Open Access Article
Open Access ArticleQuantifying the level of nanoparticle uptake in mammalian cells using flow cytometry†
HyeRim
Shin
a,
Minjeong
Kwak
b,
Tae Geol
Lee
b and
Ji Youn
Lee
 *a
*a
aCenter for Bioanalysis, Division of Chemical and Medical Metrology, Korea Research Institute of Standards and Science, 267 Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea. E-mail: jylee@kriss.re.kr; Fax: +82-42-868-5801; Tel: +82-42-868-5363
bCenter for Nano-Bio Measurement, Division of Industrial Metrology, Korea Research Institute of Standards and Science, 267 Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea
First published on 2nd July 2020
Abstract
Reliable quantification of nanoparticle uptake in mammalian cells is essential to study the effects of nanoparticles in the fields of medicine and environmental science. Most conventional quantification methods, such as electron microscopy or confocal imaging, are laborious and semi-quantitative and therefore not readily applicable to routine analyses. Here, we developed assays to quantify fluorescently labelled nanoparticle uptake in mammalian cells using a flow cytometer. The first approach was to measure the percentage of nanoparticle-containing cells based on a cutoff fluorescence intensity as set from a histogram of control cells, which is a quick and easy way to relatively compare nanoparticle uptake in the same set of experiments. The second approach was to measure the calibrated fluorescence intensity of the nanoparticle-treated cells in molecules of equivalent soluble fluorophore (MESF) values using calibration beads, which allows for comparisons between different sets of experiments. We successfully applied the developed assays to more readily measure fluorescence-labelled silica nanoparticle uptake in A549 lung carcinoma cells in a quantitative rather than semi-quantitative manner. We further tested the assays with nine different types of mammalian cells and investigated the correlation between cell type/size and nanoparticle uptake.
Introduction
Nanoparticles have drawn tremendous interest in diverse fields including medicine,1 foods,2 environmental science,3 and cosmetics4 because of their potential in related technologies. Whether it is biomedical studies to verify the efficacy of nanomedicines or environmental studies to investigate the safety of engineered nanomaterials, quantifying the cellular uptake of nanoparticles is prerequisite to understanding the underlying mechanisms.5,6–9 The cellular uptake of nanoparticles depends on physical properties such as size10–13 and shape,14,15 chemical properties such as surface charge16 and modification,17,18 and dispersion media characteristics such as the presence or absence of serum proteins,19 heat inactivated serum,20 and corona composition.21,22 Temperature also plays an important role in nanoparticle uptake because the process is active and energy-dependent.23 The molecular mechanisms for uptake vary by the physicochemical properties of the nanoparticles,24,25 among which surface charge is an important factor that regulates cellular interactions with the nanoparticles. The regulation of cellular uptake is especially important in the delivery of nanomedicine, where researchers try to reduce non-specific uptake by precise surface modification.26The analytical techniques employed to measure nanoparticle uptake are quite diverse7,27 including transmission electron microscopy (EM),28–30 inductively-coupled plasma mass spectrometry (ICP-MS),31 ICP-atomic emission spectrometry,26 microplate reader,32,33 flow cytometry,5,29,34–38 confocal microscopy,5 transmission X-ray microscopy,39 whole cell tomography,40 confocal Raman microscopy,41 and hyperspectral imaging.42,43 These can be broadly classified into three approaches: EM techniques such as transmission EM (TEM) and scanning EM (SEM), ICP-based techniques, and fluorescence-based techniques such as microplate reader, flow cytometry, and confocal imaging.
Among them, TEM and confocal microscopy are the most widely used for nanoparticle uptake analysis. They have good spatial resolution but poor throughput, making them difficult to apply widely. While the ICP-based techniques have also been chosen for their more quantitative results, they are destructive, not easily accessible, and above all limited to metal particles. At present, there is no primary method to measure nanoparticle uptake, and therefore orthogonal methods often need to be complemented to achieve reliable results.9 For example, Gottstein et al. combined confocal microscopy with flow cytometry to precisely quantify the uptake of polystyrene particles,5 and Valero et al. used flow cytometry and imaging44 to identify molecular targets of drugs.
A flow cytometer is a device that measures the scattering and fluorescence of single particles (e.g. cells) in stream. Unlike other fluorescence-based methods such as microplate readers, it is non-destructive and measures individual cells rather than averaged signals from a population of cells. Forward and side scattering vary upon the size and complexity of the particles, respectively. The introduction of nanoparticles, such as TiO2, ZnO, SiO2, and Ag, to cells significantly increases side scatter, leading to side scattering often being used to quantify nanoparticle uptake in mammalian cells35,45,46 and bacteria.47 In addition, other cellular statuses, such as reactive oxygen species production and genotoxicity, can be measured in parallel using fluorescence channels.38 Since flow cytometry is a non-destructive analysis method, if necessary, the analyzed cells can be recovered using a cell sorter and used in other conventional analyses.
In this work, we provide a novel quantitative analysis approach to measure the level of nanoparticle uptake in mammalian cells via flow cytometry. Uptake is measured in two different ways: (1) by the percentage of cells containing nanoparticles, and (2) by the calibrated median fluorescence intensity of the cells. First, we demonstrate our method with a human lung carcinoma cell line, A549, and then apply the method to other cell lines from different tissue origins.
