Open Access Article
Open Access ArticleInter-particle biomolecular reactivity tuned by surface crowders†
M. R. W.
Scheepers
 ab,
S. R. R.
Haenen
a,
J. M.
Coers
a,
L. J.
van IJzendoorn
ab and
M. W. J.
Prins
ab,
S. R. R.
Haenen
a,
J. M.
Coers
a,
L. J.
van IJzendoorn
ab and
M. W. J.
Prins
 *abc
*abc
aEindhoven University of Technology, Department of Applied Physics, PO Box 513, 5600 MB Eindhoven, The Netherlands
bEindhoven University of Technology, Institute for Complex Molecular Systems, PO Box 513, 5600 MB Eindhoven, The Netherlands
cEindhoven University of Technology, Department of Biomedical Engineering, PO Box 513, 5600 MB Eindhoven, The Netherlands. E-mail: m.w.j.prins@tue.nl
First published on 15th June 2020
Abstract
The rate at which colloidal particles can form biomolecular bonds controls the kinetics of applications such as particle-based biosensing, targeted drug delivery and directed colloidal assembly. Here we study how the reactivity of the particle surface depends on its molecular composition, quantified by the inter-particle rate of aggregation in an optomagnetic cluster experiment. Particles were functionalized with DNA or with proteins for specific binding, and with polyethylene glycol as a passive surface crowder. The data show that the inter-particle binding kinetics are dominated by specific interactions, which surprisingly can be tuned by the passive crowder molecules for both the DNA and the protein system. The experimental results are interpreted using model simulations, which show that the crowder-induced decrease of the particle surface reactivity can be described as a reduced reactivity of the specific binder molecules on the particle surface.
Introduction
The behaviour of biofunctionalized colloidal particles in complex biological systems is determined by both specific and nonspecific interactions, each conferred to the particle by different molecular means. Specific binding functionality is conferred to the particles by coupling macromolecules with specific binding affinities to the particle surface, such as antibodies,1,2 oligonucleotides,3,4 or aptamers.5,6 Nonspecific interactions can be controlled by providing e.g. charges7 or hydrophilic polymers8,9 on the particle surface, in order to stabilize the particles and achieve low biofouling properties.The specific and nonspecific binding properties of biofunctionalized particles have been studied in biochemical assays as well as in biophysical experiments. An example of a biophysical experiment is colloidal probe atomic force microscopy (AFM), to measure force–distance curves in liquids between a single micrometer sized colloidal probe and a surface or a second particle.10 The potential energy landscape of functionalized particles above a substrate has been explored using total internal reflection microscopy (TIRM),11–13 and the interactions between two functionalized particles using optical tweezers.14 However, these methods cannot quantify the biomolecular reactivity of colloidal particles.
In this paper we quantify the biomolecular reactivity of functionalized colloidal particles in an optomagnetic cluster (OMC) experiment.15 The OMC experiment was originally developed for rapid and sensitive biomarker quantification by measuring the equilibrium aggregation state of a colloidal solution.1 Ranzoni et al. showed subpicomolar detection of specific proteins in buffer and in blood plasma using antibody-coated particles. Recently, we demonstrated that the OMC experiment allows quantification of the nonspecific kinetic rate of dimer formation kagg within an ensemble of biofunctionalized particles.15 The parameter kagg represents the rate at which biochemical inter-particle aggregation occurs, for particles that are in a well-defined state of proximity. In the OMC experiment, the proximal state is created and controlled by an attractive magnetic inter-particle force. The aggregation rate kagg is a measure of the inter-particle surface reactivity and represents the particle equivalent of the molecular association rate kon.
The aggregation rate of colloidal particles has contributions from specific and nonspecific interactions, which are often studied independently. For example, passive hydrophilic molecules have been applied in order to form a crowded layer that generates steric repulsion between particles.8,9 In the study of this paper, we hypothesize that the steric repulsion by hydrophilic surface crowders might affect not only nonspecific interactions between particles, but may also have an effect on specific inter-particle interactions. To test this hypothesis, particles were provided with a coating consisting of specific binders (antibodies or DNA oligonucleotides) and passive polyethylene glycol (PEG) molecules of different molecular weights. The dependence of the particle surface reactivity on the density of specific binders was studied in a sandwich-type aggregation assay, where aggregation is induced using a specific bridging molecule (a protein or an oligonucleotide) called the analyte, see Fig. 1. The data show that the specific aggregation rate is reduced, for all analyte concentrations, by the presence of PEG molecules on the particle surface, as schematically shown in Fig. 1c. Simulations were performed to interpret the data in terms of the underlying molecular parameters. The paper concludes with an outlook on reactivity modulations that may be achieved by further surface engineering.
