown by TEM, and these defects were also seen by AFM. To confirm that the two systems were functional, we monitored NO and peroxynitrite production based on real-time chemiluminescence and fluorescence detection, respectively. PAPA NONOate induced a robust and stable NO signal, and the addition of X/XO was able to decrease the NO signal, likely due to the rapid reaction between NO and the superoxide radicals (Fig. 5a). Additionally, continuous treatment of GO revealed that PAPA NONOate alone was able to decrease the optical density of the GO suspensions, indicative of GO degradation, while X/XO alone did not exert observable effects (Fig. S2a†). Raman analysis confirmed these results (Fig. S2b†). On the other hand, the peroxynitrite donor SIN-1 did not generate a NO signal (at 300 μM), though there was a slight increase when a higher dose (900 μM) was applied (Fig. 5b). Instead, the fluorescence intensity of DAX-J2 PON Green increased in a time- and dose-dependent manner upon reacting with SIN-1, clearly showing that peroxynitrite was formed, while PAPA NONOate (an NO donor) generated a negligible signal (Fig. 5c). Collectively, these results suggest that the degradation of GO by PAPA NONOate-X/XO may be attributed to NO, but not to superoxide radicals, and that the decomposition of SIN-1 produces superoxide and NO resulting in rapid formation of ONOO−, which ‘digests’ GO. One may ask what the potential advantages or disadvantages would be of NO-driven degradation of GO. One important aspect is that NO-driven degradation does not require direct interaction with the participating enzymes which means that cellular uptake of the nanomaterial is not required. Instead, NO released from gastrointestinal cells may diffuse to the gut lumen and ‘attack’ GO, leading to its degradation. On the other hand, a possible limitation is that the kinetics of NO-driven degradation are evidently slower than those of MPO.4
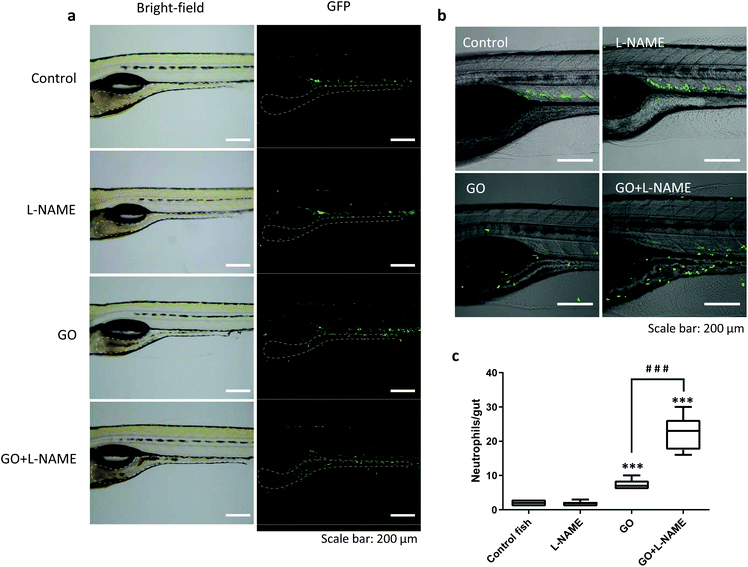
Only a few attempts have been made to assess the biological consequences of nanomaterial degradation or the lack thereof. Kagan et al.21 found that ex vivo degraded CNTs induced less pulmonary inflammation when aspirated into the lungs of mice whereas non-degraded CNTs triggered granuloma formation. In a subsequent study, inflammatory and fibrotic responses to CNTs were aggravated in MPO-deficient mice when compared to wild-type mice.22 We recently reported that MPO-degraded GO was non-cytotoxic and did not exert DNA damage in cultured human lung cells,4 but in vivo responses were not evaluated in the latter study. Here we asked whether biodegradation would alter inflammatory responses induced by GO in the GI tract. To address this, we employed transgenic zebrafish, Tg(mpx:eGFP), expressing fluorescent GFP under a neutrophil-specific promoter to visualize neutrophil infiltration,23 an acknowledged marker of inflammation. As illustrated in Fig. 6a, the GFP-positive neutrophils were mostly located in the blood vessels in control fish. Following GO exposure, neutrophils were found to infiltrate the GI tract (Fig. 6a). Interestingly, in zebrafish exposed to GO in the presence of L-NAME, significantly more neutrophils migrated towards the GI tract as compared to fish exposed to GO alone (Fig. 6b and c). However, L-NAME alone had no impact on neutrophil migration (Fig. 6c). These results suggest that NO-dependent degradation of GO which is suppressed by L-NAME serves to reduce the inflammatory response triggered by ingested GO. We found that the recruited neutrophils were mainly located in the lamina propria (beneath the epithelium) while very few GFP-positive cells infiltrated into the gut lumen (Fig. S3†). Therefore, direct interactions between neutrophils and GO seem unlikely.
