Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Guided by evolution: from biology oriented synthesis to pseudo natural products
Gregor S.
Cremosnik†
 a,
Jie
Liu†
a,
Jie
Liu†
 ab and
Herbert
Waldmann
ab and
Herbert
Waldmann
 *ab
*ab
aDepartment of Chemical Biology, Max-Planck-Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany. E-mail: herbert.waldmann@mpi-dortmund.mpg.de
bFaculty of Chemistry and Chemical Biology, TU Dortmund University, 44227 Dortmund, Germany
First published on 6th October 2020
Abstract
Covering: up to 2020
Natural products (NPs) provide inspiration for the design of biologically active compounds and libraries. In this review, we cover several experimental and in silico approaches, which have been used to simplify NPs and guide NP-based library design. Earlier approaches, like the structural classification of natural products (SCONP) and biology-oriented synthesis (BIOS), focus on the identification of activity determining scaffolds and the synthesis of corresponding compound collections. More recently, NP fragments identified by means of cheminformatic analysis of the Dictionary of Natural Products (DNP) have been combined in unprecedented fashions to yield pseudo natural products (pseudo NPs), which show biological activities unrelated to the guiding NPs. Each approach was also the source of chemical innovation, in which synthetic methods were established for the rapid assembly of NP-inspired compounds and libraries.
1 Introduction
Harnessing nature's pool of biologically active molecules has been a central topic of chemical biology and drug discovery programs.1,2 Natural products (NPs) offer highly three-dimensional scaffolds, which have been pre-validated by nature.1 Therefore, NPs have served as versatile platforms for the discovery of novel biologically active compounds. Advances in synthetic methodology and total synthesis have enabled access to complex scaffolds and the chemoselective derivatisation thereof for structure–activity relationship (SAR) studies.3–7 In addition, through elegant distortions of the NPs core-scaffold, unprecedented structural motives can be accessed. This approach was termed “Complexity-to-Diversity” (CtD) and enabled the discovery of novel biologically active molecules from terpenes,8,9 steroids8 and alkaloids.8,10–14 NPs have also been used to generate small molecule libraries for drug discovery. For example, researchers at Novartis have assembled a 150-membered fragment library from four NPs (tacrolimus, sanglifehrin A, cytochalasin E and massarigenin C) using degradation and diversification reactions previously employed in structure elucidation.15Associated with the synthetic challenges involved in NP total synthesis, methods and approaches have also focused on reducing structural complexity of NPs while keeping biological relevance. Such design approaches may arrive at NP-inspired compounds which still represent the kind of bioactivity encoded by the guiding NPs, or they may yield classes of NP-inspired compounds that display novel bioactivities.
Here we review two concepts for the design and synthesis of novel NP-inspired compound classes. In Biology Oriented Synthesis (BIOS), NP scaffolds are simplified in an iterative process arriving at series of structurally simplified, NP-inspired compound classes. Extension of the logic underlying BIOS and combining it with fragment-based compound development led to the concept of pseudo natural products (pseudo-NPs) in which NP fragments are combined in arrangements not available via biosynthesis.16,17
2 Structural classification of natural products (SCONP)
In general the bioactivity of NPs can be related to structural features.18 In an attempt to identify structurally simplified compound classes that still resemble the underlying structures of NPs, and thereby their biological relevance and activity, a cheminformatic method for structural classification of natural products (SCONP) was developed.To this end the CRC Press Dictionary of Natural Products (DNP, 190![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 939 compounds at the time) was employed as an initial dataset.18In silico fragmentation of the NPs and subsequent building of a scaffold tree based on substructure identification enabled a visual representation of scaffold relationships (Fig. 1). In the scaffold tree, more complex scaffolds are arranged on the outside and become simpler towards the centre.18
939 compounds at the time) was employed as an initial dataset.18In silico fragmentation of the NPs and subsequent building of a scaffold tree based on substructure identification enabled a visual representation of scaffold relationships (Fig. 1). In the scaffold tree, more complex scaffolds are arranged on the outside and become simpler towards the centre.18
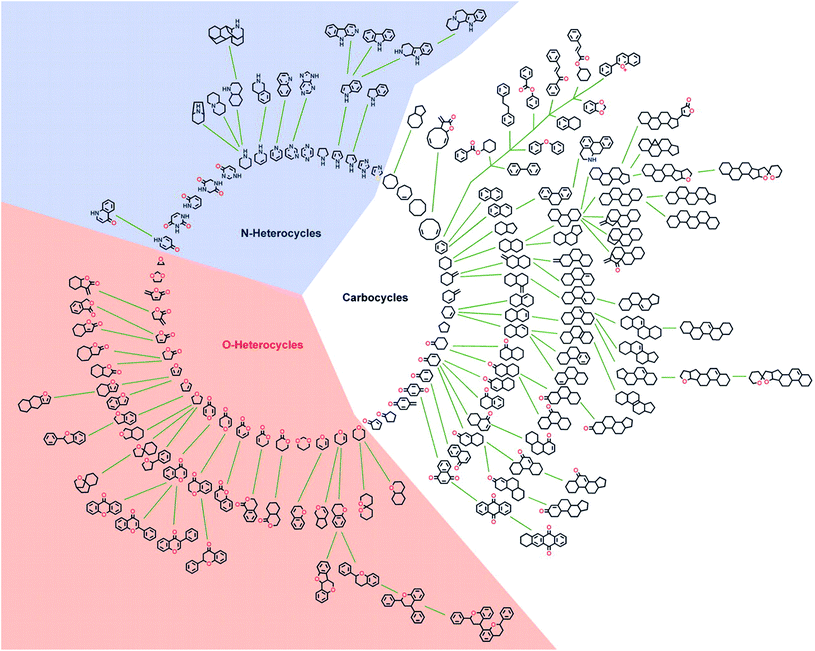 | ||
| Fig. 1 Graphical star-like representation of the NP scaffold tree. For clarity of the graphic illustration, only scaffolds are shown that represent cumulatively at least 0.2% of the NP population in the DNP. Reprinted from ref. 18, Copyright (2005) National Academy of Sciences. | ||
The SCONP concept was further validated using protein structure similarity clustering (PSSC) as an independent guiding principle (Scheme 1A) for the discovery of a selective 11β-hydroxysteroid dehydrogenase type 1 (11βHSD1) inhibitor.18 PSSC builds structure similarity clusters of proteins with similar ligand-sensing domains in which cluster members are expected to have similar ligands. 11βHSDs were found in the same structure similarity cluster as the Cdc25A phosphatase and acetylcholinesterase. Through fragmentation according to SCONP, dysidiolide, a CdC25A inhibitor, could be related to glycyrrhetinic acid (GA), a naturally occurring unselective ligand for 11βHSD1 and 11βHSD2. Based on their common octahydronaphthalene scaffold 1, a library of 162 compounds was prepared of which 28 were confirmed as selective inhibitors of 11βHSD1 over 11βHSD2 (compounds 1a–1c, Scheme 1B).
