Open Access Article
Open Access ArticleBoosting nanotoxicity to combat multidrug-resistant bacteria in pathophysiological environments†
Dana
Westmeier‡
 a,
Svenja
Siemer‡
a,
Cecilia
Vallet
b,
Jörg
Steinmann
c,
Dominic
Docter
a,
Jan
Buer
c,
Shirley K.
Knauer
a,
Svenja
Siemer‡
a,
Cecilia
Vallet
b,
Jörg
Steinmann
c,
Dominic
Docter
a,
Jan
Buer
c,
Shirley K.
Knauer
 *b and
Roland H.
Stauber
*b and
Roland H.
Stauber
 *a
*a
aENT Department, University Medical Center Mainz, Langenbeckstrasse 1, 55131 Mainz, Germany. E-mail: rstauber@uni-mainz.de
bDepartment of Molecular Biology II, Center for Medical Biotechnology/Nanointegration (ZMB/CENIDE), University Duisburg-Essen, Universitätsstrasse 5, 45117 Essen, Germany
cInstitute of Medical Microbiology, University Hospital Essen, University Duisburg-Essen, Hufelandstrasse 55, 45112 Essen, Germany
dInstitute for Biotechnology, Shanxi University, No. 92 Wucheng Road, 030006 Shanxi, China
First published on 23rd October 2020
Abstract
Nanomaterials are promising novel antibiotics, but often ineffective. We found that nanomaterial–bacteria complex formation occurred with various nanomaterials. The bactericidal activity of NMs strongly depends on their physical binding to (multidrug-resistant) bacteria. Nanomaterials' binding and antibiotic effect was reduced by various pathophysiological biomolecule coronas strongly inhibiting their antibiotic effects. We show from analytical to in vitro to in vivo that nanomaterial-based killing could be restored by acidic pH treatments. Here, complex formation of negatively-charged, plasma corona-covered, nanomaterials with bacteria was electrostatically enhanced by reducing bacteria's negative surface charge. Employing in vivo skin infection models, acidic pH-induced complex formation was critical to counteract Staphylococcus aureus infections by silver nanomaterials. We explain why nano-antibiotics show reduced activity and provide a clinically practical solution.
Introduction
Besides the wide use of engineered nanomaterials (NMs) in technical products, their applications are not only increasing in biotechnology and biomedicine, but also in the environment, including potential antimicrobial approaches, such as fighting recent pandemics as exemplified by COVID-19.1–4 Particularly, the availability of nanomaterials (NMs) has increased in size-range, complexity, and functionality, potentially allowing control over microbial pathogens by novel NM-based antimicrobial therapies, harnessing the nano-characteristics of the different materials, which mostly are different compared to their bulk counterparts.4–10 Nevertheless, besides their desired benefits, extended exposure to NMs might also result in nanotoxicity and thus, potential risks for human health and ecosystems.2,11,12 Hence, over the last decade, numerous comprehensive studies focused on interactions of NMs with eukaryotic cells, and their consequences at nano-bio interfaces.11,12 For further details on mechanisms of nanotoxicity on eukaryotic cells as well as potential involved molecular signaling pathways. We refer the reader to the pertinent literature. While the physico-chemical properties and the behavior of NMs can be engineered and characterized under idealized conditions, this is no longer the case in complex physiological or natural environments.11,13 Here, proteins and other biomolecules rapidly bind to NMs, forming the protein/biomolecule corona, which critically affects the NM's (patho)biological, biotechnical and therapeutic identity.2,11,13–22 As the corona impacts in vitro and/or in vivo NM applications in humans and ecosystems, a mechanistic understanding of its relevance and the biophysical forces regulating corona formation is mandatory, but still incomplete. Rational design of NMs to reduce their potential toxicity requires knowledge of how NMs cause toxic effects. The mechanisms of toxicity include oxidative stress, membrane damage, inflammasome activation, genotoxicity and fibrosis.2,11,12 Here, the mechanisms induced by a specific NM are highly dependent on the NM's physicochemical characteristics as well as the (patho)physiological environment.2,11,12,22 However, surprisingly little is known about the crosstalk of NMs with socio-economical highly relevant pests, including viruses, (pathogenic) fungi or the plethora of bacteria. Besides being crucial to maintain ecological homeostasis, bacteria are associated with a wide spectrum of diseases, highly relevant for humans or animals but also for agriculture.22–26Not only the increasing cases of multidrug-resistant (MDR) bacteria but also the lack of still effective and novel antibiotics represent an increasing socio-economic problem.27–29 Here, hospital-acquired MDR bacteria such as Methicillin-resistant Staphylococcus aureus (MRSA), Salmonella enterica or Pseudomonas aeruginosa, and extended-spectrum β-lactamase-producing Escherichia coli (ESBL-EC) are problematic in the clinics even in highly developed countries.10,27,30–36 Consequently, NMs are investigated as tools for diagnosis of bacteria, as antibiotic drug carriers and/or based on their antimicrobial activity, directly tested as novel nano-antibiotics.4,6–10,27,29,35,37 Antibacterial nano-therapeutics so far mainly focused on a variety of semiconductor, metal, and metal oxide NMs.4,10,22,27,29,38,39 Here, their antibacterial properties seem to be mainly due to the release of toxic metal ions, physical damage of bacterial membranes, the generation of free radicals and reactive species, as well as by local heating through surface plasmon resonance effects.10,29,38–40 Moreover, anti-microbial nano-peptide polymers have been reported, potentially acting via membrane destabilization and damage.10,32,41,42 As not all NMs are antibiotics and not all antibiotics are NMs, we here wish to define the term ‘nano-antibiotics’ as any nano-sized antibacterial material. These include NMs that are already antibacterial, but also any nano-sized carrier for antibiotic drugs.
Interestingly, nano-antibiotics often fail in practical clinical or biotechnological applications and the details are not yet understood.4,6–10,29,35 ‘Classical’ antimicrobial resistances are mainly based on a pathogen's ability to stop the antibiotic from reaching its target at a high enough concentration and/or to modify or bypass the target that the antibiotic acts on. In contrast, besides these known ‘classical’ resistance mechanisms, potentially relevant though not specific for nanoantibiotics, there is an ongoing discussion if nano-size specific effects could also contribute to limited practical applications, i.e. causing resistance.4,69 The blood-system, the oro-gastrointestinal tract, lung as well as (skin) wound applications of NMs represent main targets for nano-antibiotics.4,6–10,27,29,35–37 Notably, such (patho)physiological systems are often highly dynamic and NMs are expected to contact bacteria only for a short period of time. However, most studies investigated rather artificial static and constant treatment scenarios so far, often neglecting the dynamics of bacteria–NM interaction.10,27,29,35,39 Moreover, (patho)physiological systems contain various molecules, quickly assemble on the NM's surface and form biomolecule coronas of various identities and complexities.4,10–12,22,29,35,38–40,43–53 These coronas may modulate the physico-chemical and (patho)biological identity of NMs via different mechanisms, including their impact on the NM's colloidal stability, dissolution, and/or zeta potential.4,10,11,22,29,35,38–40,44,45,53,54 Although one may hypothesize that corona formation may potentially affect ‘nano-antibiotics’ activity, its detailed (practical) impact has not been comprehensively investigated before.
In our study, we therefore investigated these ‘neglected’ factors employing well-characterized NM models and found that complex formation is required for NMs' efficient antibacterial effects. Subsequently, from analytical to in situ to in vitro to in vivo, we provide convincing evidence that biomolecule coronas acts as a ‘resistance modulator’ to NMs, and a simple and effective way to overcome this effect to enhance the activity of nano-antibiotics relevant for practical applications of medical and biotechnological nanotechnology.