Results and discussion
Characterization of FITC-labeled silica nanoparticles (FITC-SiO2NPs)
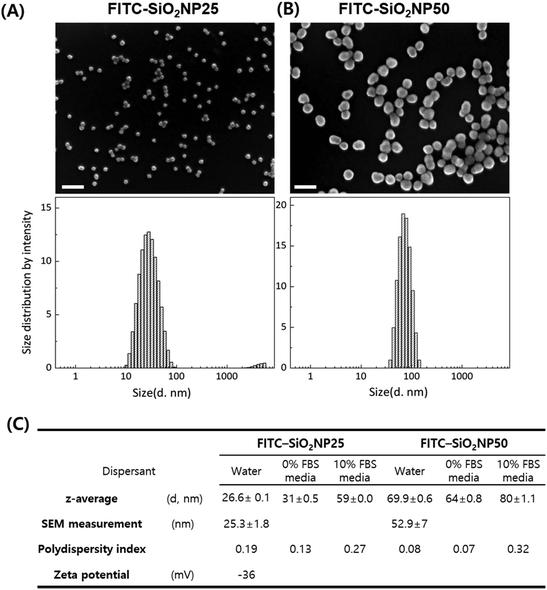
We synthesized and characterized fluorescein 5(6)-isothiocyanate-(FITC-) labeled SiO2 nanoparticles (NPs) of two different sizes, 25 and 50 nm. The nanoparticles were slightly angled rather than perfectly spherical, and their sizes were measured by SEM and dynamic light scattering (DLS, explanation on z-average and polydiversity index) as shown in Fig. 1. The measured size was slightly smaller with SEM but the difference was acceptable.The mechanisms for dye-doped silica nanoparticle synthesis have been well-documented,48,49 where silane-conjugated dye molecules are incorporated into silica nanoparticles covalently when a silica matrix is produced from the precursor, tetraethyl-orthosilicate (TEOS). Since FITC molecules are covalently incorporated into the silica matrix during synthesis, rather than being modified after nanoparticle synthesis, we anticipate that dye molecules will present both the inside and outside of nanoparticles. To confirm how stable the fluorescent molecules of the nanoparticles are, the fluorescence intensity of different concentrations of nanoparticle solutions was measured. We suppose that if nanoparticles were leaked out, the self-quenching effect between fluorescent molecules disappears and the fluorescence intensity changes. However, we observed no significant fluorescence decrease or change over 72 h. Considering the negligible deviation of fluorescence intensity among wells (≤5%), we could conclude that FITC molecules did not leach out from the silica nanoparticles (ESI Fig. S1†).
Quantification of nanoparticle uptake in A549 cells
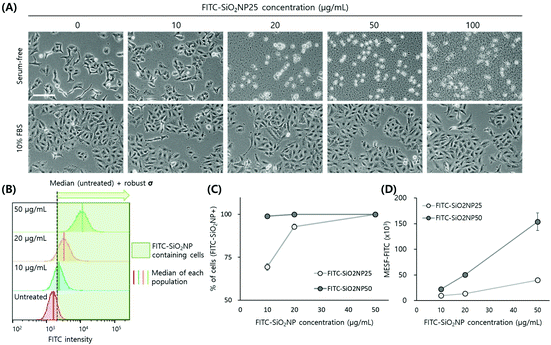
We choose the human lung carcinoma cell line A549 as a pulmonary exposure-mimicking in vitro model system to develop a nanoparticle uptake quantification method. We tested nanoparticle treatment ranging from 10 to 100 ng mL−1 under both serum-free and serum-containing conditions. The nanoparticle treatment duration was fixed at 24 h because in many standard protocols, such as ISO documents for nanoparticle-induced cytotoxicity and reactive oxygen species assays, this duration is employed to eliminate cell proliferation effects. It is important to minimize the dilution of intracellular nanoparticles due to cell cycling, because the doubling time of most mammalian cell lines is longer than 24 h. Muraca et al. employed a three-dimensional spheroid culture enabling the long-term monitoring of nanomaterials interacting with organisms, where cell cycling is effectively modulated and cells enter the quiescent (G0) phase.50A549 cells cultured under serum-free conditions for 24 h underwent morphological changes: they lost cell–cell contact and rolled up rather than spreading out on the culture dish. Cell proliferation was inhibited and fewer cells were observed via a micrograph even with the lowest tested nanoparticle concentration, 10 ng mL−1 FITC-SiO2NP25. Most cells detached and died when they were treated with FITC-SiO2NP25 at concentrations higher than 20 ng mL−1 possibly due to the adhesion of bare nanoparticles to the cell membrane. As Lesniak et al. explained,23 bare nanoparticles tend to adsorb to cell membranes to lower their surface energy, and this tendency accelerates as particle concentration increases.51 On the other hand, in the case of A549 cells in culture media containing 10% fetal bovine serum (FBS), the presence of nanoparticles, both NP25 and NP50 at concentrations as high as 100 ng mL−1, did not affect cell morphology, adhesion, or proliferation, with viability remaining high throughout the culture period (NP25 in Fig. 2(A), and NP50 in ESI Fig. S2†). Therefore, we decided to use the serum-containing medium in subsequent nanoparticle uptake analyses.