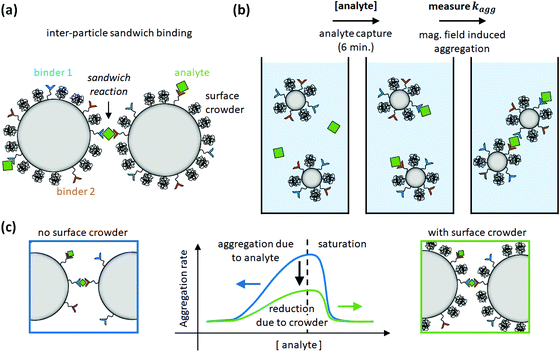 | ||
| Fig. 1 Study of inter-particle biomolecular reactivity as a function of surface-crowding PEG molecules. (a) Particle dimers are formed in a sandwich-type aggregation assay, with specific binder molecules on the particle surface (binder 1 and binder 2) and specific bridging molecules called analyte. The particle is also functionalized with PEG crowders, to investigate the influence of the PEG molecules on the specific particle aggregation process. (b) The assay consists of an analyte capture phase of 6 min, and subsequently an aggregation rate measurement using optomagnetic detection.15 (c) The aggregation rate is measured as a function of analyte concentration. The experiments show that PEG molecules on the particle surface reduce the specific aggregation rate. | ||
Results and discussion
Specific particle aggregation quantified for DNA and protein systems
The specific particle aggregation rate is first investigated for the DNA system and the protein system without passive surface crowders. The inter-particle aggregation rate is quantified using the OMC experiment. This methodology is summarized in section S1 of the ESI.†In the DNA sandwich system, see Fig. 2a, streptavidin coated particles were functionalized with biotinylated DNA docking strands and DNA filler strands. The docking DNA consists of a 20 bp dsDNA with a 15 nt ssDNA overhang. The filler strands are similar to the docking strands, but do not have the 15 nt ssDNA overhang. The maximum docking strand density per particle was quantified using an indirect supernatant assay, giving σdock,max = (2.0 ± 0.4) × 104 μm−2. By varying the concentration of docking DNA and filler DNA strands, the docking strand coverage was controlled (functionalization protocol is described in Materials and methods section). A complete description of the DNA strands and the supernatant assay is given in sections S2 and S3 of the ESI,† respectively.
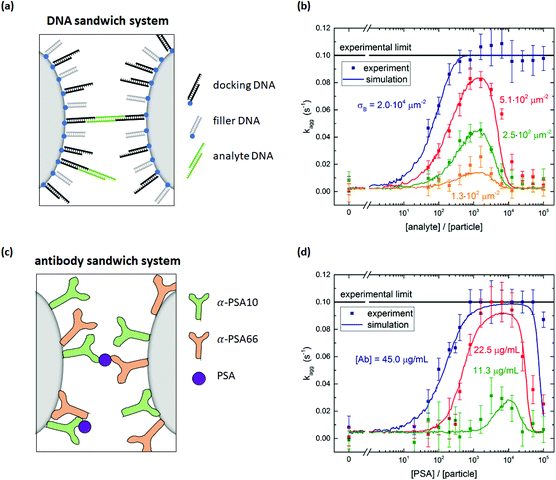 | ||
| Fig. 2 Specific particle aggregation rates measured with the optomagnetic cluster experiment. (a) DNA sandwich system consisting of streptavidin coated particles functionalized with biotinylated DNA docking and filler strands. Specific binding is induced by a symmetric DNA analyte molecule. (b) Measured aggregation rate as a function of the DNA analyte concentration for particles with different densities of DNA docking strands. The lines through the data points represent simulation results. The input parameters of these simulations can be found in ESI section S6.† (c) Antibody sandwich system consisting of carboxylic acid coated particles functionalized with a matching pair of anti-PSA antibodies. The PSA molecules form specific molecular bridges between the particles. (d) Measured aggregation rate as a function of the PSA concentration for particles functionalized with different concentrations of PSA antibodies. The lines through the data points represent simulation results. The input parameters of these simulations can be found in ESI section S6.† | ||
The DNA analyte is symmetric, with a central 20 bp dsDNA fragment and on both ends a 15 nt ssDNA overhang that can bind to the docking DNA on the particles, in this way forming a specific molecular bridge between two particles. The 15 bp complementarity ensures that this bond is stable throughout the complete experiment (∼10 min). The DNA coated particles were incubated with a controlled concentration of analyte DNA for six minutes. After five minutes of incubation, the OMC actuation protocol was started, such that during the last minute of the incubation the first measurement phase was executed. Subsequently the actuation phase, waiting phase and second measurement phase were performed. The aggregation rate kagg was then determined using eqn (S1).†
Fig. 2b shows the measured aggregation rate as a function of the analyte concentration scaled to the particle concentration, for four different docking strand coverages on the particles. Without analyte, the observed aggregation rate is much lower than the rates observed in the presence of analyte, demonstrating the high molecular control of the system. For analyte-to-particle-ratios below 103, the aggregation rate increases with analyte concentration by two effects. First, having more analytes per particle increases the probability that an analyte molecule is present in the interaction area of a dimer, allowing for specific aggregation of the two particles. Second, for even more analytes per particle, all dimers have analytes in their interaction area and an increase in the number of analytes in the interaction area increases the particle reactivity. For analyte-to-particle-ratios around 103, the aggregation rate reaches a maximum value. At this point the system is in its most reactive state, which implies that half of the docking strands is covered with an analyte strand. For analyte-to-particle-ratios higher than 103, the aggregation rate decreases because the docking strands on the particle surface become saturated with analyte strands, so there are no free docking strands on the particles anymore. Note that the actual number of analytes per particle may be lower than the indicated analyte-to-particle ratio, due to the short incubation time of six minutes.