The fact that the major organ systems are conserved in zebrafish18 provides an opportunity to decipher potential health effects of nanomaterials in humans. Specifically, zebrafish may serve as an alternative to mammalian models for the study of inflammation, as the cellular and humoral machinery that regulates immune responses is conserved. In the early stages of development, zebrafish rely on the innate immune system to protect against infections or injuries. The present study shows that the presence of GO is ‘sensed’ in the gut leading zebrafish larvae to mount a response to the offending agent. However, the implications of the present findings for other species including humans can only be speculated upon at this stage. Indeed, the overall role of NO in inflammation in mammals remains unresolved as both pro- and anti-inflammatory effects of NO and related nitrogen species have been reported.24,25 Anti-inflammatory effects of NO in mammals involve, for instance, inhibition of leucocyte activation and recruitment by endothelial cell NO.26 It should be noted that we used both pharmacological and genetic approaches to inhibit or silence iNOS in the zebrafish model. This, coupled with our in vitro results, has revealed a role for NO in the degradation of GO. However, we have not explored the different cell populations that may potentially produce NO, though iNOS is known to be expressed in many cell types in humans. We presumed that NO is generated by gastrointestinal cells in the present model and that NO (produced by iNOS) together with superoxide radicals (produced by NADPH oxidase) combined to produce the highly reactive peroxynitrite anion (ONOO−).
Conclusions
In conclusion, we have shown that GO can be degraded in vitro and in vivo in an NO-dependent manner. This study is the first to demonstrate biodegradation of GO in zebrafish. Furthermore, while many deleterious reactions have been attributed to peroxynitrite,27 the present results suggest that this potent oxidant may also contribute to the innate defense against foreign intrusion insofar as it is shown here to participate in the degradation of GO. We also found that the degradation of GO reduces the inflammation (evidenced as neutrophil infiltration) triggered upon ingestion of GO. These findings shed new light on the biological fate of GO and further support the view that GO is a biodegradable material.28Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Jens Sommertune (RISE Research Institute of Sweden, Stockholm) for assistance with the confocal Raman analysis, and Dr Neus Lozano (ICN2, Barcelona) for assistance with the analysis of the material characterization data. We also thank Dr Ahmed Sultan (Karolinska Institutet, Stockholm) for generously providing the neutrophil reporter line, and the KI zebrafish core facility for assistance with fish husbandry. This work was supported by the European Commission through the Graphene Flagship (grant no. 785219).References
- B. Fadeel, C. Bussy, S. Merino, E. Vázquez, E. Flahaut, F. Mouchet, L. Evariste, L. Gauthier, A. J. Koivisto, U. Vogel, C. Martín, L. G. Delogu, T. Buerki-Thurnherr, P. Wick, D. Beloin-Saint-Pierre, R. Hischier, M. Pelin, F. Candotto Carniel, M. Tretiach, F. Cesca, F. Benfenati, D. Scaini, L. Ballerini, K. Kostarelos, M. Prato and A. Bianco, ACS Nano, 2018, 12, 10582–10620 CrossRef CAS.
- G. P. Kotchey, B. L. Allen, H. Vedala, N. Yanamala, A. A. Kapralov, Y. Y. Tyurina, J. Klein-Seetharaman, V. E. Kagan and A. Star, ACS Nano, 2011, 5, 2098–2108 CrossRef CAS PubMed.
- R. Kurapati, J. Russier, M. A. Squillaci, E. Treossi, C. Ménard-Moyon, A. E. Del Rio-Castillo, E. Vazquez, P. Samorì, V. Palermo and A. Bianco, Small, 2015, 11, 3985–3994 CrossRef CAS PubMed.
- S. P. Mukherjee, A. R. Gliga, B. Lazzaretto, B. Brandner, M. Fielden, C. Vogt, L. Newman, A. F. Rodrigues, W. Shao, P. M. Fournier, M. S. Toprak, A. Star, K. Kostarelos, K. Bhattacharya and B. Fadeel, Nanoscale, 2018, 10, 1180–1188 RSC.
- C. Martín, A. Ruiz, S. Keshavan, G. Reina, D. Murera, Y. Nishina, B. Fadeel and A. Bianco, Adv. Funct. Mater., 2019, 29, 1–11 Search PubMed.
- R. Kurapati, S. P. Mukherjee, C. Martín, G. Bepete, E. Vázquez, A. Pénicaud, B. Fadeel and A. Bianco, Angew. Chem., Int. Ed., 2018, 11722–11727 CrossRef CAS PubMed.
- B. L. Allen, P. D. Kichambare, P. Gou, I. I. Vlasova, A. A. Kapralov, N. Konduru, V. E. Kagan and A. Star, Nano Lett., 2008, 8, 3899–3903 CrossRef CAS.
- J. Russier, C. Ménard-Moyon, E. Venturelli, E. Gravel, G. Marcolongo, M. Meneghetti, E. Doris and A. Bianco, Nanoscale, 2011, 3, 893–896 RSC.
- K. Bhattacharya, R. El-Sayed, F. T. Andón, S. P. Mukherjee, J. Gregory, H. Li, Y. Zhao, W. Seo, A. Fornara, B. Brandner, M. S. Toprak, K. Leifer, A. Star and B. Fadeel, Carbon, 2015, 91, 506–517 CrossRef CAS.