Before proceeding to the uptake analysis, we performed DLS measurements of the nanoparticles in the culture media because it has been well documented that proteins in serum rapidly adhere to nanoparticles to form a corona layer, which, consequently, changes the particle size and biological effects.33 As Lara et al. reported, certain proteins in the biomolecular corona are identified as key players in nanoparticle recognition and uptake.22 In the case of NP25, the size doubled and the z-average diameter increased to 59 nm. The size of NP50 also increased by the addition of 10% FBS, but by less than 15%, with the z-average diameter increasing to 80 nm. The z-average diameter of the nanoparticles was found to change over time-a 30% increase for NP25 and a 30% decrease for NP50 after 3 weeks (ESI Fig. S3†)-when the solution was stored at 4 °C. Therefore, we used freshly prepared nanoparticle solutions.
For the quantification of nanoparticle uptake, we adapted the flow cytometry method reported by Choi et al.,35 where the authors utilized changes in side scattering as induced by the uptake of silica nanoparticles. Their proposed method is simple and straightforward but the change in side scattering was insignificant with negatively charged particles due to the negative charge of the cell membrane. In our study, the side scattering of A549 cells did not change by the addition of silica nanoparticles presumably because of the negative surface charge and small particle size, while the fluorescence intensity of the nanoparticle-treated cells increased dramatically, with both cases ranging from 10 to 100 ng mL−1 (ESI Fig. S4†). Accordingly, we decided to use a fluorescence channel instead of a side scatter channel to quantify nanoparticle uptake.
We used two different approaches, as illustrated in Fig. 2(B). First, we quantified the percentage of cells containing FITC-SiO2NPs, as assigned by the cells with higher FITC intensities than the threshold intensity. The threshold value was the sum of the median intensity and robust standard deviation (SD) of control (i.e. untreated) cells. We used median and robust SD to obtain statistically meaningful measurements by excluding outliers in the broad histograms. Second, we calculated molecules of equivalent soluble fluorophore (MESF)52 values of the nanoparticle-treated cells. While the fluorescence intensity as measured using flow cytometers is an arbitrary value changed by instrument settings, MESF is a calibrated value that can be easily compared directly with the results of other experiments.
Following nanoparticle treatment, the resulting histogram from the FITC channel shifted to larger intensities in a dose-dependent manner, while other scattering parameters remained unchanged (ESI Fig. S2†). The percentage of cells containing FITC-SiO2NPs also increased in a dose-dependent manner, with most cells appearing to contain nanoparticles following treatment at concentrations higher than 20 ng mL−1 (Fig. 2(C)). In the case of NP50, the histogram shift was large, even at the lowest concentration of 10 ng mL−1. Many studies have reported that 50 nm is the optimum size for cellular uptake.16,53 However, since we do not have information on the FITC content inside the nanoparticles or the size-dependent difference in cellular uptake between the tested nanoparticles, the exact reason for the histogram shift cannot be specified in the present work.
To obtain MESF values for the nanoparticle-treated cells, we employed commercial hard-dyed beads (Rainbow Calibration Particles). We used six-peak particles dyed to six different fluorescence intensities and adjusted the voltage of the FITC channel to include all six peaks in the histogram. Reproducibility was reasonably sufficient to use the same calibration curve for different experiments performed over a period of about 3 months throughout the entire study (ESI Fig. S5†). Even with different voltage settings, correlations between MESF and FITC intensities did not change as long as all six peaks of the calibration particles were fit within the detection range. We subtracted the basal fluorescence level of the untreated cells from the MESF of the nanoparticle-containing cells. Similar to the percentage of cells containing FITC-SiO2NPs, the MESF increase following nanoparticle uptake was dose-dependent, with the increase by NP50 treatment greater than that by NP25 treatment when the cells were treated with the same concentrations (Fig. 1(D)).
Localization of nanoparticles in A549 cells
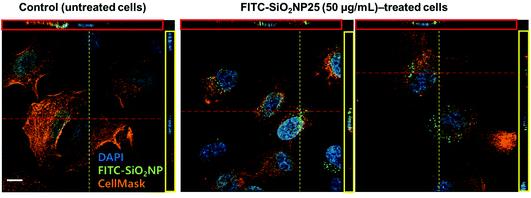
To verify the presence and location of the nanoparticles inside the cells, we imaged nanoparticle-treated cells with a confocal microscope. Often, the location of the nanoparticles (whether inside or on the surface of the cells) is not differentiated, with the amount of nanoparticles measured as a whole and referred to as cell-associated nanoparticles; however, the location is often important, especially when specific nanoparticle functionality is expected. Cell membranes were stained to differentiate the nanoparticles inside the cells from those adhered to the cell surface. Most nanoparticles existed in an aggregated form located near the nucleus rather than scattered throughout the cytoplasm (Fig. 3), clearly indicating that the nanoparticles near the nucleus were located inside the cells, as confirmed with confocal microscopy.Quantification and comparison of nanoparticle uptake in different cell types
Research has shown that the level of polystyrene nanoparticle uptake differs in different cell types,12 with other literature studies demonstrating that macrophages show much higher uptake levels as compared to other cell types.15,33 Other than these few studies though, the number of studies on nanoparticle uptake by cell type is limited.To demonstrate the versatility of the developed method, we quantified nanoparticle uptake in various cell lines from different organs, namely lungs, skin, intestine, and blood, which can be primarily exposed to nanoparticles. The list includes Beas-2B, A549, HCC827, and H1975 as lung cell lines, HaCaT, MNT1, and Hs294 T as skin cell lines, Caco2 as an intestinal cell line, and THP-1 cells as an immune cell line. Their representative micrographs are shown in ESI Fig. S6.† For THP-1 cells, we used both naïve and phorbol 12-myristate 13-acetate (PMA)-activated cells, referred to as THP-1 (M0).