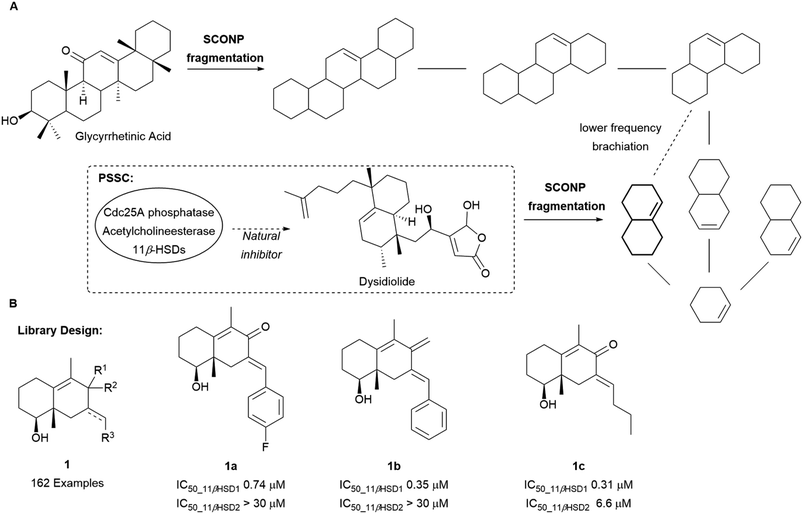 | ||
| Scheme 1 (A) Brachiation of glycyrrhetinic acid and dysidiolide to identify a common fragment. (B) Examples of 11βHSD1 selective inhibitors synthesised. | ||
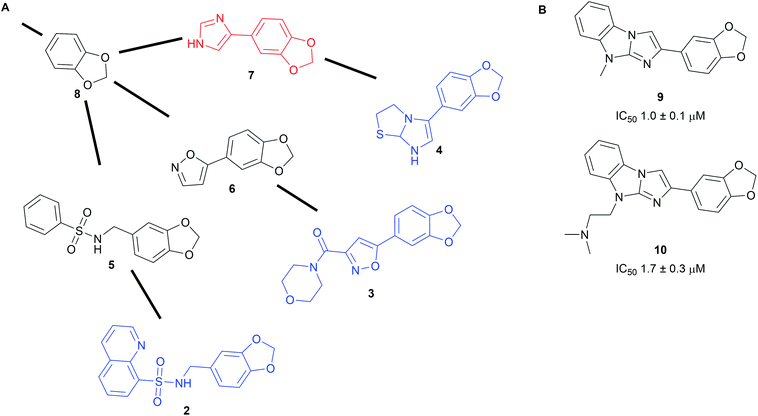
To harness the power of the SCONP approach and facilitate the building of scaffold trees, the open source software Scaffold Hunter was developed.19,20 The software uses complex data from e.g. biological screening sets as input and extracts chemically meaningful scaffolds sharing biological activity. From those it builds a scaffold tree linking related scaffolds but it also generates virtual scaffolds, where links between tested molecules are missing. The virtual scaffolds represent unexplored chemical space expected to share the bioactivity of the input and can serve as starting points for novel synthesis efforts.19 Prospective exploration using Scaffold Hunter was demonstrated with data from a pyruvate kinase screen available on PubChem (Scheme 2A).19 The program built a scaffold tree containing 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 868 scaffolds (for example fragments 2–4) from the 51
868 scaffolds (for example fragments 2–4) from the 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 415 unique molecules used in the assay. Subsequently, novel virtual scaffolds (fragments 5–7) were predicted to share the biological activity of the tested molecules. A library of 107 compounds embodying these virtual scaffolds was obtained and their half-maximal inhibitory concentrations (IC50) were determined. Nine novel pyruvate kinase inhibitors or activators with IC50 values between 1 and 10 μM were identified (selected examples 9 and 10, Scheme 2B). This corresponds to a hit rate of nearly 10% which is substantially higher than the average hit rate in PubChem high-throughput screening campaigns (0.34%).
415 unique molecules used in the assay. Subsequently, novel virtual scaffolds (fragments 5–7) were predicted to share the biological activity of the tested molecules. A library of 107 compounds embodying these virtual scaffolds was obtained and their half-maximal inhibitory concentrations (IC50) were determined. Nine novel pyruvate kinase inhibitors or activators with IC50 values between 1 and 10 μM were identified (selected examples 9 and 10, Scheme 2B). This corresponds to a hit rate of nearly 10% which is substantially higher than the average hit rate in PubChem high-throughput screening campaigns (0.34%).
The SCONP tree provides a systematisation to aid compound library design. In combination with a second independent criterion, SCONP can provide readily accessible NP scaffolds retaining affinity to proteins and biological activity. Even though the generated libraries were comparably small, high hit rates were obtained.19,21,22 It should be noted that with increasing deviation from the parent scaffold, e.g. to simpler scaffolds, biological similarity will decline.
3 Natural product fragments
While building the scaffold tree, parts of the scaffold are systematically removed to ultimately arrive at fragment-sized scaffolds. These fragments might offer valuable attachment points for efficient growth of the fragment. This was addressed in a subsequent analysis of the DNP (183![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 molecules, Scheme 3A).23 The fragmentation was performed following a set of rules that builds on the SCONP analysis, yet retains side-chains, functional groups, hybridisation states and stereochemistry (Scheme 3B). All obtained fragments were kept (751
704 molecules, Scheme 3A).23 The fragmentation was performed following a set of rules that builds on the SCONP analysis, yet retains side-chains, functional groups, hybridisation states and stereochemistry (Scheme 3B). All obtained fragments were kept (751![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 577 fragments) and further filtered through the application of an HTS filter to remove potentially unstable, toxic and undesirable fragments. The dataset was further tailored by removing macrocycles and using settings inspired by an adaptation of the original ‘rule of three’ (i.e. molecular weight < 300 Da, fewer than three hydrogen bond donors and acceptors, Clog
577 fragments) and further filtered through the application of an HTS filter to remove potentially unstable, toxic and undesirable fragments. The dataset was further tailored by removing macrocycles and using settings inspired by an adaptation of the original ‘rule of three’ (i.e. molecular weight < 300 Da, fewer than three hydrogen bond donors and acceptors, Clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 3)24,25 to NP structure (i.e. molecular weight < 350 Da, at most 3 hydrogen bond donors, at most 6 hydrogen bond acceptors, at most 6 rotatable bonds and Alog
P < 3)24,25 to NP structure (i.e. molecular weight < 350 Da, at most 3 hydrogen bond donors, at most 6 hydrogen bond acceptors, at most 6 rotatable bonds and Alog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 3.5) to afford a set of 110
P < 3.5) to afford a set of 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 485 fragments. Subsequent pharmacophore clustering generated 2000 clusters representing groups of related fragments. Comparing the molecular properties (e.g. O/N-count, Fsp3, number of rings) of this set to fragments obtained from the ZINC and SynLib libraries by principal component analysis (PCA) showed distinct differences in the covered chemical space. The NP-fragments in general showed a higher average Fsp3 content and O-count, with fewer aromatic rings present. The biological relevance of the obtained clusters was tested by searching for novel p38α MAP kinase inhibitors. Nine fragments out of 193 tested cluster centres were found to bind in various modes, as observed by X-ray crystallography. The screening revealed that sp3-rich fragments derived from cytisine and sparteine are novel allosteric inhibitors and stabilise the DFG motive in its inactive form (e.g.11 and 12 in Scheme 3C). The optimisation of binding was guided by employing structurally similar clusters to increase the diversity of the library.
