 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correlative cathodoluminescence electron microscopy bioimaging: towards single protein labelling with ultrastructural context
Kerda
Keevend
 ab,
Toon
Coenen
c and
Inge K.
Herrmann
ab,
Toon
Coenen
c and
Inge K.
Herrmann
 *ab
*ab
aLaboratory for Particles Biology Interactions, Swiss Federal Laboratories for Materials Science and Technology (Empa), Lerchenfeldstrasse 5, CH-9014, St Gallen, Switzerland. E-mail: inge.herrmann@empa.ch; Tel: +41 (0)58 765 7153
bNanoparticle Systems Engineering Laboratory, Department of Mechanical and Process Engineering, ETH Zurich, Sonneggstrasse 3, 8092 Zurich, Switzerland. E-mail: ingeh@ethz.ch
cDelmic B.V., CL solutions, Kanaalweg 4, 2628 EB, Delft, The Netherlands
First published on 10th July 2020
Abstract
The understanding of living systems and their building blocks relies heavily on the assessment of structure–function relationships at the nanoscale. Ever since the development of the first optical microscope, the reliance of scientists across disciplines on microscopy has increased. The development of the first electron microscope and with it the access to information at the nanoscale has prompted numerous disruptive discoveries. While fluorescence imaging allows identification of specific entities based on the labelling with fluorophores, the unlabelled constituents of the samples remain invisible. In electron microscopy on the other hand, structures can be comprehensively visualized based on their distinct electron density and geometry. Although electron microscopy is a powerful tool, it does not implicitly provide information on the location and activity of specific organic molecules. While correlative light and electron microscopy techniques have attempted to unify the two modalities, the resolution mismatch between the two data sets poses major challenges. Recent developments in optical super resolution microscopy enable high resolution correlative light and electron microscopy, however, with considerable constraints due to sample preparation requirements. Labelling of specific structures directly for electron microscopy using small gold nanoparticles (i.e. immunogold) has been used extensively. However, identification of specific entities solely based on electron contrast, and the differentiation from endogenous dense granules, remains challenging. Recently, the use of correlative cathodoluminescence electron microscopy (CCLEM) imaging based on luminescent inorganic nanocrystals has been proposed. While nanometric resolution can be reached for both the electron and the optical signal, high energy electron beams are potentially damaging to the sample. In this review, we discuss the opportunities of (volumetric) multi-color single protein labelling based on correlative cathodoluminescence electron microscopy, and its prospective impact on biomedical research in general. We elaborate on the potential challenges of correlative cathodoluminescence electron microscopy-based bioimaging and benchmark CCLEM against alternative high-resolution correlative imaging techniques.
Introduction
Microscopy has enabled some of the most disruptive scientific discoveries, including the discovery of pathogenic microbes and viruses. Despite rapid developments in the last 50 years in the field of electron and scanning probe microscopy, most of the microscopy investigations in biology still rely on light microscopy (LM). LM is instrumental for biology, as it is the only microscopic method enabling fast routine live-cell monitoring. The non-invasive nature, simplicity, the ease of sample preparation and the optical transparency of cells make LM particularly attractive for a variety of experimental investigations in the fields of biology and biomedicine.1 Fluorescence microscopy (FM) in particular allows simultaneous visualization of several cellular entities, including nucleic acids, proteins, and lipids by using fluorescent labels, such as organic fluorophores, semiconductor nanocrystals, and fluorescent proteins.2 FM has become a genuine workhorse technique in the overwhelming majority of biological research labs. However, the spatial resolution of traditional fluorescence microscopy is limited by Abbe's diffraction limit due to the wave nature of the light.1,2 As a consequence, nanoscale light-emitting objects are detected as a finite-sized spot, also known as the point-spread function (PSF). The PSF is a Gaussian-like intensity distribution of small (point-like) objects in the image and its width is quantified by the full-width-half-maximum (FWHM). The FWHM in a traditional lens-based system is typically 200–300 nm in the focal plane and strongly increases, reaching around 500–800 nm, in the axial plane.3 Biomolecules are however, in the range of a couple of nanometers,4 and as such cannot be resolved by traditional fluorescence microscopy.1–3Pushing the limits: sub-diffraction optical imaging of biological samples
For decades, researchers tried to overcome the diffraction limit by using near-field optical microscopy (NOM) methods based on the detection of non-propagating (evanescent) light waves close to the sample surface by using nano-sized tips.1,5 However, NOM is mostly surface sensitive and cannot visualize internal subcellular structures. Therefore, in biology, far-field techniques are preferred and have given rise to the development of super-resolution (SR) techniques, also known as optical nanoscopy, which aim to improve the resolution of traditional lens-based fluorescence microscopy.1 Super-resolution techniques can be broadly divided into two groups:6 (i) methods that use patterned illumination to control the emission behavior of fluorescent molecules, like Structured Illumination Microscopy (SIM)7 and Stimulated Emission Depletion (STED) microscopy,8 and (ii) methods that use photo-switching or other mechanisms to stochastically activate single molecules at different times and reconstruct images of measured positions of individual fluorophores, like Stochastic Optical Reconstruction Microscopy (STORM),9,10 Photo-Activated Localization Microscopy (PALM)11 and Stochastic Optical Fluctuation Imaging (SOFI).12,13 Super-resolution techniques have led to significant improvements in resolution, readily achieving lateral resolution of ≈10–40 nm and axial resolution well below 100 nm, in some cases even ≈50 nm.1,3 Balzarotti et al.14 recently demonstrated ‘MINIFLUX’, which allows localization of fluorescently labelled molecules with 6 nm lateral resolution and established three-dimensional (3D) and multi-color imaging with the same system.15 SR-FM requires specific fluorescent properties, like photoactivation, stability, high quantum yield and brightness, which have been extensively studied and optimized for hydrated conditions.16–18 While conventional FM is routinely employed for dynamic live cell imaging, SR-FM is mostly performed on fixed samples due to high laser power, long image acquisition times and lack of suitable probes.10,19 The most fundamental limitation of optical nanoscopy is the achievable labelling density with respect to the required resolution. Often the required labelling densities are not achievable in biological samples without altering their function.3,20 Nevertheless, commercially available super-resolution setups are widely adopted in biomedical research. Super-resolution techniques are discussed in more detail in recent reviews.2,3,21While fluorescence microscopy is an irreplaceable method for functional studies, the context is often missing as the majority of cell constituents remain unlabeled.22 Therefore, insight into cellular ultrastructure plays a crucial role in understanding and contextualizing the LM data. Electron microscopy (EM) is a powerful technique for visualizing the cellular ultrastructure. Electron microscopes operate in high vacuum conditions and biological samples must undergo laborious sample preparation routine (fixation, dehydration, staining, resin embedding) to make them compatible for such harsh conditions. In the conventional sample preparation route, (large) biological samples are chemically fixed using strong fixatives that cross-link the biomolecules. This is followed by the staining with heavy metals that result in non-specific membrane staining, dehydration and resin embedding, i.e. impregnation with liquid (typically hydrophobic) resin, which is then polymerized into a solid block. The cured polymer blocks are trimmed and sectioned by ultramicrotomy before subjecting the samples to EM investigations (Fig. 1).17,23–26 The localization of specific biomolecules, e.g. lipids and proteins, is only achievable using additional electron dense labels, such as immunogold.22,27 Immunogold consists of small functionalized gold nanoparticles, which are conjugated to antibodies for specific targeting and is nowadays widely adopted in electron microscopy. Immunolabeling has been demonstrated both in pre28- and post-embedding (on section)27 protocols. Even though the immunogold method has been around for decades, it still faces challenges. Despite fascinating achievements and localization of multiple targets based on differently sized and shaped immunolabels,29,30 immunogold has limited feasibility for co-localization studies (i.e. simultaneous observation of several epitopes)28,31 due to variations in sample penetration properties. This limitation is partially overcome by using quantum dot-based immunolabelling, which show superior tissue penetration.30,32 Additionally, the differentiation of the small gold nanoparticles from other (endogenous) electron dense structures is problematic.33
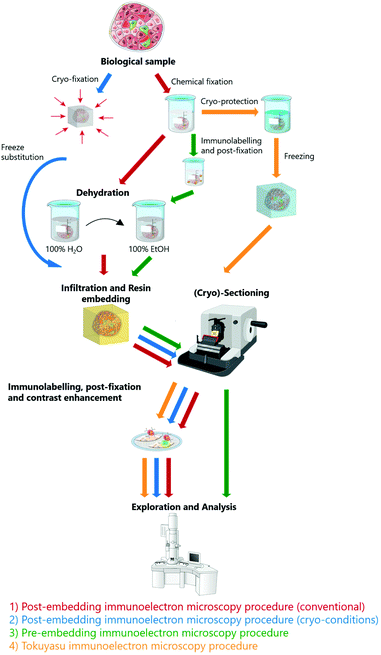 | ||
| Fig. 1 The most commonly used electron microscopy sample preparation techniques including immunolabeling protocols. Protocols adapted from Guo et al.24 | ||
EM sample preparation requires careful balance between structural integrity and antigenicity.26 The Tokuyasu method is often employed for preparation of EM immunolabeled samples. The live samples are chemically fixed, dehydrated using cryoprotection (sucrose) and frozen for cryo-sectioning. Subsequently, the obtained sections are thawed or further embedded and immunolabeled using external antibody-based labels.22,34 However, chemical fixation is slow and associated with structural changes35,36 and more often than not, alternatives such as fast (plunge) freezing (vitrification) are preferred to preserve a sample in its near-native state.37 Small samples (protein solutions, purified virus samples or cellular monolayers) applied directly to the EM grid are plunged into liquid ethane, which quickly freezes the sample and embeds it to amorphous ice.25 For larger samples, high-pressure freezing (HPF) becomes more beneficial, as the living samples are simultaneously exposed to high pressure (2100 bar) and rapidly cooled to liquid nitrogen temperature. The samples can be further freeze-substituted and embedded to resin at low temperature.38,39 HPF excellently preserves biological ultrastructure, but fixation of larger samples (>200 μm thickness) is limited by the formation of ice crystals, which hinders whole tissue investigations.25,38
Combining light and electron microscopy data enable to gain true insight into functional context.40 While traditional correlative light and electron microscopy (CLEM) suffers from the two orders of magnitude resolution mismatch between the two methods, correlative super-resolution light and electron microscopy (CSREM) partially overcomes this gap thanks to significantly improved resolution of the optical signal.23,27,41