Results and discussion
NMs' physico-chemical characteristics co-determine complex formation with bacteria
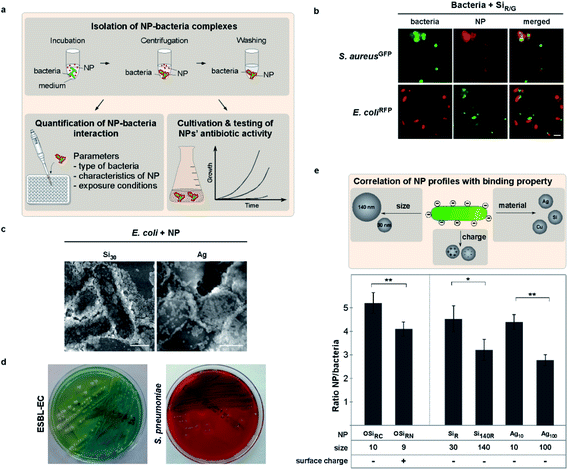
Although not tested in detail before, one may expect that the effects of NMs on bacteria are dependent on their physical contact, the type of NM and bacteria, as well as by how these attractions are potentially modulated by the (patho)physiological environment. Clearly, NMs' physicochemical characteristics define their biological behavior, though it is impossible to test all types of NMs. Hence, we here employed NM models combined with an experimental pipeline to uncover NM characteristics-function correlations. We tested NMs varying in size and/or surface functionalization, including NMs that have been proven as reliable models in previous studies, to define determinants for bacteria–NM interactions and antibiotic effects (Table 1).13,35,55–58 For one, we first analyzed the interaction of various fluorescent polymer and silica NMs with auto-fluorescent pathogenic as well as apathogenic bacterial genera in situ using mainly live cell microscopy as a non-invasive method (Fig. 1a, b and e).| TEM diameter in dry state ± s. d. [nm] | Hydrodynamic diameter in water ± s. d. [nm] | Zeta potential ± s.d. [mV] | Bacteria binding | Reduced binding by biomolecule coronas | |
|---|---|---|---|---|---|
| a NP–bacteria interaction was verified by the indicated methods: FM: (*quantitative) fluorescence microscopy, EM: electron microscopy (SEM); EDX: energy dispersive X-ray spectroscopy. Fluorescent labels (R = rhodamine; G = FITC). n.d.: not determined. | |||||
| Metal-based NMs | |||||
| Ag10 | 10.3 ± 2.2 | 12.4 ± 0.5 | −43 ± 3 | +EM/EDX/SEM; * | + |
| Ag100 | 95.6 ± 6.9 | 118.4 ± 4.5 | −36 ± 4 | +EM/EDX/SEM; * | + |
| CuO | 55.2 ± 3.6 | 488.3 ± 12 | −4.5 ± 0.5 | +EM/EDX/SEM | + |
| ZnO | 20.2 ± 2.0 | 158 ± 11 | +14 ± 3 | +SEM/EDX | + |
| CeO2 | 28.4 ± 10.4 | 135 ± 4 | +49 ± 2 | +SEM/EDX | + |
| CdTe (2.4 eV) | 3.1 ± 0.4 | 3.0 ± 0.5 | −30 ± 3 | +SEM | + |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Silica NMs | |||||
| Si30 | 31.6 ± 5.8 | 33 ± 7 | −15 ± 2 | +EM/EDX | + |
| Si140G/R | 140.8 ± 8.0 | 142.4 ± 6 | −20 ± 3 | +FM*/EM/EDX | + |
| Si140 | 141 ± 6.0 | 141 ± 6 | −21 ± 3 | +EM/EDX | + |
| SiR | 30.6 ± 6.8 | 33.6 ± 8 | −14 ± 2 | +FM*/SEM | + |
| SiG | 30.8 ± 6.4 | 34.0 ± 7.6 | −15 ± 2 | +FM* | + |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Polymer NMs | |||||
| OSiRC | 9.1 ± 1.8 | 14.9 ± 0.09 | −32 ± 2 | +FM* | + |
| OSiRN | 10.2 ± 1.9 | 15.7 ± 0.09 | +24 ± 5 | +FM* | + |
| OSiRPEG | 10.4 ± 1.7 | 22.1 ± 0.09 | −14 ± 1 | −FM* | + |
| OSiRPEtO | 11.8 ± 2.0 | 26.0 ± 0.19 | −5 ± 1 | −FM* | + |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Microparticles | |||||
| MP–Si | 3012 ± 113 | n.d. | −36.1 ± 2 | — | — |
| MP–Ag | 1012 ± 56 | n.d. | −28.3 ± 1 | —; * | — |
| MP–Ag2 | 2102 ± 98 | n.d. | −29.2 ± 2 | —; * | n.d. |
| MP–Cu | 2207 ± 213 | n.d. | −39.1 ± 2 | — | — |
To study and mimic exposure and contact times simulating realistic dynamic pathophysiological environments, we exposed bacteria to NMs under controlled conditions, allowing to vary potentially relevant parameters such as temperature, biomolecules, time or pH (Fig. 1a, ESI Fig. S1†). NM–bacteria complexes were subsequently recovered by mild centrifugation and washed to remove unbound NMs. We found by fluorescence microscopy that all tested NMs rapidly bound to all examined bacteria, including Shigella flexneri, enteropathogenic strains of E. coli as well as Listeria monocytogenes (Fig. 1b–d; ESI Fig. S2a;†Table 1).
Moreover, kinetic analyses showed that NM adsorption to bacteria occurred rapidly (∼30 s) and complex stability was not affected by variations in temperatures (8–42 °C) (ESI Fig. S1d and e†). Of note, as Brownian motion, which is directly affected by temperature, is the reason for the NMs' mobility also driving their adsorption to bacteria.
Observed NM–interactions could also be verified by independent methods, including energy-dispersive X-ray spectroscopy (EDX) or electron microscopy approaches (Table 1; Fig. 1c, 3b; ESI Fig. S1a†). Building upon the results and insights from these though non-antibacterial NM models (even when used in high concentrations – data not shown), we were able to confirm the results also for clinically and biotechnologically relevant anti-bacterial NMs, such as ZnO, CuO, and Ag (Table 1; Fig. 1f, 2a; ESI Fig. S2a†). EM showed a NM-coating of the bacteria's surface with silica NMs (Si30) (Fig. 1c), whereas NMs that already tend to aggregate in physiological buffers, such as ZnO or CeO2, mainly bound as NM clusters (data not shown). The relevance of these insights is further underlined by showing that not only laboratory bacteria but also freshly isolated MDR bacteria including primary clinical isolates, such as extended-spectrum β-lactamase-producing E. coli (ESBL-EC), MRSA as well as Salmonella enterica, efficiently bound to all investigated NMs (Fig. 1d and e; Table 1; ESI Fig. S2a†).
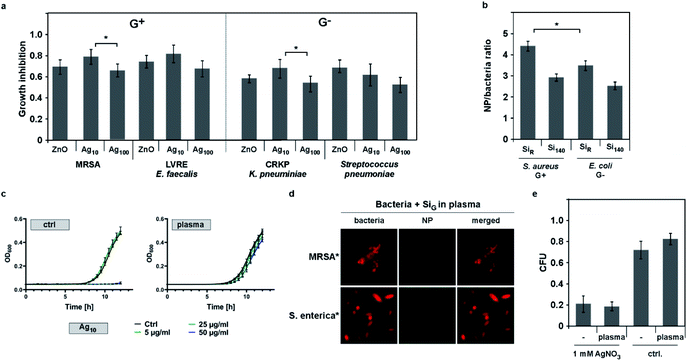
We moreover discovered that self-assembly of NMs on bacteria cannot be explained by the rules of electrostatics alone. Using NMs varying only in distinct parameters, fluorescence-microscopy automated quantification of complex formation revealed enhanced binding for negatively (OSiRC, ζ = −32 mV) compared to positively (OSiRN, ζ = +24 mV) charged polymer NMs of similar size (Table 1; Fig. 1e). Notably, all tested bacteria displayed an overall negative zeta potential, but also various negatively charged NMs, including silica as well as antibiotic Ag NMs (Ag10), bound efficiently (Fig. 1; Table 1). Binding also occurred under ion-free conditions, such as in deionized water, excluding the potential influence of counter ions (ESI Fig. S2c†). Interestingly, also protein corona formation cannot be predicted by electrostatics alone as proteins displaying an overall negative charge constitute the majority of all corona proteins, irrespective of the NM's initial surface charge (Fig. 4a; ESI Table S2†).13,55,59 Our data further indicates that NM–bacteria complex formation is enhanced by small NM size rather than charge or material. Small silica (ᴓ ∼ 30 nm) NMs bound more efficiently compared to larger ones (ᴓ ∼ 140 nm), despite their similar negative surface charge (Table 1; Fig. 1f, 2b; ESI Fig. S2c†). These NMs seem to establish more contact points than larger NMs resulting in more efficient binding. The possibility that microparticles do not bind to bacteria in contrast to NMs due to their increased negative charge is rather unlikely as less negatively charged Ag microparticles (MP-Ag, ζ = −28 mV) did not interact at all compared to highly negative Ag NMs (ζ = −43 ± 3 mV) (Table 1). Enhanced binding of smaller antibacterial NMs to pathogens, resulted in a stronger antibiotic effect (shown for silver NMs; Fig. 2a). However, one may need to test additional NM models varying only in size in order to generalize our findings. Also, one may study identical NMs only differing in their shape, which might affect NM–pathogen interaction and/or bacterial vitality.