485 fragments. Subsequent pharmacophore clustering generated 2000 clusters representing groups of related fragments. Comparing the molecular properties (e.g. O/N-count, Fsp3, number of rings) of this set to fragments obtained from the ZINC and SynLib libraries by principal component analysis (PCA) showed distinct differences in the covered chemical space. The NP-fragments in general showed a higher average Fsp3 content and O-count, with fewer aromatic rings present. The biological relevance of the obtained clusters was tested by searching for novel p38α MAP kinase inhibitors. Nine fragments out of 193 tested cluster centres were found to bind in various modes, as observed by X-ray crystallography. The screening revealed that sp3-rich fragments derived from cytisine and sparteine are novel allosteric inhibitors and stabilise the DFG motive in its inactive form (e.g.11 and 12 in Scheme 3C). The optimisation of binding was guided by employing structurally similar clusters to increase the diversity of the library.
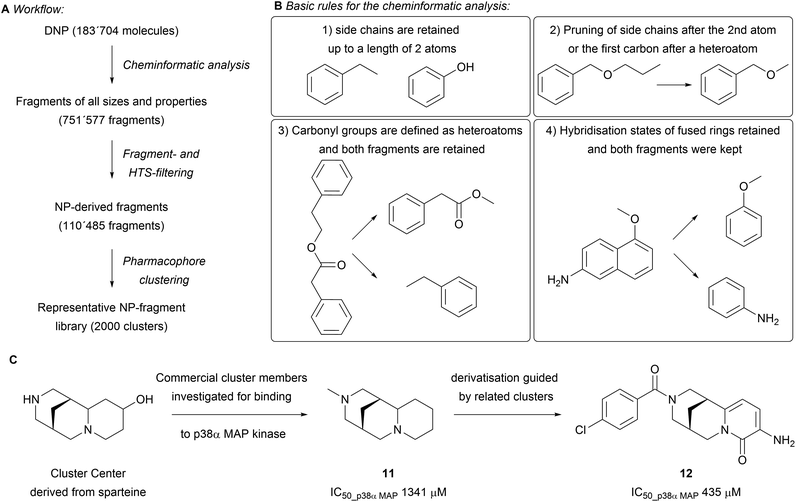 | ||
| Scheme 3 (A) Flowchart of the performed cheminformatic analysis; (B) the guiding rules for fragmentation; (C) principle for the discovery of novel p38α MAP kinase binders. | ||
This example highlights the potential of the generated clusters for development into bioactive molecules and to guide optimisation attempts. The fragment clusters retain many attributes of NPs and cover chemical space outside commercial libraries. Although the obtained fragments and cluster centres are structurally less complex than their guiding NPs, their synthesis will often require considerable effort and the development of novel methods.
In silico fragment generation from NPs with annotated biological activity or from high-throughput screening data is a powerful way to explore novel chemical space. The obtained fragments are often readily accessible, allowing for the creation of large libraries. It is important to note that fragments usually inherit the biological activity of the parent compound although they may vary in selectivity towards similar targets.20 Alternatively, fragments and structural clusters can be used as guiding structures for the rapid assessment of biological activity and subsequent development of more potent compounds.
4 Biology-oriented synthesis
The aim of biology-oriented synthesis (BIOS) is the structural simplification of NP scaffolds to obtain new compound classes, which cover biologically relevant space.1,26 Subsequently, a synthesis route is developed to give a NP-inspired compound collection for biological studies (Scheme 4). Over the past two decades, several NP scaffolds have been simplified through BIOS and have led to the discovery of novel biologically active compounds. The examples presented here were chosen due to related synthetic strategies relying on cycloaddition reactions.The 1,3-dipolar cycloaddition is a powerful method to introduce pyrrolidine moieties into scaffolds.27–29 The development of regio-, diastereo- and enantioselective 1,3-dipolar cycloadditions makes it a versatile tool for synthesis.27 Ningalin B and lamellarin D are two classes of marine natural products exhibiting interesting biological activities including antitumor activity, HIV-1 integrase inhibition and multidrug resistance reversal activity.30 The synthesis of the common pyrrolocoumarine core-scaffold was achieved through intramolecular 1,3-dipolar cycloadditions from alkynes 13 (Scheme 5).31 Cell-based screening of a collection of 43 compounds revealed Pyrcoumin as an inhibitor of canonical Wnt signaling.32 Further target identification and validation showed that it targets the nucleotide pyrophosphatase dCTPP1.33
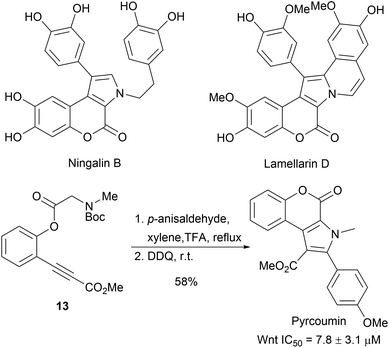 | ||
| Scheme 5 One pot synthesis of pyrcoumin. IC50 values determined by Wnt-responsive reporter gene assays in HEK293-Fz cells. | ||
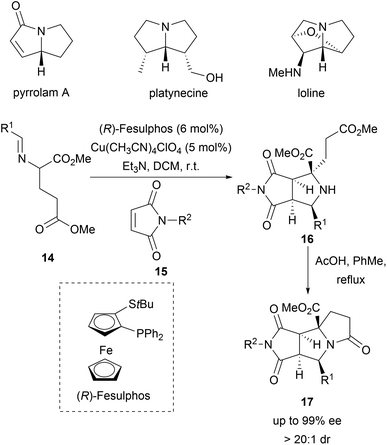
Pyrrolizidines are characteristic for a large group of alkaloids which exhibit insecticidal activity.34 A dual cyclization strategy was developed to access a pyrrolizidine-like compound collection (Scheme 6). Enantioselective Cu-catalysed 1,3-dipolar cycloadditions between azomethine ylides (14) and N-methylmaleimides (15) delivered bicyclic pyrrolidines 16 with excellent diastereoselectivity and enantioselectivity (Scheme 6).35 Subsequent intramolecular lactamisation provided the tricyclic pyrrolizidine-alkaloid analogues 17 in high yields. Of the 119 compounds, four compounds inhibited parasite growth.
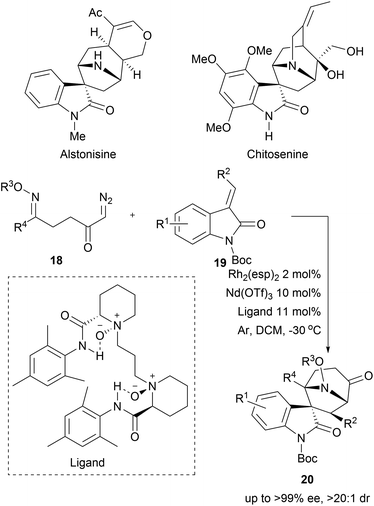
1,3-Dipolar cycloadditions have also been used to access bridged structures with high stereocomplexity. In a recent example, cyclic azomethine ylides were generated from α-diazo ketones 18 by means of rhodium catalysis. The cycloaddition between the transient cyclic azomethine ylides and the oxindole-derived alkenes 19 delivered spirotropanyl oxindole scaffolds 20 with good to excellent diastereo- and enantioselectivity (Scheme 7).36 Utilizing bimetallic relay catalysis, 24 compounds were synthesised enabling a BIOS of spirotropanyl oxindole alkaloids similar to alstonisine and chitosenine.
