Open Access Article
Open Access ArticleLocal release of siRNA using polyplex-loaded thermosensitive hydrogels†
Lies A. L.
Fliervoet
a,
Heyang
Zhang
b,
Emma
van Groesen
c,
Kyra
Fortuin
a,
Naut J. C. B.
Duin
a,
Katrien
Remaut
b,
Raymond M.
Schiffelers
 d,
Wim E.
Hennink
d,
Wim E.
Hennink
 a and
Tina
Vermonden
a and
Tina
Vermonden
 *a
*a
aDepartment of Pharmaceutics, Utrecht Institute for Pharmaceutical Sciences (UIPS), Faculty of Science, Utrecht University, PO Box 80082, 3508 TB Utrecht, the Netherlands. E-mail: T.Vermonden@uu.nl; Tel: (+31)620291631
bLaboratory of General Biochemistry and Physical Pharmacy, Faculty of Pharmaceutical Sciences, Ghent University, Ghent 9000, Belgium
cBiological Chemistry Group, Institute of Biology Leiden, Leiden University, Sylvius Laboratories, 2333 BE Leiden, the Netherlands
dDepartment of Clinical Chemistry and Haematology, University Medical Center Utrecht, Heidelberglaan 100, 3508 GA Utrecht, the Netherlands
First published on 28th April 2020
Abstract
One of the challenges for the clinical translation of RNA interference (RNAi)-based therapies concerns the deposition of therapeutically effective doses of the nucleic acids, like siRNA, at a local tissue level without severe off-target effects. To address this issue, hydrogels can be used as matrices for the local and sustained release of the siRNA cargo. In this study, the formation of polyplexes based on siRNA and poly(2-dimethylaminoethyl methacrylate) (PDMAEMA)-based polymers was investigated, followed by their loading in a thermosensitive hydrogel to promote local siRNA release. A multifunctional NPD triblock copolymer consisting of a thermosensitive poly(N-isopropylacrylamide) (PNIPAM, N), a hydrophilic poly(ethylene glycol) (PEG, P), and a cationic PDMAEMA (D) block was used to study the binding properties with siRNA taking the non-thermosensitive PD polymer as control. For both polymers, small polyplexes with sizes ranging from 10–20 nm were formed in aqueous solution (HBS buffer, 20 mM HEPES, 150 mM NaCl, pH 7.4) when prepared at a N/P charge ratio of 5 or higher. Formulating the siRNA into NPD or PD polyplexes before loading into the thermosensitive PNIPAM–PEG–PNIPAM hydrogel resulted in a more controlled and sustained release compared to free siRNA release from the hydrogel. The polyplexes were released for 128 hours in HBS, when changing the release medium twice a day, while free siRNA was completely released within 50 hours with already 40% being released after changing the release medium just once. The release of the polyplexes was dependent on the dissolution rate of the hydrogel matrix. Moreover, intact polyplexes were released from the hydrogels with a similar size as before loading, suggesting that the hydrogel material did not compromise the polyplex stability. Finally, it was shown that the released polyplexes were still biologically active and transfected FaDu cells, which was observed by siRNA-induced luciferase silencing in vitro. This study shows the development of an injectable thermosensitive hydrogel to promote local and sustained release of siRNA, which can potentially be used to deliver siRNA for various applications, such as the treatment of tumors.
1. Introduction
Since the discovery of RNA interference (RNAi) by Fire and Mello in 1998,1 many researchers have focused on using this for therapeutic purposes. RNAi is a biological process in which RNA molecules, like microRNA (miRNA) and small interfering RNA (siRNA), inhibit gene expression by binding to complementary messenger RNA (mRNA) resulting in silencing of the protein expression. The process is exploited to knock down specific genes creating many opportunities for treating diseases. Despite the high potential of these nucleic acid therapeutics, translation has been limited and only recently the first siRNA therapy was approved by the FDA.2,3 Efficient delivery of siRNA molecules in the target cells remains the major challenge for clinical translation. Nucleic acids are highly negatively charged and large hydrophilic molecules, making them unable to pass cellular membranes by passive diffusion. Moreover, siRNAs have a half-life of only a few minutes after intravenous administration due to their susceptibility to degradation by nucleases in the bloodstream and rapid renal clearance.4 Other challenges include induction of immune responses and off-target effects.5 To facilitate the intracellular delivery of siRNA, non-viral polymer-based systems have been widely studied both in vitro and in vivo.6–8 Cationic polymers form complexes (i.e. polyplexes) with the negatively charged siRNA molecules exploiting electrostatic interactions, and these polyplexes protect siRNA from degradation and promote cellular uptake. A great variety of cationic polymers have been developed as nucleic acid carriers, including polyethylenimine (PEI), poly-L-lysine (PLL), polyamidoamine (PAMAM), and poly(2-dimethylaminoethyl methacrylate) (PDMAEMA).9–11 The advantages of these polymeric deliver systems are their tailorable design, the relatively ease to scale-up the production, and they are safer as compared to viral vectors. However, one of the challenges that remains is the accumulation of therapeutically effective doses of siRNA at a local tissue level with acceptable off-target effects.12,13 To address these limitations, hydrogels can be used to facilitate local and sustained release of siRNA. Hydrogels are water-swollen cross-linked networks of hydrophilic polymers, which have been used for the delivery of various biomolecules, including siRNA.14–17 The nucleic acids are entrapped within the hydrogel either as siRNA conjugates or loaded in nanoparticles, and the release kinetics can be tuned by varying hydrogel properties such as polymer concentration and cross-link density.17,18 In this way, hydrogels assist in local retention and sustained release of siRNA to enhance in vivo efficacy and at the same time limit off-target toxicity.18While many hydrogels have been investigated for local release of therapeutics, injectable formulations are preferred over preformed hydrogels, since in situ gelation can take place under physiological conditions upon injection without the need of an invasive surgical intervention.19–21 Especially the use of stimuli-responsive polymers gained a lot of interest, as they can respond to external environmental triggers, like temperature, pH or light exposure.17 Temperature-sensitive hydrogels are one of the most studied class of stimuli-responsive polymer systems, and the self-assembly of these polymers occurs at a critical temperature, the so-called cloud point (CP) in aqueous solution. Because poly(N-isopropylacrylamide) (PNIPAM)-based polymers have a CP around 32 °C, they are very attractive materials for biomedical and pharmaceutical applications.22,23
Previously, a multifunctional triblock copolymer consisting of a thermosensitive PNIPAM, an hydrophilic poly(ethylene glycol) (PEG) and a cationic PDMAEMA block was investigated for the release of plasmid DNA (pDNA). The presented results showed that multiple functionalities can be introduced in the polymer design without compromising the interactions with pDNA and hence the polyplex structure.24 The aim of the present study is to investigate the applicability of the thermosensitive polyplexes based on the NPD (N = PNIPAM, P = PEG, D = PDMAEMA) triblock copolymer for siRNA delivery. Furthermore, the loading of these polyplexes into a thermosensitive PNIPAM–PEG–PNIPAM hydrogel was evaluated to facilitate local and sustained release of siRNA. First, the physical–chemical properties of polyplexes formed between siRNA and the multifunctional NPD polymer were evaluated taking a non-thermosensitive polymer as control. Second, the release profile of polyplexes from the thermosensitive PNIPAM–PEG–PNIPAM hydrogel was evaluated as well as the ability of the released polyplexes to deliver siRNA in vitro into cells.