Currently, supramolecular structural and/or charged domains on biomolecules are recognized and addressed as targets by the powerful field of supramolecular chemistry. It is likely that supramolecular domains of components constituting the complex architecture and molecular surface structure of bacteria may also determine the binding of NMs. Hence, targeted supramolecular chemistry may be investigated as a strategy to rationally guide design of NM–pathogen interaction. Such chemical strategies aiming at the surface of bacteria have been tested, potentially allowing to control NM–bacteria complex formation upon their integration on NM surfaces.60–63
Clearly, we feel that our findings are of general relevance, and may with caution be extrapolated to other NMs, as we could not test all existing NMs. Future work might likewise examine identical NMs only differing in their shape, although the mechanisms of how NM-shape alone could enhance their bactericidal activity needs to be determined and is controversially discussed.64
Relevance of protein coronas for NM–bacteria complex formation as a ‘resistance modulator’ to nano-antibiotics
As introduced, it is accepted in the field that biomolecule coronas rapidly form on almost all NMs in pathophysiologically relevant and complex environments. However, as their impact on NMs' antimicrobial activity has not been studied in detail and pathophysiological environments relevant for practical application include protein-rich environments, such as wounds or the blood, we focussed on the (hard) protein corona in our study. We observed that enhanced binding of NMs correlated directly with improved anti-microbial activity of metal based NMs, such as CuO, ZnO or different Ag NMs, inhibiting also the growth of MDR bacteria (Fig. 2a and b; ESI Fig. S2†). However, when we used human plasma as an additional component in our SOP, we observed that the blood protein corona mediated resistance to anti-microbial NM reducing the NMs' colloidal stability (Fig. 2c and e; Table 2; ESI Fig. S1c; ESI Table S1†), explaining the NMs' reduced antibiotic effects in physiologically relevant surroundings. Here, SDS-PAGE and LC-MS confirmed that all tested NMs developed a plasma protein corona (Fig. 4a; ESI Table S2†). Biomolecule coronas may transform NMs by also modulating their colloidal stability and/or zeta potential.4,10,11,22,29,35,38–40,44,45,47,48,54,55,65 Summarizing the results of numerous studies, (patho)biological and ecological environments seem to be facing mainly negatively-charged, protein corona-covered NMs, irrespective of the NM's initial zeta potential.46,55,59,66–68 Although we only tested a selection of relevant NMs, our data demonstrates that binding to also negatively charged bacteria occurs and thus, that binding cannot be predicted by colloidal electrostatics alone in the absence or presence of biomolecules.| Ag10 | ZnO | |||||
|---|---|---|---|---|---|---|
| H2O | SWF pH 7 | SWF pH 3 | H2O | SWF pH 7 | SWF pH 3 | |
| MRSA | 6.5 | >50 | 3.5 | 21.5 | >50 | 14.5 |
| ESBL-EC | 5.5 | >50 | 1.5 | 24.0 | >50 | 12.0 |
| *Si30 | *OSiRC | ||||||
|---|---|---|---|---|---|---|---|
| H2O | SWF pH 7 | SWF pH 3 | H2O | SWF pH 7 | SWF pH 3 | ||
| MRSA | >150 | >150 | >150 | >150 | >150 | >150 | |
| ESBL-EC | >150 | >150 | >150 | >150 | >150 | >150 | |
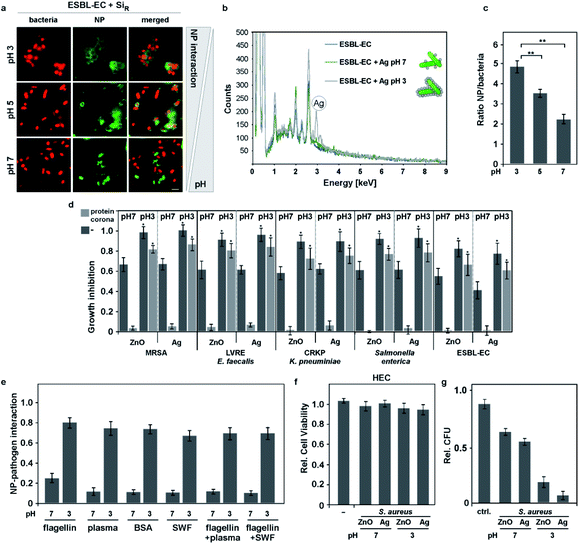
Moreover, NM exposure to simulated wound fluid (SWF; ESI Table S3†) equally resulted in resistance of Gram+ and Gram− model bacteria as well as freshly isolated multi-drug resistant primary clinical isolates (Gram+ and Gram−) (Fig. 1d, 2; Table 2). However, pathogens pre-incubated even with undiluted human plasma still bound NMs (ESI Fig. S3e†), showing that the NMs' protein coronas but not proteins potentially adsorbed on microbes, are critical for the observed effects. Also, no protein corona-mediated resistance was evident for free silver ions, demonstrating the nano-size relevance of the identified mechanism (Fig. 2e).
In a recent study, it was suggested that flagellin confers resistance to Ag NMs.4,69 Under laboratory conditions also flagellin formed a protein corona on anti-microbial NMs, thereby conferring bacterial resistance by inhibiting NM–bacteria complex formation (Fig. 3e). In contrast, in pathophysiological realistic environments already containing high protein concentrations, such as wound fluid or human blood plasma, adding flagellin did not further increase bacterial resistance. In conclusion, it seems that not flagellin's biological activity but its mere physical binding to NMs is relevant for resistance by inhibiting NM–pathogen complex formation (Fig. 3e).
In addition, we examined whether the reduction of antibiotic activity mediated by protein coronas is caused by reduced liberation of toxic ions due to high protein concentrations or pH-mediated dissolution. Here, we studied antibiotic quantum dots (QDs; 2.4 eV CdTe QDs; 150 nM), for which the antibiotic activity is not dependent on the release of toxic ions, but on the generation of photo-activated charge carriers.29 As expected, we found that light-activated inhibition of bacterial growth was also inhibited by protein coronas (ESI Fig. S2b†) and improved by low pH (light-activated inhibition of S. aureus MIC [μg mL−1]: pH 7 = 130 nM; pH 3 = 55 nM).
Collectively, this data further supports our main finding that protein corona formation is most likely the main resistance modulator mechanism to nano-antibiotics in general and that complex formation of nano-antibiotics with bacteria seems essential for an optimal therapeutic effect, independent of the NMs' mode of activity, such as the release of toxic ions (Table 3).