Apart from the synthesis of NP scaffolds by novel methodologies, established multistep syntheses of NP fragments have been used to access simplified scaffolds of NPs, resembling a function-oriented synthesis (FOS)37,38 approach but with library synthesis in mind. Withanolides are a family of at least 300 steroids exhibiting diverse bioactivities, including potent anti-inflammatory effects or modulation of the mTOR and Wnt pathways.39 Comparison of the structural features of withanolides showed that many retained the trans-hydrindane unit (C/D rings) linked to an α,β-unsaturated lactone (Scheme 8). The A/B rings of the steroid scaffold on the other hand showed variability. This observation led to the assumption that the retained structural features of the C/D ring may be responsible for common biological activity along with the modification of the dehydro-δ-lactone moiety.40 A compound collection was assembled based on these observations using a multi-step sequence and late-stage derivatization.40 Prins reaction of olefin 21 with paraformaldehyde afforded alcohol 22 as a single stereoisomer. Diastereoselective hydrogenation of the double bond introduced another stereocenter. Swern oxidation followed by a diastereoselective Brown allylation gave the homoallylic alcohol 23. After removing the acetal protecting group, an appendage was incorporated into the scaffold through an esterification reaction. Ring closing metathesis furnished the dehydro-δ-lactone moiety. This core scaffold (24) was further derivatised to obtain a diverse library (examples 25 and 26). Biological evaluation of the compound class revealed novel modulators of the hedgehog signalling pathway targeting the Smoothened (Smo) protein.41 Notably, the specific bioactivity was modulated by the appendages on the C ring. Withanolides have not been identified as hedgehog pathway inhibitors before.
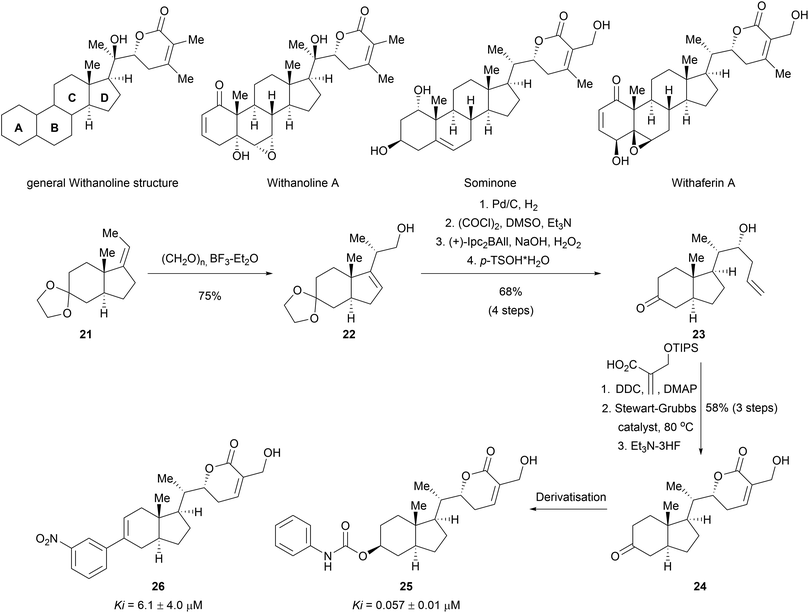 | ||
| Scheme 8 Biology-oriented synthesis of withanolid-inspired compound collection. Ki value for smoothened binding. | ||
BIOS provides a strategy to access biologically active compounds based on natural products through truncations and appendage modifications. This has also been exploited to assemble libraries fit for drug discovery.42,43 Natural products were truncated to their core scaffold while keeping functionalities intact, which could be used for derivatisation and growing of the scaffold.42 The obtained libraries were used in a rescaffolding approach for the discovery of novel inhibitors. Interactions of known inhibitors were analysed based on crystal structures and provided the basis for scaffold selection. In the example of the caspase-1 inhibitor PGE-3935199, it was found important that keeping the biaryl- and lactone side chains intact is crucial (Scheme 9A).42 Truncation between C10 and C11 as well as C20 and N23 enabled screening for a replacement of the 5,8-fused bicyclic core of PGE-3935199. A screening through the assembled virtual library identified 16 NP scaffolds fitting the descriptors. Out of these the bridged 2-azabicyclo[2.2.2]octane (2-ABO), a core scaffold of several marine NPs, offered a compelling solution to the problem as docking studies predicted an inhibitor in the nM affinity range. Synthesis and biological testing of CD10847 confirmed these predictions and an IC50 17 nM was obtained in cell-based assays.
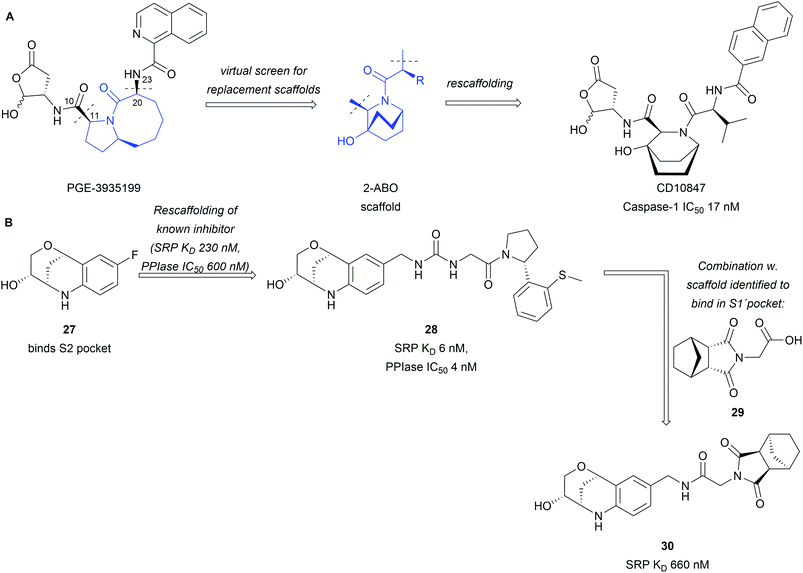 | ||
| Scheme 9 Rescaffolding of known inhibitors with BIOS inspired scaffolds: (A) rescaffolding of a caspase-1 inhibitor; (B) BIOS inspired CypD inhibitors. | ||
The same library was also employed in the discovery of Cyclophilin D (CypD) inhibitors through high throughput screening combined with X-ray crystallography.43 Tetrahydroquinoline derivative 27 was identified to bind into the S2 pocket (Scheme 9B). Rescaffolding of a known inhibitor led to compound 28 with significant increase in potency and affinity to the protein. In the same campaign succinimide 29 was found to bind into the S1′ pocket of the target protein. The two scaffolds were subsequently combined (30) with a fitting linker but no further improvement in potency was observed.