- F. T. Andôn, A. A. Kapralov, N. Yanamala, W. Feng, A. Baygan, B. J. Chambers, K. Hultenby, F. Ye, M. S. Toprak, B. D. Brandner, A. Fornara, J. Klein-Seetharaman, G. P. Kotchey, A. Star, A. A. Shvedova, B. Fadeel and V. E. Kagan, Small, 2013, 9, 2721–2729 CrossRef PubMed.
- V. E. Kagan, A. A. Kapralov, C. M. St. Croix, S. C. Watkins, E. R. Kisin, G. P. Kotchey, K. Balasubramanian, I. I. Vlasova, J. Yu, K. Kim, W. Seo, R. K. Mallampalli, A. Star and A. A. Shvedova, ACS Nano, 2014, 8, 5610–5621 CrossRef CAS PubMed.
- D. Elgrabli, W. Dachraoui, C. Ménard-Moyon, X. J. Liu, D. Bégin, S. Bégin-Colin, A. Bianco, F. Gazeau and D. Alloyeau, ACS Nano, 2015, 9, 10113–10124 CrossRef CAS PubMed.
- K. Bhattacharya, C. Sacchetti, P. M. Costa, J. Sommertune, B. D. Brandner, A. Magrini, N. Rosato, N. Bottini, M. Bottini and B. Fadeel, Adv. Healthcare Mater., 2018, 7, 1–11 Search PubMed.
- C. Szabó, H. Ischiropoulos and R. Radi, Nat. Rev. Drug Discovery, 2007, 6, 662–680 CrossRef PubMed.
- I. S. Sohal, K. S. O'Fallon, P. Gaines, P. Demokritou and D. Bello, Part. Fibre Toxicol., 2018, 15, 29 CrossRef PubMed.
- A. Pietroiusti, E. Bergamaschi, M. Campagna, L. Campagnolo, G. De Palma, S. Iavicoli, V. Leso, A. Magrini, M. Miragoli, P. Pedata, L. Palombi and I. Iavicoli, Part. Fibre Toxicol., 2017, 14, 47 CrossRef PubMed.
- J. O. Lundberg, E. Weitzberg and M. T. Gladwin, Nat. Rev. Drug Discovery, 2008, 7, 710 CrossRef CAS.
- L. I. Zon and R. T. Peterson, Nat. Rev. Drug Discovery, 2005, 4, 35–44 CrossRef CAS PubMed.
- C. M. Girish, A. Sasidharan, G. S. Gowd, S. Nair and M. Koyakutty, Adv. Healthcare Mater., 2013, 2, 1489–1500 CrossRef CAS PubMed.
- D. Guarnieri, P. Sánchez-Moreno, A. E. Del Rio Castillo, F. Bonaccorso, F. Gatto, G. Bardi, C. Martín, E. Vázquez, T. Catelani, S. Sabella and P. P. Pompa, Small, 2018, 14, 1800227 CrossRef.
- V. E. Kagan, N. V. Konduru, W. Feng, B. L. Allen, J. Conroy, Y. Volkov, I. I. Vlasova, N. A. Belikova, N. Yanamala, A. Kapralov, Y. Y. Tyurina, J. Shi, E. R. Kisin, A. R. Murray, J. Franks, D. Stolz, P. Gou, J. Klein-Seetharaman, B. Fadeel, A. Star and A. A. Shvedova, Nat. Nanotechnol., 2010, 5, 354–359 CrossRef CAS PubMed.
- A. A. Shvedova, A. A. Kapralov, W. H. Feng, E. R. Kisin, A. R. Murray, R. R. Mercer, C. M. St. Croix, M. A. Lang, S. C. Watkins, N. V. Konduru, B. L. Allen, J. Conroy, G. P. Kotchey, B. M. Mohamed, A. D. Meade, Y. Volkov, A. Star, B. Fadeel and V. E. Kagan, PLoS One, 2012, 7, e30923 CrossRef CAS PubMed.
- S. A. Renshaw, C. A. Loynes, D. M. I. Trushell, S. Elworthy, P. W. Ingham and M. K. B. Whyte, Blood, 2006, 108, 3976–3978 CrossRef CAS.
- J. O. Lundberg, J. M. Lundberg, K. Alving and E. Weitzberg, Nat. Med., 1997, 3, 30–31 CrossRef CAS PubMed.
- P. Kubes, Gut, 2000, 47, 6–9 CrossRef CAS PubMed.
- P. Kubes, M. Suzuki and D. N. Granger, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 4651–4655 CrossRef CAS PubMed.
- S. Pfeiffer, B. Mayer and B. Hemmens, Angew. Chem., Int. Ed., 1999, 38, 1714–1731 CrossRef.
- K. Bhattacharya, S. P. Mukherjee, A. Gallud, S. C. Burkert, S. Bistarelli, S. Bellucci, M. Bottini, A. Star and B. Fadeel, Nanomedicine, 2016, 12, 333–351 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr03675g |
| This journal is © The Royal Society of Chemistry 2020 |