| NM size | (Potentially) highly relevant; improved binding for smaller NMs, correlating with enhanced antibacterial effects (shown for Ag NMs) |
| NM charge | Potentially less relevant (binding of negatively or positively charged NMs) |
| Material | Less relevant (binding of various NMs) |
| Biomolecule coronas | Concentration-dependent inhibition of complex formation by various types of coronas |
| Exposure time | Rapid binding of NMs to bacteria |
| Type of bacteria | Potentially less relevant (although a limited number of bacteria have been tested) |
| Antibacterial activity | Antibiotic activity of the NM is required; increased upon NM–pathogen complex formation |
Strategies to overcome protein corona-mediated NM resistance
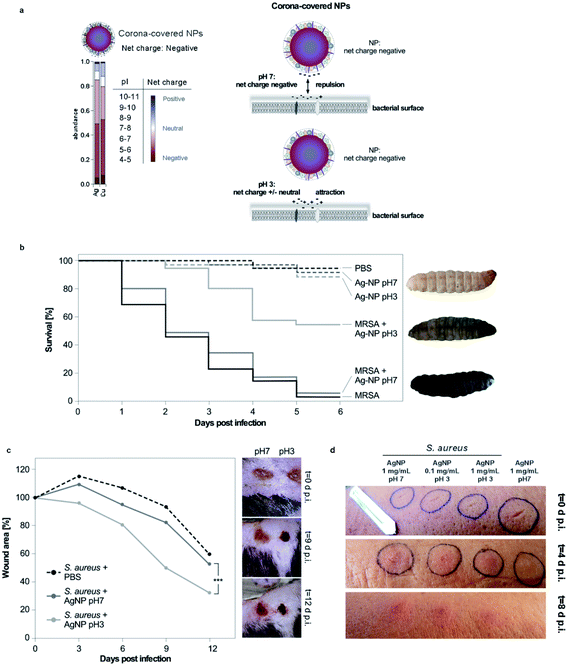
As outlined above, supramolecular structural and/or charged domains on biomolecules are recognized and addressed as targets by the field of supramolecular chemistry. One may speculate that targeted supramolecular chemistry may also be exploited in the future as a strategy to increase NM–pathogen complex formation and thus, the NMs' bactericidal activity, even in practical environments and applications. Clearly, our data indicates that approaches restoring NM–bacteria complex formation even in protein corona forming environments are expected to boost NMs' antibiotic efficacy. Therefore, we tested environmental parameters, which may influence the bacteria's cell surface charge. The field has shown that the dense glycocalyx, containing numerous deprotonated carbohydrates, is mainly responsible for the overall negative bacterial surface charge. We speculated that variations in pH may lower (pH < 7) or increase (pH > 7) this negative surface charge, based on protonation or deprotonation of molecules. Indeed, our zeta potential analyses showed that the overall negative surface-charge of Gram-pathogens (E. coli; pH 7; ζ = −74.2 mV) or of Gram+ bacteria (S. aureus; pH 7; ζ = −12.8 mV) became less negative (E. coli, pH 3; ζ = −6.2 mV; S. aureus; pH 3; ζ = −1.3 mV) under acidic pH conditions. Interestingly, our analyses revealed that NM–bacteria complex formation was enhanced in acidic surroundings (pH 3–5) for Gram− as well as for Gram+ (MDR) bacteria (Fig. 3a–c; ESI Fig. S3†). Notably, even a short acidic pulse (<1 min) enhanced complex formation, which remained stable under subsequent transfer to neutral pH conditions (ESI Fig. S3a and b†). Moreover, we observed that when bacteria, which have been pretreated at pH 3, were mixed with bacteria pretreated at pH 7, the tested NMs mainly adsorbed to pH 3-pretreated bacteria (ESI Fig. S3c†). These results indicate that pH-induced effects on the bacterial cell rather than at the NMs' surfaces are determining likelihood and strength of interaction.Building upon the obtained results, we further hypothesized that lowering the negative surface charge of bacteria by acidic pH increases the electrostatic attraction even of protein corona-covered NMs to pathogens, which was confirmed by zeta potential analyses, measured 10 min post exposure in plasma (Cu NM: ζ = −0.6 ± 0.8 mV; AgNM: ζ = −33 ± 6 mV).
Furthermore, we used LC-MS based proteomics to profile the number and identity of human plasma proteins adsorbing on anti-microbial Cu or silver NMs (ESI Table S2†). Analyses and bioinformatic classification of the identified corona components (>200 proteins) could demonstrate that mainly proteins with an overall negative-charge composed the majority of the protein corona (Fig. 4a). As (patho)biological environments seem to be facing mainly negatively-charged, protein corona-covered NMs, reducing the bacteria's surface charge by acidic pH would mechanistically explain enhanced complex formation and improved anti-microbial activity promoted by the induced electrostatic attractions (Fig. 4a). It is suffice that by enhancing the physical binding of NMs with pathogens' cell surface the nanotoxic effects mediated by light-activated free radicals and/or the release of toxic ions is increased and may also enhance the impact of bactericidal chemicals, which are released from engineered NMs.
In order to probe the relevance of our discovery for realistic treatments, we subsequently examined the effects of lowering the pH of the applied NMs in in vitro scenarios such as simulated wound fluids or human blood plasma. Notably, resistance to anti-microbial NMs' mediated by protein coronas could be overcome in these relevant environments by acidic pH treatments (Fig. 3d; Table 2), allowing even the killing of clinical MDR isolates, including MRSA, ESBL-EC or CRKP (Fig. 3d).
To further examine the relevance of the findings for practical (clinical) applications, primary human epithelial cells (HEC) were exposed to bacteria in combination with anti-microbial silver or ZnO NMs or inert fluorescent Si NMs at different pH conditions (pH 7 or 3). NM–bacteria complexes were stable and detectable at the cell membrane (ESI Fig. S3f†). Notably, these treatments did not affect cellular vitality but the bacterial growth was efficiently inhibited mainly at pH 3 (Fig. 3f and g).
Long-term acidic pH conditions have been used as food preservation method for centuries and are present in the human stomach.70 To investigate if acidic pH and not enhanced interaction of bacteria with antibiotic NMs is key for the observed enhanced killing of (MDR) bacteria, bacteria were exposed to non-antibiotic NMs (silica or polymer NMs) at various pH conditions. However, we did not observe an antibiotic effect of acidic pH with these non-bactericidal NMs (Table 2). Previous studies also suggested that highly acidic pH can promote dissolution of (metal)-based NMs.71–73 Thus, we performed DLS measurements, which confirmed that the increased antibiotic effect of Ag or Cu NMs at pH 3 conditions was not merely mediated by enhanced NM dissolution under our experimental conditions and exposure time points (NMs' hydrodynamic diameter: Cu NM: H2O ± s. d. [nm], t = 0 h → 488.3 ± 12; t = 3 h → 479.5 ± 10; SWF pH 3 ± s. d. [nm], t = 0 h → 491.3 ± 14; t = 3 h → 475.3 ± 13; Ag NM: H2O ± s. d. [nm], t = 0 h → 12.4 ± 0.5; t = 3 h → 12.3 ± 0.3; SWF pH 3 ± s. d. [nm], t = 0 h → 12.1 ± 0.4; t = 3 h → 10.4 ± 0.5). Although less important for most biomedical applications, it is likely that acidic pH will promote NM dissolution upon prolonged incubation, such as days or month in ecologically and agriculturally relevant environments, which may influence desired or undesired nanotoxicology for microbial habitats.4,11,35 In addition, EDX analysis showed improved binding of silver NMs to (MDR) E. coli following 1 h exposure at pH 3, arguing against a major dissolution-mediated antibiotic impact of Ag NMs (Fig. 3b).
Collectively, our data supports our conclusion that resistance to nano-antibiotic NMs is mediated by pathophysiological protein coronas reducing NMs' binding to bacteria. We thus provide a nanosize-specific explanation why the activity of current nano-antibiotics is often impaired under clinically relevant conditions. Also, we found that low pH treatment seems to be a promising approach to override the observed protein corona-mediated resistance by restoring NM–bacteria complex formation.
Practical translation: applying acidic-pH NM formulations to treat bacterial infections of the skin
Following our tiered experimental pipeline, we subsequently tested the relevance of the encouraging in vitro data by using in vivo models of growing complexity and (pre)clinical relevance. Clearly, in contrast to the intravenous systemic use of nanoformulations, the topical application of nanoformulations is currently more accepted by the field, expected to minimize potential adverse nanotoxicological complications. The pH of wounds during infection, treatments, and healing seems relevant though complex topic involving multiple signaling pathways in both, the wound microenvironment, as well as in microbes.74,75 Although not fully understood, acidosis of the wound supports oxygenation, macrophage activity, collagen formation, angiogenesis as well as the proliferation of fibroblasts, whereas alkalization is often favorable for the growth of microbes.74,75 Hence, we chose topical wound infections as a relevant model to test our anti-bacteria nanoformulations, protein corona effects, and the impact of pH modulation on improving NM–pathogen complex formation, and thus, increasing the NMs' antibiotic effect. Of note, our single topical ointment treatments did not allow to modulate the wound pH for a prolonged period.Currently, research heavily relies on murine models for studying the activity of (nano-based) anti-microbials.12,35 However, there is a growing public consciousness demanding alternative experimental models as initial proof-of-concept screening systems, as there are ethical, financial, and logistical problems connected with the use of mammals. The invertebrate insect Galleria mellonella can be infected by numerous microorganisms including bacteria, and thus is suggested to be highly useful to investigate bacterial infections and their therapies.76G. mellonella larvae can be easily and inexpensively obtained in large numbers, and have a short life cycle. Though lacking a typical vertebrate adaptive immune response, insects possess well-developed innate immune responses with remarkable similarities to vertebrates. In particular, their anti-bacterial immunity also involves cellular and humoral components.77–79 The hemolymph system contains different types of hemocytes, important for recognition and elimination of pathogenic microorganisms.77–79 Moreover, insects and humans have evolutionary conserved anti-microbial peptides, including defensins.22,77–79 Here, G. mellonella larvae were scratched with a lancet in the left proleg and infected with MRSA. Animals were then treated by topical administration of pH 7 or pH 3 AgNM formulations. Compared to PBS-treated controls, the treatment with pH 3 AgNMs prevented bacterial growth and animal death, whereas pH 7 formulations were less effective (Fig. 4a). As an additional control for pH-regulated effects in the wound microenvironment, using non-antibiotic silica NMs even at pH 3 did not counteract MRSA infection and animal death, excluding again a major relevance of acidic pH alone, independent of nanoantibiotics (ESI Fig. S4b†).