5 Pseudo natural products
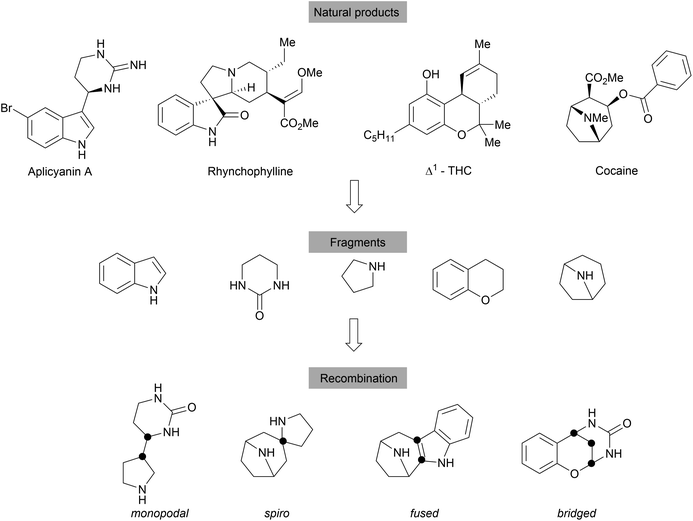
With an emphasis on the synthesis of specific NP scaffolds, BIOS only explores the chemical and biological space covered by the guiding NPs. This biased approach limits the exploration of the wider chemical- and biological space, i.e. new chemical entities and mechanisms of actions (MoAs). By analogy, a retrospective analysis on the discovery of NPs shows that the NP space is largely limited and the number of fundamentally novel scaffolds discovered from nature is decreasing.44,45 To overcome such limitations, biosynthesis may combine two unrelated synthetic routes to generate hybrid NPs, for example hybrid PKS-NRPS,46 terpenoid alkaloids47 and meroterpenoids.48 However, the biosynthetic efficiency is limited by the accessible precursors among microorganisms and plants and the available biosynthetic machinery.Thus, the chemical combination of two biosynthetically unrelated NP fragments would yield novel compound classes still endowed with biological relevance and would cover novel chemical and potentially biological space (Scheme 10).16,49 Such hybrid compounds may be regarded as pseudo natural products (pseudo NPs).16 Similar to diversity oriented synthesis (DOS)50 libraries, the fragment based design principle can lead to compound collections with high chemical diversity. However, pseudo NPs are biologically prevalidated by the choice of fragments directly derived from natural products, while DOS libraries mostly are built with chemical and structural considerations as predominating arguments.16 Compounds synthesised according to DOS can be considered NP-like as they encompass many attributes of NPs (e.g. high Fsp3 content) but their biological relevance is initially unclear.16 Pseudo NP library synthesis also does not necessarily follow the build/couple/pair strategy,51 which was very successfully applied in DOS. Inspiration for suitable fragments to combine was found in the fragment clusters from the cheminformatic analysis of the DNP.23 The identified 2000 cluster centres (see above), most of which are mono- or bicyclic scaffolds, offer a large variety of basic fragments for recombination. To increase complexity, NPs or their derivatives can serve as suitable platforms for combination with simpler fragments. Different connection types and positions lead to topologically diverse structures that may also differ in biological activities. According to the number of shared atoms between the fragments, recombined structures can be divided into four main groups, including monopodal connections (0 atom), spirocycles (1 atom), fused rings (2 atoms) and bridged scaffolds (≥3 atoms). Determination of the NP-likeness score52 of different fragment combinations showed that pseudo NPs are distinctly different from NPs and BIOS compounds but overlap with the space covered by drugs.16,49,53,54
Differing from BIOS, where the guiding NPs can suggest possible biological targets, pseudo NPs are expected to exhibit different biological activity than their individual fragments. To biologically characterize these compound collections, target-agnostic screening methods can be applied.55–58 The “Cell Painting” assay has shown great potential in characterizing pseudo NPs. Through staining of different cellular compartments and structures, phenotypic changes induced upon compound treatment are observed by microscopy. The obtained morphological profiles (fingerprints) can be compared to reference compounds with known biological activity to arrive at a target hypothesis, which has to be further confirmed in more specific assays. The outlined strategy has led to the discovery of several novel biologically active compound classes with different molecular topologies, which will be discussed below. The focus is set on the employed chemistry as well as the biological characterisation, and is categorised by fragment recombination patterns.
5.1 Bridged recombination
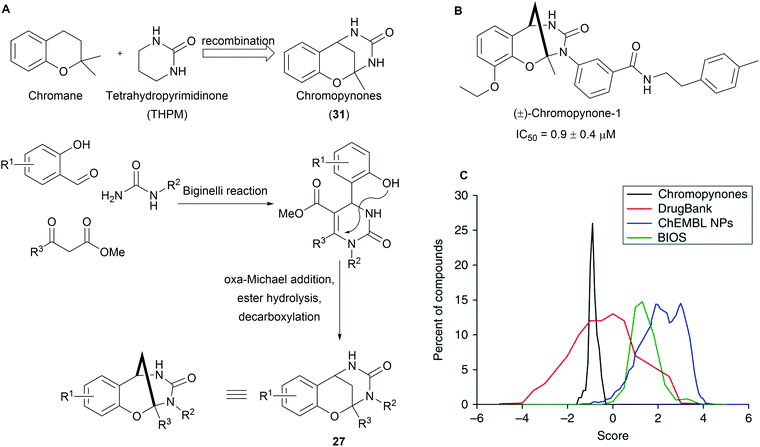
Chromopynones (31) combine biosynthetically unrelated chromane and tetrahydropyrimidinone (THPM) fragments and were synthesized by means of a Biginelli reaction followed by an intramolecular cyclization (Scheme 11A).49 The obtained library was investigated in several cell-based assays and an inhibition of glucose uptake in HCT116 cells was observed (Scheme 11B). Further biological investigations revealed that chromopynones are selective inhibitors of the glucose transporters GLUT-1 and -3, and inhibit cancer cell growth.59 Furthermore, neither chromane- nor THPM fragments exhibit any activity towards glucose transporters; therefore the combination of chromane and THPM is essential for the biological activity. The NP-likeness scores of the synthesised chromopynones are comparable with synthetic drugs but distinctly differ from the scores calculated for NPs and BIOS collections (Scheme 11C).Tropane and indole are two common fragments found in NPs. The recombination of these two fragments afford highly sp3-rich indotropane pseudo NPs (32, Scheme 12).60 Their enantioselective synthesis was achieved via 1,3-dipolar cycloaddition of imine 33 and nitroalkenes 34. This scaffold can also be seen as a bridged recombination of tryptoline and pyrrolidine. Phenotypic screening of the obtained collection resulted in the discovery of myokinasib, which induces striking changes in mouse L fibroblast morphology indicating an involvement in cytokinesis.53 Kinase screening and target validation revealed that myokinasib is a mixed-type myosin light chain kinase 1 (MLCK1) inhibitor. This chemotype was unprecedented for kinase inhibitors.