2. Materials and methods
2.1. Materials
All materials were obtained from Sigma-Aldrich (Zwijndrecht, the Netherlands) and used as received unless noted otherwise. Acetonitrile (ACN) and N,N-dimethylformamide (DMF) were purchased from Biosolve (Valkenswaard, the Netherlands) and DMF was dried by molecular sieves for 24 hours before use. To remove the inhibitor from 2-(dimethylamino)ethyl methacrylate (DMAEMA), the monomer was passed over a column of alumina prior to use. Slide-A-lyzer™ Dialysis cassettes (Molecular weight cut-off, MWCO: 10 kDa) were obtained from Thermo Fisher Scientific (Bleiswijk, the Netherlands). Agarose multi-purpose was purchased from Roche Molecular Biochemicals (Mannheim, Germany) and Midori Green DNA gel stain was obtained from Nippon Genetics (Düren, Germany). Human epithelial hypopharyngeal carcinoma cells stably expressing luciferase (FaDu-luc) were obtained from Erasmus Medical Center (Rotterdam, the Netherlands).25 Linear polyethylenimine (l-PEI, Mw 25 kDa) was obtained from Polysciences (Hirschberg an der Bergstraße, Germany). Lipofectamine 3000, alamarBlue™ Cell Viability Reagent and Quant-iT™ RiboGreen™ RNA Assay Kit were purchased from Thermo Fisher Scientific and Luciferase assay kit was obtained from Promega (Leiden, the Netherlands).The siRNA specifically targeting firefly luciferase and the negative control siRNA were provided by GlaxoSmithKline (Stevenage, UK) and their sequences are shown in Table 1. siRNA labeled with Alexa 647 was used for fluorescence correlation spectroscopy (FCS) experiments and this Alexa 647 labeled siRNA has the negative control sequence with the label present at the 3′ end of the sense strand.
| siRNA | Sequence |
|---|---|
| Underlined bases indicate a 2′O-methyl modification. dT indicates a deoxyribonucleic acid base having phosphorothioate (PS) bond. | |
| Luciferase siRNA (siRNA-luc) | Sense 5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) -A -A![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) G- G-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) G-AG G-AG![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) -A -A![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) - -![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) G-AdTdT-3′ G-AdTdT-3′ |
| Antisense 5′-UCG-AAG-UAC-UCA-GCG-UAA-GdTdT-3′ | |
| Negative control siRNA (siRNA-nc) | Sense 5′-AU![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) -G -G![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) A- A-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) G G![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) -A -A![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) C-G C-G![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) C-G C-G![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) A- A-![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) dTdT-3′ dTdT-3′ |
Antisense 5′-A![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) A- A-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GA- GA-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GG- GG-![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) A A![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) -G -G![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) A- A-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GA- GA-![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) dTdT-3′ dTdT-3′ |
|
2.2. Synthesis and characterization of polymers
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture was added. The reaction mixture was flushed with nitrogen and placed in an ice bath while stirring until all compounds were completely dissolved. Subsequently, 2 equiv. of Me6TREN was added and the polymerization reaction was carried out for one hour on ice. The polymer solution was dialyzed (MWCO: 10 kDa) against water for 48 hours at 4 °C, while changing the dialysate three times a day and subsequently lyophilized.
1) mixture was added. The reaction mixture was flushed with nitrogen and placed in an ice bath while stirring until all compounds were completely dissolved. Subsequently, 2 equiv. of Me6TREN was added and the polymerization reaction was carried out for one hour on ice. The polymer solution was dialyzed (MWCO: 10 kDa) against water for 48 hours at 4 °C, while changing the dialysate three times a day and subsequently lyophilized.
2.3. Polyplex preparation
Polymer stock solutions (ranging from 110–1370 μg mL−1, depending on polymer type and intended N/P ratio), and siRNA stock solution (30 μM) were prepared in HBS buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) and cooled at 0 °C. Next, the polymer solution was added to the siRNA solution in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio (typically 400
1 volume ratio (typically 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 μL), and the mixture was vortexed for 10 seconds. The formed polyplexes were incubated at 0 °C for 30 minutes before further use.
200 μL), and the mixture was vortexed for 10 seconds. The formed polyplexes were incubated at 0 °C for 30 minutes before further use.
2.4. Polyplex characterization
 | (1) |
In which D is the diffusion coefficient, kB the Boltzmann's constant, T the absolute temperature, η the viscosity, and RH the hydrodynamic radius of the particle.
2.5. Hydrogel formation and polyplex/siRNA release
The NPN polymers (final concentration of 15% w/w) were dissolved in the NPD or PD polyplexes dispersions (N/P 5 mol/mol, final siRNA concentration of 500 μM) overnight at 4 °C. Free siRNA (thus not formulated as polyplexes; same final concentration) was included as a control. Next, the glass vials were placed at 37 °C and gels (100 μL) were allowed to form. After 30 minutes, 500 μL of prewarmed release buffer (HBS, 20 mM HEPES, 150 mM NaCl, pH 7.4) of 37 °C was added on top of the hydrogels. Twice a day, 450 μL of the release medium was replaced with fresh prewarmed release buffer and the concentration of siRNA in the samples was determined using the RiboGreen Assay (section 2.6).2.6. Quantification of siRNA with Quant-iT™ RiboGreen™ assay
The amount of siRNA released from the thermosensitive hydrogel was quantified using an intercalating reagent that produces a fluorescent signal. First, release samples were diluted 100 times and 40 μL was transferred into a 96-well plate. Next, 55 μL of heparin solution (4.5 mg mL−1) was added to the release samples and incubated for 1 hour at RT to destabilize the polyplexes. Next, the commercially available Quant-iT™ RiboGreen® RNA Assay Kit was used according to the manufacturer's protocol and the ‘high range assay’ (20 ng mL−1 to 1 μg mL−1) was selected. The siRNA's with the same sequences were used for calibration and the fluorescence signal was measured at 484/520 nm using a Jasco FP-8300 spectrophotometer (JASCO, Easton, MD).2.7. Characterization of released polyplexes by fluorescence correlation spectroscopy (FCS)
The size of the released polyplexes from the thermosensitive hydrogel was determined by FCS measurements. Polyplexes containing Alexa 647 labeled siRNA were prepared as described in section 2.3 and subsequently loaded in thermosensitive hydrogels as described in section 2.5. Release medium was refreshed twice a day and at predetermined timepoints (8 and 72 hours) 40 μL of the release medium was transferred onto a coverslip. In parallel, to destabilize the polyplexes, 90 μL of the release medium was mixed with 10 μL heparin sodium salt solution (20 mg mL−1) and incubated for 1 hour at RT. Afterwards, 40 μL of each sample was transferred onto a coverslip. The coverslips were placed in a temperature controlled plate to maintain the temperature at 37 °C. FCS measurements were further performed as described in section 2.4.3.2.8. In vitro transfection activity of polyplexes