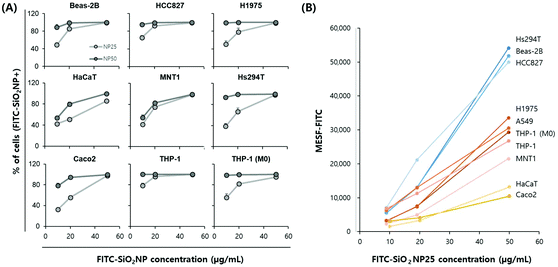
First, we analyzed the percentage of cells containing FITC-SiO2NPs in the nine tested cell lines (Fig. 4A). Lung cell lines and THP-1 cells, both naïve and M0, showed a similar pattern as A549 cells: a high uptake percentage for NP50 and a dose-dependent uptake increase for NP25. In general, the level of nanoparticle uptake was lower in skin and intestinal cells than in lung and immune cells. However, Hs294 and Caco2 cells treated with NP50 displayed uptake percentages comparable to the lung cell lines. Therefore, we could not conclude that the level of nanoparticle uptake can be classified according to the organ from which the cell line was derived.
Then, we analyzed MESF–FITC values from the median fluorescence intensity of an ensemble of cells treated with FITC-SiO2NP25 (Fig. 4B). Since the background level of each cell line in the FITC channel was not negligible and different for each cell line (ESI, Fig. S7†), we subtracted the background values for an objective comparison. Here, we present only the NP25 results because, for some cell lines, the FITC histogram for the highest nanoparticle concentration was truncated due to a high level of fluorescence. Cell lines were roughly divided into three groups based on the MESF level of the 50 ng mL−1 NP25-treated samples. The highest MESF-level group includes a skin cell line (Hs294T) and two lung cell lines (Beas-2B and HCC629); the middle includes two lung cell lines (H1975 and A549), immune cell lines (THP-1 and THP-1 (M0)), and a skin cell line (MNT1); and the lowest includes a skin cell line (HaCaT) and the intestinal cell line (Caco2). In general, lung cells showed higher MESF values while skin and intestinal cells showed lower MESF values; however, exceptions were observed, similar to the uptake percentage results. Notably, the three skin cell lines, Hs294T, MNT1, and HaCaT, all presented different levels of MESF values, with Hs294T showing very high results.
Since one of the main parameters affecting the cellular uptake of nanoparticles is cell size, and we already have that information in the form of forward scatter (FSC) from the flow cytometer data, we investigated the correlation between cell size and MESF value. Because FSC reflects the size of cells in the form of spheres and may not directly reflect the area of the cells in culture, we further estimated the cell areas of the eight adherent types of cells (except THP-1, which is a suspension type) using a cytoplasm staining dye, carboxyfluorescein diacetate succinimidyl ester (CFSE). The FSC (i.e. FSC-A) and CFSE-area results showed good linear correlation (R2 = 0.7851, ESI, Fig. S8A†), although the measured area depends somewhat on the flatness of the cells. For example, Caco2 cells maintained a round shape with a convex edge, and thus displayed a smaller attached area than other cell lines, resulting in a relatively lower CFSE-area value compared to FSC. In contrast, Beas-2B cells tended to spread out over the surface and remained flat, resulting in relatively a higher CFSE-area value compared to FSC (Fig. S8B†).
Interestingly, when we plot the cell size, both FSC and CFSE-area, versus MESF–FITC, the two parameters displayed a meaningful correlation, with the tested cell lines again roughly dividing into three groups (Fig. 5). The Hs294 T, Beas-2B, and HCC827 cells showed the largest area and the highest MESF–FITC; the A549, H1975, THP-1 (M0), THP-1, and MNT1 cells showed moderate area and MESF–FITC; and the Caco2 and HaCaT cells showed the smallest area and the lowest MESF–FITC. It is well known that nanoparticle uptake depends on both the physicochemical properties of the nanoparticles, such as size, surface charge, and shape, and the cells, such as tissue origin15,33 and cellular mechanical state.54 Our test results from numerous cell types also suggest that the surface area of the cells plays an important role in determining the level of nanoparticle uptake, regardless of the cell type, presumably due to the increased frequency of interaction between nanoparticles and cells.