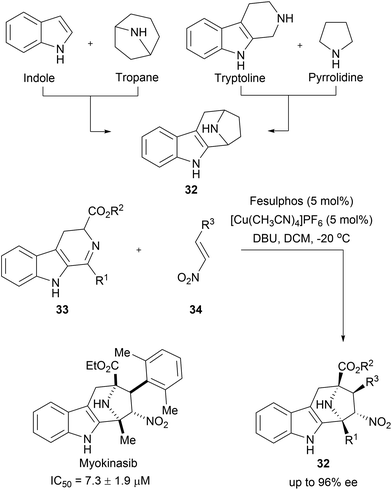 | ||
| Scheme 12 Enantioselective synthesis of indotropanes (32). IC50 data represents the ability to inhibit MLCK1. | ||
5.2 Fused recombination
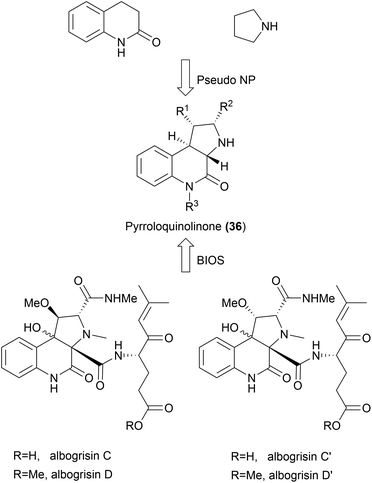
The 1,3-dipolar cycloaddition also proved to be applicable for the synthesis of a variety of pseudo NPs with fused fragment combination (Scheme 13). A combination of pyrrolidine- and quinolinone fragments was obtained from amide 35 through an enantioselective intramolecular 1,3-dipolar cycloaddition to afford pyrroloquinolones (36, Scheme 13A).61 The high selectivity of the method could furthermore be used for the kinetic resolution of racemic tropanes 37 to afford a rich collection of enantiopure pyrrotropanes (38, Scheme 13B).62 Finally, pyrriridoids (39) were assembled from pyrone fragments 40 through 1,3-dipolar cycloadditions affording highly stereogenic compounds with five contiguous stereocenters (Scheme 13C).63 Several members of the obtained libraries showed promising activities in Wnt- and Hedgehog reporter-gene assays.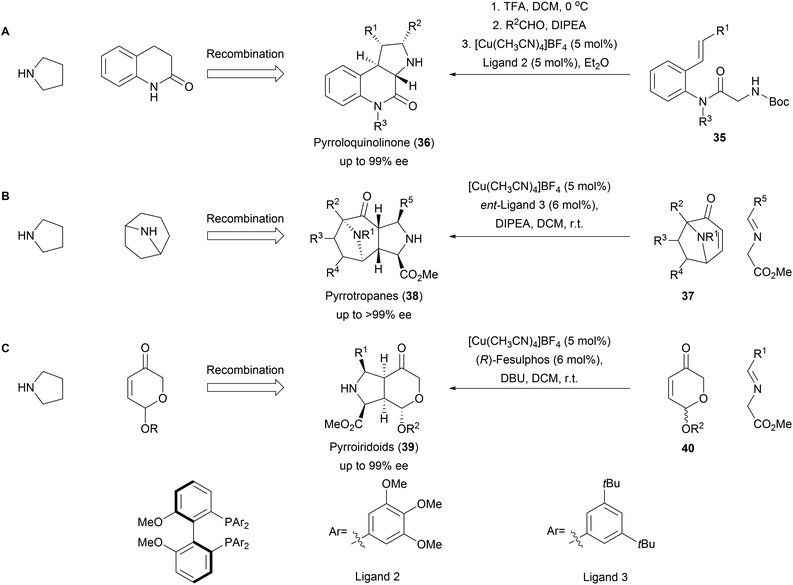 | ||
| Scheme 13 Asymmetric fusion of pyrrolidine moiety into diverse NP fragments. Synthesis of the pseudo NP classes pyrroloquinolinone (A), pyrrotropanes (B) and pyrroiridoids (C). | ||
While the aim of the pseudo NP strategy is to combine fragments in a manner not yet observed in nature, it is possible that certain scaffolds may exist in nature but may not have been isolated from natural sources yet. In 2015, pyrroloquinolinones were synthesized as one of the first examples of pseudo NPs by combining pyrrolidine and quinolinone fragments (36, Scheme 14).61 More recently, this scaffold has been isolated from Streptomyces albogriseolus MGR072.64 While pyrroloquinolinones 36 cannot be considered pseudo NPs anymore, they would still fit the BIOS principle. More examples like this will emerge with the discovery of novel NPs. Such discoveries highlight the strength of the pseudo NP principle in providing biologically active compounds.
Indole is frequently present in naturally occurring bioactive molecules. Owing to the practicability of indole synthesis methodologies,65 indole fragments can be readily combined with other fragments to afford large indole-based pseudo NP libraries. For example, sp3-rich morphan fragments 41 were subjected to Fischer-indole synthesis to efficiently gain access to an indomorphan collection (42, Scheme 15).66 Biological screening of this 63-membered library resulted in the discovery of glupin, another potent GLUT-1 and GLUT-3 inhibitor. In further studies, glupin was systematically truncated to validate the necessity of the recombination of these two fragments.
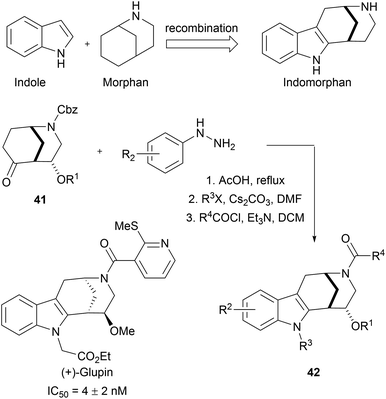 | ||
| Scheme 15 Fusion of indole moiety with morphan fragment. IC50 values determined for the inhibition of 2DG uptake in MDA-MB-231 cells. | ||
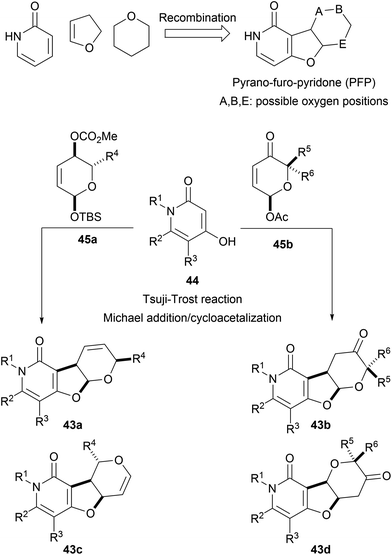
The pseudo NP principle can be extended to more than two fragments. For instance, pyridone-, dihydrofuran- and pyran fragments were combined to form pyrano-furo-pyridones (43, PFP). Fusion of pyridines 44 and functionalized pyrans 45 through Tsuji–Trost reactions or Michael additions afforded the desired tricyclic cores as different regioisomers (43a–d, Scheme 16).54 Biological analysis of the PFPs by means of the cell painting assay showed high biosimilarities with aumitin, a known inhibitor of mitochondrial respiration.67 Further investigations validated this hypothesis and proved that PFPs were a novel chemotype for the inhibition of mitochondrial respiration by targeting mitochondrial complex I.