The docking strand surface density was also varied in this experiment. The total number of DNA strands on the particles was kept constant by saturating the surface with filler strands, in order to keep the electrostatic repulsion constant. For each non-zero analyte concentration, the particles with a higher docking strand coverage aggregate faster. For a docking strand surface density σdock ≤ 5.1 × 102 μm−2, the aggregation rate is measurable for the complete analyte concentration range. For a binder density of σdock = 2.0 × 104 μm−2 and analyte-to-particle-ratios above 4 × 102, the aggregation rate is equal to the experimental limit of 0.1 s−1. Under these conditions, the particle surface is so reactive that all magnetic dimers chemically aggregate during the interaction time in the experiment.
In the antibody sandwich system, see Fig. 2c, carboxylic acid coated Ademtech Masterbeads were functionalized with a matching pair of anti-PSA antibodies (α-PSA10 and α-PSA66) via EDC-NHS chemistry (complete functionalization protocol is described in Materials and methods section). Note that each particle contains both types of antibodies. In the presence of PSA, the particles can specifically aggregate by sandwiching a PSA molecule between an α-PSA10 antibody on one particle and an α-PSA66 antibody on the other particle, or vice versa. The particle surface reactivity was varied by varying the antibody concentration during the particle functionalization and the incubation with the PSA analyte. Fig. 2d shows the measured aggregation rate as a function of the PSA concentration scaled to the particle concentration, for three different antibody concentrations. The PSA incubation time was six minutes.
The measured aggregation rate of the antibody coated particles shows a similar analyte concentration dependence as the DNA sandwich system. For increasing PSA concentrations the aggregation rate initially increases w.r.t. the nonspecific aggregation rate. For PSA-to-particle-ratios between 103 and 104, the aggregation rate reaches its maximum value and for the highest antibody concentration reaches the experimental limit. For PSA-to-particle-ratios higher than 104, the aggregation rate decreases due to saturation of the antibodies on the particle surface. A control experiment was performed in which the aggregation rate was measured for particles functionalized with only a single type of antibody. Fig. S4† shows that the aggregation rate of the control does not depend on PSA concentration, thus confirming that the PSA induced binding is only possible for a matching antibody pair.
The particle aggregation process is not only determined by the surface reactivity of the particles, but also by the particle encountering process. Previous measurements performed by Ranzoni et al.1 on a comparable antibody sandwich system showed that the equilibrium particle cluster concentration as a function of PSA concentration is sensitive in the range of 0.2–200 PSA per particle. The aggregation rate measurements described here show a PSA dependence in the range of 300–3000 PSA per particle. The difference in PSA dynamic range is caused by the different particle encountering processes. Ranzoni et al. used multiple short magnetic field pulses, whereby magnetic dimers were brought in close proximity many times for a short interaction time. Each time a different interaction area on the particle was probed, such that even particles with a single PSA molecule are at some point in the correct orientation for binding. For a PSA concentration that is much larger than the particle concentration, all particles can aggregate and the equilibrium state is always the fully clustered state. In the experiments of this paper, we measure the aggregation rate by bringing dimers in close proximity only once for a longer interaction time. For [PSA] ≈ [particle], most magnetic dimers will not have their binding sites aligned, and therefore the aggregation rate cannot be distinguished from the nonspecific interaction. For [PSA] ≫ [particle], all magnetic dimers can aggregate if the antibody surface density is sufficiently high.
Simulating specific particle aggregation
To interpret the measured aggregation rates, a stochastic binding simulation was developed which mimics the specific aggregation process in the OMC experiment. Briefly, during an actuation time tact magnetic dimers are formed at a constant rate (as shown in section S1 of the ESI†). Each dimer has a probability to chemically aggregate which depends on several parameters that are included in the simulation: the binder and analyte density on the particle surface, the intrinsic molecular analyte-binder binding rate, and the interaction time of the magnetic dimer. For all magnetic dimers it is checked if chemical aggregation occurs, after which the average aggregation rate of the ensemble of particles is calculated according to eqn (S1).† The resulting aggregation rates are then compared to the experimental results.In the simulation, particles are modelled as spheres with radius R and a binder surface density σB (either DNA docking strands or antibodies) where the number of binders per particle follows a Poisson distribution. For a given analyte concentration [A] and a given binder areal surface density σB, the rate at which analytes from the solution bind to the binders on the particle surface during the incubation time is described by eqn (1).
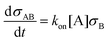 | (1) |
Here, σAB represents the particle surface density of bound analytes. kon is the association rate between analyte and binder, for a binder on a particle surface and analyte in solution. Using eqn (1) we can numerically calculate the analyte surface density after the incubation time tinc as a function of the analyte concentration in solution.
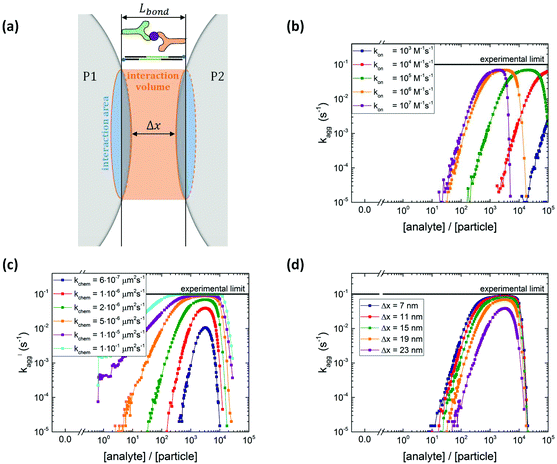
During the actuation time, magnetic dimers are created at a constant encounter rate kenc, neglecting monomer depletion and the formation of larger clusters (as justified in section S1† and ref. 15). For two particles that are magnetically attracted to become a dimer, we define an interparticle distance Δx with a cylindrical interaction volume defined as schematically shown in Fig. 3a. The length of the cylindrical volume is limited by the specific bond length of the interaction, Lbond. For the DNA sandwich system this is taken as the length of the hybridized docking–analyte–docking DNA complex (∼31 nm), whereas for the antibody system it is taken as the length of the antibody–antigen–antibody sandwich (∼35 nm).