Building upon the effects observed in the invertebrate system, we next investigated the pH-dependent antibiotic effect of AgNMs in a preclinical mouse wound infection model. Here, we generated full-thickness skin wounds on the back of mice, which were infected with S. aureus bacterial suspensions. Wounds were treated by topically administering AgNMs in pH 7 or pH 3 formulations or using saline as controls for one time only. We monitored wound healing macroscopically over time and found that mainly the application of pH 3 silver NMs effectively inhibited the bacterial infection and promoted wound healing (Fig. 4b), supporting our conclusions obtained from employing our in vitro data and the data acquired in the invertebrate wound infection model. Of note, when monitoring treatment-induced changes in wound pH, we observed that ten minutes after treatments, the wound fluids' pH correlated with the pH of the topical ointments (pH < 5 for Ag10 NMs in pH 3; pH 7 for Ag10 NMs in pH 7). Already two hours later, the pH of recovered wound fluid was similar and around pH 7 for both treatments, thus neutralized by the fluid's physiological buffering capacity. Due to wound healing/closure and ethical reasons, we could not recover wound fluids at later time points. Though, based on the current literature, there are no reasons to expect that acidification of wounds will occur at later time points, particularly without additional treatments.74,75 Hence, as NM–pathogen complex formation occurs rapidly and was mimicked by our experimental procedures from analytical to in vitro as well as in vivo, our data strongly indicate that complex formation of nano-antibiotics with bacteria seems essential for an optimal therapeutic effect, rather then a low-pH triggered rapid dissolution of metal-based NMs.
As a final proof-of-concept experiment, we tested our pH-switch to treat human skin wounds infected with S. aureus in a single healthy volunteer (R.S.). Topical application of silver NMs (pH 3), even when diluted 10-fold higher than the other treatments, reduced infection and increased wound healing more effectively than treatment with silver NMs at pH 7 (Fig. 4d). Treatments were applied for one time only. No signs of skin irritation or inflammation were observed by administering silver NMs alone (Fig. 4d).
Experimental
Reagents
If not stated otherwise, reagents and chemicals were purchased from Merck, MSC UG, Invitrogen, and Promega.NM synthesis and characterization
Silica, polymer, and metal-based NMs as well as various microparticles were synthesized and provided by the DENANA/NanoBEL Research consortium, purchased (Merck, Invitrogen, MSC UG) or synthesized as indicated.22,29,35,55–57,80–82 NMs were characterized by EM, DLS, and zeta potential measurements as reported.55,57,83Clinical isolates and cultivation of bacteria
MDR bacteria were isolated and respective resistance phenotypes characterized at the University Medical Centers of Essen and Mainz using microbial methods as described.22,32,33,81 Identity confirmation and susceptibility testing were performed by the semi-automated systems VITEK 2 (bioMérieux, Germany) and MicroScan WalkAway (Siemens, Germany) or by MALDI Biotyper® System (Bruker, Germany).33,84 For microscopy, bacteria strains, expressing red (tdTomato) or green fluorescent protein (GFP) were used.35 Bacteria were grown in liquid cultures overnight at 37 °C and 140 rpm or on agar plates in their respective media.Characterization of NM–bacteria complexes
To form NM–bacteria complexes, bacteria from an overnight culture were washed five times before incubation with NMs in the indicated media, temperature, and time points. NM–bacteria complexes were harvested by mild centrifugation (10 min, 3000 rpm, 20 °C), the pellet was resuspended in PBS buffer and two more washing steps were performed (10 min, 3000 rpm, 20 °C) before resuspension in PBS buffer, counting and subsequent use. Binding of NMs was analysed by independent methods, including fluorescence microscopy, automated high content microscopy analysis, EM, and EDX.Biocorona profiling
Label free quantitative liquid chromatography-mass spectrometry (LC-MS)-based profiling of human plasma coronas on Ag and Cu NMs and bioinformatic analyses were performed as reported.55,83 Briefly, NMs were incubated in human plasma and then separated from unbound plasma molecules via a centrifugation-based protocol. Proteins were eluted from the NMs with the help of lysis buffer and quantified by LC-MS.Microscopy
Confocal laser scanning microscopy, fluorescence microscopy, and automated high content microscopy analysis were used to visualize and quantify NM–bacteria complexes in situ and to show their cellular localisation.55 The ArrayScanVTI automated HTS microscopy platform was employed for quantification of NM–bacteria complexes.55 A minimum of 1 × 103 NM–bacteria complexes/well was analysed. A binary image mask was created from the GFP signal of the bacteria (XF93 GFP 475/515). In the second channel (XF32 TRITC 547/570), this circular mask was dilated by one pixel to quantify the adhered NMs on the bacterial surface in the Target Activation mode.57 All samples were imaged unfixed.Growth curve analysis of bacteria
Bacterial growth was analysed as optical density (OD600) using the Infinite® 200 PRO Tecan reader (Tecan Group) at time points after exponential growth of control samples or a Tecan Spark® (Tecan Group) for continuous measurements while being incubated at 37 °C and 162 rpm (3.5 mm) in a humidity chamber. Growth inhibition was defined as 1 − (OD600(treated)/OD600(untreated)).For quantum dot (QD) inhibition studies, bacteria–QD complexes were illuminated using a filtered halogen lamp (GE 35200-EKE) as described.29 When applicable, a colony forming unit (CFU) assay was performed additionally.33,35,81,85
Cell and cytotoxicity experiments
Primary human epithelial skin cells (HEC) were purchased from Provitro AG and cultured for up to five passages in Epithelial Cell Medium (Provitro AG) as described elsewhere.86 For cytotoxicity studies, cells were seeded in 96-well cell culture plates. After incubation for 24 h, cells were exposed to the respective samples (bacteria preincubated ± NMs; 90 min 37 °C, 5% CO2). Cells were washed three times with PBS buffer to remove unattached and non-phagocytosed bacteria and viability measured using the CellTiter-Glo® Cell Viability Assay (Promega).Galleria mellonella skin infections
Larvae of G. mellonella (MSC UG) weighing 250–300 mg were randomly selected and scratched with a lancet in the left proleg. Both treatment and control group were hereafter infected with MRSA (5 μL; 4.0 × 107 CFU mL−1). They were then treated with AgNMs in pH 3 solution (5 μL; 0.1 mg mL−1; 1% PEG600, 1 mM citrate), pH 7 solution (5 μL; 0.1 mg mL−1; 1% PEG600 PBS) or PBS alone (5 μL; 1% PEG600 PBS) for one time only. Infected and control larvae were housed under air conditioned S1-safety requirements at 20 °C/55% humidity for 96 h in the dark. Survival was recorded by visual inspection and testing signs of vitality by brief touching every 24 h.Murine skin infections
CD-1 mice (Charles River Laboratories) were housed and used for infection experiments according to locally approved German Animal Welfare Guidelines at the University of Mainz and Essen as previously described.32,87 Briefly, full-thickness skin wounds were induced on the back of anesthetized mice and infected with S. aureus (10 μL; 3.0 × 107 CFU mL−1). Animals were treated in three groups of n = 5 with Ag10 NMs in pH 3 solution (10 μL, Ag10 NMs – 0.1 mg mL−1; 1% PEG600, 1 mM citrate), pH 7 solution (10 μL, pH 7 Ag10 NMs – 0.1 mg mL−1; 1% PEG600 PBS), or as control with PBS (10 μL, PBS, 1% PEG600 PBS). NM-formulations were topically administered on wound beds immediately post infection for one time only. Wound fluids were collected using glass microcapillaries (Fisherbrand™ Color-Coded Capillary Tubes; Fisher Scientific) and pH determined qualitatively by using pH indicator stripes (Merck AG). The mean wound area was macroscopically monitored over time and visualized by photography on days 0, 3, 6, 9, and 12 after infection. The remaining wound area was used as a read-out for wound healing.Human skin infections