Since the first demonstration of CSREM,11 microscopy hardware has rapidly developed for both imaging modalities leading to fascinating new imaging capabilities, that are extensively reviewed by Hauser et al.17
The main remaining challenges are thus oftentimes linked to the incompatibility of LM and EM sample preparation protocols.23,42 On one hand, fluorescence microscopy samples require mild chemical fixation and permeabilization of the cells prior to the labelling, which diminishes ultrastructure preservation.22,42,43
On the other hand, EM sample preparation requires strong fixatives (like glutaraldehyde and osmium tetroxide), which obscures epitopes for antibody labelling and quenches fluorescence.43,44 Therefore, overcoming these challenges necessitates careful strategies and non-traditional probes.23,43,45 For example, recent developments in genetically encoded tags developed for CLEM/CSREM allow use of the conventional EM sample preparation route because of new osmium resistant photoactivatable fluorescent proteins, which enable super resolution microscopy on resin embedded sections.44,46 Many (SR)-CLEM methods require fluorescent imaging prior to the EM sample preparation,47 while others have adapted EM sample preparation protocols by reducing the amount of strong fixatives in order to preserve fluorescence.39,43 However, the modifications on the embedding protocols oftentimes result in critical structural alterations and poorly preserved ultrastructure, which makes correlation between LM and EM images even more problematic.23 Cryo-fixation is a superior method for ultrastructure preservation, but cryo-SR-FM is still challenging. First, cryo-SR-FM requires special fluorophores to exploit blinking phenomena at low temperature.48 Although cryogenic conditions increase fluorophore photostability and dark state lifetime enabling high contrast,49,50 the underlying photophysics of SR fluorophores in cryo-conditions is poorly understood. Second, the most used single molecule localization techniques require high laser powers, causing local devitrification of the samples, which may result in sample damage.51,52 The vitrification can be alleviated by lowering the laser intensity52 or introducing cryoprotectants.51 Additionally, only the lower numerical aperture (NA) air lens could be used and it must be separated from the cryo-sample resulting in reduced light collection and resolution.23,37,53 Wang et al.54 recently overcame this problem by using solid immersion lens for multi-color STORM demonstrating single molecule resolution (12 nm) in cryo-conditions. Hoffman et al.50 combined cryo-SR imaging prior to EM sample preparation and demonstrated whole cell 3D CSREM using 3D-SIM and FIB/SEM techniques. However, performing FL imaging prior to the EM sample preparation could cause difficulties in image alignment and critical structural alterations as the EM sample preparation causes nonlinear and spatially inhomogeneous sample deformations.50 Instead of performing cryo-SR-FM before the EM sample preparation, freeze-substituted samples enable SR-FM imaging in ambient temperatures using high NA objectives.55,56 Nevertheless, SR probes are optimized for hydrated conditions and their fluorescence emission after dehydration and resin embedding is usually compromised.17,39
Generating light by focused electron beams: cathodoluminescence electron microscopy
Rather than collecting optical information with super-resolution light microscopy based techniques, sub-diffraction resolution can also be obtained by measuring photon emission resulting from the interactions of the samples with accelerated electrons, a phenomenon known as cathodoluminescence (CL).57,58 The primary electron beam generates various signals upon interaction with the sample, including secondary (SE) and backscattered electrons (BSE), characteristic X-rays (EDX), and luminescence (Fig. 2a). The aforementioned signals can be used for imaging biological samples. Depending on the sample preparation and imaging conditions, biological specimens can be imaged in either ambient or cryogenic conditions using scanning or scanning transmission mode (Fig. 2b). Cathodoluminescence is usually collected by a parabolic mirror and detected by a photodetector or spectrometer. Collection of all the emitted signals by the sample in response to electron beam interactions allows simultaneous analysis of sample morphology, composition, and luminescence properties. CL has been implemented to scanning electron microscopes (SEM) (Fig. 2c and d)59–61 and scanning transmission electron microscopes (STEM) (Fig. 2e).57,62–64 Recently, systems equipped with a pulsed electron beam65 (Fig. 2d) in conjunction with time-resolved CL detection66 or integrated light-electron microscopes65,67,68 (Fig. 2f) have gained attention, as they allow studies on complex dynamics of the materials.66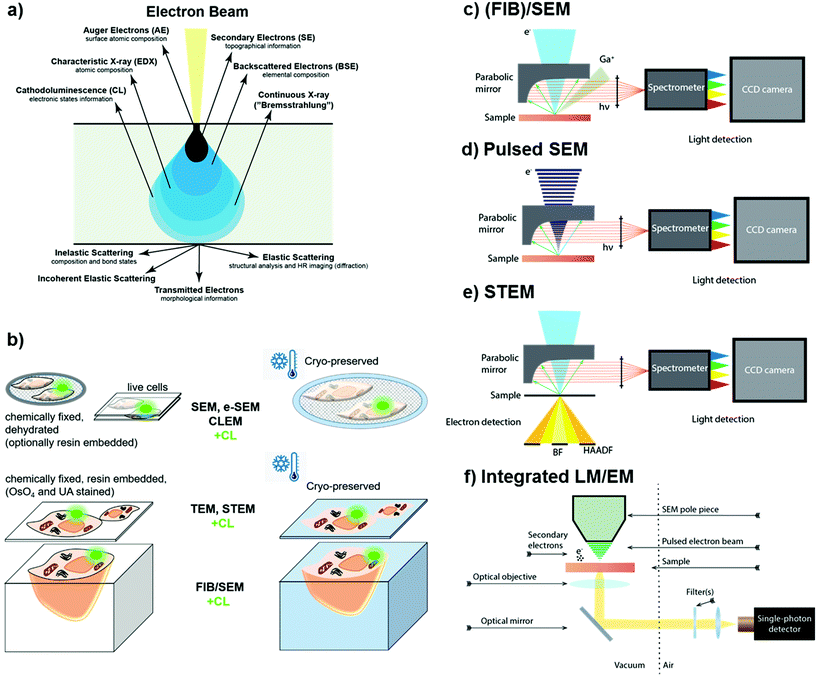 | ||
| Fig. 2 (a) Generation of various signals via electron beam – matter interactions, including secondary (SE), backscattered (BSE), and Auger (AE) electrons, characteristic X-rays (EDX) as well as cathodoluminescence (CL). (b) Different sample geometries and preparation methods suitable for the different electron microscopy configurations. CL acquisition setups using (c) a scanning electron microscope (SEM) including focused ion beam (FIB)/SEM,58,59 (d) a pulsed SEM,66 (e) a scanning transmission electron microscope (STEM),63 and (f) an integrated light and electron microscope.65 | ||
Cathodoluminescence can be generated by distinct physical processes that are commonly divided into two main categories: incoherent and coherent CL emission.58,69 Incoherent emission is typically generated within the medium and is incoherent, unpolarized and isotropic. Although this excitation mechanism for cathodoluminescence is comparable to other luminescence phenomena, e.g. photoluminescence,70 electron beam excitation generally leads to emission by all the luminescence mechanisms present in the material since the electrons act as a supercontinuum source.58,70 In photoluminescence, the emission is strongly dependent on the excitation energy hν.70 Systems containing impurities and defects, such as rare-earth ion doped nanocrystals and nitrogen-vacancy centers in diamonds, usually produce incoherent CL with multiple emission peaks.58 Coherent CL emission on the other hand has a fixed phase relation with the incoming electrons and is strongly polarized. Coherent transition radiation emission is always generated when there is a refractive index contrast with vacuum. Although coherent CL is generated in all materials, the intensity is usually orders of magnitude weaker compared to incoherent CL. Typically, (metallic) plasmonic systems are characterized by their coherent CL emission, as incoherent CL in these materials is diminished by non-radiative processes.58 This review focuses on the use of incoherent CL in biological samples. More comprehensive overviews on CL in general can be found in the literature.58,69,70
The advantage of CL over traditional fluorescence methods is its spatial resolution in nanometer range due to the highly localized excitation, which is mainly driven by three factors: (i) the electron beam spot size, (ii) the generation volume of the signal, and (iii) the propagation of generated secondary charge carriers within the sample.70 However, depending on the imaging method, e.g. SEM or STEM, all the above mentioned factors contribute differently to the resolution. An electron beam can be focused down to a nanometer scale beam waist in modern electron microscopes. Note that scanning transmission electron microscopes normally operate with smaller spot size compared to scanning electron microscopes. The spot size of an electron beam is mostly determined by the beam current; a smaller spot size is achieved by using lower electron beam current.71 At a given current, higher acceleration voltages results in smaller spot sizes, however, lead to increased CL generation volumes in bulk samples.70,71 Therefore, in SEM the resulting increased interaction volume and lateral spread significantly reduces CL spatial resolution when operating at high voltages.61 In STEM, the generation volume is primarily determined by the thickness of the sample, which is typically between 70 and 100 nm.57 Calculations have shown that the diffusion length of the secondary charge carriers in biological samples is small and the light generation predominantly occurs within the electron beam excitation volume. This implies that the resolution of the CL in biological samples does not hinge on the carrier diffusion length.70 Depending on the sample, the chosen instrumentation, and the imaging conditions, CL spatial resolution well below the Abbe diffraction limit are well within reach.61,72
(Single protein) labels: nanocrystals and nanodiamonds
Historically, the CL phenomenon has mostly been exploited for the analysis of minerals and semiconductors, even though first reports on CL in biological samples date back to the 1970s, where Pease et al.73 enhanced the contrast of spinach leaves by absorbing thioflavin dyes into cell walls (Fig. 3a). This early study gave rise to follow-up studies exploring potential cathodoluminescence dyes.74,75 However, CL emission from these samples was poor and CL microscopy was constrained to niche applications in soft matter and life science. Only with the emergence of luminescent inorganic nanocrystals featuring bright and narrow emission, correlative cathodoluminescence electron microscopy (CCLEM) bioimaging has gained increasing attention.59,63 Especially the prospect that CL spectra can be acquired across a wide spectral range to enable identification of specific protein labels directly based on spectral signatures (Fig. 3b) rather than their electron density and size (as in the case of immunogold labelling) gives significant appeal to CL bioimaging.76,77 However, CCLEM still faces obstacles related to poor CL signal as well as the electron beam damage to the biological samples78 and the labels themselves74 due to relatively high energies required for the excitation of existing luminescent labels. There is an urgent need for bright and stable imaging probes for molecular labelling applications in CCLEM setting.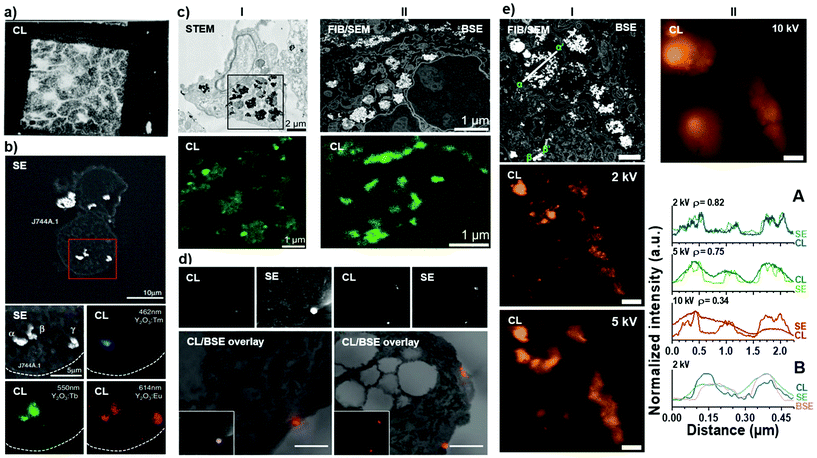 | ||
| Fig. 3 Seminal examples of CL bioimaging. (a) First demonstration of cathodoluminescence on biological samples by Pease et al.73 demonstrating CL emission from thioflavin dyes absorbed into spinach leaves. Reprinted by permission from Nature/Springer: Nature (Scanning Electron Microscopy of Biological Material, Pease et al. 1966). (b) The first demonstration of multi-color CL of biological sample using rare-earth element doped nanocrystals by Niioka et al.77 Secondary electron and CL images of Y2O3 nanoparticles doped with Tm (blue), Tb (green) and Eu (red) ions in mouse macrophage-like cell line (J744A.1). Reprinted by permission from ©2011 The Japan Society of Applied Physics. (c) The first demonstration of CL and full ultrastructural analysis from (I) STEM and (II) FIB/SEM mode on biological samples by Keevend et al.59 (I) STEM bright-field image of resin-embedded human cells incubated with green CL-emitting LaF3:Tb3+ nanoparticles. (II) BSE image of the same sample imaged using FIB/SEM mode revealing well-preserved cellular ultrastructure and CL emission from LaF3:Tb3+ nanoparticles. Reproduced by permission of The Royal Society of Chemistry. (d) The first demonstration of immunolabelling on biological samples using CL-based label detection by Hemelaar et al.67 CL and SE images (top) demonstrating presence of bioconjugated 70 nm FND on the surface of human epithelial colon carcinoma cell clusters (HT29) and (bottom) the corresponding CL overlaid with BSE images. Scale bars: 1 μm. (e) Demonstration of cathodoluminescence resolution dependence of the electron beam acceleration voltage in FIB/SEM setting – reprinted with permission from ref. 61. (I) BSE image of YVO4:Bi3+,Eu3+ nanocrystal containing human cells in a FIB cross-section. CL images acquired with 2 and 5 kV. Co-localization studies of BSE, SE, and CL signals in two regions and the co-localization studies have been indicated (low magnification, α–α′ and high magnification, β–β′). (II) CL image acquired with 10 kV acceleration voltage and the corresponding line profiles of SE and CL signals with different acceleration voltages for low-magnification (α–α′, A) and high-magnification (β–β′, B) settings. Scale bars: 1 μm. Copyright 2019 American Chemical Society. | ||