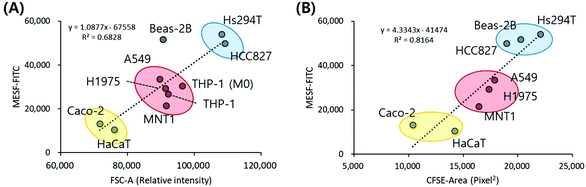 | ||
| Fig. 5 Correlation between cell size and nanoparticle uptake. Plots between median MESF values with (A) FSC-A obtained by flow cytometry and (B) CFSE-area obtained by image analysis. | ||
Lastly, we investigated the uptake dynamics for a few cell lines, namely A549, HCC827, HaCaT, and THP-1. Sample preparation for flow cytometry is relatively simple and fast, making it suitable for monitoring nanoparticle uptake dynamics. For A549 cells, a steady increase of nanoparticle uptake over time until 5 h (ref. 19) and until 24 h (ref. 53) has previously been reported. We observed a similar steady linear increase of nanoparticle uptake until 24 h (ESI, Fig. S9A–C†). Interestingly, while all tested adherent cells showed a linear increase in MESF–FITC values over time until 24 h, THP-1 cells showed an immediate uptake of nanoparticles upon nanoparticle exposure (Fig. S9D†). Even after 5 min, the minimal time to transfer and load a sample for flow cytometer measurement, after treatment, the level of MESF–FITC was similar to that of the 1 h sample. Macrophages are known to engulf foreign substances by their nature, and are known to act as scavengers for the delivery of nanoparticles using the blood stream.18 While a detailed investigation into the nanoparticle uptake mechanism in THP-1 cells is not the scope of this paper, our proposed method can be useful in analyzing nanoparticle uptake dynamics because the sample preparation time is significantly shorter than other assays.
In summary, we developed flow cytometry-based assays to quantify the level of nanoparticle uptake in mammalian cells and validated the assays with different types of cell lines. In-house synthesized FITC-labeled silica nanoparticles of two different sizes, average diameter 25 nm and 50 nm, were employed. The first approach to measure nanoparticle uptake simply quantified the percentage of nanoparticle-containing cells based on the median fluorescence intensity of the control cell population. Although this method allows for a rapid assessment of nanoparticle uptake, it does not reflect the amount of nanoparticles present in the cell. The second approach, therefore, measured the fluorescence intensity of a population of nanoparticle-treated cells with values normalized to calibration particles, MESF. This quantitative measure also provides information about the amount of nanoparticles in the cells, and enables the comparison of experiments from different setups. Assays with a flow cytometer are high-throughput measurements providing extensive information on thousands of individual cells. By using multiple fluorescence channels, numerous cellular states such as cell cycle, oxidative stress, and genotoxicity can be simultaneously monitored to investigate the correlation between cellular status and nanoparticle uptake. In addition, the assays are non-destructive and the cells can be recovered, allowing for further downstream analysis of the nanoparticle-treated cells. We examined our assays with A549 cells and other cell lines from different tissue origins, and found that the level of nanoparticle uptake differed among the tested cell lines and largely depended on cell shape and adhered surface area rather than tissue origin. One of the important factors involved in nanoparticle uptake, but not covered in detail in the current study, is corona proteins. Corona proteins are frontline molecules interacting with the cellular membrane during uptake and their crucial role has been highlighted in multiple articles. Recent studies focus on not only analyzing the composition and amount of the corona layer using mass spectrometry,19 but also understanding the underlying mechanism of nanoparticle uptake by cells through specific proteins in the corona layer. We believe that our quantification assays could accelerate the understanding of the interactions of nanoparticles with organisms along with those pioneering studies.
Methods
Nanoparticle synthesis and characterization
Fluorescein 5(6)-isothiocyanate (FITC, >90%, HPLC grade), tetraethyl orthosilicate (TEOS, >98%), 3-aminopropyl triethoxysilane (APTES, >99%), butanol (>99%), ethanol (anhydrous, >99.5%), ammonia (anhydrous, >99.98%), L-arginine (>98%), and cyclohexane (anhydrous, >99.5%) were provided from Sigma-Aldrich and all of the chemicals were analytical reagent grade. Ultrapure Milli-Q water (18.2 MΩ cm−1) was used for all the procedures.All procedures for FITC-SiO2NP synthesis were conducted while minimizing exposure to light. For 25 nm nanoparticle synthesis, we modified the method from Quan et al.55 In brief, an APTES–FITC mixture was prepared by mixing 2 mg of FITC, 400 μl of butanol, and 2 drops of APTES for 24 h. The APTES–FITC mixture was then mixed with 70 ml of 0.82 μM L-arginine solution, 12 ml of butanol, and 18 ml of TEOS, and allowed to react in a round-bottom flask under vigorous stirring at 65 °C for 18 hours. The aqueous phase, which contains highly monodisperse FITC-SiO2NPs, was obtained after removing the organic phase using a glass funnel. For 50 nm nanoparticle synthesis, we modified the method from Imhof et al.56 In brief, an APTES–FITC mixture was prepared by mixing 12 μg of FITC, 10 ml of ethanol, and 14 μg of APTES for 24 hours. The APTES–FITC mixture was mixed with 44.25 ml of ethanol, 2.1 ml of ammonia, and 0.75 ml of TEOS, and then kept in a round-bottom flask under moderate stirring at room temperature for 20 hours. The solvent was exchanged with deionized water using microcentrifugation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) and washed twice.
000 rpm, 10 min) and washed twice.
The morphology and size information of the FITC-SiO2NPs were obtained by SEM and DLS. SEM images of the 25 nm and 50 nm FITC-SiO2NPs were obtained using a HITACHI S-4800 and ZEISS GeminiSEM 500, respectively. The average particle size was obtained by measuring at least 100 single particles using ImageJ software. The hydrodynamic size and zeta potential were determined using a Zetasizer Nano ZSP (Malvern). DLS results are reported as the average of three runs.
Cell culture
A549 and Caco2 cells were maintained in minimum essential medium (MEM) supplemented with 10% FBS, 200 units per mL penicillin, 200 units per mL streptomycin, 2 mM L-glutamine; Beas-2B, HCC827, HaCaT, and MNT1 cells were maintained in DMEM supplemented with 10% FBS, 200 units per mL penicillin, and 200 units per mL streptomycin; H1975, Hs294 T, THP-1, and activated THP-1 cells were maintained in RPMI supplemented with 10% FBS, 200 units per mL penicillin, and 200 units per mL streptomycin. All cells were incubated at 37 °C in a humidified 5% CO2 atmosphere. THP-1 cells were activated by stimulating them with 320 nM PMA for 24 h. MNT1 and HS294 T cells were obtained from KCLB (Korea Cell Line Bank) and other cell lines were obtained from ATCC (American Type Culture Collection). Phase contrast images of the cells were obtained using an inverted microscope (Olympus IX71). All culture supplies were obtained from Thermo Fisher Scientific, unless otherwise indicated.Nanoparticle treatment
Cells were seeded in 6-well or 12-well plates at a density of 2–2.5 × 104 cells per cm2 and cultured for 24 h. The cells were washed with fresh media and incubated with various concentrations of nanoparticle solutions prepared in the culture media. After the treatment, cells were washed twice with 1× PBS to remove residual nanoparticles both in culture media and on the cell surfaces. The cells were kept at 4 °C until flow cytometry analysis.Nanoparticle uptake analysis using flow cytometry
Cells were analyzed using FACSVerse (BD Biosciences). In brief, the live single-cell population was gated in a plot of FSC versus SSC after excluding cell debris and doublets, and a histogram from the FITC channel for the single-cell population was obtained and analyzed using FlowJo (version X, FlowJo LLC). Each time the cells were measured, Rainbow Calibration Particles (6-peak, Spherotech) were run, which served as a quality control and later as a FITC intensity calibrator.Visualization of nanoparticle location using fluorescence microscopy
Nanoparticle-treated cells were harvested and re-plated on a coverslip. The next day, the cells were fixed with 4% paraformaldehyde and 0.1% Triton-X-100 for 2 min, and then post-fixed with 4% paraformaldehyde for 15 min. Cell membranes were stained with CellMask™ Orange for 30 min at room temperature, followed by nucleus counterstaining with 4,6-diamidino-2-phenylindole (DAPI) for 5 min. ProLong® Gold antifade reagent was used for mounting the coverslips. The cells were imaged and analyzed with an LSM 810 confocal microscope (ZEISS).Cell-area estimation by ImageJ
Cells were labeled using a CellTrace™ CFSE Cell Proliferation Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Fluorescence images of cells were obtained using an epifluorescence microscope (Olympus IX71) and analyzed using ImageJ. Briefly, images were first converted to 8-bit greyscale and then converted to binary images with a threshold value set to include the maximum cell attachment area. The area parameter for the binary mask was analyzed using the ‘Analyze Particles’ function.Statistical analysis
Experiments were independently repeated at least in triplicate. Error bars in the graphical data represent standard deviations. A two-tailed t-test was used for statistical analysis in Excel 2013 (Microsoft), and statistical significance was claimed when the p-value was lower than 0.05.Author contributions
Idea and study design: H.S. and J.Y.L. Data generation: H.S. and M.J.K. Data analysis: H.S., M.J.K., and J.Y.L. Manuscript writing: H.S., T.G.L., and J.Y.L.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Nano Material Technology Development Program (No. 2016M3A7B6908929) of the National Research Foundation (NRF) and KRISS-GP2018-0006-04 from the Korea Research Institute of Standards and Science from the Ministry of Science and ICT.References
- Z. Cheng, A. Al Zaki, J. Z. Hui, V. R. Muzykantov and A. Tsourkas, Multifunctional Nanoparticles: Cost Versus Benefit of Adding Targeting and Imaging Capabilities, Science, 2012, 338, 903–910 CrossRef CAS PubMed.
- P. Sanguansri and M. A. Augustin, Nanoscale materials development - a food industry perspective, Trends Food Sci. Technol., 2006, 17, 547–556 CrossRef CAS.
- B. Nowack and T. D. Bucheli, Occurrence, behavior and effects of nanoparticles in the environment, Environ. Pollut., 2007, 150, 5–22 CrossRef CAS PubMed.
- J. W. Wiechers and N. Musee, Engineered inorganic nanoparticles and cosmetics: Facts, issues, knowledge gaps and challenges, J. Biomed. Nanotechnol., 2010, 6, 408–431 CrossRef CAS PubMed.
- C. Gottstein, G. Wu, B. J. Wong and J. A. Zasadzinski, Precise quantification of nanoparticle internalization, ACS Nano, 2013, 7, 4933–4945 CrossRef CAS PubMed.
- H. Peuschel, T. Ruckelshausen, C. Cavelius and A. Kraegeloh, Quantification of Internalized Silica Nanoparticles via STED Microscopy, BioMed Res. Int., 2015, 2015, 1–16 CrossRef PubMed.
- A. Elsaesser, et al., Quantification of nanoparticle uptake by cells using microscopic and analytical techniques, Nanomedicine, 2010, 5, 1447–1457 CrossRef CAS PubMed.
- D. Vanhecke, F. Blank and A. Petri-fink, Quantification of nanoparticles at the single-cell level: an overview about state-of-the-art techniques and their limitations, Nanomedicine, 2014, 9, 1885–1900 CrossRef CAS PubMed.
- B. Drasler, D. Vanhecke, L. Rodriguez-Lorenzo, A. Petri-Fink and B. Rothen-Rutishauser, Quantifying nanoparticle cellular uptake: which method is best?, Nanomedicine, 2017, 12, 1095–1099 CrossRef CAS PubMed.
- S. D. Conner and S. L. Schmid, Regulated portals of entry into the cell, Nature, 2003, 422, 37–44 CrossRef CAS PubMed.
- A. Lesniak, et al., Rapid Growth Cone Uptake and Dynein-Mediated Axonal Retrograde Transport of Negatively Charged Nanoparticles in Neurons Is Dependent on Size and Cell Type, Small, 2018, 1803758, 1–10 Search PubMed.
- T. Dos Santos, J. Varela, I. Lynch, A. Salvati and K. A. Dawson, Quantitative assessment of the comparative nanoparticle-uptake efficiency of a range of cell lines, Small, 2011, 7, 3341–3349 CrossRef CAS PubMed.
- M. V. D. Z. Park, et al., The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles, Biomaterials, 2011, 32, 9810–9817 CrossRef CAS PubMed.
- S. Xiao, et al., Morphological and mechanical determinants of cellular uptake of deformable nanoparticles, Nanoscale, 2018, 10, 11969–11979 RSC.
- H. Herd, et al., Nanoparticle geometry and surface orientation influence mode of cellular uptake, ACS Nano, 2013, 7, 1961–1973 CrossRef CAS PubMed.
- P. Foroozandeh and A. A. Aziz, Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles, Nanoscale Res. Lett., 2018, 13, 1–12 CrossRef CAS PubMed.
- A. Verma and F. Stellacci, Effect of surface properties on nanoparticle-cell interactions, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. W. Chan, Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- A. Lesniak, et al., Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- A. Lesniak, et al., Serum heat inactivation affects protein corona composition and nanoparticle uptake, Biomaterials, 2010, 31, 9511–9518 CrossRef CAS PubMed.
- V. Francia, et al., Corona Composition Can Affect the Mechanisms Cells Use to Internalize Nanoparticles, ACS Nano, 2019, 13, 11107–11121 CrossRef CAS PubMed.
- S. Lara, et al., Identification of Receptor Binding to the Biomolecular Corona of Nanoparticles, ACS Nano, 2017, 11, 1884–1893 CrossRef CAS PubMed.
- A. Lesniak, et al., Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- S. Behzadi, et al., Cellular uptake of nanoparticles: Journey inside the cell, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- A. E. Nel, et al., Understanding biophysicochemical interactions at the nano–bio interface, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- J. Xie, C. Xu, N. Kohler, Y. Hou and S. Sun, Controlled PEGylation of monodisperse Fe3O4 nanoparticles for reduced non-specific uptake by macrophage cells, Adv. Mater., 2007, 19, 3163–3166 CrossRef CAS.
- A. Ivask, A. J. Mitchell, A. Malysheva, N. H. Voelcker and E. Lombi, Methodologies and approaches for the analysis of cell–nanoparticle interactions, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10, 1–22 Search PubMed.
- Z. Chu, Y. Huang, Q. Tao and Q. Li, Cellular uptake, evolution, and excretion of silica nanoparticles in human cells, Nanoscale, 2011, 3, 3291–3299 RSC.
- A. Kumar, A. K. Pandey, S. S. Singh, R. Shanker and A. Dhawan, A flow cytometric method to assess nanoparticle uptake in bacteria, Cytometry, Part A, 2011, 79 A, 707–712 Search PubMed.
- H. D. Summers, et al., Quantification of nanoparticle dose and vesicular inheritance in proliferating cells, ACS Nano, 2013, 7, 6129–6137 CrossRef CAS PubMed.
- X. Wei, et al., High-Throughput/High-Precision Sampling of Single Cells into ICP-MS for Elucidating Cellular Nanoparticles, Anal. Chem., 2018, 90, 13543–14550 Search PubMed.
- S. Jeon, et al., Surface Charge-Dependent Cellular Uptake of Polystyrene Nanoparticles, Nanomaterials, 2018, 8, 1028 CrossRef PubMed.
- M. Claudia, Ö. Kristin, O. Jennifer, R. Eva and F. Eleonore, Comparison of fluorescence-based methods to determine nanoparticle uptake by phagocytes and non-phagocytic cells in vitro, Toxicology, 2017, 378, 25–36 CrossRef CAS PubMed.
- X.-Y. Sun, Q.-Z. Gan and J.-M. Ouyang, Size-dependent cellular uptake mechanism and cytotoxicity toward calcium oxalate on Vero cells, Sci. Rep., 2017, 7, 1–12 CrossRef PubMed.