6 Conclusion and outlook
We have reviewed complementary principles for the design of biologically active NP-inspired compound classes and the evolution of the underlying logic. In silico fragmentation of NPs has been highlighted as a guiding tool for compound design. Combined with independent guiding principles, structural fragments with biological relevance can be extracted from complex NPs.18 These fragments are typically readily accessible and libraries with high biological relevance were obtained. Through the hierarchical sorting of fragments, new and unexplored scaffolds were identified and were used for library design.19 Biology-oriented synthesis uses a similar approach but reduces NP scaffolds to readily accessible yet highly stereogenic scaffolds.1 In BIOS, biological space covered is restricted to NP structure and activity, i.e. biological activities of the novel entities may be similar to the guiding NPs. This challenge could be addressed by the pseudo NP concept. In a cheminformatic analysis of the DNP, 2000 NP fragment clusters were identified.23 Members of these were combined in a fashion not observed in nature, creating scaffolds with unprecedented biological activity.Since there is a vast number of possible fragment combinations, connection types and regioisomers, it is anticipated that the concept of pseudo NPs will inspire the design and synthesis of numerous compound classes with novel biological activity. A systematic analysis of these molecules is therefore highly desirable. The pseudo NP concept benefits from unbiased phenotypic screening techniques, like cell painting. They may provide target hypotheses and mode of action for novel scaffolds unprecedented in nature. The generated target hypotheses could be further confirmed in specific cell-based assays.
7 Conflicts of interest
There are no conflicts to declare.8 Acknowledgements
GSC in grateful for the financial support provided by the Swiss National Science Foundation (Early PostDoc.Mobility, P2SKP2_184064) and the European Union's Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie grant agreement No. 836947. Open Access funding provided by the Max Planck Society.9 References
- S. Wetzel, R. S. Bon, K. Kumar and H. Waldmann, Angew. Chem., Int. Ed., 2011, 50, 10800–10826 CrossRef CAS.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS.
- K. C. Nicolaou, J. S. Chen and S. M. Dalby, Bioorg. Med. Chem., 2009, 17, 2290–2303 CrossRef CAS.
- K. C. Nicolaou, C. R. H. Hale, C. Nilewski and H. A. Ioannidou, Chem. Soc. Rev., 2012, 41, 5185–5238 RSC.
- C. A. Kuttruff, M. D. Eastgate and P. S. Baran, Nat. Prod. Rep., 2014, 31, 419–432 RSC.
- O. Robles and D. Romo, Nat. Prod. Rep., 2014, 31, 318–334 RSC.
- B. J. Huffman and R. A. Shenvi, J. Am. Chem. Soc., 2019, 141, 3332–3346 CrossRef CAS.
- K. C. Morrison and P. J. Hergenrother, Nat. Prod. Rep., 2014, 31, 6–14 RSC.
- R. J. Rafferty, R. W. Hicklin, K. A. Maloof and P. J. Hergenrother, Angew. Chem., Int. Ed., 2014, 53, 220–224 CrossRef CAS.
- N. G. Paciaroni, R. Ratnayake, J. H. Matthews, V. M. t. Norwood, A. C. Arnold, L. H. Dang, H. Luesch and R. W. Huigens III, Chemistry, 2017, 23, 4327–4335 CrossRef CAS.
- A. Garcia, B. S. Drown and P. J. Hergenrother, Org. Lett., 2016, 18, 4852–4855 CrossRef CAS.
- S. Z. Tasker, A. E. Cowfer and P. J. Hergenrother, Org. Lett., 2018, 20, 5894–5898 CrossRef CAS.
- L. Laraia, K. Ohsawa, G. Konstantinidis, L. Robke, Y.-W. Wu, K. Kumar and H. Waldmann, Angew. Chem., Int. Ed., 2017, 56, 2145–2150 CrossRef CAS.
- L. Laraia, G. Garivet, D. J. Foley, N. Kaiser, S. Müller, S. Zinken, T. Pinkert, J. Wilke, D. Corkery, A. Pahl, S. Sievers, P. Janning, C. Arenz, Y. Wu, R. Rodriguez and H. Waldmann, Angew. Chem., Int. Ed., 2020, 59, 5721–5729 CrossRef CAS.
- H. Prescher, G. Koch, T. Schuhmann, P. Ertl, A. Bussenault, M. Glick, I. Dix, F. Petersen and D. E. Lizos, Bioorg. Med. Chem., 2017, 25, 921–925 CrossRef CAS.
- G. Karageorgis, D. J. Foley, L. Laraia and H. Waldmann, Nat. Chem., 2020, 12, 227–235 CrossRef CAS.
- M. Grigalunas, A. Burhop, A. Christoforow and H. Waldmann, Curr. Opin. Chem. Biol., 2020, 56, 111–118 CrossRef CAS.
- M. A. Koch, A. Schuffenhauer, M. Scheck, S. Wetzel, M. Casaulta, A. Odermatt, P. Ertl and H. Waldmann, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17272–17277 CrossRef CAS.
- S. Wetzel, K. Klein, S. Renner, D. Rauh, T. I. Oprea, P. Mutzel and H. Waldmann, Nat. Chem. Biol., 2009, 5, 581–583 CrossRef CAS.
- S. Renner, W. A. van Otterlo, M. Dominguez Seoane, S. Mocklinghoff, B. Hofmann, S. Wetzel, A. Schuffenhauer, P. Ertl, T. I. Oprea, D. Steinhilber, L. Brunsveld, D. Rauh and H. Waldmann, Nat. Chem. Biol., 2009, 5, 585–592 CrossRef CAS.
- A. Schuffenhauer, P. Ertl, S. Roggo, S. Wetzel, M. A. Koch and H. Waldmann, J. Chem. Inf. Model., 2007, 47, 47–58 CrossRef CAS.
- S. Wetzel, A. Schuffenhauer, S. Roggo, P. Ertl and H. Waldmann, Chimia, 2007, 61, 355–360 CrossRef CAS.
- B. Over, S. Wetzel, C. Grutter, Y. Nakai, S. Renner, D. Rauh and H. Waldmann, Nat. Chem., 2013, 5, 21–28 CrossRef CAS.
- M. Congreve, R. Carr, C. Murray and H. Jhoti, Drug Discovery Today, 2003, 8, 876–877 CrossRef.
- H. Koster, T. Craan, S. Brass, C. Herhaus, M. Zentgraf, L. Neumann, A. Heine and G. Klebe, J. Med. Chem., 2011, 54, 7784–7796 CrossRef.
- H. van Hattum and H. Waldmann, J. Am. Chem. Soc., 2014, 136, 11853–11859 CrossRef CAS.
- J. Adrio and J. C. Carretero, Chem. Commun., 2019, 55, 11979–11991 RSC.
- T. Hashimoto and K. Maruoka, Chem. Rev., 2015, 115, 5366–5412 CrossRef CAS.
- R. Narayan, M. Potowski, Z.-J. Jia, A. P. Antonchick and H. Waldmann, Acc. Chem. Res., 2014, 47, 1296–1310 CrossRef CAS.
- H. Fan, J. Peng, M. T. Hamann and J.-F. Hu, Chem. Rev., 2008, 108, 264–287 CrossRef CAS.
- S. R. Vidadala and H. Waldmann, Tetrahedron Lett., 2015, 56, 3358–3360 CrossRef CAS.
- R. Nusse and H. Clevers, Cell, 2017, 169, 985–999 CrossRef CAS.