The well-established fluorescence microscopy markers, including organic fluorophores, fluorescent proteins, and semiconductor quantum dots, are not suitable for CCLEM since their emission stability and brightness are severely limited.61,79 Organic molecules bleach rapidly under accelerated electrons due to structural damage introduced by the primary beam.74,79,80 Semiconductor quantum dots (QDs), such as CdSe/ZnS structures, which have gained increasing importance as probes in traditional fluorescence microscopy, suffer from electron ionization, followed by trapping of the ejected electrons at deep traps inside their matrix.79,81
As alternatives, rare-earth element (REE) doped nanoparticles59,64,82 (Fig. 3c) and fluorescent nanodiamonds (FNDs)65,76,83 (Fig. 3d) have been proposed as multi-color cathodoluminescence labels. In the case of the nanodiamonds a range of (nitrogen-related) radiative defects can be employed for this purpose. REE3+ doped (nano)particles can be synthesized with different lanthanide species, each with a distinct emission spectrum. REE3+ doped nanocrystals have been reported to show minimum bleaching when irradiated with high-energy electrons, even after long-term exposure.62 However, decreasing CL intensity using low acceleration voltages has been reported.59,61 Loss in emission intensity in REE3+ doped nanocrystals can be attributed to knock-on damage79,84 and the accumulation of active quenchers.61,62,76 Nonetheless, the stability of inorganic nanocrystals is still significantly higher compared to widely adopted fluorescence microscopy labels. The emission stability of nanodiamonds has not been studied in detail, but STEM studies report that they can be detected even after 2–3 hours of continuous beam exposure.85
Both REE3+ doped nanocrystals and nanodiamonds show high cytocompatibility,86,87 high emission stability, and have been employed as potential labels in fluorescence87,88 and cathodoluminescence67,77 bioimaging. The emission spectrum can be tailored, enabling multi-color imaging, by introducing impurities (e.g. REE3+)62 or engineered defects89 in the nanocrystals. Multi-color imaging is an attractive feature of CL microscopy, especially since current protein labelling techniques in EM are significantly limited in the simultaneous detection of several epitopes as a result of the diffusivity differences of differently sized gold nanoparticles.28,31,90 Thus, creation of labels with similar size but distinct emission lines helps to overcome the major immunogold limitation. Another advantage of the proposed labels is their stable luminescence emission in spite of the osmium staining or resin embedding,59,61,67 in sharp contrast to conventional labels used for CLEM and CSREM.42,44,53 This is also an attractive feature for in situ CLEM (i.e. same instrument for FL and EM data acquisition91,92), although the majority of REE3+-based nanocrystals require deep-UV excitation, which is poorly compatible with conventional imaging systems.88 CL-based multi-target imaging can alternatively be achieved by selecting labels based on their distinctly different emission lifetimes in combination with time-resolved and/or gated CL imaging.65,83 Additionally, other sources of CL, such as autofluorescence, can be filtered out based on distinctly different lifetimes.
The main challenges of the novel labels lie in their synthesis, colloidal stability, and surface functionalization. The potential markers must be in the size range of a typical protein (2–6 nm (ref. 4)), show bright emission, and have high colloidal stability. Synthesis of REE3+ doped nanocrystals has been attempted by various methods, including the flame aerosol technique,88,93 homogeneous precipitation,64 solvothermal,94 thermal decomposition,95 and microwave-assisted hydrothermal route.96,97 While many methods yield in highly crystalline nanoparticles, they also involve either collection of particles in dry state88,93 or high temperature annealing.64 As a result, nanocrystals oftentimes show comparatively poor colloidal stability,64,86 which may be improved by post-synthesis surface functionalization.86 Even though post-functionalization may significantly increase colloidal stability, it remains challenging to obtain fully monodisperse colloids, especially when the materials are irreversibly sintered.86 Synthesis in high boiling point solvents typically results in small (≤10 nm), near-perfect single crystals with narrow size distribution. However, the obtained particles are generally only dispersible in organic solvents and need further modification for actual cell targeting applications.82,95,96 Furthermore, small nanoparticles typically suffer from low brightness, as they have a high surface-to-volume ratio and a large portion of dopant is located on the surface, which is more prone to quenching (surface impurities, ligands, and solvents).95 Overcoming these drawbacks requires careful nanoparticle engineering, such as the creation of core/shell structures,98,99 to enhance surface passivation and luminescence brightness.
Additionally, luminescence emission can be enhanced by sensitizer ions, which absorb the excitation energy and transfer it to the emitting ions.95,100 For example, highly efficient energy transfer between Ce3+ and Tb3+ ions takes place as Ce3+ ions have allowed f–d absorption and broad emission band, which overlaps with Tb3+ absorption band, resulting in energy transfer and enhanced photoluminescence emission.100 These techniques can be implemented to REE3+ based nanocrystal synthesis and functionalization routes, to further increase the cathodoluminescence efficiency.
Nanodiamond-based labels may be used as an alternative to REE3+ doped nanocrystals. Initially, nanodiamonds were produced by detonation, but a wide variety of methods are used nowadays in research and industrial laboratories. These include laser ablation, plasma-assisted chemical vapor deposition (CVD), high-energy ball-milling of high-pressure high-temperature (HPHT) diamond microcrystals, as well as hydrothermal synthesis routes.101 While the combustion method yields in clusters of nanodiamonds, several methods have been developed, which result in monodisperse nanoparticles with 3–4 nm particle size, which may be of interest for bioimaging.101,102 The fluorescence emission from nanodiamonds mainly originates from nitrogen-vacancy (NV) centers, which are created by irradiation and annealing processes. NV centers occur in two charge states, neutral (NV0) and negatively charged (NV−), contributing both to the fluorescence emission spectra, while in CL NV− centers are reversibly converted to NV0 leading to NV0 dominated CL emission spectra.67,101,103 Although fluorescence emission of nanodiamonds proves to be photo-stable,104 small-sized nanodiamonds (≤40 nm) typically contain a high fraction of non-emitting particles and those that do emit suffer from limited brightness, which critically hampers specific labelling applications.67 Despite the limitations for very small nanodiamonds, there are many surface functionalization methods available. Primary surface functionalization protocols for nanodiamonds are based on versatile wet-chemistry methods, which enable carboxylation, halogenation, Diels–Alder reactions, and diazonium chemistry.101,104
For both REE3+ doped nanocrystals and nanodiamonds, single protein labelling could be achieved by using functionalization techniques similar to immunogold. Prior to that, biologically relevant functional groups need to be attached on the nanoparticles, which can be performed in two consecutive steps: first introducing primary functional group with silanes105,106 or polymers107 and then adding a bioactive molecule, which acts as a targeting moiety, such as a small molecule87,108 or an antibody.106,109 While most of the advancements in surface functionalization have been currently achieved on lanthanide nanoparticles based on a fluoride matrix, adaptation of these functionalization methods may enable surface modifications of other matrices as well. Versatile coatings allow a wide variety of targeting applications, which have been extensively reviewed by Dong et al.110
Next to the instrument and sample properties, CL resolution is intimately linked to the optical properties of the label, including brightness, stability, and emission lifetime as these dictate imaging conditions. Bright and stable labels allow usage of low acceleration voltages and beam currents (Fig. 3e), while having sufficient signal-to-noise (SN) ratio.61 Prigozhin et al.82 have concluded that an optimum SN ratio can be achieved if the electron interaction volume is matched to the nanoparticle size (0.75–1 keV electron landing energies for nanoparticles with a core size of 15–20 nm). Xu et al.111 have also found that the energy of ∼1 keV offers optimal trade-off between the BSE contrast and the axial resolution in biological SEM. The luminescence lifetime of the labels affects the electron beam scanning parameters and must be chosen based on the application. Smearing strongly reduces the image quality and occurs when the dwell time is shorter than the fluorescence emission lifetime.83 Rare earth elements typically have lifetimes in the range of milliseconds88,93 (with exceptions, such as Ce3+ ions with lifetimes of ∼60 ns (ref. 112)), while organic fluorophores, nanodiamonds and semiconductor quantum dots exhibit lifetimes from a few nanoseconds113 to a few tens of nanoseconds.83,113,114 Relatively long emission lifetimes of rare-earth element-based labels (typically Tm, Tb, Eu, Er), which otherwise fulfil the requirements for stability and brightness, hinder the acquisition of fast CCLEM. Therefore, label lifetimes are currently limiting the (large-scale) image acquisition times to values in the same order as alternative techniques, e.g. energy dispersive X-ray analysis (EDX)/electron energy loss spectroscopy (EELS) (see section “Alternative Methods: EDX and EELS Mapping” below) (Table 1). Recent developments of labels unifying sufficiently high brightness and stability with short lifetimes (such as Ce-based nanocrystals and nanodiamonds) pave the way for faster CL data acquisition. Next to emission lifetime, emission brightness and stability also dictate the scanning parameters. The electron dose, i.e. the number of electrons per unit area of sample, depends on electron beam current, dwell time and scanning area and determines the extent of electron beam damage to the samples.78–80 The optimal dose can be obtained by minimizing the dose to a value where the CL generation volume is minimal, but an acceptable S/N ratio can be obtained. Therefore, bright labels with short emission lifetime allow usage of low electron beam currents and fast scanning speeds, which will reduce locally accumulated dose and potential beam damage compared to dimmer nanoparticles.