In vitro studies were performed according to recommendations as previously described.33 FaDu cells which stably express firefly luciferase were cultured in Dulbecco's Modified Eagle's Medium (DMEM) with high glucose (4.5 g L−1 glucose) supplemented with 10% FBS (referred to as full medium) at 37 °C in a humidified atmosphere containing 5% CO2. Transfection studies were done in 96-well plates, with FaDu-luc cells seeded at a density of 7000 cells per well 24 hours before transfections. At the day of the experiment, cells were washed once with PBS and incubated with 150 μL of various siRNA formulations (anti-luciferase or negative control) in complete medium for 6 hours at 37 °C. For the experiments with fresh polyplexes, particles were prepared on the same day as described in section 2.3 and subsequently diluted in full medium (100 or 500 nM siRNA). Commercially available transfection agents, l-PEI (25 kDa) and Lipofectamine 3000, were included as positive controls and complexes were prepared according to manufacturer's protocol. Free siRNA, without a transfection agent, was included as negative control. Cells treated with only medium were used as negative control and each condition was measured in octuple. For the released polyplexes, the supernatant from the hydrogels collected at various timepoints (8, 24, 48 and 72 hours) was diluted with full medium to yield a final siRNA concentration of 100 or 500 nM. Each condition was measured in quadruple. All transfection mixtures were replaced after 6 hours incubation by 200 μL fresh medium and the plates were incubated for another 24 hours at 37 °C. After 16 hours, medium was replaced with 100 μL fresh medium containing 1× alamarBlue (50 nM) and incubated for 4 hours at 37 °C to assess the cytotoxicity of the tested formulations. From each well, 80 μL was transferred into a new 96-well plate and the absorbance at 570 nm (taking 630 nm as a reference) was measured using a BMG SPECTROstar Nano wellplate reader (BMG Labtech, de Meern, the Netherlands). Medium containing 1× alamarBlue was included as blank. Data analysis was performed using MARS Data analysis software version 2.22 (BMG Labtech) and cell viability was calculated relative to untreated cells. To evaluate siRNA-induced silencing, a Luciferase Assay was performed. Medium was removed and the cells were lysed with 100 μL lysis buffer (25 mM tris(hydroxymethyl)aminomethane (Tris), 2 mM dithiothreitol (DTT), 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid (DCTA), 1% Triton X-100, 10% glycerol) on a shaking board at RT for 15 minutes. Thereafter, 50 μL of lysate was transferred into a white luminescence plate and 50 μL of Luciferase Assay Reagent was injected using a FLUOstar OPTIMA microplate reader (BMG Labtech, Ortenberg, Germany) equipped with an injection pump. After two seconds, luminescence was measured for ten seconds according to supplier's recommendation.3. Results and discussion
3.1. Polymer synthesis and characterization
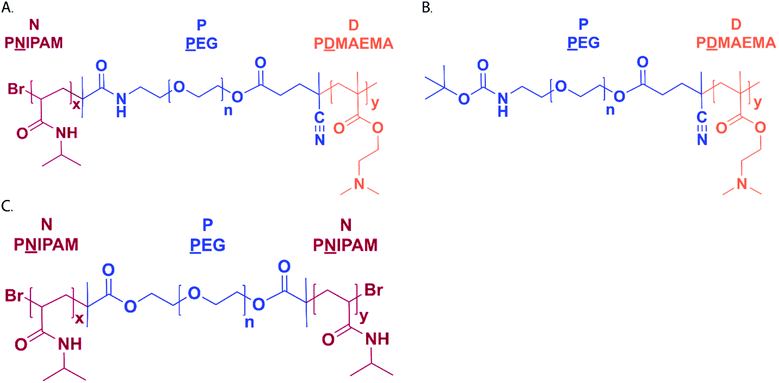
The NPD triblock copolymer consisting of a PEG midblock (P), flanked by blocks of PNIPAM (N) and PDMAEMA (D) to introduce thermosensitive and cationic properties into the polymer structure, respectively, was synthesized and characterized (Fig. 1A and Table 2). The polymer synthesis followed a two-step synthesis route using a hetero-functional PEG macroinitiator as reported before (Scheme S1†).26 In the first step, NIPAM was polymerized by ATRP and the corresponding NPPN polymer (N = PNIPAM, P = PEG) was obtained with a monomer conversion of 92% based on 1H-NMR analysis and a yield of 90% (Table 2). Subsequently, classical free radical polymerization was used to polymerize DMAEMA to yield the final NPD triblock copolymer. DMAEMA conversion was 79% as determined with 1H-NMR and the polymer was obtained in a yield of 71% (Table 2, Fig. S1†). A non-thermosensitive PD diblock copolymer lacking the PNIPAM block was also synthesized (Fig. 1B) and a similar monomer conversion of DMAEMA (77%) and polymer yield (68%) were obtained as for the NPD polymer (Table 2, Fig. S2†). A NPN (N = PNIPAM, P = PEG) triblock copolymer (Fig. 1C) was synthesized according to a previously reported method27 and its characteristics are summarized in Table 2. The monomer conversion based on 1H-NMR analysis was 100% (Fig. S3†), and the obtained polymer had a rather narrow polydispersity index (PDI 1.2), which is expected for polymers synthesized by controlled radical polymerization. The cloud points of the synthesized polymers (NPD and NPN) were 34 and 35 °C, respectively (Table 2, Fig. S4†). The rheological properties of the temperature dependent gelation behavior of NPN, as well as representative images of the thermosensitive hydrogel before and after gelation are shown in the ESI (Fig. S5 and S6†). The injectability as previously shown by De Graaf et al.,27 was confirmed by passing the solution through a 23 G needle.![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) IPAM, P =
IPAM, P = ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) EG, D = P
EG, D = P![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) MAEMA)
MAEMA)
| Name | Feed initiator![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) monomer ratio (mol/mol) monomer ratio (mol/mol) |
Monomer conversiona (%) | M n N blocka (kDa) | M n P blocka (kDa) | M n D blocka (kDa) | Total Mn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (kDa) a (kDa) |
Total Mn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b (kDa) b (kDa) |
PDIb | Cloud pointc (°C) |
|---|---|---|---|---|---|---|---|---|---|
| a Determined by 1H-NMR. b Determined by GPC. c Determined by light scattering at 550 nm. n.d. = not determined, n.a. = not applicable. | |||||||||
| NPPN | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 283 283 |
92 | 29 | 10 (2 × 5) | n.a. | 39 | 64 | 1.6 | n.d. |
| NPD | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790 790 |
79 | 15 | 5 | 49 | 69 | 51 | 1.9 | 34 |
| PD | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
77 | n.a. | 5 | 47 | 52 | 32 | 2.1 | n.d. |
| NPN | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302 302 |
100 | 34 | 6 | n.a. | 40 | 45 | 1.2 | 35 |
3.2. Polyplex characterization