Skin wounds were induced on the arm of a healthy volunteer (author RS) using a lancet and infected by adding 3 μL of Staphylococcus aureus bacterial suspension (3.0 × 107 CFU mL−1) or saline control. Subsequently, 5 μL of AgNMs pH 3 (1 mg mL−1 or 0.1 mg mL−1; 1% PEG600, 1 mM citrate) or AgNMs pH 7 (1 mg mL−1; 1% PEG600 PBS) formulations were topically applied immediately post infection for one time only. Infection and wound healing was macroscopically monitored. All experiments were performed in accordance with in compliance with relevant laws, the University Medical Center Mainz Guidelines, and approved by the institutional ethics committee at the University Medical Center Mainz. Informed consents were obtained from human participants of this study.Statistical analysis
Statistical significance was determined by using the Mann–Whitney test or paired t-test assuming significance at *P: 0.05; **P: 0.01; ***P: 0.005 as described previously.46,55,83 Columns show the mean ± s.d. from three independent experiments, unless stated otherwise.Conclusions
Despite the initial enthusiasm for exploiting NMs to combat MDR infections worldwide, nano-antibiotics were often mainly tested under artificial laboratory conditions and often fail in practical clinical applications. Clearly, the combined complexity of pathophysiology, microbiology, chemistry, and material sciences explains why the details are not yet understood. Hence, we did not investigate known resistance mechanisms here, which are not specific for nano-antibiotics, but rather focused on the relevance of biomolecule coronas forming in (patho)physiological systems, which has been neglected so far. Employing comprehensive experimental approaches from analytical to in situ to in vitro to in vivo, we found that physical complex formation of anti-microbial NMs with bacteria is required to achieve an optimal therapeutic effect, which is promoted by small NM size. We also found that resistance to antimicrobial NMs is mediated by protein coronas forming in pathophysiologically relevant environments, such as blood or wounds, reducing the binding of NMs' to bacteria. Based on our data, we strongly feel we uncovered a nanosize-specific mechanism explaining why the antibiotic activity of NMs in general is often reduced under pathophysiologically relevant conditions. We though wish to emphasize that additional, yet unknown (NM-specific) effects may contribute to nano-antibiotic resistance.Furthermore, we discovered a simple and effective way to overcome protein corona-mediated resistance by employing an acidic pH-mediated mechanism, electrostatically restoring complex formation and NMs' antibiotic activity also against highly pathogenic and MDR bacteria.
As previous studies and our own data showed that (most) protein corona-covered NMs in pathophysiological environments are negatively charged, lowering bacteria's cell surface charge by acidic pH may be used to boost the anti-microbial activity of nano-antibiotics in general. Though, we emphasize that our insights may need to be confirmed in detail for alternative antibacterial nanoformulations. In addition to lowering bacteria's cell surface charge, acidic pH treatments may also induce additional structural and chemical changes favouring NM–pathogen complex formation. Whether such effects may be exploited to design nano-antibiotics specific for Gram-positive or Gram-negative pathogens, needs to be explored.
Due to the complexity of pH-dependent effects during wound healing, acidosis of wounds seems to enhance healing although it was reported that the effectiveness of silver antiseptics and gentamycin was reduced in acidic wounds.74,75 Moreover, as depicted in the graphical abstract, wounds infected by bacteria are often alkalescent, counteracting NMs' binding to pathogens.74,75 Hence, lowering the pH by acidic NM formulations could prove relevant for improving nano-antibiotic effects in general, as shown for silver NMs here.
Our proof-of-concept study further proposes to employ our invertebrate insect model to study the therapeutic and/or toxicological activity of nanoformulations in vivo prior to performing extensive studies in mammals. Although we also tested our pH-switch to treat human skin wound infections, we are fully aware that well-controlled clinical trials are required prior to using acidic silver NM formulations in the clinical routine. Also, our insights need to be confirmed for alternative factors causing resistance to antibacterial nanoformulations. Moreover, one might test alternative approaches, such as pre-covering NMs with a widely effective protease (e.g., proteinase K) to reduce protein corona formation and sustain prolonged antibiotic activity. Collectively, we feel that our findings may stimulate the development of more effective antibacterial NM-containing treatments for broad clinical applications as well to rationally guide bacteria–NM interaction in biotechnology.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by NatMedFZ, DFGDO/SR10902, and foreign experts project of Shanxi Provincial ‘100 Talents Plan’. We also wish to acknowledge the kind support from the Mainz imaging facility (Dr H. Goetz), Dr J. Eckrich for support with animal experiments, Dr S. Tenzer for support with proteomics, Dr C. Seckert for support with pathogens, Dr M. Barz and the DENANA/NanoBEL BMBF-research consortium for NM synthesis and financial support.References
- G. Nikaeen, S. Abbaszadeh and S. Yousefinejad, Nanomedicine, 2020, 15(15), 1501–1512 CrossRef CAS.
- D. Westmeier, R. H. Stauber and D. Docter, Toxicol. Appl. Pharmacol., 2016, 299, 53–57 CrossRef CAS.
- S. Siemer, D. Westmeier, C. Vallet, S. Becker, J. Voskuhl, G. B. Ding, E. Thines, R. H. Stauber and S. K. Knauer, ACS Appl. Mater. Interfaces, 2019, 11, 104–114 CrossRef CAS.
- R. H. Stauber, S. Siemer, S. Becker, G. B. Ding, S. Strieth and S. K. Knauer, ACS Nano, 2018, 12, 6351–6359 CrossRef CAS.
- A. Machelart, G. Salzano, X. Li, A. Demars, A. S. Debrie, M. Menendez-Miranda, E. Pancani, S. Jouny, E. Hoffmann, N. Deboosere, I. Belhaouane, C. Rouanet, S. Simar, S. Talahari, V. Giannini, B. Villemagne, M. Flipo, R. Brosch, F. Nesslany, B. Deprez, E. Muraille, C. Locht, A. R. Baulard, N. Willand, L. Majlessi, R. Gref and P. Brodin, ACS Nano, 2019, 13, 3992–4007 CrossRef CAS.
- X. Yang, J. Yang, L. Wang, B. Ran, Y. Jia, L. Zhang, G. Yang, H. Shao and X. Jiang, ACS Nano, 2017, 11, 5737–5745 CrossRef CAS.
- H. A. Hemeg, Int. J. Nanomed., 2017, 12, 8211–8225 CrossRef CAS.
- M. AlMatar, E. A. Makky, I. Var and F. Koksal, Curr. Drug Deliv., 2018, 15, 470–484 CrossRef CAS.
- P. V. Baptista, M. P. McCusker, A. Carvalho, D. A. Ferreira, N. M. Mohan, M. Martins and A. R. Fernandes, Front. Microbiol., 2018, 9, 1441 CrossRef.
- S. J. Lam, N. M. O'Brien-Simpson, N. Pantarat, A. Sulistio, E. H. Wong, Y. Y. Chen, J. C. Lenzo, J. A. Holden, A. Blencowe, E. C. Reynolds and G. G. Qiao, Nat. Microbiol., 2016, 1, 16162 CrossRef CAS.
- M. Markiewicz, J. Kumirska, I. Lynch, M. Matzke, J. Koser, S. Bemowsky, D. Docter, R. Stauber, D. Westmeier and S. Stolte, Green Chem., 2018, 20, 4133–4168 RSC.
- D. Docter, S. Strieth, D. Westmeier, O. Hayden, M. Gao, S. K. Knauer and R. H. Stauber, Nanomedicine, 2015, 10, 503–519 CrossRef CAS.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2015, 44, 6094–6121 RSC.
- Z. S. Al-Ahmady, M. Hadjidemetriou, J. Gubbins and K. Kostarelos, J. Contr. Release, 2018, 276, 157–167 CrossRef CAS.