| Sample | Instrument | Image acquisition conditions | Specimen | Ref. | ||
|---|---|---|---|---|---|---|
| Acceleration voltage (kV) | Beam current (nA) | Dwell time (ms) | ||||
| Y2O3:Ln3+ (Ln = Eu, Tm, Tb) | SEM | 10 | 0.47 | 50 | Eukaryotic cells (J744A.1) | 77 |
| Nanodiamonds and LuAG:Ce3+ | SEM | 5 | 1.2 | 0.09 | No cells | 76 |
| Y2O3:Zn,Eu3+ | SEM | 3 | 0.053 | 100 | Eukaryotic cells (HeLa) | 126 |
| Y2O3:Tm3+,Yb3+ | SEM | 3 | N/A | 100 | Eukaryotic cells (HeLa) | 127 |
| Nanodiamonds | SEM | 5 | 0.895 | 0.106 | Eukaryotic cells (HeLa) | 89 |
| Y2O3:Ln3+ (Ln = Eu, Tb) | STEM | 80 | 2 | 100 | Eukaryotic cells (HeLa) | 128 |
| Y2O3:Ln3+,Yb3+ (Ln = Tm, Er) | STEM | 80 | N/A | 500 | Eukaryotic cells (HeLa) | 64 |
| Gd2O3:Ln3+,Yb3+ (Ln = Tm, Ho, Er) | STEM | 80 | N/A | N/A | Eukaryotic cells (HeLa) | 129 |
| Enhanced green fluorescent protein | TEM | 200 | 0.1–0.3 | N/A | No cells | 68 |
| LaF3:Tb3+ nanocrystals | SEM/STEM | 2/5 | 0.4 | 50 | Eukaryotic cells (A549) | 59 |
| Nanodiamonds | STEM | 60 | N/A | N/A | Eukaryotic cells (A673) | 63 |
| Nanodiamonds | SEM | 3 | 0.8 | N/A | Eukaryotic cells (J744A.1/HT29) | 67 |
| YVO4:Bi3+,Eu3+ and Y2O3:Tb3+ | SEM | 2 | 0.4 | 1/5 | Eukaryotic cells (HUVEC) | 61 |
| Auto-cathodoluminescence | SEM | 8 | N/A | 20 | Animal/human collagen tissues | 118 |
| NaGdF4:Eu3+ | SEM | 1.5–20 | 0.3–0.5 | 2 | No cells | 82 |
Alternative methods: EDX and EELS mapping
The identification of biomolecules is strongly dependent on available labels in fluorescence as well as in electron microscopy. Alternative to label-based methods, label-free approaches for identification of proteins, amino acids, and sugars in the cells have gained increasing attention. CCLEM could also be employed as label-free method, when imaging auto-cathodoluminescence of tissues and cells.115–118 “ColorEM” or analytical electron microscopy converts grey-scale analytical information into false color-coded images, and can be used to identify biological features based on elemental composition.119,120 ColorEM techniques include energy dispersive X-ray analysis, electron energy loss spectroscopy, and correlative cathodoluminescence electron microscopy, which are reviewed in great detail by Pirozzi et al.120The interaction between an incident electron beam and the sample generates additional signals, as described previously. Elemental analysis in electron microscope can be achieved by using EDX or EELS. EDX analyses specific energetic electromagnetic radiation produced by primary electrons, which interact with core electrons from atoms in the material. In case of collision, created electron vacancies are being filled with electrons from the higher levels, leading to the emission of characteristic X-ray radiation. As the produced radiation is specific to the atom, energy dispersive X-ray analysis allows elemental composition studies. EDX systems are nowadays also able to measure light elements (down to Li) with acceptable signal-to-noise ratio.121 It has been extensively used in geology and material science; however, has not yet been widely adopted in life sciences. Scotuzzi et al.122 have demonstrated detection of commonly used EM-immunolabels (CdSe, Au) in rat pancreatic samples. Additionally, they have identified different cell types and peptides purely based on their elemental composition using EDX mapping. Similar to CL imaging, EDX resolution is strongly dependent on acceleration voltage and the material density.123,124 Currently, EDX mapping typically requires acquisition times ≈10× longer than CL for a comparable signal/noise ratio.61,122
Another imaging modality that has been suggested for elemental mapping is electron energy loss spectroscopy. EELS analyses inelastic scattering of primary electrons after interaction with ultrathin samples (preferred thickness ≤100 nm) using electron beams with high incident energy (≥100 keV). Transmitted electrons are separated based on their kinetic energy and analyzed with a high-resolution electron spectrometer, producing an electron energy-loss spectrum. The primary electron can ionize an atom in the sample and loses energy approximate to the binding energy of the dislodged electron. This can reveal elemental composition, but also physical and chemical properties of the material.125 EELS determines single elements (like in case of carbon, nitrogen, and oxygen), with careful analysis of valence electron energy-loss spectrum; it also reveals chemical fingerprints of biological molecules, like nucleic acids and amino acids in proteins.
EELS can also detect exogenous elements, such as gold and cadmium, which may be employed for immunolabeling.130 Instead of applying nanoparticle-based labels, Adams et al.131 described multi-target labeling in cells and tissues by selectively precipitating and detecting diaminobenzidine (DAB)-conjugates bound to lanthanide chelates. The resolution of EELS is determined by the electron beam size and interaction volume. With modern STEM instruments, the electron beam can be focused down to well below a 1 nm and EELS is usually measured on ultrathin samples to reduce the interaction between the sample and the high energy electron beam (≥100 keV). Therefore, theoretically STEM EELS enables atomic resolution, however, in biological samples it is compromised by the radiation damage on the sample and therefore reaches maximal resolutions of around 50 nm.130
The choice of the imaging method depends on the scientific question to be addressed, as all aforementioned methods have their advantages and limitations. All the above described methods are based on primary electron beam interaction with the sample and have a potential for label-free detection.118,120,122 In combination with STEM, all the methods have nanometric spatial resolution and complement each other, as the resolution is mostly determined by the sample thickness and electron beam spot size.120,130 High-energy STEM-EELS imaging can induce significant beam damage,132 while EDX and CL are less invasive due to the lower beam energies. EELS must be acquired from ultrathin samples,120,130 whereas CL and EDX can be collected both from bulk and ultrathin samples.59,120 X-rays are generated in a (slightly) smaller generation volume compared to CL at the given acceleration voltage. However, beam energies required for generating EDX signal are typically ≥5 kV and collection times oftentimes exceed several hours.122,124 Meanwhile, CL can be generated from heavy elements with lower acceleration voltages, routinely reaching sub-diffraction limit resolution and the acquisition of EELS and CL maps is achievable within (tens of) minutes.59,61,130 EELS and EDX are able to identify currently available immunolabels, e.g. based on gold and quantum dots. CL on the other hand requires novel light-emitting labels with high brightness, short emission lifetime and stability.120,130 Yet, despite their different physical origin, all the above mentioned methods can complement each other, and offer a comprehensive multi-modal imaging approach also for biological samples.
Towards live cell imaging and nanotomography
Provided that suitable labels become available in the near future, additional imaging modalities may combine well with the new opportunities given by multi-color imaging directly in the electron microscope. Additionally, correlative CL and electron microscopy may be extended to live cell imaging.89,117,133 Inami et al.134 developed an electron beam excitation assisted optical (EXA) microscope, which combines SEM and a conventional optical microscope by creating membranes with thin luminescent films. The achievable resolution is below 100 nm and strongly dependent on the luminescent film.135Generally, electron-transparent membranes allow imaging of hydrated116 and live cells.133 The EXA microscope has been employed for imaging fluorescent nanodiamonds in live cells89 and cellular constituents in a label-free manner.117,133 However, live-cell imaging suffers from low signal-to-noise ratio (low autofluorescence signal intensity133) and electron beam damage, which limits prolonged investigations.89,133 Cathodoluminescence can also be collected under cryogenic conditions, which typically yields narrow(er) emission lines and enhanced CL emission intensity.136,137 Cryogenic sample preparation becomes useful when attempting to preserve auto-cathodoluminescence signals of biological samples, which are typically quenched by osmium tetroxide, and study dynamic processes in biological samples.115,138 Despite these advantages, cryogenic sample preparation in the biological CCLEM field is not as widely adopted (yet) as the imaging of resin-embedded samples.
A somewhat similar approach exploiting CL for nanoscale bio-imaging is cathodoluminescence-activated imaging by resonant energy transfer (CLAIRE).139 In this technique a thin-film phosphor material is used in which CL is efficiently generated in a subwavelength volume. The locally generated CL is then used to probe an underlying material layer, which can be solid or a liquid cell environment, after which any resulting fluorescence emission is collected by a conventional parabolic mirror. The phosphor material absorbs the majority of the incoming electrons, thereby protecting the underlying material, and can act as a (partial) liquid cell seal at the same time. However, the supercontinuum excitation character of the electron beam is lost because the excitation is mediated by the phosphor, although different phosphors can be used to match particular absorption bands. The electron beam should not penetrate the layer of interest in CLAIRE, therefore simultaneous acquisition of SE and BSE signals from the layer of interest is impossible. The optical CLAIRE data can only be correlated with SE/BSE images acquired at a higher acceleration voltage, which results in the electron beam penetration into the layer of interest, but this is a sequential approach.
3D imaging, e.g. tomography has continuously gained importance in microscopy. Three-dimensional imaging aids a better understanding of cellular features and physical interaction between them. Transmission electron microscopy (TEM) is nowadays a standard tool for ultrastructural imaging, which also enables 3D tomography. Typically, images are acquired from thin sections (thickness ≤100 nm) to assure single-scattering events for the accelerated electron. Tomographic images can be constructed from the series of TEM images collected from a range of angles.31 Alternatively, array tomography (AT) has been developed to study large volumes of biological samples in 3D.140 In this case, serial sections are cut using an ultramicrotome, placed as an array on a conductive substrate and subsequently imaged using SEM.23 The main limitation in AT lays in the sectioning quality, as deformations on the sections (e.g. wrinkles) can ruin the whole data set.141 While array tomography is offering a potential also in CL bioimaging,142 CL has not been combined with it yet. Another technique, where CL could be employed is serial block-face electron microscopy, where the slicing is performed with an integrated ultramicrotome directly in the electron microscope.143 However, next to lack of novel labels, integrating the CL collection optics with the slicing hardware could be challenging due to space constraints and therefore has not been reported.
Alternatively, 3D information can be acquired using FIB/SEM, which allows precise sectioning and enables nanometric resolution in x–y–z direction.144,145 Accelerated ions (typically gallium) are bombarded onto the sample surface to sputter away atoms and expose inner structures of the sample. Sputtering depth and section size are adjusted by altering the spot size of the ion beam.60 The exposed structures are subsequently scanned with a focused electron beam using low acceleration voltages. High-resolution CL tomography requires optimal milling and imaging conditions. The FIB slicing thickness is dependent on the electron beam acceleration voltage used for imaging, since it is preferred to avoid overlapping signals between the slices. Cathodoluminescence bioimages with nanometric resolution from FIB sectioned samples have been acquired from nanoparticle exposed mammalian cells.59,61 However, to our best knowledge, in situ 3D FIB-SEM-CL has only been demonstrated on diamond60 and not yet on biological samples. 3D CL tomography on diamond sample revealed growth patterns within the material illustrating the potential of gaining mechanistic insights. However, the achieved spatial (≈300 nm) and axial resolutions (≈650 nm) did not reach the attainable resolution in FIB-SEM.60,111 The size of the electron beam interaction volume is strongly dependent on the material and therefore requires further tweaking for specific applications. Additionally, FIB-SEM-CL can only be acquired in manual slicing mode in the commercially available microscopes.59–61
CL tomography has also been demonstrated without focused ion beam. Recently, Atre et al.146 established CL tomography on metal-dielectric crescent in SEM by studying differently oriented particles and collecting corresponding coherent CL signals, locating regions of high CL intensity at each wavelength in 3D.
The main limitation in adoption of FIB-SEM-CL tomography in biological research is the strong need for bright and stable labels, which are critical in order to minimize the CL generation volume and give access to high resolution in x–y–z direction. Additionally, workflows in current imaging systems must improve drastically; and depend on fully automated FIB milling, and subsequent rapid acquisition of CL and electron micrographs.
The future of CCLEM bioimaging
Cathodoluminescence has a great potential to be adopted by biologists for routine analysis: it can be acquired simultaneously in scanning or transmission electron microscopes on live cells, histology sections, cryopreserved or resin embedded samples. The main advantage of cathodoluminescence over existing super-resolution methods is that sub-wavelength optical information can be collected directly in the electron microscope, along with the sample ultrastructure. The CL signal is in perfect registry with the other EM signals, contrary to SR or other correlative approaches, which are prone to misalignments and distortions. CCLEM in turn allows imaging of the wider context and enables structure–function studies. Ideally, CCLEM-based microscopy offers a route to luminescence imaging approaching the resolution of conventional electron microscopy, enabling ultimate correlative microscopy with no resolution mismatch. However, this heavily relies on the development of suitable labels and the sample properties. Current CCLEM studies on biological samples indicate a resolution of approximately 20–50 nm,61,67,118 in the order of a typical nanoparticle-label size82 and on par with several of the SR microscopy techniques.Despite the promising prospects, wide-spread adoption of CCLEM by the biomedical community likely depends on the availability of off-the-shelf CL-labels.120 Critical steps towards unlocking of the full potential of CL include:
• Development of bright and stable molecular labels based on REE3+ doped nanocrystals or fluorescent nanodiamonds. This has the prospect to enable labelling at low electron beam energy thus giving access to single molecule localization studies directly in EM.