The thermosensitive NPD and the non-thermosensitive PD polymers (Table 2) were used to prepare siRNA-containing polyplexes at various N/P ratios. The N/P ratio is defined as the molar ratio between the amine groups (N) on the cationic polymers and the phosphate groups (P) on the siRNA. Increasing the N/P molar ratio from 1 to 5 resulted in a significant decrease in NPD polyplex size from 108 ± 1 to 23 ± 1 nm (Fig. 2A). A further increase of the N/P ratio to 10 did not result in a further decrease in the size of the polyplexes. The same trend was observed for the PD polyplexes, for which a decrease in polyplex size from 158 ± 1 to 23 ± 1 nm (N/P 1 and 5, respectively) was found upon increasing N/P ratio (Fig. 2A). Interestingly, polyplexes prepared with N/P 1 had very narrow size distributions (PDI < 0.1), while PDIs of polyplexes with higher N/Ps increased but were still within an acceptable range (<0.3) (Fig. 2B). The polyplexes prepared at an N/P ratio of 1 had a ζ-potential of −3 ± 1 mV and −1 ± 1 mV (NPD and PD polyplexes, respectively) indicating incomplete shielding of the siRNA molecules (Fig. 2C). As expected, with an increasing N/P ratio of the formulation, due to the excess of cationic groups, the polyplexes showed an increase in ζ-potential (up to + 9 ± 1 mV at an N/P ratio of 10 for PD polyplexes). Slightly higher ζ-potentials were found for PD polyplexes as compared to NPD polyplexes ( + 9 ± 1 mV and 5 ± 1 mV at N/P 10, respectively), which can be explained by the additional shielding of the hydrophilic PNIPAM block (below the CP) in the NPD polyplexes. Similar findings were also reported before for polyplexes containing pDNA.24 The ability of the cationic polymers to complex with siRNA was evaluated by an agarose gel retardation assay. Even at low N/P ratios, no free siRNA was detected in the both types of polyplex dispersions, indicating that all siRNA molecules complexed with the cationic polymers (Fig. 2D). To show the reversibility of the electrostatic interactions, strongly negatively charged heparin was added to the polyplexes, which resulted in release of siRNA (Fig. 2D).3.3. Polyplex stability at 37 °C
Since the NPD polyplexes contain thermosensitive blocks, the effect of temperature changes above (37 °C) and below (10 °C) the CP was evaluated by DLS. For polyplexes prepared at N/P 1 and 3, increasing the temperature to 37 °C resulted in polyplexes with larger sizes (298 ± 3 at 37 °C and 362 ± 12 at 37 °C, respectively) and broader size distributions (Fig. 2A and B). A similar effect was observed for PD polyplexes with the same N/P ratio, suggesting that the observed changes for NPD polyplexes with temperature are not caused by the thermosensitive PNIPAM block. Additionally, the increase in size at 37 °C was not completely reversible after cooling down the polyplexes again to 10 °C (Fig. 2A). The size of NPD polyplexes at N/P1 increased from 108 ± 1 to 298 ± 3 nm upon heating from 10 to 37 °C, whereas upon cooling to 10 °C the size was 304 ± 6 nm. Fig. 2A also shows that for the NPD polyplexes prepared at N/P 5 and 10, a significant increase in particle size (from ∼23 nm to ∼170 nm) was observed when heated to 37 °C, which was completely reversible after cooling down to 10 °C. The particle size of PD polyplexes with N/P 5 and 10 remained unchanged during the same temperature changes. At N/P 5 and 10, a large excess of polymer is added to the siRNA, making it likely that free polymer chains are present in the polyplex dispersion. In addition, the free polymer chains can self-assemble into micellar structures at 37 °C, because of the presence of the thermosensitive PNIPAM blocks. Indeed, the control sample containing only NPD polymer, showed the presence of particles with a similar size (∼165 nm) at 37 °C (Fig. 2A). In addition, the ζ-potential of NPD polyplexes slightly increased at 37 °C which can indicate the presence of positively charged micellar structures (Fig. 2C). The stability of the polyplexes was studied in HBS buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) over time. DLS measurements showed that the size of PD polyplexes (N/P 5) remained constant (∼30 nm) after incubation for 16 hours at 37 °C (Fig. 3B). Again, at 37 °C the size of the NPD polyplexes (N/P 5) significantly increased in time (from 25 to 120 nm), which was similar to the micelle size formed by the NPD polymer alone (Fig. 3A and C). Cooling down the polyplexes to 10 °C after 16 hours incubation at 37 °C, resulted again in only the small sized polyplexes (23 nm). Polyplexes prepared at N/P ratios of 1 and 3 showed significant increase in size and PDI over time (Fig. S7†). From literature it is known that among others particle size is a very important factor for the uptake efficiency of complexes. For example, it has been shown that the uptake efficiency of 20 and 40 nm nanoparticles was 5–10 times greater than that of 100 nm particles.34 Not only the efficiency of the uptake but also the type of uptake pathway by which the particles are internalized, is influenced by the size of the particles.35Fluorescence correlation spectroscopy (FCS), a light microscopy based technique used to study interactions on the single molecule level, was exploited to further characterize the thermosensitive polyplexes at 37 °C. FCS showed that free siRNA in buffer and at 37 °C has a diffusion coefficient of 181 ± 7 μm2 s−1 (Fig. 4A) which using the Stokes–Einstein equation corresponds to a size of 2 nm, that is in line with earlier reported sizes for siRNA molecules (Fig. 4B).36,37 FCS measurements showed that the diffusion coefficient for both NPD and PD polyplexes prepared at N/P ratio of 1 and 3 was ∼3 μm2 s−1, which corresponds to a particle size of ∼100 nm (Fig. 4A and B). On the other hand, the diffusion coefficients of polyplexes prepared at higher N/P ratios 5 and 10 were around 10 times larger (∼30 μm2 s−1) for both types of polymers. This diffusion coefficient corresponds to a polyplex size of ∼10 nm, which is in the same range as the small particle size found by DLS at 10 °C (Fig. 2A and Fig. 4B). These results confirm the presence of micellar structures in the NPD polyplex dispersions, which dominate the DLS measurement of NPD polyplexes (N/P 5 and 10) at 37 °C. Particles in a colloid dispersion scatter an incident laser beam, and the intensity of this scattered light is proportional to the 6th power of the diameter of the particles.38 As a consequence, the excessive scattering of bigger particles can mask the low intensity scattering from smaller particles.39
Polyplexes with comparable small sizes (10–30 nm) have been reported in literature before using various block copolymers.40–45 For example, Hayashi et al. used poly(ethylene glycol)-b-poly(L-lysine) (PEG–PLL) block copolymers to complex siRNA and reported sizes of around 10 nm for polyplexes based on this block copolymer.42 The authors attribute this small size to a monomolecular assembly of the positively charged polymer chain and the negatively charged siRNA molecule. In other words, it is considered that each particle consists of a single siRNA molecule complexed with one PEG–PLL chain via electrostatic interactions. In the present study, it is shown that the polyplex size is dependent on the N/P ratio, where the small polyplexes are only observed at higher N/P ratios (N/P ≥ 5). At higher N/P ratios, the siRNA molecules become more diluted over the polymer chains making it possible that at some point the self-assembly behavior resembles that of PEG–PLL, in which each polyplex contains a single siRNA molecule. From the FCS data, an estimation of the number of siRNA molecules per complex can be calculated by normalizing the fluorescent particle concentration of the polyplexes to that of free siRNA,46 assuming that no quenching occurs. If quenching would occur to a large extent, the peaks in the FCS time traces are likely to disappear,47 which was not observed (Fig. S8A, C, E and G†). It is determined that indeed polyplexes prepared at N/P 5 and 10 contain between 1–3 siRNA molecules per complex compared to >100 siRNAs for polyplexes at N/P 1 and 3 (Table 3).