- R. Garcia-Alvarez, M. Hadjidemetriou, A. Sanchez-Iglesias, L. M. Liz-Marzan and K. Kostarelos, Nanoscale, 2018, 10, 1256–1264 RSC.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Buggio, J. Swift and K. Kostarelos, Biomaterials, 2019, 188, 118–129 CrossRef CAS.
- M. Hadjidemetriou, Z. Al-Ahmady and K. Kostarelos, Nanoscale, 2016, 8, 6948–6957 RSC.
- M. Hadjidemetriou and K. Kostarelos, Nat. Nanotechnol., 2017, 12, 288–290 CrossRef CAS.
- M. Hadjidemetriou, S. McAdam, G. Garner, C. Thackeray, D. Knight, D. Smith, Z. Al-Ahmady, M. Mazza, J. Rogan, A. Clamp and K. Kostarelos, Adv. Mater., 2019, 31, e1803335 CrossRef.
- M. Mazza, H. Ahmad, M. Hadjidemetriou, G. Agliardi, O. N. Pathmanaban, A. T. King, B. W. Bigger, S. Vranic and K. Kostarelos, Nanomedicine, 2019, 14, 3127–3142 CrossRef CAS.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Mazza, R. F. Collins, K. Dawson and K. Kostarelos, ACS Nano, 2015, 9, 8142–8156 CrossRef CAS.
- D. Westmeier, D. Solouk-Saran, C. Vallet, S. Siemer, D. Docter, H. Gotz, L. Mann, A. Hasenberg, A. Hahlbrock, K. Erler, C. Reinhardt, O. Schilling, S. Becker, M. Gunzer, M. Hasenberg, S. K. Knauer and R. H. Stauber, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 7087–7092 CrossRef.
- L. P. Erwig and N. A. Gow, Nat. Rev. Microbiol., 2016, 14, 163–176 CrossRef CAS.
- L. Romani, Nat. Rev. Immunol., 2011, 11, 275–288 CrossRef CAS.
- R. Dean, J. A. Van Kan, Z. A. Pretorius, K. E. Hammond-Kosack, A. Di Pietro, P. D. Spanu, J. J. Rudd, M. Dickman, R. Kahmann, J. Ellis and G. D. Foster, Mol. Plant Pathol., 2012, 13, 414–430 CrossRef.
- V. Aimanianda, J. Bayry, S. Bozza, O. Kniemeyer, K. Perruccio, S. R. Elluru, C. Clavaud, S. Paris, A. A. Brakhage, S. V. Kaveri, L. Romani and J. P. Latge, Nature, 2009, 460, 1117–1121 CrossRef CAS.
- G. Ghodake, M. Kim, J. S. Sung, S. Shinde, J. Yang, K. Hwang and D. Y. Kim, Nanomaterials, 2020, 10(2) DOI:10.3390/nano10020360.
- J. M. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. Piddock, Nat. Rev. Microbiol., 2015, 13, 42–51 CrossRef CAS.
- C. M. Courtney, S. M. Goodman, J. A. McDaniel, N. E. Madinger, A. Chatterjee and P. Nagpal, Nat. Mater., 2016, 15, 529–534 CrossRef CAS.
- B. M. Hover, S. H. Kim, M. Katz, Z. Charlop-Powers, J. G. Owen, M. A. Ternei, J. Maniko, A. B. Estrela, H. Molina, S. Park, D. S. Perlin and S. F. Brady, Nat Microbiol, 2018, 3, 415–422 CrossRef CAS.
- J. Y. H. Lee, I. R. Monk, A. Goncalves da Silva, T. Seemann, K. Y. L. Chua, A. Kearns, R. Hill, N. Woodford, M. D. Bartels, B. Strommenger, F. Laurent, M. Dodemont, A. Deplano, R. Patel, A. R. Larsen, T. M. Korman, T. P. Stinear and B. P. Howden, Nat Microbiol, 2018, 3, 1175–1185 CrossRef CAS.
- B. D. Henry, D. R. Neill, K. A. Becker, S. Gore, L. Bricio-Moreno, R. Ziobro, M. J. Edwards, K. Muhlemann, J. Steinmann, B. Kleuser, L. Japtok, M. Luginbuhl, H. Wolfmeier, A. Scherag, E. Gulbins, A. Kadioglu, A. Draeger and E. B. Babiychuk, Nat. Biotechnol., 2015, 33, 81–88 CrossRef CAS.
- M. Krull, I. Klare, B. Ross, R. Trenschel, D. W. Beelen, D. Todt, E. Steinmann, J. Buer, P. M. Rath and J. Steinmann, Antimicrob. Resist. Infect. Contr., 2016, 5, 31 CrossRef CAS.
- J. Steinmann, A. Hamprecht, M. J. Vehreschild, O. A. Cornely, D. Buchheidt, B. Spiess, M. Koldehoff, J. Buer, J. F. Meis and P. M. Rath, J. Antimicrob. Chemother., 2015, 70, 1522–1526 CrossRef CAS.
- D. Westmeier, A. Hahlbrock, C. Reinhardt, J. Frohlich-Nowoisky, S. Wessler, C. Vallet, U. Poschl, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2018, 47, 5312–5337 RSC.
- A. Zipperer, M. C. Konnerth, C. Laux, A. Berscheid, D. Janek, C. Weidenmaier, M. Burian, N. A. Schilling, C. Slavetinsky, M. Marschal, M. Willmann, H. Kalbacher, B. Schittek, H. Brotz-Oesterhelt, S. Grond, A. Peschel and B. Krismer, Nature, 2016, 535, 511–516 CrossRef CAS.
- A. N. Brown, K. Smith, T. A. Samuels, J. Lu, S. O. Obare and M. E. Scott, Appl. Environ. Microbiol., 2012, 78, 2768–2774 CrossRef CAS.
- M. J. Hajipour, K. M. Fromm, A. A. Ashkarran, D. Jimenez de Aberasturi, I. R. de Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 30, 499–511 CrossRef CAS.
- X. Li, S. M. Robinson, A. Gupta, K. Saha, Z. Jiang, D. F. Moyano, A. Sahar, M. A. Riley and V. M. Rotello, ACS Nano, 2014, 8, 10682–10686 CrossRef CAS.
- A. J. Huh and Y. J. Kwon, J. Contr. Release, 2011, 156, 128–145 CrossRef CAS.
- N. Abed, F. Said-Hassane, F. Zouhiri, J. Mougin, V. Nicolas, D. Desmaele, R. Gref and P. Couvreur, Sci. Rep., 2015, 5, 13500 CrossRef CAS.
- M. Karavolos and A. Holban, Pharmaceuticals, 2016, 9(4) DOI:10.3390/ph9040062.
- B. J. Teubl, B. Stojkovic, D. Docter, E. Pritz, G. Leitinger, I. Poberaj, R. Prassl, R. H. Stauber, E. Frohlich, J. G. Khinast and E. Roblegg, Clin. Oral Invest., 2018, 22, 929–940 CrossRef.
- K. R. Raghupathi, R. T. Koodali and A. C. Manna, Langmuir, 2011, 27, 4020–4028 CrossRef CAS.
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 27, 76–83 CrossRef CAS.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS.
- J. E. Riviere, C. Scoglio, F. D. Sahneh and N. A. Monteiro-Riviere, Comput. Sci. Discov., 2013, 6, 014005 CrossRef.
- E. Alarcon, C. Bueno-Alejo, C. Noel, K. Stamplecoskie, N. Pacioni, H. Poblete and J. C. Scaiano, J. Nanoparticle Res., 2013, 15, 1–14 CrossRef.
- M. T. Ortega, J. E. Riviere, K. Choi and N. A. Monteiro-Riviere, Toxicol. Vitro, 2017, 42, 150–160 CrossRef CAS.
- J. Li, H. Sang, H. Guo, J. T. Popko, L. He, J. C. White, O. Parkash Dhankher, G. Jung and B. Xing, Nanotechnology, 2017, 28, 155101 CrossRef.
- K. Choi, J. E. Riviere and N. A. Monteiro-Riviere, Nanotoxicology, 2017, 11, 64–75 CrossRef CAS.
- P. Chandran, J. E. Riviere and N. A. Monteiro-Riviere, Nanotoxicology, 2017, 11, 507–519 CrossRef CAS.