- S. Y. Choi, N. Yang, S. K. Jeon and T. H. Yoon, Semi-quantitative estimation of cellular SiO 2 nanoparticles using flow cytometry combined with X-ray fluorescence measurements, Cytometry, Part A, 2014, 85, 771–780 CrossRef PubMed.
- R. M. Zucker, E. J. Massaro, K. M. Sanders, L. L. Degn and W. K. Boyes, Detection of TiO2 nanoparticles in cells by flow cytometry, Cytometry, Part A, 2010, 77, 677–685 CrossRef CAS PubMed.
- H. Suzuki, T. Toyooka and Y. Ibuki, Simple and easy method to evaluate uptake potential of nanoparticles in mammalian cells using a flow cytometric light scatter analysis, Environ. Sci. Technol., 2007, 41, 3018–3024 CrossRef CAS PubMed.
- Y. Toduka, T. Toyooka and Y. Ibuki, Flow cytometric evaluation of nanoparticles using side-scattered light and reactive oxygen species-mediated fluorescence-correlation with genotoxicity, Environ. Sci. Technol., 2012, 46, 7629–7636 CrossRef CAS PubMed.
- H. H. Chen, et al., Quantitative analysis of nanoparticle internalization in mammalian cells by high resolution X-ray microscopy, J. Nanobiotechnol., 2011, 9, 14 CrossRef CAS PubMed.
- A. W. Sanders, K. M. Jeerage, C. L. Schwartz, A. E. Curtin and A. N. Chiaramonti, Gold Nanoparticle Quantitation by Whole Cell Tomography, ACS Nano, 2015, 9, 11792–11799 CrossRef CAS PubMed.
- N. B. Shah, J. Dong and J. C. Bischof, Cellular uptake and nanoscale localization of gold nanoparticles in cancer using label-free confocal Raman microscopy, Mol. Pharm., 2011, 8, 176–184 CrossRef CAS PubMed.
- M. Vetten and M. Gulumian, Differences in uptake of 14 nm PEG-liganded gold nanoparticles into BEAS-2B cells is dependent on their functional groups, Toxicol. Appl. Pharmacol., 2018, 363, 131–141 CrossRef PubMed.
- P. Vallotton, B. Angel, M. Mccall, M. Osmond and J. Kirby, Imaging nanoparticle-algae interactions in three dimensions using Cytoviva microscopy, J. Microsc., 2015, 257, 166–169 CrossRef CAS PubMed.
- T. Valero, et al., Drug ‘clicking’ on cell-penetrating fluorescent nanoparticles for in cellulo chemical proteomics, Bioconjugate Chem., 2018, 29, 3154–3160 CrossRef CAS PubMed.
- N. Bohmer, et al., Interference of engineered nanomaterials in flow cytometry: A case study, Colloids Surf., B, 2018, 172, 635–645 CrossRef CAS PubMed.
- X. Zhao and Y. Ibuki, Evaluating the toxicity of silver nanoparticles by detecting phosphorylation of histone h3 in combination with flow cytometry side-scattered light, Environ. Sci. Technol., 2015, 49, 5003–5012 CrossRef CAS PubMed.
- Y. Ibuki and T. Toyooka, Nanoparticle Uptake Measured by Flow Cytometry, Nanotoxicity, 2012, 926, 157–166 CAS.
- V. Gubala, G. Giovannini, F. Kunc, M. P. Monopoli and C. J. Moore, Dye-doped silica nanoparticles: Synthesis, surface chemistry and bioapplications. Cancer Nanotechnology, Springer, Vienna, 2020, vol. 11 Search PubMed.
- B. Korzeniowska, R. Nooney, D. Wencel and C. McDonagh, Silica nanoparticles for cell imaging and intracellular sensing, Nanotechnology, 2013, 24, 1–20 CrossRef PubMed.
- F. Muraca, et al., A Three-Dimensional Cell Culture Platform for Long Time-Scale Observations of Bio-Nano Interactions, ACS Nano, 2019, 13, 13524–13536 CrossRef CAS PubMed.
- J. S. Chang, K. L. B. Chang, D. F. Hwang and Z. L. Kong, In vitro cytotoxicitiy of silica nanoparticles at high concentrations strongly depends on the metabolic activity type of the cell line, Environ. Sci. Technol., 2007, 41, 2064–2068 CrossRef CAS PubMed.
- A. Schwartz, et al., Quantitating fluorescence intensity from fluorophore: The definition of MESF assignment, J. Res. Natl. Inst. Stand. Technol., 2002, 107, 83 CrossRef CAS PubMed.
- K. Shapero, et al., Time and space resolved uptake study of silica nanoparticles by human cells, Mol. BioSyst., 2011, 7, 371–378 RSC.
- X. Wang, et al., Influence of cell size on cellular uptake of gold nanoparticles, Biomater. Sci., 2016, 4, 970–978 RSC.
- B. Quan, C. Lee, J. S. Yoo and Y. Piao, Facile scalable synthesis of highly monodisperse small silica nanoparticles using alkaline buffer solution and their application for efficient sentinel lymph node mapping, J. Mater. Chem. B, 2017, 5, 586–594 RSC.
- A. Imhof, et al., Spectroscopy of Fluorescein (FITC) Dyed Colloidal Silica Spheres, J. Phys. Chem. B, 2002, 103, 1408–1415 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr01627f |
| This journal is © The Royal Society of Chemistry 2020 |