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) IPAM, P =
IPAM, P = ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) EG, D = P
EG, D = P![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) MAEMA). Mean ± SD (n = 3)
MAEMA). Mean ± SD (n = 3)
| Polymer | N/P ratio | Polyplex size (nm) | Number of siRNA molecules per complex |
|---|---|---|---|
| NPD | 1 | 149 ± 9 | 102 ± 2 |
| 3 | 88 ± 13 | 128 ± 9 | |
| 5 | 10 ± 1 | 3 ± 1 | |
| 10 | 12 ± 1 | 2 ± 1 | |
| PD | 1 | 100 ± 16 | 132 ± 32 |
| 3 | 137 ± 22 | 109 ± 5 | |
| 5 | 12 ± 1 | 2 ± 1 | |
| 10 | 12 ± 1 | 1 ± 1 |
Polyplexes prepared with both types of polymers at different N/P ratios were incubated with or without fetal bovine serum (FBS). A significant increase in polyplex size (from 149 ± 9 to 866 ± 204 nm) was observed for NPD polyplexes with N/P 1 when incubated without FBS for 6 hours at 37 °C, which is in line with the DLS results (Fig. 2A and 4B). In contrast, the observed sizes of the polyplexes at N/P 1 and 3 did not change after incubation with FBS at 37 °C. However, the FCS time traces showed a significant decrease in the count rate (Fig. S8†), which might indicate aggregation and subsequent sedimentation of the particles. Interestingly, FCS analysis showed that the size of NPD and PD polyplexes prepared at higher N/P ratios of 5 and 10 did not change significantly when incubated at 37 °C for 6 hours without FBS (Fig. 4B). However, in the presence of serum, the originally small sized NPD polyplexes (size ∼10 nm; N/P 5 and 10) aggregate to bigger particles of around 200 nm after 6 hours incubation (Fig. 4B). It should be noted that such a polyplex size is still generally considered acceptable to achieve efficient transfection.11,48 The same trend was observed for the PD polyplexes (N/P 5 and 10) which increased slightly more in size to around 400 nm after incubation with FBS for 6 hours at 37 °C To evaluate whether siRNA loaded in NPD polyplexes is protected against degradation by RNases present in FBS, an agarose retardation assay was performed (Fig. S9†). Compared to the observed band for free siRNA in buffer (shown at N/P 0), a time-dependent decrease in intensity was clearly seen when siRNA was incubated with serum, indicating the presence of nucleases in FBS. When the siRNA was complexed with polymers, only the polyplexes with N/P 1 showed a significant decrease in siRNA signal in the gel, while all other formulations obviously protected the loaded siRNA against degradation.
3.4. Hydrogel formation and siRNA release
Polyplexes prepared at N/P 5 were selected for further studies because of their small size and excellent colloidal stability in buffer and good stability in serum. These polyplexes were loaded in a thermosensitive hydrogel based on the PNIPAM–PEG–PNIPAM (NPN) triblock polymer by mixing NPD or PD polyplexes (N/P 5) with a NPN solution at 4 °C and subsequently increasing the temperature to 37 °C enabling gel formation. Free siRNA was quantitatively released from the hydrogels in 50 hours (Fig. 5, circles). Moreover, a rapid release was observed for this formulation, since 40% of the siRNA was released after changing the release medium just once. When siRNA was embedded in the hydrogel as polyplexes, a significant slower and more controlled release was observed. For both type of polyplexes (NPD and PD polyplexes), siRNA was quantitatively released during 128 hours (Fig. 5, squares and triangles). Furthermore, the amount of siRNA released was rather similar per time point, which suggests that the release of the polyplexes is dependent on the dissolution rate of the hydrogel system. This is not completely surprising, since the hydrogel is physically cross-linked and it has been reported before that the equilibrium between the hydrogel and free polymer chains in solution influences the dissolution rate of such systems.27,49 The NPN triblock copolymers are known to form micelles in aqueous solutions above their CP, and at high concentrations the micelles are closely packed into a hydrogel hold together by “bridging” chains. When this hydrogel is immersed into an aqueous medium, an equilibrium between bridged micelles in the gel, free micelles and unimers in solution is established.27,50 As a result, the frequency with which the medium is refreshed determines the dissolution rate of the hydrogel, and consequently the release of the entrapped polyplexes. As a consequence, the release profiles in vivo may be substantially different from the ones observed in vitro studies because of a more dynamic environment in vivo. The rationale for comparing the thermosensitive NPD polyplexes with the non-thermosensitive PD polyplexes was based on the hypothesis that, in the case of the NPD polyplexes, besides dissolution of the hydrogel also interactions between the PNIPAM blocks in the polyplexes and those present in the hydrogel may play a role in the kinetics of polyplex release. However, no significant differences in release kinetics between the NPD and PD polyplexes were observed (Fig. 5), indicating that the release of the polyplexes is governed by the dissolution rate of the hydrogel. Hydrogels can be physically cross-linked by noncovalent interactions or chemically cross-linked by covalent bond formation, or via a combination of both. In this study, the thermosensitive physical cross-linking causes the polyplex release to be dependent on dissolution of the hydrogel matrix and not on degradation of the chemical cross-links, which could offer an alternative strategy to tune the release kinetics. | ||
| Fig. 5 Release of luciferase siRNA (siRNA-luc) from a thermosensitive hydrogel in its free form, or formulated as NPD or PD polyplexes (N/P 5) at 37 °C. The values are given as the mean ± SD (n = 3). | ||
It is important to note that the siRNA quantification measurements were performed after the addition of heparin to dissociate the siRNA from the cationic polymers enabling intercalating of the RiboGreen reagent. This means that the detection does not discriminate between siRNA released in the form of polyplexes or free siRNA. Moreover, analyzing the release medium with dynamic light scattering does not give insight whether polyplexes were released since this technique detects both polyplexes and nanoparticulate fragments of the hydrogel matrix. Therefore, FCS was used to evaluate whether in the release samples free siRNA was present or siRNA in the form of polyplexes. Since only the fluorescent signal of Alexa 647 labeled siRNA was tracked, the presence of other particles derived from degradation of the hydrogel did not influence the measurement. The diffusion coefficient of free siRNA in buffer was slightly higher than that of free siRNA in release medium (181 ± 7 μm2 s−1 and 154 ± 17 μm2 s−1, respectively), indicating only a minor effect of the NPN polymers on the viscosity of the release medium (Fig. 6A). More importantly, FCS measurements showed that the diffusion coefficients of NPD and PD polyplexes released after 8 hours were comparable to those before loading in the thermosensitive hydrogel (Fig. 6A), demonstrating that intact polyplexes were released from the thermosensitive hydrogel. The released polyplexes were similar in size as the ones before encapsulation (Fig. 6B). Even after 72 hours, the diffusion coefficient, and correspondingly the size, of the released NPD polyplexes were similar to freshly prepared polyplexes. For the PD polyplexes, the diffusion coefficient after 72 hours was slightly decreased, which also translated into larger particles (Fig. 6B). This might be explained by aggregation of the released polyplexes over time in the release medium. To confirm that indeed polyplexes were released, the strongly negatively charged heparin biopolymer was added to the release medium (at 8 hours) and the resulting samples were analyzed by FCS (Fig. 6C and D). For both type of polyplexes (NPD and PD polyplexes), the addition of this biopolymer resulted in an increase in diffusion coefficient from ∼38 μm2 s−1 to ∼165 μm2 s−1 (Fig. 6C). The larger diffusion coefficient correlates perfectly with the value for free siRNA, demonstrating destabilization of the polyplexes after addition of heparin. To further support the finding that intact polyplexes were released, agarose gel assay was performed which showed the absence of free siRNA (Fig. 7). Again, the addition of heparin to the release medium resulted in dissociation of the polyplexes, demonstrating also the reversibility of the complexation. As expected, free siRNA was detected in the release medium when incorporated into the hydrogel without the presence of cationic polymers (Fig. 7).