The interaction volume defines an interaction area at the particle surfaces in which the analytes on one particle can form bonds with the binders on the other particle, and vice versa. The specific particle aggregation rate depends on the surface reactivity of the particles and can be expressed in terms of the analyte areal density σAB (analyte A bound to binder B) and the free binder areal density σB. The rate of formation of chemical bonds between two interacting surfaces is described by kchem, expressed in unit μm2 s−1. This describes the rate of bond formation between one particle with unit binder density and another particle with unit analyte density, averaged over all possible configurations of the binders and analytes in the interaction area on the particles. The rate kchem depends on the molecular interaction and the distance between the interacting surfaces. For two particles, the interacting surfaces are curved and the interaction distance is a function of the position in the interaction area.
The density of bonds that is formed between analytes on particle 1 and binders on particle 2 and vice versa during the interaction time tint is numerically calculated using eqn (2).
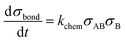 | (2) |
The absolute number of bonds between the particles in the dimer is calculated by multiplying the bond density by the interaction area, see eqn (3).
| Nbonds = σbondAint | (3) |
In case Nbonds ≥ 1, the dimer is marked as a chemical dimer. In addition to the specific aggregation a constant nonspecific aggregation rate kchem,ns is included in the simulation. Similar to the experiment, the ensemble average aggregation rate kagg is calculated, see eqn (S1).†
A parameter scan has been performed to investigate the effect of several input parameters on the simulated particle aggregation rate kagg. In each scan all other parameters were kept constant at the default value shown in Table S4.†Fig. 3b–d show the simulated aggregation rates as a function of the analyte-to-particle concentration ratio. The simulated curves resemble the trends observed in the measurements: for low analyte concentration the aggregation rate increases with analyte concentration, for intermediate analyte concentration the aggregation rate reaches a maximum value and for high analyte concentration the aggregation rate decreases. The position of the maximum represents the situation where 50% of the binders is covered by an analyte.
Fig. 3b shows the simulated aggregation rate for different analyte association rates kon. For increasing kon, the analyte coverage on the particle is higher at the same analyte concentration, shifting the curves to lower analyte concentrations. In the limit of an infinite association rate, the analyte coverage equals the analyte-to-particle concentration ratio until the surface is saturated with analytes. Fig. S4a† shows that for increasing binder density σB the aggregation rate increases and the position of the maximum shifts to higher analyte concentrations.
The simulated particle aggregation rate kagg depends strongly on the intrinsic molecular aggregation rate kchem, see Fig. 3c. For increasing kchem, bond formation occurs faster which makes the particle surface more reactive, and thus the aggregation rate increases. The x-position of the maximum of the curve depends only on σB, tinc and kon and does not change with kchem. However, the height of the maximum does increase with kchem. Fig. 3d shows that increasing the interparticle distance Δx leads to a decrease in the particle aggregation rate which can be attributed to a decrease in the interaction area and the accompanying decrease in the absolute number of interacting analytes and binders.
The parameter scan for the experimental settings, magnetic dimer formation rate kenc (Fig. S5b†) and the actuation time tact (Fig. S5c†) are relatively modest and are explained in more detail in section S5 of the ESI.† Adding a nonspecific aggregation rate to the simulation, kchem,ns = 0.1 s−1, leads to a non-zero background aggregation, see Fig. S5d.†
The experimentally measured aggregation rates of Fig. 2b and d are accompanied by simulated aggregation rate curves. Apart from σB and the parameters relevant for the molecular system such as Lbond and kchem,ns, all other input parameters are kept constant in the simulation. Particle radius R, incubation time tinc and actuation time tact are known in the experiment and the interparticle distance is estimated to be equal to the bond length minus the thickness of the two coatings on the particles (Δx ∼19 nm). A complete list of the values of the input parameters for these simulations is given in section S6 of the ESI.†
The simulated aggregation rate curves for the DNA sandwich system in Fig. 2b closely resemble the experimental data. From this simulation two unknown parameters are obtained: the analyte association rate kon = 1.5 × 106 M−1 s−1 and the intrinsic chemical aggregation rate kchem = 5.0 × 10−4 μm2 s−1. The association rate obtained here, is similar in magnitude to the association rate measured for similar sized DNA oligonucleotides bound on the surface of a graphene field-effect transistor.16 For the highest possible DNA docking strand density on the particle, the analyte association rate seems to be lower. Fig. S7† shows the measured aggregation rate for σdock = 2.0 × 104 μm−2 accompanied by simulations using kon = 1.5 × 106 M−1 s−1 and kon = 1.0 × 105 M−1 s−1. The data and simulation agree best for the lower association rate. At this high DNA docking strand density, association of the negatively charged DNA analyte from the solution may be hindered by the negative charge on the particle.