• Improvement of single nanoparticle emission properties, i.e. shorter lifetimes, higher emission stability, and enhanced quantum yield to enable faster image acquisition times. First fascinating steps have been taken towards this by creating complex core/shell structures for upconversion lanthanide nanoparticles,98,99 which may be tailored for CCLEM.
• Demonstration of multi-color single molecule labelling. Despite recent reports in developing near-monodisperse nanocrystals with nine different emissions,82 multiple epitope labelling using small nanoparticles (<50 nm) has yet to be demonstrated. Once established, this technology can be used for simultaneous labelling of multiple epitopes by using labels with distinctly different optical emissions thereby overcoming major limitations of the established immunogold method.
• Development of post-functionalization protocols to bridge the gap between precision nanoparticle synthesis protocols in organic environments and successful phase transfer. First promising steps into the direction of efficient phase transfer have recently been published by Dragoman et al.147 Alternative routes based on untemplated synthesis also are increasingly explored.
• Demonstration of robust and versatile bio-functionalization strategies.
• For truly unleashing the potential of the developed labels, epitope labeling efficiency must be assessed compared to established immunolabels, such as immunogold and quantum dots.
The present minireview reports key developments related to label development, imaging conditions and instrument adjustments, which pave the way for CCLEM-based immunolabelling (immunoCCLEM). Importantly, the use of bright inorganic nanocrystals allows the use of straightforward, well-established and robust sample preparation methods in an unaltered way, since the label emission is neither affected by traditional fixatives nor heavy metal contrasting. These developments in turn pave the way for 3D correlative multi-color cathodoluminescence electron microscopy upon integration of CL detectors into (S)TEM and FIB/SEM. Making high-quality near-monodisperse nanocrystals with specific binding moieties available for use in aqueous environments and the increasing availability of CL detectors as well as creating automated imaging systems in electron microscopy facilities is expected to significantly boost the adoption of CL by the biologist community as an attractive alternative to super-resolution microscopy and traditional CLEM.
Conflicts of interest
Toon Coenen is an employee of Delmic B.V., a company that develops and sells the cathodoluminescence system that was used in some of the work presented in this paper.Acknowledgements
We thank Dr Job Fermie and Dr Sangeetha Hari for their critical revision of the review. We have used icons from Smart Servier Medical Art for the preparation of Fig. 1. We acknowledge funding from the Swiss National Science Foundation (SNSF, Spark grant no. 190832).References
- S. W. Hell, Far-Field Optical Nanoscopy, Science, 2007, 316(5828), 1153 CrossRef CAS PubMed.
- C. A. Combs and H. Shroff, Fluorescence Microscopy: A Concise Guide to Current Imaging Methods, Curr. Protoc. Neurosci., 2017, 79(1), 2.1.1–2.1.25 Search PubMed.
- L. Schermelleh, A. Ferrand, T. Huser, C. Eggeling, M. Sauer, O. Biehlmaier and G. P. C. Drummen, Super-resolution microscopy demystified, Nat. Cell Biol., 2019, 21(1), 72–84 CrossRef CAS PubMed.
- H. P. Erickson, Size and Shape of Protein Molecules at the Nanometer Level Determined by Sedimentation, Gel Filtration, and Electron Microscopy, Biol. Proced. Online, 2009, 11(1), 32 CrossRef CAS PubMed.
- L. Novotony and B. Hecht, Principles of nano-optics, Cambridge, New York, 2006 Search PubMed.
- B. Huang, H. Babcock and X. Zhuang, Breaking the Diffraction Barrier: Super-Resolution Imaging of Cells, Cell, 2010, 143(7), 1047–1058 CrossRef CAS PubMed.
- M. G. L. Gustafsson, Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy, J. Microsc., 2000, 198(2), 82–87 CrossRef CAS PubMed.
- K. I. Willig, S. O. Rizzoli, V. Westphal, R. Jahn and S. W. Hell, STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis, Nature, 2006, 440(7086), 935–939 CrossRef CAS PubMed.
- M. J. Rust, M. Bates and X. Zhuang, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods, 2006, 3, 793 CrossRef CAS PubMed.
- S. A. Jones, S.-H. Shim, J. He and X. Zhuang, Fast, three-dimensional super-resolution imaging of live cells, Nat. Methods, 2011, 8, 499 CrossRef CAS PubMed.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Imaging Intracellular Fluorescent Proteins at Nanometer Resolution, Science, 2006, 313(5793), 1642–1645 CrossRef CAS PubMed.
- T. Dertinger, R. Colyer, G. Iyer, S. Weiss and J. Enderlein, Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI), Proc. Natl. Acad. Sci. U. S. A., 2009, 106(52), 22287–22292 CrossRef CAS PubMed.
- T. Dertinger, M. Heilemann, R. Vogel, M. Sauer and S. Weiss, Superresolution Optical Fluctuation Imaging with Organic Dyes, Angew. Chem., 2010, 122(49), 9631–9633 CrossRef.
- F. Balzarotti, Y. Eilers, K. C. Gwosch, A. H. Gynnå, V. Westphal, F. D. Stefani, J. Elf and S. W. Hell, Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes, Science, 2017, 355(6325), 606–612 CrossRef CAS PubMed.
- K. C. Gwosch, J. K. Pape, F. Balzarotti, P. Hoess, J. Ellenberg, J. Ries and S. W. Hell, MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells, Nat. Methods, 2020, 17(2), 217–224 CAS.
- M. Fernández-Suárez and A. Y. Ting, Fluorescent probes for super-resolution imaging in living cells, Nat. Rev. Mol. Cell Biol., 2008, 9(12), 929–943 CrossRef PubMed.
- M. Hauser, M. Wojcik, D. Kim, M. Mahmoudi, W. Li and K. Xu, Correlative Super-Resolution Microscopy: New Dimensions and New Opportunities, Chem. Rev., 2017, 117(11), 7428–7456 CrossRef CAS PubMed.
- L. Nahidiazar, A. V. Agronskaia, J. Broertjes, B. van den Broek and K. Jalink, Optimizing Imaging Conditions for Demanding Multi-Color Super Resolution Localization Microscopy, PLoS One, 2016, 11(7), e0158884 CrossRef PubMed.
- Y. Chen, W. Liu, Z. Zhang, C. Zheng, Y. Huang, R. Cao, D. Zhu, L. Xu, M. Zhang, Y.-H. Zhang, J. Fan, L. Jin, Y. Xu, C. Kuang and X. Liu, Multi-color live-cell super-resolution volume imaging with multi-angle interference microscopy, Nat. Commun., 2018, 9(1), 4818 CrossRef PubMed.
- E. Wegel, A. Göhler, B. C. Lagerholm, A. Wainman, S. Uphoff, R. Kaufmann and I. M. Dobbie, Imaging cellular structures in super-resolution with SIM, STED and Localisation Microscopy: A practical comparison, Sci. Rep., 2016, 6, 27290 CrossRef CAS PubMed.
- J. Vangindertael, R. Camacho, W. Sempels, H. Mizuno, P. Dedecker and K. P. F. Janssen, An introduction to optical super-resolution microscopy for the adventurous biologist, Methods Appl. Fluoresc., 2018, 6(2), 022003 CrossRef CAS PubMed.
- P. de Boer, J. P. Hoogenboom and B. N. G. Giepmans, Correlated light and electron microscopy: ultrastructure lights up!, Nat. Methods, 2015, 12(6), 503–513 CrossRef CAS PubMed.
- T. Ando, S. P. Bhamidimarri, N. Brending, H. Colin-York, L. Collinson, N. De Jonge, P. J. de Pablo, E. Debroye, C. Eggeling, C. Franck, M. Fritzsche, H. Gerritsen, B. N. G. Giepmans, K. Grunewald, J. Hofkens, J. P. Hoogenboom, K. P. F. Janssen, R. Kaufmann, J. Klumpermann, N. Kurniawan, J. Kusch, N. Liv, V. Parekh, D. B. Peckys, F. Rehfeldt, D. C. Reutens, M. B. J. Roeffaers, T. Salditt, I. A. T. Schaap, U. S. Schwarz, P. Verkade, M. W. Vogel, R. Wagner, M. Winterhalter, H. Yuan and G. Zifarelli, The 2018 correlative microscopy techniques roadmap, J. Phys. D: Appl. Phys., 2018, 51(44), 443001 CrossRef PubMed.
- F. Guo and B. Q. Huang, Immunogold Labeling for Electron Microscopy: Strategy and Problem Solving, in Plant Microtechniques and Protocols, ed. E. C. T. Yeung, C. Stasolla, M. J. Sumner and B. Q. Huang, Springer International Publishing, Cham, 2015, pp. 225–249 Search PubMed.
- Y. S. Bykov, M. Cortese, J. A. G. Briggs and R. Bartenschlager, Correlative light and electron microscopy methods for the study of virus–cell interactions, FEBS Lett., 2016, 590(13), 1877–1895 CrossRef CAS PubMed.
- C. E. Sarraf, Immunolabeling for Electron Microscopy, in Diagnostic and Therapeutic Antibodies, ed. A. J. T. George and C. E. Urch, Humana Press, Totowa, NJ, 2000, pp. 439–452 Search PubMed.
- D. M. Van Elsland, E. Bos, J. B. Pawlak, H. S. Overkleeft, A. J. Koster and S. I. Van Kasteren, Correlative light and electron microscopy reveals discrepancy between gold and fluorescence labelling, J. Microsc., 2017, 267(3), 309–317 CrossRef CAS PubMed.
- R. C. N. Melo, E. Morgan, R. Monahan-Earley, A. M. Dvorak and P. F. Weller, Pre-embedding immunogold labeling to optimize protein localization at subcellular compartments and membrane microdomains of leukocytes, Nat. Protoc., 2014, 9(10), 2382–2394 CrossRef CAS PubMed.
- V. V. Philimonenko, A. A. Philimonenko, I. Šloufová, M. Hrubý, F. Novotný, Z. Halbhuber, M. Krivjanská, J. Nebesářová, M. Šlouf and P. Hozák, Simultaneous detection of multiple targets for ultrastructural immunocytochemistry, Histochem. Cell Biol., 2014, 141(3), 229–239 CrossRef CAS PubMed.
- B. N. G. Giepmans, T. J. Deerinck, B. L. Smarr, Y. Z. Jones and M. H. Ellisman, Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots, Nat. Methods, 2005, 2(10), 743–749 CrossRef CAS PubMed.
- D. S. Lidke and K. A. Lidke, Advances in high-resolution imaging – techniques for three-dimensional imaging of cellular structures, J. Cell Sci., 2012, 125(11), 2571–2580 CrossRef CAS PubMed.
- B. M. Humbel, M. D. M. de Jong, W. H. Müller and A. J. Verkleij, Pre-embedding immunolabeling for electron microscopy: An evaluation of permeabilization methods and markers, Microsc. Res. Tech., 1998, 42(1), 43–58 CrossRef CAS PubMed.
- C. Brandenberger, M. J. Clift, D. Vanhecke, C. Mühlfeld, V. Stone, P. Gehr and B. Rothen-Rutishauser, Intracellular imaging of nanoparticles: Is it an elemental mistake to believe what you see?, Part. Fibre Toxicol., 2010, 7(1), 15 CrossRef PubMed.