3.5. In vitro transfection activity of polyplexes
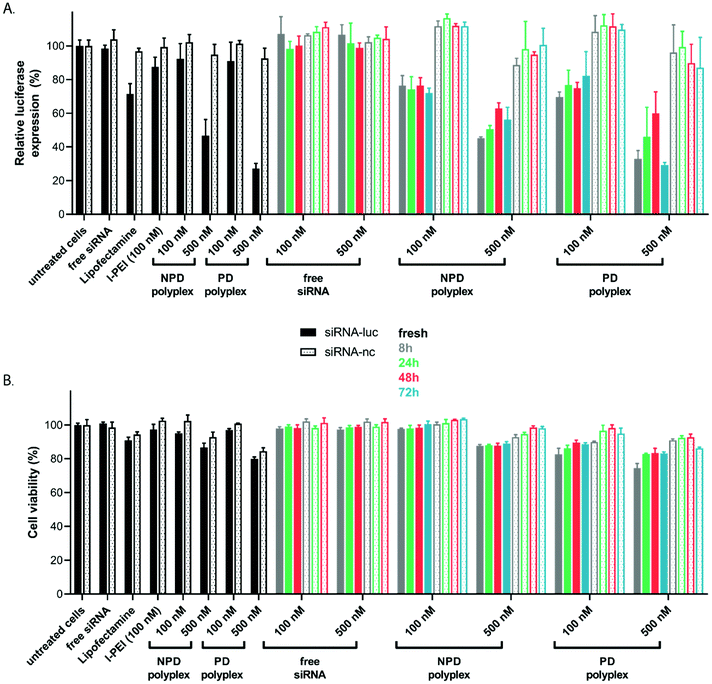
The transfection efficiency and cytotoxicity of polyplexes based on NPD and PD were evaluated in the presence of serum using FaDu-luc cells (Fig. 8). For the freshly prepared NPD and PD polyplexes, increasing the dose from 100 to 500 nM siRNA resulted in a lower luciferase expression and thus increased silencing (Fig. 8A). This increase in polyplex dose was associated with a slight drop in cell viability, which can be explained by the higher concentration of the cationic polymer to which the cells were exposed (Fig. 8B). It has been reported in many papers that cationic polymers adversely affect the viability of living cells.51,52 Furthermore, comparison of the silencing effects between luciferase siRNA (Fig. 8A, closed bars) and the non-coding siRNA (Fig. 8A, open bars) supports that the observed reduced luminescence observed upon incubation of the cells with the polyplex formulations was because of specific silencing of the luciferase gene and not related to their cytotoxicity. The commercially available transfection reagent Lipofectamine was included as control, and resulted in ∼30% knockdown of the luciferase gene. This observed silencing effect of Lipofectamine is lower compared to the ∼80% luciferase silencing as reported before.43,53 This however can be explained by differences in experimental conditions as in this study transfections were performed in full medium supplemented with FBS compared to serum-free conditions in other studies. Polyplexes prepared with l-PEI (25 kDa) performed in a similar way as the NPD and PD polyplexes with the same siRNA dose. Incubation of the cells with siRNA in its free form did not result in any luciferase gene silencing, which confirms the requirement of a delivery system for these molecules. The ability to induce siRNA-mediated gene silencing for polyplexes after being released at different time points from the hydrogel (Fig. 5 and Fig. S10†) was examined. Cells incubated with the supernatant from the hydrogels containing polyplexes showed a decrease in luciferase signal for all release samples tested (8, 24, 48 and 72 hours) with a gene knockdown ranging between 30–70% (Fig. 8A). These results further support the finding that intact polyplexes were released from the hydrogel, since siRNA released in its free form from the hydrogel was not able to induce silencing of luciferase (Fig. 8A). Additionally, limited cytotoxicity was observed for all released formulations (Fig. 8B). Similar to the freshly prepared polyplexes, increasing the polyplex dose from 100 to 500 nM siRNA resulted in an increased luciferase silencing with mild cytotoxicity (Fig. 8A and B). No significant differences in gene silencing were observed between the NPD and PD polyplexes, however, PD polyplexes (100 nM) were slightly cytotoxic whereas NPD polyplexes were not (∼86% and ∼100% cell viability respectively). Similar results were reported before for pDNA polyplexes, where NPD polyplexes showed a better cytocompatibility than PD polyplexes with similar efficiencies of delivering its cargo into HeLa cells.24 Together, these data show that the released siRNA polyplexes are able to enter cells and induce gene silencing.4. Conclusion
This study shows the feasibility of an injectable thermosensitive hydrogel to promote local and sustained delivery of siRNA. Both the NPD and PD polymers formed small nanosized polyplexes (10–20 nm) with siRNA when prepared at a N/P ratio of 5 or higher which were stable under physiological conditions. Formulating siRNA into polyplexes using either the NPD or PD polymer before loading into the thermosensitive hydrogel resulted in a more controlled and sustained release compared to free siRNA. The results presented here also demonstrate that intact polyplexes were released suggesting that the hydrogel material does not compromise the polyplex stability. Importantly, transfection experiments demonstrated that the released polyplexes were still active by promoting the cellular uptake and luciferase gene silencing in FaDu cells. This polyplex-hydrogel delivery approach could potentially be used to deliver siRNA for various applications, such as the treatment of tumors, where an injectable hydrogel is valuable for localized retention of the nucleic acid cargo.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Netherlands Organization for Scientific Research (NWO/VIDI 13457 and NWO/Aspasia 015.009.038) is acknowledged for funding. The authors thank Cristina Casadidio for her contribution to this project.References
- A. Fire, S. Q. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature, 1998, 391(6669), 806–811 CrossRef CAS PubMed.