- J. S. Gebauer, M. Malissek, S. Simon, S. K. Knauer, M. Maskos, R. H. Stauber, W. Peukert and L. Treuel, Langmuir, 2012, 28, 9673–9679 CrossRef CAS.
- Z. Gao, T. Ma, E. Zhao, D. Docter, W. Yang, R. H. Stauber and M. Gao, Small, 2016, 12, 556–576 CrossRef CAS.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS.
- R. Holm, B. Weber, P. Heller, K. Klinker, D. Westmeier, D. Docter, R. H. Stauber and M. Barz, Macromol. Biosci., 2017, 17 DOI:10.1002/mabi.201600514.
- D. Westmeier, G. Posselt, A. Hahlbrock, S. Bartfeld, C. Vallet, C. Abfalter, D. Docter, S. K. Knauer, S. Wessler and R. H. Stauber, Nanoscale, 2018, 10, 1453–1463 RSC.
- J. Kasper, M. I. Hermanns, C. Bantz, M. Maskos, R. Stauber, C. Pohl, R. E. Unger and J. C. Kirkpatrick, Part. Fibre Toxicol., 2011, 8, 6 CrossRef CAS.
- C. D. Walkey, J. B. Olsen, F. Song, R. Liu, H. Guo, D. W. Olsen, Y. Cohen, A. Emili and W. C. Chan, ACS Nano, 2014, 8, 2439–2455 CrossRef CAS.
- M. Lan, J. Wu, W. Liu, W. Zhang, J. Ge, H. Zhang, J. Sun, W. Zhao and P. Wang, J. Am. Chem. Soc., 2012, 134, 6685–6694 CrossRef CAS.
- J. J. Cid Martin, M. Assali, E. Fernandez-Garcia, V. Valdivia, E. M. Sanchez-Fernandez, J. M. Garcia Fernandez, R. E. Wellinger, I. Fernandez and N. Khiar, J. Mater. Chem. B, 2016, 4, 2028–2037 RSC.
- K. P. Yu, Y. T. Huang and S. C. Yang, J. Hazard. Mater., 2013, 261, 155–162 CrossRef CAS.
- B. Schmidt, S. Sankaran, L. Stegemann, C. A. Strassert, P. Jonkheijm and J. Voskuhl, J. Mater. Chem. B, 2016, 4, 4732–4738 RSC.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS.
- J. E. Riviere, M. Jaberi-Douraki, J. Lillich, T. Azizi, H. Joo, K. Choi, R. Thakkar and N. A. Monteiro-Riviere, Nanotoxicology, 2018, 12, 1093–1112 CrossRef CAS.
- S. Siemer, D. Westmeier, C. Vallet, J. Steinmann, J. Buer, R. H. Stauber and S. K. Knauer, Mater. Today, 2018, 26, 19–29 CrossRef.
- C. D. Walkey and W. C. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- L. Treuel, D. Docter, M. Maskos and R. H. Stauber, Beilstein J. Nanotechnol., 2015, 6, 857–873 CrossRef CAS.
- A. Panacek, L. Kvitek, M. Smekalova, R. Vecerova, M. Kolar, M. Roderova, F. Dycka, M. Sebela, R. Prucek, O. Tomanec and R. Zboril, Nat. Nanotechnol., 2018, 13, 65–71 CrossRef CAS.
- S. M. Tennant, E. L. Hartland, T. Phumoonna, D. Lyras, J. I. Rood, R. M. Robins-Browne and I. R. van Driel, Infect. Immun., 2008, 76, 639–645 CrossRef CAS.
- M. Breisch, K. Loza, K. Pappert, A. Rostek, C. Rurainsky, K. Tschulik, M. Heggen, M. Epple, J. C. Tiller, T. A. Schildhauer, M. Koller and C. Sengstock, Nanotechnology, 2020, 31, 055703 CrossRef CAS.
- Y. Kirmanidou, M. Sidira, A. Bakopoulou, A. Tsouknidas, O. Prymak, R. Papi, T. Choli-Papadopoulou, M. Epple, N. Michailidis, P. Koidis and K. Michalakis, Dent. Mater., 2019, 35, e220–e233 CrossRef CAS.
- T. Tenkumo, K. Ishiyama, O. Prymak, K. Nakamura, M. Shirato, T. Ogawa, M. Miyashita, M. Takahashi, M. Epple, T. Kanno and K. Sasaki, Sci. Rep., 2020, 10, 8553 CrossRef CAS.
- P. G. Bowler, B. I. Duerden and D. G. Armstrong, Clin. Microbiol. Rev., 2001, 14, 244–269 CrossRef CAS.
- S. L. Percival, S. Finnegan, G. Donelli, C. Vuotto, S. Rimmer and B. A. Lipsky, Crit. Rev. Microbiol., 2016, 42, 293–309 CAS.
- C. J. Tsai, J. M. Loh and T. Proft, Virulence, 2016, 7, 214–229 CrossRef CAS.
- L. Jiang, M. K. Greene, J. L. Insua, J. S. Pessoa, D. M. Small, P. Smyth, A. P. McCann, F. Cogo, J. A. Bengoechea, C. C. Taggart and C. J. Scott, J. Contr. Release, 2018, 279, 316–325 CrossRef CAS.
- A. Gomez-Lopez, A. Forastiero, E. Cendejas-Bueno, L. Gregson, E. Mellado, S. J. Howard, J. L. Livermore, W. W. Hope and M. Cuenca-Estrella, Med. Mycol., 2014, 52, 311–319 CrossRef CAS.
- N. Ramarao, C. Nielsen-Leroux and D. Lereclus, J. Vis. Exp., 2012, 70, e4392 Search PubMed.
- O. Koshkina, D. Westmeier, T. Lang, C. Bantz, A. Hahlbrock, C. Wurth, U. Resch-Genger, U. Braun, R. Thiermann, C. Weise, M. Eravci, B. Mohr, H. Schlaad, R. H. Stauber, D. Docter, A. Bertin and M. Maskos, Macromol. Biosci., 2016, 16, 1287–1300 CrossRef CAS.
- S. Siemer, A. Hahlbrock, C. Vallet, D. J. McClements, J. Balszuweit, J. Voskuhl, D. Docter, S. Wessler, S. K. Knauer, D. Westmeier and R. H. Stauber, NPJ Sci. Food, 2018, 2, 22 CrossRef.
- J. Xiao, A. Kuc, S. Pokhrel, L. Madler, R. Pottgen, F. Winter, T. Frauenheim and T. Heine, Chemistry, 2013, 19, 3287–3291 CrossRef CAS.
- D. Docter, U. Distler, W. Storck, J. Kuharev, D. Wunsch, A. Hahlbrock, S. K. Knauer, S. Tenzer and R. H. Stauber, Nat. Protoc., 2014, 9, 2030–2044 CrossRef CAS.
- M. Kaase, S. Schimanski, R. Schiller, B. Beyreiss, A. Thurmer, J. Steinmann, V. A. Kempf, C. Hess, I. Sobottka, I. Fenner, S. Ziesing, I. Burckhardt, L. von Muller, A. Hamprecht, I. Tammer, N. Wantia, K. Becker, T. Holzmann, M. Furitsch, G. Volmer and S. G. Gatermann, Int. J. Med. Microbiol., 2016, 306, 415–420 CrossRef CAS.
- S. Siemer, D. Westmeier, M. Barz, J. Eckrich, D. Wunsch, C. Seckert, C. Thyssen, O. Schilling, M. Hasenberg, C. Pang, D. Docter, S. K. Knauer, R. H. Stauber and S. Strieth, Biomaterials, 2019, 192, 551–559 CrossRef CAS.
- K. Engels, S. K. Knauer, S. Loibl, V. Fetz, P. Harter, A. Schweitzer, A. Fisseler-Eckhoff, F. Kommoss, L. Hanker, V. Nekljudova, I. Hermanns, H. Kleinert, W. Mann, A. du Bois and R. H. Stauber, Cancer Res., 2008, 68, 5159–5166 CrossRef CAS.
- R. H. Stauber, S. K. Knauer, N. Habtemichael, C. Bier, B. Unruhe, S. Weisheit, S. Spange, F. Nonnenmacher, V. Fetz, T. Ginter, S. Reichardt, C. Liebmann, G. Schneider and O. H. Kramer, Oncotarget, 2012, 3, 31–43 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00644k |
| ‡ Equal contributor. |
| This journal is © The Royal Society of Chemistry 2020 |