The simulations of the antibody sandwich system are shown in Fig. 2d. The data give kon = 1.9 × 105 M−1 s−1 and kchem = 9.5 × 10−6 μm2 s−1. The association and the intrinsic chemical binding rate for the PSA antibody system are lower compared to the DNA sandwich system. Apparently, DNA hybridization is faster than antibody–antigen binding, which is probably due to multiple orientations in which DNA oligos can start hybridizing, while the reaction between an antibody and an antigen is orientationally much more restricted.
Tuning the specific aggregation rate using passive surface crowders
In this section we investigate if it is possible to tune the specific reactivity of particles by incorporating PEG molecules on the particle surface. For the DNA sandwich system the particles were coated with a lower density of biotinylated DNA docking strands and the remainder of the surface was saturated with biotin-PEG, see Fig. 4a. The DNA docking strand density was quantified to be σdock = (5.6 ± 0.5) × 103 μm−2, so 28% of the docking strand capacity, leaving room for immobilization of PEG molecules. After saturation of the particles with PEG, multiple magnetic washing steps are performed, so there is no free PEG in the solution during the aggregation rate measurements.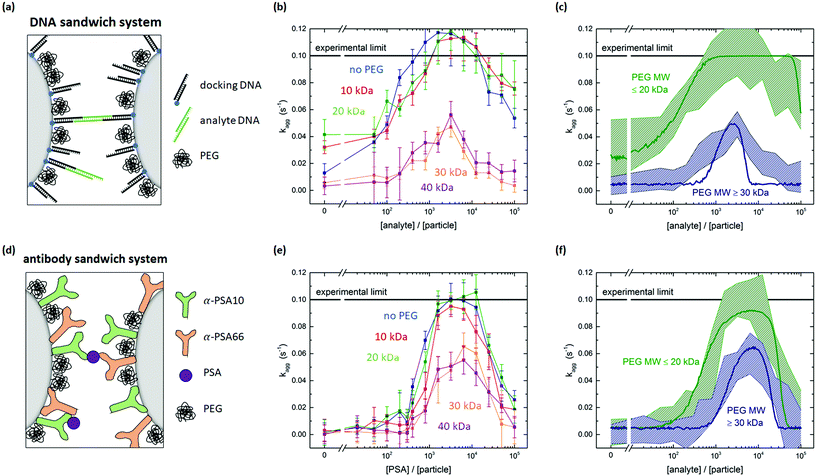 | ||
| Fig. 4 Tuning particle surface reactivity using PEG surface crowders. (a) DNA sandwich system including a PEG coating. (b) Measured aggregation rate for the DNA sandwich system for particles coated with biotinylated docking strands and biotin-PEG of different molecular weights. For PEG MW ≥30 kDa, the aggregation rates clearly decrease. (c) Experimental data plotted in two bands. A low MW PEG band (≤20 kDa) and a high MW PEG band (≥30 kDa). The curves represent simulations using input parameters shown in Table S8.† (d) Antibody sandwich system including a PEG coating. (e) Measured aggregation rate for the antibody sandwich system, for particles coated with a matching pair of PSA antibodies and amine-PEG of different molecular weights. For PEG MW ≥30 kDa the aggregation rates clearly decrease. (f) Experimental data plotted in two bands. A low MW PEG band (≤20 kDa) and a high MW PEG band (≥30 kDa). The curves represent simulations using input parameters shown in Table S8.† | ||
Fig. 4b shows the measured aggregation rate as a function of the analyte-to-particle ratio, for DNA particles coated with 10, 20, 30 or 40 kDa PEG or without PEG. The 10 and 20 kDa PEG curves are very similar to the measurement without PEG, with a small shift of the 10 and 20 kDa curves to higher analyte concentrations. Such a shift of the curve along the concentration axis has been observed in the parameter scan of the simulation for a decreasing association rate kon. Consequently, one might hypothesize that the presence of the PEG close to the docking strands may hinder the association of analytes on the docking strands.
The nonspecific interaction (at zero analyte concentration) is higher for the 10 and 20 kDa PEG coated particles compared to the 0, 30 and 40 kDa PEG coated particles, possibly caused by nonspecific PEG-DNA interaction.17 The Flory radius of a 10 kDa PEG molecule (2RF = 18 nm) is similar to the length of the docking strands (Ldock ≈ 16 nm) on the particle, which makes interaction between the PEG on one particle and the DNA on the other particle possible. For the 20 kDa PEG (2RF = 28 nm), the 30 kDa PEG (2RF = 36 nm) and the 40 kDa PEG (2RF = 42 nm), the Flory radius significantly exceeds the length of the docking strand, such that steric repulsion between the PEG molecules may prevent nonspecific aggregation. However, for the 20 kDa PEG the nonspecific aggregation rate is still high. The confinement of the particles due to the attractive magnetic dipole–dipole force may deform the steric barrier and still allow for nonspecific interactions between the particles in a dimer.
The specific aggregation rate significantly decreases for the 30 and 40 kDa PEG, see Fig. 4b. By adding the PEG molecules to the particle surface, steric hindrance occurs and the aggregation rate between the particles decreases. The reduction of the specific inter-particle binding rate represents a reduction of the specific inter-particle surface reactivity. We attribute the reduced particle reactivity to the steric hindrance caused by the PEG molecules. The steric hindrance itself is a nonspecific effect, but it has important consequences for the specific binding, because the hindrance reduces the overlap probability of the complementary molecules on the respective particles.