- S. M. Hoy, Patisiran: First Global Approval, Drugs, 2018, 78(15), 1625–1631 CrossRef CAS PubMed.
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope, T. D. Madden, B. L. Mui, S. C. Semple, Y. K. Tam, M. Ciufolini, D. Witzigmann, J. A. Kulkarni, R. van der Meel and P. R. Cullis, The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs, Nat. Nanotechnol., 2019, 14(12), 1084–1087 CrossRef CAS PubMed.
- C. F. Bennett and E. E. Swayze, RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 259–293 CrossRef CAS PubMed.
- K. A. Whitehead, J. E. Dahlman, R. S. Langer and D. G. Anderson, Silencing or stimulation? siRNA delivery and the immune system, Annu. Rev. Chem. Biomol. Eng., 2011, 2, 77–96 CrossRef CAS PubMed.
- E. Wagner, Biomaterials in RNAi therapeutics: quo vadis?, Biomater. Sci., 2013, 1(8), 804–809 RSC.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Non-viral vectors for gene-based therapy, Nat. Rev. Genet., 2014, 15, 541 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Delivery materials for siRNA therapeutics, Nat. Mater., 2013, 12(11), 967–977 CrossRef CAS PubMed.
- S. Agarwal, Y. Zhang, S. Maji and A. Greiner, PDMAEMA based gene delivery materials, Mater. Today, 2012, 15(9), 388–393 CrossRef CAS.
- E. Wagner, Polymers for nucleic acid transfer-an overview, Adv. Genet., 2014, 88, 231–261 CAS.
- U. Lachelt and E. Wagner, Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond), Chem. Rev., 2015, 115(19), 11043–11078 CrossRef PubMed.
- V. J. Venditto and F. C. Szoka Jr., Cancer nanomedicines: so many papers and so few drugs!, Adv. Drug Delivery Rev., 2013, 65(1), 80–88 CrossRef CAS PubMed.
- J. L. Au, B. Z. Yeung, M. G. Wientjes, Z. Lu and M. G. Wientjes, Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities, Adv. Drug Delivery Rev., 2016, 97, 280–301 CrossRef CAS PubMed.
- M. D. Krebs, O. Jeon and E. Alsberg, Localized and Sustained Delivery of Silencing RNA from Macroscopic Biopolymer Hydrogels, J. Am. Chem. Soc., 2009, 131(26), 9204–9206 CrossRef CAS PubMed.
- M. K. Nguyen, O. Jeon, M. D. Krebs, D. Schapira and E. Alsberg, Sustained localized presentation of RNA interfering molecules from in situ forming hydrogels to guide stem cell osteogenic differentiation, Biomaterials, 2014, 35(24), 6278–6286 CrossRef CAS PubMed.
- L. L. Wang, J. N. Sloand, A. C. Gaffey, C. M. Venkataraman, Z. C. Wang, A. Trubelja, D. A. Hammer, P. Atluri and J. A. Burdick, Injectable, Guest-Host Assembled Polyethylenimine Hydrogel for siRNA Delivery, Biomacromolecules, 2017, 18(1), 77–86 CrossRef CAS PubMed.
- L. A. L. Fliervoet, J. F. J. Engbersen, R. M. Schiffelers, W. E. Hennink and T. Vermonden, Polymers and hydrogels for local nucleic acid delivery, J. Mater. Chem. B, 2018, 6(36), 5651–5670 RSC.
- L. L. Wang and J. A. Burdick, Engineered Hydrogels for Local and Sustained Delivery of RNA-Interference Therapies, Adv. Healthcare Mater., 2017, 6(1), 1601041 CrossRef PubMed.
- J. E. Mealy, C. B. Rodell and J. A. Burdick, Sustained Small Molecule Delivery from Injectable Hyaluronic Acid Hydrogels through Host-Guest Mediated Retention, J. Mater. Chem. B, 2015, 3(40), 8010–8019 RSC.
- K. N. Minh and D. S. Lee, Injectable Biodegradable Hydrogels, Macromol. Biosci., 2010, 10(6), 563–579 CrossRef PubMed.
- M. Norouzi, B. Nazari and D. W. Miller, Injectable hydrogel-based drug delivery systems for local cancer therapy, Drug Discovery Today, 2016, 21(11), 1835–1849 CrossRef CAS PubMed.
- M. Najafi, E. Hebels, W. E. Hennink and T. Vermonden, Poly(N-isopropylacrylamide): Physicochemical Properties and Biomedical Applications, in Temperature-Responsive Polymers: Chemistry, Properties, and Applications, ed. V. V. Khutoryanskiy and T. K. Georgiou, John Wiley & Sons, 2018, pp. 3–34 Search PubMed.
- H. G. Schild, Poly(N-isopropylacrylamide): experiment, theory and application, Prog. Polym. Sci., 1992, 17(2), 163–249 CrossRef CAS.
- L. A. L. Fliervoet, C. F. van Nostrum, W. E. Hennink and T. Vermonden, Balancing hydrophobic and electrostatic interactions in thermosensitive polyplexes for nucleic acid delivery, Multifunct. Mater., 2019, 2(2), 024002 CrossRef.
- S. Keereweer, I. M. Mol, A. L. Vahrmeijer, P. B. Van Driel, R. J. Baatenburg de Jong, J. D. Kerrebijn and C. W. Lowik, Dual wavelength tumor targeting for detection of hypopharyngeal cancer using near-infrared optical imaging in an animal model, Int. J. Cancer, 2012, 131(7), 1633–1640 CrossRef CAS PubMed.
- L. A. L. Fliervoet, M. Najafi, M. Hembury and T. Vermonden, Heterofunctional Poly(ethylene glycol) (PEG) Macroinitiator Enabling Controlled Synthesis of ABC Triblock Copolymers, Macromolecules, 2017, 50(21), 8390–8397 CrossRef CAS PubMed.
- A. J. de Graaf, S. Azevedo Prospero dos II, E. H. Pieters, D. T. Rijkers, C. F. van Nostrum, T. Vermonden, R. J. Kok, W. E. Hennink and E. Mastrobattista, A micelle-shedding thermosensitive hydrogel as sustained release formulation, J. Controlled Release, 2012, 162(3), 582–590 CrossRef CAS PubMed.
- O. Soga, C. F. van Nostrum and W. E. Hennink, Poly(N-(2-hydroxypropyl) methacrylamide mono/di lactate): A new class of biodegradable polymers with tuneable thermosensitivity, Biomacromolecules, 2004, 5(3), 818–821 CrossRef CAS PubMed.
- D. Neradovic, W. L. J. Hinrichs, K.-v. d. Bosch, J. J. Hennink and W. E, Poly(N-isopropylacrylamide) with hydrolyzable lactic acid ester side groups: a new type of thermosensitive polymer, Macromol. Rapid Commun., 1999, 20(11), 577–581 CrossRef CAS.
- H. Zhang, S. C. De Smedt and K. Remaut, Fluorescence Correlation Spectroscopy to find the critical balance between extracellular association and intracellular dissociation of mRNA complexes, Acta Biomater., 2018, 75, 358–370 CrossRef CAS PubMed.