- K. T. Tokuyasu, A Technique For Ultracryotomy Of Cell Suspensions And Tissues, J. Cell Biol., 1973, 57(2), 551–565 CrossRef CAS PubMed.
- U. Schnell, F. Dijk, K. A. Sjollema and B. N. G. Giepmans, Immunolabeling artifacts and the need for live-cell imaging, Nat. Methods, 2012, 9, 152 CrossRef CAS PubMed.
- C. K. E. Bleck, A. Merz, M. G. Gutierrez, P. Walther, J. Dubochet, B. Zuber and G. Griffiths, Comparison of different methods for thin section EM analysis of Mycobacterium smegmatis, J. Microsc., 2010, 237(1), 23–38 CrossRef CAS PubMed.
- G. Wolff, C. Hagen, K. Grünewald and R. Kaufmann, Towards correlative super-resolution fluorescence and electron cryo-microscopy, Biol. Cell, 2016, 108(9), 245–258 CrossRef CAS PubMed.
- D. Studer, S. Zhao, X. Chai, P. Jonas, W. Graber, S. Nestel and M. Frotscher, Capture of activity-induced ultrastructural changes at synapses by high-pressure freezing of brain tissue, Nat. Protoc., 2014, 9(6), 1480–1495 CrossRef CAS PubMed.
- C. J. Peddie, K. Blight, E. Wilson, C. Melia, J. Marrison, R. Carzaniga, M.-C. Domart, P. O'Toole, B. Larijani and L. M. Collinson, Correlative and integrated light and electron microscopy of in-resin GFP fluorescence, used to localise diacylglycerol in mammalian cells, Ultramicroscopy, 2014, 143(100), 3–14 CrossRef CAS PubMed.
- M. R. G. Russell, T. R. Lerner, J. J. Burden, D. O. Nkwe, A. Pelchen-Matthews, M.-C. Domart, J. Durgan, A. Weston, M. L. Jones, C. J. Peddie, R. Carzaniga, O. Florey, M. Marsh, M. G. Gutierrez and L. M. Collinson, 3D correlative light and electron microscopy of cultured cells using serial blockface scanning electron microscopy, J. Cell Sci., 2017, 130(1), 278–291 CrossRef CAS PubMed.
- B. G. Kopek, G. Shtengel, C. S. Xu, D. A. Clayton and H. F. Hess, Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(16), 6136–6141 CrossRef CAS PubMed.
- S. W. Hell, S. J. Sahl, M. Bates, X. Zhuang, R. Heintzmann, M. J. Booth, J. Bewersdorf, G. Shtengel, H. Hess, P. Tinnefeld, A. Honigmann, S. Jakobs, I. Testa, L. Cognet, B. Lounis, H. Ewers, S. J. Davis, C. Eggeling, D. Klenerman, K. I. Willig, G. Vicidomini, M. Castello, A. Diaspro and T. Cordes, The 2015 super-resolution microscopy roadmap, J. Phys. D: Appl. Phys., 2015, 48(44), 443001 CrossRef.
- S. Watanabe, A. Punge, G. Hollopeter, K. I. Willig, R. J. Hobson, M. W. Davis, S. W. Hell and E. M. Jorgensen, Protein localization in electron micrographs using fluorescence nanoscopy, Nat. Methods, 2010, 8, 80 CrossRef PubMed.
- M. G. Paez-Segala, M. G. Sun, G. Shtengel, S. Viswanathan, M. A. Baird, J. J. Macklin, R. Patel, J. R. Allen, E. S. Howe, G. Piszczek, H. F. Hess, M. W. Davidson, Y. Wang and L. L. Looger, Fixation-resistant photoactivatable fluorescent proteins for CLEM, Nat. Methods, 2015, 12, 215 CrossRef CAS PubMed.
- J. Kuipers, T. J. van Ham, R. D. Kalicharan, A. Veenstra-Algra, K. A. Sjollema, F. Dijk, U. Schnell and B. N. G. Giepmans, FLIPPER, a combinatorial probe for correlated live imaging and electron microscopy, allows identification and quantitative analysis of various cells and organelles, Cell Tissue Res., 2015, 360(1), 61–70 CrossRef CAS PubMed.
- Z. Fu, D. Peng, M. Zhang, F. Xue, R. Zhang, W. He, T. Xu and P. Xu, mEosEM withstands osmium staining and Epon embedding for super-resolution CLEM, Nat. Methods, 2020, 17(1), 55–58 CrossRef CAS PubMed.
- D. Kim, T. J. Deerinck, Y. M. Sigal, H. P. Babcock, M. H. Ellisman and X. Zhuang, Correlative Stochastic Optical Reconstruction Microscopy and Electron Microscopy, PLoS One, 2015, 10(4), e0124581 CrossRef PubMed.
- P. D. Dahlberg, A. M. Sartor, J. Wang, S. Saurabh, L. Shapiro and W. E. Moerner, Identification of PAmKate as a Red Photoactivatable Fluorescent Protein for Cryogenic Super-Resolution Imaging, J. Am. Chem. Soc., 2018, 140(39), 12310–12313 CrossRef CAS PubMed.
- W. Li, S. C. Stein, I. Gregor and J. Enderlein, Ultra-stable and versatile widefield cryo-fluorescence microscope for single-molecule localization with sub-nanometer accuracy, Opt. Express, 2015, 23(3), 3770–3783 CrossRef CAS PubMed.
- D. P. Hoffman, G. Shtengel, C. S. Xu, K. R. Campbell, M. Freeman, L. Wang, D. E. Milkie, H. A. Pasolli, N. Iyer, J. A. Bogovic, D. R. Stabley, A. Shirinifard, S. Pang, D. Peale, K. Schaefer, W. Pomp, C.-L. Chang, J. Lippincott-Schwartz, T. Kirchhausen, D. J. Solecki, E. Betzig and H. F. Hess, Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells, Science, 2020, 367(6475), eaaz5357 CrossRef CAS PubMed.
- Y.-W. Chang, S. Chen, E. I. Tocheva, A. Treuner-Lange, S. Löbach, L. Søgaard-Andersen and G. J. Jensen, Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography, Nat. Methods, 2014, 11(7), 737–739 CrossRef CAS PubMed.
- M. W. Tuijtel, A. J. Koster, S. Jakobs, F. G. A. Faas and T. H. Sharp, Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins, Sci. Rep., 2019, 9(1), 1369 CrossRef PubMed.
- E. Johnson, E. Seiradake, E. Y. Jones, I. Davis, K. Grünewald and R. Kaufmann, Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins, Sci. Rep., 2015, 5, 9583 CrossRef CAS PubMed.
- L. Wang, B. Bateman, L. C. Zanetti-Domingues, A. N. Moores, S. Astbury, C. Spindloe, M. C. Darrow, M. Romano, S. R. Needham, K. Beis, D. J. Rolfe, D. T. Clarke and M. L. Martin-Fernandez, Solid immersion microscopy images cells under cryogenic conditions with 12 nm resolution, Commun. Biol., 2019, 2(1), 74 CrossRef PubMed.
- C. J. Peddie, M.-C. Domart, X. Snetkov, P. O'Toole, B. Larijani, M. Way, S. Cox and L. M. Collinson, Correlative super-resolution fluorescence and electron microscopy using conventional fluorescent proteins in vacuo, J. Struct. Biol., 2017, 199(2), 120–131 CrossRef CAS PubMed.
- T. K. Tsang, E. A. Bushong, D. Boassa, J. Hu, B. Romoli, S. Phan, D. Dulcis, C. Y. Su and M. H. Ellisman, High-quality ultrastructural preservation using cryofixation for 3D electron microscopy of genetically labeled tissues, eLife, 2018, 7, e3552 CrossRef PubMed.
- Z. Mahfoud, A. T. Dijksman, C. Javaux, P. Bassoul, A.-L. Baudrion, J. Plain, B. Dubertret and M. Kociak, Cathodoluminescence in a Scanning Transmission Electron Microscope: A Nanometer-Scale Counterpart of Photoluminescence for the Study of II–VI Quantum Dots, J. Phys. Chem. Lett., 2013, 4(23), 4090–4094 CrossRef CAS.
- T. Coenen and N. M. Haegel, Cathodoluminescence for the 21st century: Learning more from light, Appl. Phys. Rev., 2017, 4(3), 031103 Search PubMed.
- K. Keevend, M. Stiefel, A. L. Neuer, M. T. Matter, A. Neels, S. Bertazzo and I. K. Herrmann, Tb3+-doped LaF3 nanocrystals for correlative cathodoluminescence electron microscopy imaging with nanometric resolution in focused ion beam-sectioned biological samples, Nanoscale, 2017, 9(13), 4383–4387 RSC.
- D. A. M. De Winter, M. N. Lebbink, D. F. W. De Vries, J. A. Post and M. R. Drury, FIB-SEM cathodoluminescence tomography: practical and theoretical considerations, J. Microsc., 2011, 243(3), 315–326 CrossRef CAS PubMed.
- K. Keevend, L. Puust, K. Kurvits, L. R. H. Gerken, F. H. L. Starsich, J.-H. Li, M. T. Matter, A. Spyrogianni, G. A. Sotiriou, M. Stiefel and I. K. Herrmann, Ultrabright and Stable Luminescent Labels for Correlative Cathodoluminescence Electron Microscopy (CCLEM) Bioimaging, Nano Lett., 2019, 19(9), 6013–6018 CrossRef CAS PubMed.
- T. Furukawa, S. Fukushima, H. Niioka, N. Yamamoto, J. Miyake, T. Araki and M. Hashimoto, Rare-earth-doped nanophosphors for multicolor cathodoluminescence nanobioimaging using scanning transmission electron microscopy, J. Biomed. Opt., 2015, 20(5), 056007–056007 CrossRef PubMed.
- S. Nagarajan, C. Pioche-Durieu, L. H. G. Tizei, C.-Y. Fang, J.-R. Bertrand, E. Le Cam, H.-C. Chang, F. Treussart and M. Kociak, Simultaneous cathodoluminescence and electron microscopy cytometry of cellular vesicles labeled with fluorescent nanodiamonds, Nanoscale, 2016, 8(22), 11588–11594 RSC.
- S. Fukushima, T. Furukawa, H. Niioka, M. Ichimiya, T. Sannomiya, N. Tanaka, D. Onoshima, H. Yukawa, Y. Baba, M. Ashida, J. Miyake, T. Araki and M. Hashimoto, Correlative near-infrared light and cathodoluminescence microscopy using Y2O3:Ln,Yb (Ln = Tm, Er) nanophosphors for multiscale, multicolour bioimaging, Sci. Rep., 2016, 6, 25950 CrossRef CAS PubMed.
- M. W. H. Garming, I. G. C. Weppelman, P. de Boer, F. P. Martínez, R. Schirhagl, J. P. Hoogenboom and R. J. Moerland, Nanoparticle discrimination based on wavelength and lifetime-multiplexed cathodoluminescence microscopy, Nanoscale, 2017, 9(34), 12727–12734 RSC.
- S. Meuret, M. Solà Garcia, T. Coenen, E. Kieft, H. Zeijlemaker, M. Lätzel, S. Christiansen, S. Y. Woo, Y. H. Ra, Z. Mi and A. Polman, Complementary cathodoluminescence lifetime imaging configurations in a scanning electron microscope, Ultramicroscopy, 2019, 197, 28–38 CrossRef CAS PubMed.
- S. R. Hemelaar, P. de Boer, M. Chipaux, W. Zuidema, T. Hamoh, F. P. Martinez, A. Nagl, J. P. Hoogenboom, B. N. G. Giepmans and R. Schirhagl, Nanodiamonds as multi-purpose labels for microscopy, Sci. Rep., 2017, 7(1), 720 CrossRef CAS PubMed.
