ZnO nanowire-integrated bio-microchips for specific capture and non-destructive release of circulating tumor cells†
Abstract
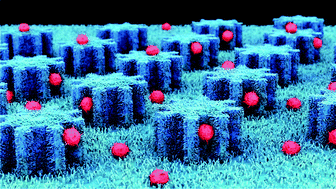
Circulating tumor cells (CTCs) are one type of significant biomarker in cancer patients’ blood that have been attracting attention from researchers for decades, and their efficient and viable isolation is of vital importance in cancer prevention and treatment. However, the development of efficient and low-cost bio-microchips still faces significant challenges. In this paper, we construct a novel three-dimensional micro-nano bio-microchip that has dual functions of specifically capturing and non-destructively releasing cancer cells. ZnO nanowire arrays were vertically grown on the surface of a polydimethylsiloxane (PDMS) pillar substrate with a gear structure (ZnO-coated G-PDMS pillar microchips). The gear structure provides more binding sites for antibodies and target cancer cells, while ZnO nanowires provide a rough surface for CTC attachment and size-specific effects for retaining CTCs. For subsequent culture and bioanalysis, the captured CTCs can be non-destructively released with high efficiency and good viability using a mild acidic solution treatment. Furthermore, the manufacturing process of the G-PDMS pillar microchips is convenient and low-cost, and the preparation approach of the ZnO nanowire is mature and simple to operate. In particular, the bio-microchips showed high capture efficiency (91.11% ± 5.53%) and excellent cell viability (96%) using a spiked cell sample. Moreover, we successfully achieved the specific fluorescent labeling of CTCs in 9 clinical breast cancer patients’ samples. The ZnO-coated G-PDMS pillar microchips not only have great potential for new target drug development for cancer stem cells but also open up new opportunities for individualized treatment.


 Please wait while we load your content...
Please wait while we load your content...