- E. L. Elson and D. Magde, Fluorescence correlation spectroscopy. I. Conceptual basis and theory, Biopolymers, 1974, 13(1), 1–27 CrossRef CAS.
- K. Remaut, B. Lucas, K. Braeckmans, N. N. Sanders, S. C. De Smedt and J. Demeester, FRET-FCS as a tool to evaluate the stability of oligonucleotide drugs after intracellular delivery, J. Controlled Release, 2005, 103(1), 259–271 CrossRef CAS PubMed.
- E. V. van Gaal, R. van Eijk, R. S. Oosting, R. J. Kok, W. E. Hennink, D. J. Crommelin and E. Mastrobattista, How to screen non-viral gene delivery systems in vitro?, J. Controlled Release, 2011, 154(3), 218–232 CrossRef CAS PubMed.
- Z. Wang, C. Tiruppathi, R. D. Minshall and A. B. Malik, Size and dynamics of caveolae studied using nanoparticles in living endothelial cells, ACS Nano, 2009, 3(12), 4110–4116 CrossRef CAS PubMed.
- S. Xiang, H. Tong, Q. Shi, J. C. Fernandes, T. Jin, K. Dai and X. Zhang, Uptake mechanisms of non-viral gene delivery, J. Controlled Release, 2012, 158(3), 371–378 CrossRef CAS PubMed.
- A. Schroeder, C. G. Levins, C. Cortez, R. Langer and D. G. Anderson, Lipid-based nanotherapeutics for siRNA delivery, J. Intern. Med., 2010, 267(1), 9–21 CrossRef CAS PubMed.
- K. Raemdonck, K. Remaut, B. Lucas, N. N. Sanders, J. Demeester and S. C. De Smedt, In Situ Analysis of Single-Stranded and Duplex siRNA Integrity in Living Cells, Biochemistry, 2006, 45(35), 10614–10623 CrossRef CAS PubMed.
- C. E. Barnett, Some Applications of Wave-length Turbidimetry in the Infrared, J. Phys. Chem., 1942, 46(1), 69–75 CrossRef CAS.
- S. Bhattacharjee, DLS and zeta potential – What they are and what they are not?, J. Controlled Release, 2016, 235, 337–351 CrossRef CAS PubMed.
- R. J. Christie, Y. Matsumoto, K. Miyata, T. Nomoto, S. Fukushima, K. Osada, J. Halnaut, F. Pittella, H. J. Kim, N. Nishiyama and K. Kataoka, Targeted polymeric micelles for siRNA treatment of experimental cancer by intravenous injection, ACS Nano, 2012, 6(6), 5174–5189 CrossRef CAS PubMed.
- S. Florinas, M. Liu, R. Fleming, L. Van Vlerken-Ysla, J. Ayriss, R. Gilbreth, N. Dimasi, C. Gao, H. Wu, Z.-Q. Xu, S. Chen, A. Dirisala, K. Kataoka, H. Cabral and R. J. Christie, A Nanoparticle Platform To Evaluate Bioconjugation and Receptor-Mediated Cell Uptake Using Cross-Linked Polyion Complex Micelles Bearing Antibody Fragments, Biomacromolecules, 2016, 17(5), 1818–1833 CrossRef CAS PubMed.
- K. Hayashi, H. Chaya, S. Fukushima, S. Watanabe, H. Takemoto, K. Osada, N. Nishiyama, K. Miyata and K. Kataoka, Influence of RNA Strand Rigidity on Polyion Complex Formation with Block Catiomers, Macromol. Rapid Commun., 2016, 37(6), 486–493 CrossRef CAS PubMed.
- B. Lou, N. Beztsinna, G. Mountrichas, J. B. van den Dikkenberg, S. Pispas and W. E. Hennink, Small nanosized poly(vinyl benzyl trimethylammonium chloride) based polyplexes for siRNA delivery, Int. J. Pharm., 2017, 525(2), 388–396 CrossRef CAS PubMed.
- M. Miteva, K. C. Kirkbride, K. V. Kilchrist, T. A. Werfel, H. Li, C. E. Nelson, M. K. Gupta, T. D. Giorgio and C. L. Duvall, Tuning PEGylation of mixed micelles to overcome intracellular and systemic siRNA delivery barriers, Biomaterials, 2015, 38, 97–107 CrossRef CAS PubMed.
- C. Dufès, W. N. Keith, A. Bilsland, I. Proutski, I. F. Uchegbu and A. G. Schätzlein, Synthetic Anticancer Gene Medicine Exploits Intrinsic Antitumor Activity of Cationic Vector to Cure Established Tumors, Cancer Res., 2005, 65(18), 8079–8084 CrossRef PubMed.
- H. J. Kim, H. Takemoto, Y. Yi, M. Zheng, Y. Maeda, H. Chaya, K. Hayashi, P. Mi, F. Pittella, R. J. Christie, K. Toh, Y. Matsumoto, N. Nishiyama, K. Miyata and K. Kataoka, Precise engineering of siRNA delivery vehicles to tumors using polyion complexes and gold nanoparticles, ACS Nano, 2014, 8(9), 8979–8991 CrossRef CAS PubMed.
- K. Remaut, B. Lucas, K. Raemdonck, K. Braeckmans, J. Demeester and S. C. De Smedt, Protection of Oligonucleotides against Enzymatic Degradation by Pegylated and Nonpegylated Branched Polyethyleneimine, Biomacromolecules, 2007, 8(4), 1333–1340 CrossRef CAS.
- Y. Liu, O. Samsonova, B. Sproat, O. Merkel and T. Kissel, Biophysical characterization of hyper-branched polyethylenimine-graft- polycaprolactone-block-mono-methoxyl-poly(ethylene glycol) copolymers (hy-PEI-PCL-mPEG) for siRNA delivery, J. Controlled Release, 2011, 153(3), 262–268 CrossRef CAS PubMed.
- C. L. McCormick, B. S. Sumerlin, B. S. Lokitz and J. E. Stempka, RAFT-synthesized diblock and triblock copolymers: thermally-induced supramolecular assembly in aqueous media, Soft Matter, 2008, 4(9), 1760–1773 RSC.
- A. N. Semenov, J. F. Joanny and A. R. Khokhlov, Associating polymers: Equilibrium and linear viscoelasticity, Macromolecules, 1995, 28(4), 1066–1075 CrossRef CAS.
- D. Fischer, Y. Li, B. Ahlemeyer, J. Krieglstein and T. Kissel, In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis, Biomaterials, 2003, 24(7), 1121–1131 CrossRef CAS PubMed.
- P. van de Wetering, J. Y. Cherng, H. Talsma and W. E. Hennink, Relation between transfection efficiency and cytotoxicity of poly(2-(dimethylamino)ethyl methacrylate)/plasmid complexes, J. Controlled Release, 1997, 49(1), 59–69 CrossRef CAS.
- E. E. Oude Blenke, J. van den Dikkenberg, B. van Kolck, A. Kros and E. Mastrobattista, Coiled coil interactions for the targeting of liposomes for nucleic acid delivery, Nanoscale, 2016, 8(16), 8955–8965 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr03147j |
| This journal is © The Royal Society of Chemistry 2020 |