To interpret the effect of the PEG molecules on particle surface reactivity, the molecular rate of bond formation kchem was varied in the simulation. In the simulations, the kon and kchem values that were obtained in the previous section are used as the basis for the low MW data. The nonspecific aggregation rate is obtained from the measurements of Fig. 4b and the association rate is chosen to be kon = 1.5 × 105 M−1 s−1 to match with the experimental data. Table S8† shows an overview of the used simulation parameters. The experimental data of the low molecular weight PEG (≤20 kDa) and high molecular weight PEG (≥30 kDa) is matched with the simulations, see Fig. 4c. The green and blue bands show the error margin of the experimental data and the green and blue lines are simulated aggregation rate curves. The simulated curve for the high MW data is obtained from the low MW simulation by only changing the molecular rate of bond formation from kchem = 5.0 × 10−4 μm2 s−1 to kchem = 6.5 × 10−7 μm2 s−1. This indicates that the effect of the PEG coating on the particle can be described as an effective decrease of the intrinsic molecular binding rate between the DNA molecules, by almost three orders of magnitude.
For the antibody sandwich system, the antibody concentration during functionalization was kept constant at 11.25 μg mL−1. The simulations of the previous section showed that for this antibody concentration the antibody density is σAB = 3.8 × 103 μm−2. This corresponds to 19–57% of the geometrically expected maximum surface coverage, for upright or side-on orientation respectively. The remainder of the surface was blocked with amine-PEG molecules. Fig. 4e shows the measured aggregation rate as a function of the PSA-to-particle ratio, for particles coated with 10, 20, 30 or 40 kDa PEG or without PEG. The nonspecific interaction is lower compared to the DNA sandwich system and does not depend on the molecular weight of the PEG. The curves show a similar trend with PEG molecular weight as was observed for the DNA sandwich system. PEG molecular weights ≥30 kDa cause a significant decrease in the aggregation rate. This observation is supported by the maximum bond length being similar in both systems: Lbond ≈ 31 nm for the docking-analyte-docking complex and Lbond ≈ 35 nm for the antibody sandwich.
Fig. 4f shows the experimental bands and the simulated curves for the low MW PEG (≤20 kDa) and the high MW PEG (≥30 kDa) for the antibody sandwich system. Again, by changing the molecular rate of bond formation kchem, the simulation can be matched with the experimental data. The effective decrease in molecular binding rate for the antibody sandwich system is from kchem = 9.5 × 10−6 μm2 s−1 to kchem = 1.4 × 10−6 μm2 s−1, which is a significantly lower decrease than for the DNA sandwich system. This might be explained by a significantly lower PEG density is on the DNA coated particles compared to the antibody coated particles, originating from the different coupling chemistries. To test this hypothesis, the effect of PEG density on the reduction in particle reactivity should be studied more thoroughly.
Conclusions
We described aggregation rate measurements on biofunctionalized particles, for a DNA sandwich system and an antibody sandwich system, using an optomagnetic cluster experiment. Particles were coated with specific binders, either DNA docking strands or a matching pair of antibodies. The kinetics of particle aggregation was measured in the presence of analyte molecules, from which the particle surface reactivity was quantified.Subsequently, we studied the influence of crowder molecules on the specific interactions between particles. Particles were coated with a mixed surface functionalization of specific binder molecules and PEG as a passive surface crowder. The aggregation rates were measured for a wide range of analyte concentrations and several different molecular weights of the PEG molecules. For high MW PEG (≥30 kDa), the specific aggregation rate significantly decreases with respect to the low MW PEG (≤20 kDa), indicating that the crowder molecules tune the particle surface reactivity. Aggregation rate simulations were compared with the experimental results, showing that the decrease in particle surface reactivity by the high MW PEG molecules can be modelled as an effective decrease in the molecular rate of bond formation.
The results of this paper show that it is possible to tune the particle surface reactivity using passive surface crowders in systems with strong molecular affinity, namely 15 bp DNA hybridization and antibody–antigen interaction. The effect of the neutral crowder on the particle aggregation process will depend on the repulsive force of the crowder layer relative to the strength of the specific inter-particle binding. Further research should aim to map out what level of steric repulsion by the surface crowders is needed to affect the aggregation process of a certain biomolecular interaction, in terms of the nature and density of the molecular binders and the neutral crowders. To our knowledge, this is the first study that quantifies the effect of passive surface crowders on the specific reactivity of biofunctionalized particles. The results reveal a novel way to tune the specific reactivity, namely by adding passive surface crowder molecules on the surface, which gives a new dimension for the control of properties of colloidal particles for biological applications.
Experimental section
Materials
Carboxylated superparamagnetic Masterbeads were purchased from Ademtech SA (diameter 528 nm, coefficient of variation 25%). Streptavidin coated superparamagnetic silica particles were obtained from MircoParticles GmbH (diameter 511 nm, coefficient of variation <5%). Biotinylated DNA docking strands and DNA analyte strands were purchased from Integrated DNA Technology Inc. (IDT). For a complete list of the used DNA sequences, see ESI section S2.† Prostate specific antigen (PSA) was purchased from Abcam plc. and a matching pair of monoclonal mouse IgGs against PSA (α-PSA10 and α-PSA66) were supplied by Fujirebio Europe N.V. Amine-terminated polyethylene glycol (PEG) and biotin-terminated PEG with molecular weights of 5, 10, 20, 30 and 40 kDa were purchased from Creative PEGworks. Phosphate buffered saline (PBS) tablets, Pluronic F-127, 2-(N-morpholino)ethanesulfonic acid (MES), 1-ethyl-3-(dimethylaminopropyl)carbodiimide hydrochloride (EDC), tris(hydroxymethyl)aminomethane (tris), N-hydroxysulfosuccinimide (sulfo-NHS), bovine serum albumin (BSA, >98% pure), biotin-Atto655 and DNA LoBind and Protein LoBind Eppendorf tubes were obtained from Sigma Aldrich Inc. Borosilicate glass 3.3 cuvettes with inner dimensions of 1.00 ± 0.05 mm × 1.00 ± 0.05 mm and outer dimensions of 1.23 ± 0.05 mm × 1.23 ± 0.05 mm, and a length of 20 ± 1 mm were obtained from Hilgenberg GmbH and used in the optomagnetic cluster experiment.Particle functionalization for DNA sandwich system
Streptavidin coated silica MicroParticles were functionalized with biotinylated DNA docking and DNA filler strands and biotinylated PEG (10, 20, 30 or 40 kDa) by sequential incubation steps. First, 15 μL of the particle stock solution (10 mg mL−1) was mixed with 285 μL of DNA docking strand solution in PBS, and incubated for 60 minutes in an incubator shaker (1200 rpm, room temperature). Subsequently 2 μL of a large excess of DNA filler strands is added to saturate the particle surface with DNA, and incubated for 60 minutes in an incubator shaker (1200 rpm, room temperature). The amount of functional docking DNA during the first incubation step was varied throughout the experiments.For measuring the effect of PEG on the aggregation rate, first DNA docking strand were coated on the particle, and the docking-DNA-to-particle-ratio was kept equal to 5000. After the first incubation step, the particle solution was magnetically washed to remove unbound DNA docking strands. The particles were redispersed in a 1 mM biotin-PEG solution of a certain molecular weight and incubated for 60 minutes in an incubator shaker (1200 rpm, room temperature) to saturate the remaining streptavidin groups with biotin-PEG.
After the second incubation step, the particle solution was magnetically washed again to remove the unbound PEG. The particles were redispersed in a 10 mg mL−1 BSA in PBS solution to suppress nonspecific aggregation of the particles. The particle solution was then incubated in a sonic bath for 10 minutes and the solution was sonicated (10 × 0.5 s) to reduce the number of clusters in the solution.
Quantification of the DNA docking strand coverage
To quantify the number of DNA docking strands on the streptavidin coated Microparticles an indirect fluorescence supernatant assay is performed. First, the biotin capacity of the particles is quantified by binding increasing amounts of biotin-atto655 on the particles, during 60 minutes in an incubator shaker (1200 rpm, room temperature). The lowest b-atto655 concentration at which there is still b-atto655 left over in the supernatant after incubation is quantified using a Fluoroskan Ascent FL (λex = 646 nm, λem = 679 nm, spectral width 5 nm). The b-atto655 capacity per particle is equal to Nb-atto655 = (4.3 ± 0.5) × 104.Next, increasing amounts of biotinylated DNA are added to the particles and incubated during 60 minutes in an incubator shaker (1200 rpm, room temperature). After this incubation step, the particle solution is magnetically washed to remove unbound DNA strands. Subsequently, b-atto655 is added in a concentration that is slightly above the b-atto655 capacity of the particles. Again an incubation step is performed during 60 minutes in an incubator shaker (1200 rpm, room temperature). Particles that are not fully coated with DNA strands will bind some of the b-atto655 in the solution, the fully coated particles will not bind b-atto655. After the incubation the particle solutions are magnetically washed and the fluorescence of the supernatant is measured. The amount of b-atto655 in the supernatant for a certain DNA concentration, Is.n.([DNA]), is related to the number of DNA on the particle, NDNA, according to eqn (4).
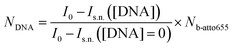 | (4) |
Here I0 is the fluorescence intensity of the b-atto655 solution that is added in the second incubation step, and Is.n.([DNA] = 0) is the fluorescence intensity of the supernatant when adding non-functionalized streptavidin Microparticles. For more details of the supernatant assay, see ESI section S3.†
Particle functionalization for the antibody sandwich system
Carboxylic acid coated Ademtech Masterbeads were functionalized through an EDC-NHS reaction with a matching pair of monoclonal PSA antibodies that target a different epitope on the PSA antigen. The particle surface area in between the antibodies area was coated with an amine-terminated PEG (10, 20, 30 or 40 kDa) and the remaining surface area was blocked with tris. All steps were performed at room temperature.First, 4 μL of particle stock solution (50 mg mL−1) was mixed with 196 μL a 100 mM MES solution (pH 5.0), 50 μL EDC in MES solution (10 mg mL−1) and 50 μL NHS in MES solution (10 mg mL−1). The particles were incubated for 30 minutes on a roller bench to activate the carboxyl groups on the particle surface.
After the activation step the particles were magnetically washed twice and redispersed in 200 μL MES solution. Subsequently the particle solution was sonicated (10 × 0.5 s) to undo the particle clustering that occurred during the magnetic washing steps. Then, 100 μL of antibody solution was added to the particles and incubated for 30 minutes on a roller bench, to covalently attach the antibodies to the particles. The antibody concentration was varied in one of the experiments. However, if not specified in the text, the antibody-to-particle ratio during incubation was kept equal to 1500.