- K. Nagayama, T. Onuma, R. Ueno, K. Tamehiro and H. Minoda, Cathodoluminescence and Electron-Induced Fluorescence Enhancement of Enhanced Green Fluorescent Protein, J. Phys. Chem. B, 2016, 120(6), 1169–1174 CrossRef CAS PubMed.
- F. J. García de Abajo, Optical excitations in electron microscopy, Rev. Mod. Phys., 2010, 82(1), 209–275 CrossRef.
- B. G. Yacobi and D. B. Holt, Cathodoluminescence Microscopy of Inorganic Solids, Springer, 1990 Search PubMed.
- A. Gustafsson and E. Kapon, Cathodoluminescence in the scanning electron microscope: application to low-dimensional semiconductor structures, Scanning Microsc., 1998, 12(2), 285–299 Search PubMed.
- T. Coenen, B. J. M. Brenny, E. J. Vesseur and A. Polman, Cathodoluminescence microscopy: Optical imaging and spectroscopy with deep-subwavelength resolution, MRS Bull., 2015, 40(4), 359–365 CrossRef CAS.
- R. F. W. Pease and T. L. Hayes, Scanning Electron Microscopy of Biological Material, Nature, 1966, 210(5040), 1049–1049 CrossRef CAS PubMed.
- M. Mets and A. Lagasse, An investigation of some organic chemicals as cathodoluminescent dyes using the scanning electron microscope, J. Microsc., 1971, 94(2), 151–156 CrossRef PubMed.
- R. Herbst and D. Hoder, Cathodoluminescence in biological studies, Scanning, 1978, 1(1), 35–41 CrossRef CAS.
- D. R. Glenn, H. Zhang, N. Kasthuri, R. Schalek, P. K. Lo, A. S. Trifonov, H. Park, J. W. Lichtman and R. L. Walsworth, Correlative light and electron microscopy using cathodoluminescence from nanoparticles with distinguishable colours, Sci. Rep., 2012, 2, 865 CrossRef CAS PubMed.
- H. Niioka, T. Furukawa, M. Ichimiya, M. Ashida, T. Araki and M. Hashimoto, Multicolor Cathodoluminescence Microscopy for Biological Imaging with Nanophosphors, Appl. Phys. Express, 2011, 4(11), 112402 CrossRef.
- R. E. Thach and S. S. Thach, Damage to Biological Samples Caused by the Electron Beam during Electron Microscopy, Biophys. J., 1971, 11(2), 204–210 CrossRef CAS PubMed.
- J. J. H. A. Van Hest, A. V. Agronskaia, J. Fokkema, F. Montanarella, A. Gregorio Puig, C. De Mello Donega, A. Meijerink, G. A. Blab and H. C. Gerritsen, Towards robust and versatile single nanoparticle fiducial markers for correlative light and electron microscopy, J. Microsc., 2019, 274(1), 13–22 CrossRef CAS PubMed.
- J.-I. Niitsuma, H. Oikawa, E. Kimura, T. Ushiki and T. Sekiguchi, Cathodoluminescence investigation of organic materials, J. Electron Microsc., 2005, 54(4), 325–330 CAS.
- J. Rodriguez-Viejo, K. F. Jensen, H. Mattoussi, J. Michel, B. O. Dabbousi and M. G. Bawendi, Cathodoluminescence and photoluminescence of highly luminescent CdSe/ZnS quantum dot composites, Appl. Phys. Lett., 1997, 70(16), 2132–2134 CrossRef CAS.
- M. B. Prigozhin, P. C. Maurer, A. M. Courtis, N. Liu, M. D. Wisser, C. Siefe, B. Tian, E. Chan, G. Song, S. Fischer, S. Aloni, D. F. Ogletree, E. S. Barnard, L. M. Joubert, J. Rao, A. P. Alivisatos, R. M. Macfarlane, B. E. Cohen, Y. Cui, J. A. Dionne and S. Chu, Bright sub-20 nm cathodoluminescent nanoprobes for electron microscopy, Nat. Nanotechnol., 2019, 14(5), 420–425 CrossRef CAS PubMed.
- H. Zhang, I. Aharonovich, D. R. Glenn, R. Schalek, A. P. Magyar, J. W. Lichtman, E. L. Hu and R. L. Walsworth, Silicon-Vacancy Color Centers in Nanodiamonds: Cathodoluminescence Imaging Markers in the Near Infrared, Small, 2014, 10(10), 1908–1913 CrossRef CAS PubMed.
- X. Sun, B. Wang, I. Kempson, C. Liu, Y. Hou and M. Gao, Restructuring and Remodeling of NaREF4 Nanocrystals by Electron Irradiation, Small, 2014, 10(22), 4711–4717 CrossRef CAS PubMed.
- L. H. G. Tizei and M. Kociak, Spectrally and spatially resolved cathodoluminescence of nanodiamonds: local variations of the NV0 emission properties, Nanotechnology, 2012, 23(17), 175702 CrossRef CAS PubMed.
- K. Keevend, G. Panzarasa, F. H. L. Starsich, M. Zeltner, A. Spyrogianni, E. Tsolaki, G. Fortunato, S. E. Pratsinis, S. Bertazzo and I. K. Herrmann, Facile meltPEGylation of flame-made luminescent Tb3+-doped yttrium oxide particles: hemocompatibility, cellular uptake and comparison to silica, Chem. Commun., 2018, 54(23), 2914–2917 RSC.
- T.-K. Ryu, S. W. Baek, G.-J. Lee, C.-K. Rhee and S.-W. Choi, Targeted Tumor Therapy Based on Nanodiamonds Decorated with Doxorubicin and Folic Acid, Macromol. Biosci., 2017, 17(2), 1600180 CrossRef PubMed.
- A. Spyrogianni, P. Tiefenboeck, F. H. L. Starsich, K. Keevend, F. Krumeich, I. K. Herrmann, J.-C. Leroux and G. A. Sotiriou, Near-UV activated, photostable nanophosphors for in vitro dosimetry and dynamic bioimaging, AIChE J., 2018, 64(8), 2947–2957 CrossRef CAS.
- Y. Nawa, W. Inami, S. Lin, Y. Kawata, S. Terakawa, C.-Y. Fang and H.-C. Chang, Multi-Color Imaging of Fluorescent Nanodiamonds in Living HeLa Cells Using Direct Electron-Beam Excitation, ChemPhysChem, 2014, 15(4), 721–726 CrossRef CAS PubMed.
- J. Roth and M. Binder, Colloidal gold, ferritin and peroxidase as markers for electron microscopic double labeling lectin techniques, J. Histochem. Cytochem., 1978, 26(3), 163–169 CrossRef CAS PubMed.
- A. C. Zonnevylle, R. F. C. Van Tol, N. Liv, A. C. Narvaez, A. P. J. Effting, P. Kruit and J. P. Hoogenboom, Integration of a high-NA light microscope in a scanning electron microscope, J. Microsc., 2013, 252(1), 58–70 CrossRef CAS PubMed.
- N. Liv, A. C. Zonnevylle, A. C. Narvaez, A. P. J. Effting, P. W. Voorneveld, M. S. Lucas, J. C. Hardwick, R. A. Wepf, P. Kruit and J. P. Hoogenboom, Simultaneous Correlative Scanning Electron and High-NA Fluorescence Microscopy, PLoS One, 2013, 8(2), e55707 CrossRef CAS PubMed.
- G. A. Sotiriou, M. Schneider and S. E. Pratsinis, Green, Silica-Coated Monoclinic Y2O3:Tb3+ Nanophosphors: Flame Synthesis and Characterization, J. Phys. Chem. C, 2012, 116(7), 4493–4499 CrossRef CAS PubMed.
- N. Pinna, G. Garnweitner, P. Beato, M. Niederberger and M. Antonietti, Synthesis of Yttria-Based Crystalline and Lamellar Nanostructures and their Formation Mechanism, Small, 2005, 1(1), 112–121 CrossRef CAS PubMed.
- G. Chen, T. Y. Ohulchanskyy, R. Kumar, H. Ågren and P. N. Prasad, Ultrasmall Monodisperse NaYF4:Yb3+/Tm3+ Nanocrystals with Enhanced Near-Infrared to Near-Infrared Upconversion Photoluminescence, ACS Nano, 2010, 4(6), 3163–3168 CrossRef CAS PubMed.
- H.-Q. Wang and T. Nann, Monodisperse Upconverting Nanocrystals by Microwave-Assisted Synthesis, ACS Nano, 2009, 3(11), 3804–3808 CrossRef CAS PubMed.
- E. V. Samsonova, A. V. Popov, A. S. Vanetsev, K. Keevend, K. Kaldvee, L. Puust, A. E. Baranchikov, A. V. Ryabova, S. G. Fedorenko, V. Kiisk, I. Sildos, J. Kikas, R. Steiner, V. B. Loschenov and Y. V. Orlovskii, Fluorescence quenching mechanism for water-dispersible Nd3+:KYF4 nanoparticles synthesized by microwave-hydrothermal technique, J. Lumin., 2016, 169, 722–727 CrossRef CAS.
- F. Vetrone, R. Naccache, V. Mahalingam, C. G. Morgan and J. A. Capobianco, The Active-Core/Active-Shell Approach: A Strategy to Enhance the Upconversion Luminescence in Lanthanide-Doped Nanoparticles, Adv. Funct. Mater., 2009, 19(18), 2924–2929 CrossRef CAS.
- M. Banski, M. Afzaal, A. Podhorodecki, J. Misiewicz, A. L. Abdelhady and P. O'Brien, Passivation of lanthanide surface sites in sub-10 nm NaYF4:Eu3+ nanocrystals, J. Nanopart. Res., 2012, 14(11), 1228 CrossRef CAS PubMed.
- H. A. A. Seed Ahmed, O. M. Ntwaeaborwa and R. E. Kroon, The energy transfer mechanism in Ce,Tb co-doped LaF3 nanoparticles, Curr. Appl. Phys., 2013, 13(7), 1264–1268 CrossRef.
- V. N. Mochalin, O. Shenderova, D. Ho and Y. Gogotsi, The properties and applications of nanodiamonds, Nat. Nanotechnol., 2011, 7, 11 CrossRef PubMed.
- O. A. Williams, J. Hees, C. Dieker, W. Jäger, L. Kirste and C. E. Nebel, Size-Dependent Reactivity of Diamond Nanoparticles, ACS Nano, 2010, 4(8), 4824–4830 CrossRef CAS PubMed.
- M. Sola-Garcia, S. Meuret, T. Coenen and A. Polman, Electron-induced state conversion in diamond NV centers measured with pump-probe cathodoluminescence spectroscopy, ACS Photonics, 2020, 7(1), 232–240 CrossRef CAS PubMed.
- C.-C. Fu, H.-Y. Lee, K. Chen, T.-S. Lim, H.-Y. Wu, P.-K. Lin, P.-K. Wei, P.-H. Tsao, H.-C. Chang and W. Fann, Characterization and application of single fluorescent nanodiamonds as cellular biomarkers, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(3), 727–732 CrossRef CAS PubMed.
- K. C. Barick, A. Sharma, N. G. Shetake, R. S. Ningthoujam, R. K. Vatsa, P. D. Babu, B. N. Pandey and P. A. Hassan, Covalent bridging of surface functionalized Fe3O4 and YPO4:Eu nanostructures for simultaneous imaging and therapy, Dalton Trans., 2015, 44(33), 14686–14696 RSC.
- J. Pichaandi, G. Zhao, A. Bouzekri, E. Lu, O. Ornatsky, V. Baranov, M. Nitz and M. A. Winnik, Lanthanide nanoparticles for high sensitivity multiparameter single cell analysis, Chem. Sci., 2019, 10(10), 2965–2974 RSC.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, PEGylation as a strategy for improving nanoparticle-based drug and gene delivery, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- A. Jain, P. G. J. Fournier, V. Mendoza-Lavaniegos, P. Sengar, F. M. Guerra-Olvera, E. Iñiguez, T. G. Kretzschmar, G. A. Hirata and P. Juárez, Functionalized rare earth-doped nanoparticles for breast cancer nanodiagnostic using fluorescence and CT imaging, J. Nanobiotechnol., 2018, 16(1), 26 CrossRef PubMed.
- G. Yi, H. Lu, S. Zhao, Y. Ge, W. Yang, D. Chen and L.-H. Guo, Synthesis, Characterization, and Biological Application of Size-Controlled Nanocrystalline NaYF4:Yb,Er Infrared-to-Visible Up-Conversion Phosphors, Nano Lett., 2004, 4(11), 2191–2196 CrossRef CAS.
- H. Dong, S.-R. Du, X.-Y. Zheng, G.-M. Lyu, L.-D. Sun, L.-D. Li, P.-Z. Zhang, C. Zhang and C.-H. Yan, Lanthanide Nanoparticles: From Design toward Bioimaging and Therapy, Chem. Rev., 2015, 115(19), 10725–10815 CrossRef CAS PubMed.
- C. S. Xu, K. J. Hayworth, Z. Lu, P. Grob, A. M. Hassan, J. G. García-Cerdán, K. K. Niyogi, E. Nogales, R. J. Weinberg and H. F. Hess, Enhanced FIB-SEM systems for large-volume 3D imaging, eLife, 2017, 6, e25916 CrossRef PubMed.
- M. Odziomek, F. Chaput, F. Lerouge, M. Sitarz and S. Parola, Highly luminescent YAG:Ce ultra-small nanocrystals, from stable dispersions to thin films, J. Mater. Chem. C, 2017, 5(47), 12561–12570 RSC.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Quantum dots versus organic dyes as fluorescent labels, Nat. Methods, 2008, 5, 763 CrossRef CAS PubMed.
- J. Storteboom, P. Dolan, S. Castelletto, X. Li and M. Gu, Lifetime investigation of single nitrogen vacancy centres in nanodiamonds, Opt. Express, 2015, 23(9), 11327–11333 CrossRef CAS PubMed.
- T. Fujimoto, T. Itoh, K. Yamamoto, K. Fujimoto, M. Sakai, T. Inoué, H. Koike, T. Sakai and K. Ogawa, Cathodoluminescence electron microscopy for cytochemical studies at the organelle level, Acta Histochem. Cytochem., 1983, 16(4), 374–380 CrossRef.
- S. Thiberge, A. Nechushtan, D. Sprinzak, O. Gileadi, V. Behar, O. Zik, Y. Chowers, S. Michaeli, J. Schlessinger and E. Moses, Scanning electron microscopy of cells and tissues under fully hydrated conditions, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(10), 3346–3351 CrossRef CAS PubMed.
- M. Fukuta, Y. Masuda, W. Inami and Y. Kawata, Label-free cellular structure imaging with 82 nm lateral resolution using an electron-beam excitation-assisted optical microscope, Opt. Express, 2016, 24(15), 16487–16495 CrossRef CAS PubMed.
- M. S. Zielinski, E. Vardar, G. Vythilingam, E.-M. Engelhardt, J. A. Hubbell, P. Frey and H. M. Larsson, Quantitative intrinsic auto-cathodoluminescence can resolve spectral signatures of tissue-isolated collagen extracellular matrix, Commun. Biol., 2019, 2(1), 69 CrossRef PubMed.
- M. M. G. Barfels, X. Jiang, H. Yew Meng, A. L. Arsenault and F. P. Ottensmeyer, Low energy loss electron microscopy of chromophores, Micron, 1998, 29(2), 97–104 CrossRef CAS PubMed.
- N. M. Pirozzi, J. P. Hoogenboom and B. N. G. Giepmans, ColorEM: analytical electron microscopy for element-guided identification and imaging of the building blocks of life, Histochem. Cell Biol., 2018, 150(5), 509–520 CrossRef CAS PubMed.
- P. Hovington, V. Timoshevskii, S. Burgess, H. Demers, P. Statham, R. Gauvin and K. Zaghib, Can we detect Li K X-ray in lithium compounds using energy dispersive spectroscopy?, Scanning, 2016, 38(6), 571–578 CrossRef CAS PubMed.
- M. Scotuzzi, J. Kuipers, D. I. Wensveen, P. de Boer, K. W. Hagen, J. P. Hoogenboom and B. N. G. Giepmans, Multi-color electron microscopy by element-guided identification of cells, organelles and molecules, Sci. Rep., 2017, 7, 45970 CrossRef CAS PubMed.
- J. J. Friel, X-Ray and Image Analysis in Electron Microscopy, Princeton Gamma-Tech, Incorporated, 1994, pp. 112 Search PubMed.
- D. Bell and A. Garratt-Reed, Energy Dispersive X-ray Analysis in the Electron Microscope, Taylor & Francis, 2003 Search PubMed.
- R. F. Egerton, Electron energy-loss spectroscopy in the electron microscope, Springer Science & Business Media, 2011 Search PubMed.
- T. Furukawa, H. Niioka, M. Ichimiya, T. Nagata, M. Ashida, T. Araki and M. Hashimoto, High-resolution microscopy for biological specimens via cathodoluminescence of Eu- and Zn-doped Y2O3 nanophosphors, Opt. Express, 2013, 21(22), 25655–25663 CrossRef PubMed.
- S. Fukushima, T. Furukawa, H. Niioka, M. Ichimiya, J. Miyake, M. Ashida, T. Araki and M. Hashimoto, Y2O3:Tm,Yb nanophosphors for correlative upconversion luminescence and cathodoluminescence imaging, Micron, 2014, 67, 90–95 CrossRef CAS PubMed.
- T. Furukawa, S. Fukushima, H. Niioka, N. Yamamoto, J. Miyake, T. Araki and M. Hasimoto, Rare-earth-doped nanophosphors for multicolor cathodoluminescence nanobioimaging using scanning transmission electron micrsocopy, J. Biomed. Opt., 2015, 20(5), 056007 CrossRef PubMed.
- D. Thi Kim Dung, S. Fukushima, T. Furukawa, H. Niioka, T. Sannomiya, K. Kobayashi, H. Yukawa, Y. Baba, M. Hashimoto and J. Miyake, Multispectral Emissions of Lanthanide-Doped Gadolinium Oxide Nanophosphors for Cathodoluminescence and Near-Infrared Upconversion/Downconversion Imaging, Nanomaterials, 2016, 6(9), 163 CrossRef PubMed.
- M. A. Aronova and R. D. Leapman, Development of electron energy-loss spectroscopy in the biological sciences, MRS Bull., 2012, 37(1), 53–62 CrossRef CAS PubMed.
- S. R. Adams, M. R. Mackey, R. Ramachandra, S. F. Palida Lemieux, P. Steinbach, E. A. Bushong, M. T. Butko, B. N. G. Giepmans, M. H. Ellisman and R. Y. Tsien, Multicolor Electron Microscopy for Simultaneous Visualization of Multiple Molecular Species, Cell Chem. Biol., 2016, 23(11), 1417–1427 CrossRef CAS PubMed.
- P. A. Midgley and M. Weyland, 3D electron microscopy in the physical sciences: the development of Z-contrast and EFTEM tomography, Ultramicroscopy, 2003, 96(3), 413–431 CrossRef CAS PubMed.
- Y. Nawa, W. Inami, A. Chiba, A. Ono, A. Miyakawa, Y. Kawata, S. Lin and S. Terakawa, Dynamic and high-resolution live cell imaging by direct electron beam excitation, Opt. Express, 2012, 20(5), 5629–5635 CrossRef PubMed.
- W. Inami, K. Nakajima, A. Miyakawa and Y. Kawata, Electron beam excitation assisted optical microscope with ultra-high resolution, Opt. Express, 2010, 18(12), 12897–12902 CrossRef CAS PubMed.
- M. Fukuta, A. Ono, Y. Nawa, W. Inami, L. Shen, Y. Kawata and S. Terekawa, Cell structure imaging with bright and homogeneous nanometric light source, J. Biophotonics, 2017, 10(4), 503–510 CrossRef CAS PubMed.
- H. Watanabe, T. Kitamura, S. Nakashima and S. Shikata, Cathodoluminescence characterization of a nitrogen-doped homoepitaxial diamond thin film, J. Appl. Phys., 2009, 105(9), 093529 CrossRef.
- N. Vaskovicova, R. Skoupy, A. Patak, K. Hrubanova and V. Krzyzanek, Cathodoluminescence Study of Microdiamonds and Improvements of Signal Detection by Lowering Temperature of the Sample, Microsc. Microanal., 2017, 23(S1), 2284–2285 CrossRef.
- C. Borges-Merjane, O. Kim and P. Jonas, Functional Electron Microscopy (“Flash and Freeze”) of Identified Cortical Synapses in Acute Brain Slices, Neuron, 2020, 105, 992–1006 CrossRef CAS PubMed.
- C. G. Bischak, R. B. Wai, C. Cherqui, J. A. Busche, S. C. Quillin, C. L. Hetherington, Z. Wang, C. D. Aiello, D. G. Schlom, S. Aloni, D. F. Ogletree, D. J. Masiello and N. S. Ginsberg, Noninvasive Cathodoluminescence-Activated Nanoimaging of Dynamic Processes in Liquids, ACS Nano, 2017, 11(10), 10583–10590 CrossRef CAS PubMed.
- K. D. Micheva and S. J. Smith, Array Tomography: A New Tool for Imaging the Molecular Architecture and Ultrastructure of Neural Circuits, Neuron, 2007, 55(1), 25–36 CAS.
- A. Burel, M.-T. Lavault, C. Chevalier, H. Gnaegi, S. Prigent, A. Mucciolo, S. Dutertre, B. M. Humbel, T. Guillaudeux and I. Kolotuev, A targeted 3D EM and correlative microscopy method using SEM array tomography, Development, 2018, 145(12), dev160879 CrossRef PubMed.
- S. J. Smith, Q&A: Array tomography, BMC Biol., 2018, 16(1), 98 CrossRef PubMed.
- W. Denk and H. Horstmann, Serial Block-Face Scanning Electron Microscopy to Reconstruct Three-Dimensional Tissue Nanostructure, PLoS Biol., 2004, 2(11), e329 CrossRef PubMed.
- K. Narayan and S. Subramaniam, Focused ion beams in biology, Nat. Methods, 2015, 12, 1021 CAS.
- J. Fermie, N. Liv, C. ten Brink, E. G. van Donselaar, W. H. Müller, N. L. Schieber, Y. Schwab, H. C. Gerritsen and J. Klumperman, Single organelle dynamics linked to 3D structure by correlative live-cell imaging and 3D electron microscopy, Traffic, 2018, 19(5), 354–369 CrossRef CAS PubMed.
- A. C. Atre, B. J. M. Brenny, T. Coenen, A. García-Etxarri, A. Polman and J. A. Dionne, Nanoscale optical tomography with cathodoluminescence spectroscopy, Nat. Nanotechnol., 2015, 10, 429 CAS.
- R. M. Dragoman, M. Grogg, M. I. Bodnarchuk, P. Tiefenboeck, D. Hilvert, D. N. Dirin and M. V. Kovalenko, Surface-Engineered Cationic Nanocrystals Stable in Biological Buffers and High Ionic Strength Solutions, Chem. Mater., 2017, 29(21), 9416–9428 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |
