An electrophoretic DNA extraction device using a nanofilter for molecular diagnosis of pathogens†
Abstract
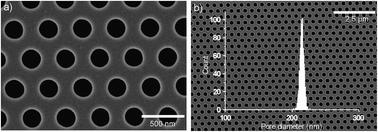
Rapid and efficient nucleic acid (NA) extraction and concentration are required for point-of-care analysis in order to prevent an epidemic/pandemic disease outbreak. Typical silica-based NA extraction methods have limitations such as being time-consuming, requiring human intervention, and resulting in a low recovery yield. In this study, we have developed a pathogenic DNA extraction device based on electrokinetic separation incorporated with a silicon nitride (SiNx) nanofilter, which expedites the DNA extraction procedure with advantages of being convenient, efficient, and inexpensive. This DNA extraction device consists of a computer numerical control (CNC) milled-Teflon gadget with a cis-chamber as a cell lysate reservoir and a trans-chamber as a elution solution reservoir, with the SiNx nanofilter being inserted between the two chambers. The SiNx nanofilter was fabricated using a photolithographic method in conjunction with nanoimprinting. Approximately 7.2 million nanopores of 220 nm diameter were located at the center of the nanofilter. When a DC electric field is applied through the nanopores, DNA is transferred from the cis-chamber to the trans-chamber to isolate the DNA from the cell debris. To demonstrate the DNA extraction performance, we measured the absorbances at 260 and 280 nm and performed a real-time polymerase chain reaction (real-time PCR) using the recovered DNA to verify its feasibility for downstream genetic analysis. Moreover, the DNA extraction device was successfully operated using a 1.5 V alkaline battery, which verifies the portability of the device for point-of-care testing. Such an advanced DNA extraction system can be utilized in various fields including clinical analysis, pathogen detection, forensic analysis, and on-site detection.
- This article is part of the themed collection: Coronavirus articles - free to access collection


 Please wait while we load your content...
Please wait while we load your content...