After the antibody coating step 50 μL of amine-terminated PEG solution (1 mM) was added to the particle solution and incubated for 180 minutes on a roller bench. To block the remaining active carboxyl groups, 100 μL of tris buffer is added (150 mM NaCl, 50 mM tris, pH 7.6) and incubated overnight on a roller bench. Finally, the particle solution is magnetically washed twice and the particle are redispersed in a 10 mg mL−1 BSA in PBS solution to suppress nonspecific particle aggregation. Finally, the particle solution was sonicated (10 × 0.5 s) to undo the particle clustering that occurred during the functionalization process.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the Peri-Operative IMPULS program for their financial support.References
- A. Ranzoni, G. Sabatte, L. J. van IJzendoorn and M. W. J. Prins, One-step homogeneous magnetic nanoparticle immunoassay for biomarker detection directly in blood plasma, ACS Nano, 2012, 6(4), 3134–3141 CrossRef CAS PubMed.
- D. R. Davies and G. H. Cohen, Interactions of protein antigens with antibodies, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 7–12 CrossRef CAS PubMed.
- A. J. Bonham, G. Braun, I. Pavel, M. Moskovits and N. O. Reich, Detection of sequence-specific protein-DNA interactions via surface enhanced resonance Raman scattering, J. Am. Chem. Soc., 2007, 129(47), 14572–14573 CrossRef CAS PubMed.
- M. D. Wang, H. Yin, R. Landick, J. Gelles and S. M. Block, Stretching DNA with optical tweezers, Biophys. J., 1997, 72(3), 1335–1346 CrossRef CAS.
- M. Jing and M. T. Bowser, Methods for measuring aptamer-protein equilibria: A review, Anal. Chim. Acta, 2011, 686, 9–18 CrossRef CAS PubMed.
- R. L. Srinivas, S. C. Chapin and P. S. Doyle, Aptamer-functionalizes microgel particles for protein detection, Anal. Chem., 2011, 83(23), 9138–9145 CrossRef CAS PubMed.
- J. Müller, K. N. Bauer, D. Prozeller, J. Simon, V. Mailänder, F. R. Wurm, S. Winzen and K. Landfester, Coating nanoparticles with tunable surfactants facilitates control over the protein corona, Biomaterials, 2017, 115, 1–8 CrossRef PubMed.
- T. Gillich, C. Acikgöz, L. Isa, A. D. Schlüter, N. D. Spencer and M. Textor, PEG-stabilized core-shell nanoparticles: Impact of linear versus dendritic polymer shell architecture on colloidal properties and the reversibility of temperature induced aggregation, ACS Nano, 2013, 7(1), 316–329 CrossRef CAS PubMed.
- L. L. Sorret, C. R. Monticello, M. A. DeWinter, D. K. Schwartz and T. W. Randolph, Steric repulsion forces contributed by PEGylation of interleukin-1 receptor antagonist reduce gelation and aggregation at the silicon oil-water interface, J. Pharm. Sci., 2019, 108, 162–172 CrossRef CAS.
- N. Helfricht, A. Mark, L. Drowling-Carter, T. Zambelli and G. Papastavrou, Extending the limits of direct force measurements: colloidal probes from sub-micron particles, Nanoscale, 2017, 9, 9491 RSC.
- P. Schein, P. Kang, D. O'Dell and D. Erickson, Nanophotonic force microscopy: Characterizing particle-surface interactions using near field photonics, Nano Lett., 2015, 15, 1414–1420 CrossRef CAS PubMed.
- K. van Ommering, M. Koets, R. Paesen, L. J. van IJzendoorn and M. W. J. Prins, Bond characterization by detection and manipulation of particle mobility in an optical evanescent field biosensor, J. Phys. D: Appl. Phys., 2010, 43, 385501 CrossRef.
- Z. Wang, C. He, X. Gong, J. Wang and T. Ngai, Measuring the surface-surface interactions induced by serum proteins in a physiological environment, Langmuir, 2016, 32, 12129–12136 CrossRef CAS PubMed.
- P. L. Biancaniello and J. C. Crocker, Line optical tweezers instrument for measuring nanoscale interactions and kinetics, Rev. Sci. Instrum., 2006, 77, 113702 CrossRef.
- M. R. W. Scheepers, A. R. Romijn, L. J. van IJzendoorn and M. W. J. Prins, Rate of dimer formation in stable colloidal solutions quantified using an attractive interparticle force, Langmuir, 2019, 35, 10533–10541 CrossRef CAS PubMed.
- S. Xu, J. Zhan, B. Man, S. Jiang, W. Yue, S. Gao, C. Guo, H. Liu, Z. Li, J. Wang and Y. Zhou, Real-time reliable determination of binding kinetics of DNA hybridization using a multi-channel graphene biosensor, Nat. Commun., 2017, 8, 14902 CrossRef CAS PubMed.
- D. B. Knowles, A. S. LaCroix, N. F. Deines, I. Shkel and M. Thomas Record Jr., Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(31), 12699–12704 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Optomagnetic cluster experiment; list of used DNA strands; supernatant assay for DNA docking strand quantification; antibody sandwich system control experiment; simulation parameter scan; input parameters for simulation of varying binder density; association rate dependence for high docking strand coverage; input parameters for simulation of varying PEG molecular weight. See DOI: 10.1039/d0nr03125a |
| This journal is © The Royal Society of Chemistry 2020 |