- A. Friese, S. Kapoor, T. Schneidewind, S. R. Vidadala, J. Sardana, A. Brause, T. Förster, M. Bischoff, J. Wagner, P. Janning, S. Ziegler and H. Waldmann, Angew. Chem., Int. Ed., 2019, 58, 13009–13013 CrossRef CAS.
- J. Robertson and K. Stevens, Nat. Prod. Rep., 2014, 31, 1721–1788 RSC.
- Z.-J. Jia, H. Takayama, Y. Futamura, H. Aono, J. O. Bauer, C. Strohmann, A. P. Antonchick, H. Osada and H. Waldmann, J. Org. Chem., 2018, 83, 7033–7041 CrossRef CAS.
- Z.-J. Jia, G. Shan, C. G. Daniliuc, A. P. Antonchick and H. Waldmann, Angew. Chem., Int. Ed., 2018, 57, 14493–14497 CrossRef CAS.
- P. A. Wender, Nat. Prod. Rep., 2014, 31, 433–440 RSC.
- P. A. Wender, V. A. Verma, T. J. Paxton and T. H. Pillow, Acc. Chem. Res., 2008, 41, 40–49 CrossRef CAS.
- L.-X. Chen, H. He and F. Qiu, Nat. Prod. Rep., 2011, 28, 705–740 RSC.
- J. Švenda, M. Sheremet, L. Kremer, L. Maier, J. O. Bauer, C. Strohmann, S. Ziegler, K. Kumar and H. Waldmann, Angew. Chem., Int. Ed., 2015, 54, 5596–5602 CrossRef.
- L. L. Rubin and F. J. de Sauvage, Nat. Rev. Drug Discovery, 2006, 5, 1026–1033 CrossRef CAS.
- A. Brethon, L. Chantalat, O. Christin, L. Clary, J. F. Fournier, M. Gastreich, C. S. Harris, T. Isabet, J. Pascau, E. Thoreau, D. Roche and V. Rodeschini, Bioorg. Med. Chem. Lett., 2017, 27, 5373–5377 CrossRef CAS.
- U. Gradler, D. Schwarz, M. Blaesse, B. Leuthner, T. L. Johnson, F. Bernard, X. Jiang, A. Marx, M. Gilardone, H. Lemoine, D. Roche and C. Jorand-Lebrun, Bioorg. Med. Chem. Lett., 2019, 29, 126717 CrossRef.
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5601–5606 CrossRef CAS.
- A. M. E. Palazzolo, C. L. W. Simons and M. D. Burke, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5564–5566 CrossRef CAS.
- L. Du, C. Sánchez and B. Shen, Metab. Eng., 2001, 3, 78–95 CrossRef CAS.
- E. C. Cherney and P. S. Baran, Isr. J. Chem., 2011, 51, 391–405 CrossRef CAS.
- Y. Matsuda and I. Abe, Nat. Prod. Rep., 2016, 33, 26–53 RSC.
- G. Karageorgis, E. S. Reckzeh, J. Ceballos, M. Schwalfenberg, S. Sievers, C. Ostermann, A. Pahl, S. Ziegler and H. Waldmann, Nat. Chem., 2018, 10, 1103–1111 CrossRef CAS.
- S. L. Schreiber, Science, 2000, 287, 1964–1969 CrossRef CAS.
- T. E. Nielsen and S. L. Schreiber, Angew. Chem., Int. Ed., 2008, 47, 48–56 CrossRef CAS.
- P. Ertl, S. Roggo and A. Schuffenhauer, J. Chem. Inf. Model., 2008, 48, 68–74 CrossRef CAS.
- T. Schneidewind, S. Kapoor, G. Garivet, G. Karageorgis, R. Narayan, G. Vendrell-Navarro, A. P. Antonchick, S. Ziegler and H. Waldmann, Cell Chem. Biol., 2019, 26, 512–523 CrossRef CAS.
- A. Christoforow, J. Wilke, A. Binici, A. Pahl, C. Ostermann, S. Sievers and H. Waldmann, Angew. Chem., Int. Ed., 2019, 58, 14715–14723 CrossRef CAS.
- S. Kremb and C. R. Voolstra, Sci. Rep., 2017, 7, 44472 CrossRef.
- V. Fetz, H. Prochnow, M. Brönstrup and F. Sasse, Nat. Prod. Rep., 2016, 33, 655–667 RSC.
- C. J. Gerry, B. K. Hua, M. J. Wawer, J. P. Knowles, S. D. Nelson Jr, O. Verho, S. Dandapani, B. K. Wagner, P. A. Clemons, K. I. Booker-Milburn, Z. V. Boskovic and S. L. Schreiber, J. Am. Chem. Soc., 2016, 138, 8920–8927 CrossRef CAS.
- M.-A. Bray, S. Singh, H. Han, C. T. Davis, B. Borgeson, C. Hartland, M. Kost-Alimova, S. M. Gustafsdottir, C. C. Gibson and A. E. Carpenter, Nat. Protoc., 2016, 11, 1757–1774 CrossRef CAS.
- C. C. Barron, P. J. Bilan, T. Tsakiridis and E. Tsiani, Metabolism, 2016, 65, 124–139 CrossRef CAS.
- R. Narayan, J. O. Bauer, C. Strohmann, A. P. Antonchick and H. Waldmann, Angew. Chem., Int. Ed., 2013, 52, 12892–12896 CrossRef CAS.
- S. R. Vidadala, C. Golz, C. Strohmann, C.-G. Daniliuc and H. Waldmann, Angew. Chem., Int. Ed., 2015, 54, 651–655 CAS.
- H. Xu, C. Golz, C. Strohmann, A. P. Antonchick and H. Waldmann, Angew. Chem., Int. Ed., 2016, 55, 7761–7765 CrossRef CAS.
- H. Takayama, Z.-J. Jia, L. Kremer, J. O. Bauer, C. Strohmann, S. Ziegler, A. P. Antonchick and H. Waldmann, Angew. Chem., Int. Ed., 2013, 52, 12404–12408 CrossRef CAS.
- D. Gao, T. Zhou, L.-T. Da, T. Bruhn, L.-L. Guo, Y.-H. Chen, J. Xu and M.-J. Xu, Org. Lett., 2019, 21, 8577–8581 CrossRef CAS.
- D. F. Taber and P. K. Tirunahari, Tetrahedron, 2011, 67, 7195–7210 CrossRef CAS.
- J. Ceballos, M. Schwalfenberg, G. Karageorgis, E. S. Reckzeh, S. Sievers, C. Ostermann, A. Pahl, M. Sellstedt, J. Nowacki, M. A. Carnero Corrales, J. Wilke, L. Laraia, K. Tschapalda, M. Metz, D. A. Sehr, S. Brand, K. Winklhofer, P. Janning, S. Ziegler and H. Waldmann, Angew. Chem., Int. Ed., 2019, 58, 17016–17025 CrossRef CAS.
- L. Robke, Y. Futamura, G. Konstantinidis, J. Wilke, H. Aono, Z. Mahmoud, N. Watanabe, Y.-W. Wu, H. Osada, L. Laraia and H. Waldmann, Chem. Sci., 2018, 9, 3014–3022 RSC.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |