Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A WS2-gold nanoparticle heterostructure-based novel SERS platform for the rapid identification of antibiotic-resistant pathogens
Avijit
Pramanik
,
Dalephine
Davis
,
Shamily
Patibandla
,
Salma
Begum
,
Priyadarshini
Ray
 ,
Kaelin
Gates
,
Ye
Gao
,
Kaelin
Gates
,
Ye
Gao
 and
Paresh
Chandra Ray
*
and
Paresh
Chandra Ray
*
Department of Chemistry and Biochemistry, Jackson State University, Jackson, MS, USA. E-mail: paresh.c.ray@jsums.edu; Fax: +1 6019793674
First published on 31st March 2020
Abstract
The emergence of antibiotic-resistant bacteria is the biggest threat to our society. The rapid discovery of drug resistant bacteria is very urgently needed to guide antibiotic treatment development. The current manuscript reports the design of a 2D–0D heterostructure-based surface enhanced Raman spectroscopy (SERS) platform, which has the capability for the rapid identification of the multidrug resistant strain of Salmonella DT104. Details of the synthesis and characterization of the heterostructure SERS platform using a two dimensional (2D) WS2 transition metal dichalcogenide (TMD) and zero dimensional (0D) plasmonic gold nanoparticles (GNPs) have been reported. The current manuscript reveals that the 2D–0D heterostructure-based SERS platform exhibits extremely high Raman enhancement capabilities. Using Rh-6G and 4-ATP probe molecules, we determined that the SERS sensitivity is in the range of ∼10−10 to 10−11 M, several orders of magnitude higher than 2D-TMD on its own (10−3 M) or 0D-GNPs on their own (∼10−6 to 10−7 M). Experimental and theoretical finite-difference time-domain (FDTD) simulation data indicate that the synergistic effect of an electromagnetic mechanism (EM) and a chemical mechanism (CM) on the heterostructure is responsible for the excellent SERS enhancement observed. Notably, the experimental data reported here show that the heterostructure-based SERS has the ability to separate a multidrug resistance strain from a normal strain of Salmonella by monitoring the antibiotic–pathogen interaction within 90 minutes, even at a concentration of 100 CFU mL−1.
Introduction
After the discovery of graphene, two-dimensional (2D) materials have drawn significant attention in the scientific community due to their excellent structural, physical, optical and electronic properties.1–5 In recent years, heterostructural building blocks using 2D, one dimensional (1D) and zero dimensional (0D) materials have opened up unique opportunities for fundamental scientific studies due to the combined advantages of the individual materials.6–12 Recent reports demonstrated that heterostructures exhibit exceptional optical and electronic properties that are not available in the individual material.4–10Due to its high sensitivity, nondestructive character and fingerprint identification capability, SERS is highly promising in biomedical technology.13–20 In the last two decades we and others have reported that extremely weak Raman signals can be enhanced significantly using a proper design of the SERS substrate via the electromagnetic mechanism (EM) and chemical mechanism (CM).16–24 Zero dimensional plasmonic nanoparticles such as gold and silver nanoparticles have been used extensively as SERS materials, where Raman intensity can be enhanced by ∼106via EM & CM.13–20 On the other hand, in the last few years SERS substrates based on two-dimensional (2D) transition-metal dichalcogenides (TMD) have been reported, where Raman intensity can be enhanced by ∼102via CM as well as due to the resonance with near- and far-field optical properties.11,16,20–24 To take the advantages of the EM and CM effect from both types of materials, in the current manuscript we have reported the development of novel heterostructure material-based SERS using 2D WS2 TMD and 0D plasmonic gold nanoparticles (GNPs). The reported data show a remarkably high SERS sensitivity of ∼10−10 to 10−11 M from the heterostructure.
Antibiotic resistant bacteria, which are a nightmare for medical treatment, are responsible for more than a million deaths in the world every year.25–30 As per the Center for Disease Control and Prevention (CDC), antibiotic resistance is one of the biggest public health challenges in the 21st century.26 As a result, there is a huge urgency in finding rapid and cost-effective assays which will have the capability for rapid and accurate determination of the appropriate antibiotic for an infection in a timely manner.31–42 Driven by this need, the current manuscript demonstrates that 0D–2D heterostructure-based SERS is capable of rapid identification of drug resistant bacteria by monitoring structural changes in the pathogen's cell wall during antibiotic treatment. Reported data show that the SERS can be used for the identification of multidrug resistant Salmonella DT104 and a normal strain Salmonella Typhi antimicrobial susceptibility test using Augmentin antibiotics, even at the concentration of 100 CFU mL−1 experimental data reported here indicates that heterostructure-based SERS has the capability of antimicrobial susceptibility testing in less than 2 hours, whereas the “gold standard” broth microdilution (BMD) test takes 16 to 20 h in clinics until the number of bacteria reaches above 105 CFU mL−1.
Experimental section
Development of the heterostructure-based SERS platform
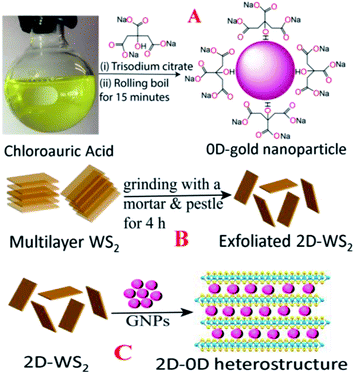
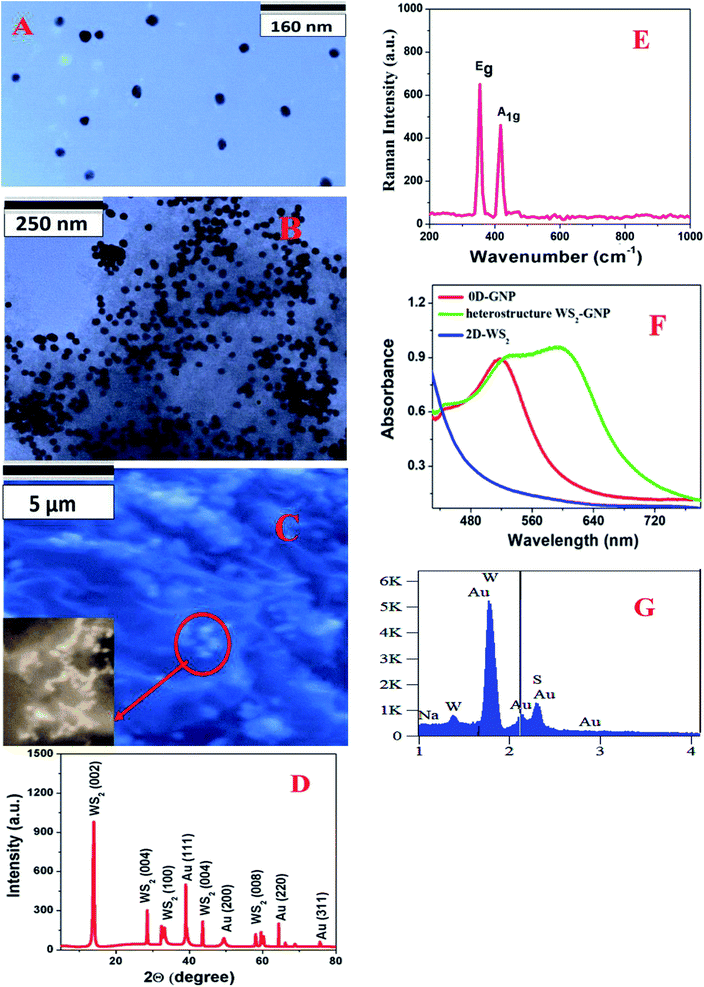
0D-plasmonic- GNPs were synthesized according to the previous work by our group18–20,24,29 using HAuCl4 and trisodium citrate dihydrate, as shown in Scheme 1A. For this purpose, 1.25 mL of 10 mM HAuCl4 solution was completely dissolved in 50 mL of deionized water. The mixture was stirred on a hot plate and brought to a rolling boil. Next, we added 2 mL of 1% trisodium citrate dihydrate to the boiling solution. After a few minutes, a colloidal solution was formed and then we continued heating for another few minutes. After that the colloidal gold nanoparticles were centrifuged at 5000 rpm for a few hours and the supernatant was removed by decantation. The purified AuNPs were kept at 4 °C for future use. After purification, the AuNPs were characterized by electron microscopy and absorption techniques. Fig. 1A shows the transmission electron microscopy (TEM) image which indicates that the size of the 0D-GNPs is ∼25 ± 5 nm. Fig. 1F shows the absorption spectra of 0D-GNPs, which clearly show a strong plasmon band at 520 nm. The observed strong plasmon band is mainly due to the localized surface plasmonic resonance (LSPR) of the GNPs induced by the incident light.13–182D-WS2 nanosheets were synthesized using a simple ultrasonication route from WS2 powder.16,20–22 Briefly, commercial WS2 bulk powder was pelletised by a grinding miller for about 4 h, as shown in Scheme 1B. In the next step, WS2 nanosheets were exfoliated through liquid exfoliation followed by intercalation. For this purpose, WS2 nanosheets were added to H2SO4 (98%) at 90 °C for 24 h.
After that WS2 nanosheets were collected after centrifugation at 5000 rpm for 1 h and kept in the fridge for future use. At the end, 2D-WS2 nanosheets were characterized by electron microscopy and absorption techniques. Fig. 1F shows the absorption spectra of 2D-WS2, which shows some absorption at 400–650 nm and it is mainly due to the A1 and B1 excitonic transitions of WS2.16,20–22 The observed exitonic transition originates from the energy split of the valence-band and spin-orbital coupling, as reported before.16,20–22
In the next step we synthesized 0D–2D heterostructures using 2D-WS2 and 0D-GNPs. For this purpose, GNP solution was dropped in freshly prepared WS2 solutions in phosphate-buffered saline. After that the 2D-WS2 and 0D-GNP mixture was sonicated for 2 h at room temperature. In the next step the 2D-WS2 and 0D-GNP mixture was continuously stirred at very low speed overnight at room temperature for the completion of the reaction. After that the product was washed with methanol three times by centrifugation at 5000 rpm for 15 min followed by decantation. Finally, the heterostructure materials were dried under vacuum at room temperature for a week. After that the heterostructure materials were characterized by TEM, SEM, absorption, energy dispersive X-ray (EDX), X-ray diffraction (XRD) and Raman spectroscopy techniques, as reported in Fig. 1C–G. Fig. 1B shows the TEM image and Fig. 1C shows the SEM image of the freshly prepared heterostructure which indicates that 0D-GNPs are on the surface of 2D-WS2. Both the TEM and inserted high-resolution SEM images indicate the formation of GNP aggregates on the 2D-WS2 surface. EDX data from the heterostructure has been reported as Fig. 1G, which clearly shows the presence of W, S, and Au. Fig. 1D shows the powder XRD data from the heterostructure, which shows the presence of (002), (004), (006) and (008) reflections for WS2, and (111), (200), (220) and (3111) reflections for GNP.17,20–22,24Fig. 1F shows the absorption spectra of the heterostructures which clearly indicate a broad plasmon band which is due to the aggregation of 0D-GNP on the surface of 2D-WS2 as shown in the TEM images reported in Fig. 1B. Both the absorption spectra and TEM/SEM images clearly indicate the formation of 0D-GNP aggregates on the surface of 2D-WS2 in the heterostructure. Fig. 1E shows the Raman spectra from the heterostructure, which indicate the presence of an in-plane (E2g) Raman band at ∼356 cm−1 and an out-of-plane (A1g) Raman band at ∼417 cm−1, which is due to the W–S vibration of WS2.17,20–22
SERS using 2D-WS2, 0D-GNPs and the heterostructure substrate
For the SERS experiment we used 1 mg mL−1 of the as-synthesized ‘2D-WS2’ and ‘2D–0D heterostructure’ samples. For the Raman experiments, we used a confocal Raman system with a laser excitation of 670 nm at ∼2 mW power. We used 100× magnification, and a numerical aperture of 0.9 for our experiment. We collected signals in a backscattering configuration. In our experiment, for the determination of the SERS enhancement factor on the 2D-WS2, 0D-GNP and heterostructure surfaces, we assumed that the analytes were distributed uniformly on the surface. For the Raman intensity measurements, the heterostructure platform was immersed in different concentrations of the analyte for two hours and the SERS substrate was dried under N2 flow. For the Raman data collection from the 2D-WS2, 0D-GNPs and heterostructure substrates with 4-ATP we used a 10 second acquisition time and 5-scan averaging, so that we could achieve a very good signal-to-noise ratio. For the Raman measurements from the bacteria, at first, we located the bacteria position using a microscope attached to a confocal Raman microscopy system, where a single bacterium or clusters of bacteria could be easily identified. After that the laser beam was focused on the bacteria through the microscope object, high quality Raman spectra from 10 to 15 spots were measured and after that the spectra were processed with a baseline removal program. In the next step, we performed averaging from the different spots, so that we could achieve a very good signal-to-noise ratio.FDTD simulations for full-field electromagnetic wave calculations
We used the finite-difference time-domain (FDTD) simulation for full-field electromagnetic wave calculations, as previously reported by our group.13,18,19,24 In this process we used numerical calculations which can be used to understand plasmon-coupling in a “hot spot”, which is responsible for the huge EM enhancement mechanism in SERS.13–20,24 In our calculations the electric field intensities were simulated by using a diameter of the 0D-GNPs of 25 nm which are decorated on the 2D-WS2 nanosheets as we observed experimentally. To keep the experimental conditions, we used an incident laser wavelength of 670 nm for our calculations. For the simulation calculations the entire process was performed with a 0.001 nm mesh resolution and 4000 fs time, as we have reported before.13,18,19,24Bacteria sample preparation
The multidrug resistant strain Salmonella DT104, the normal strain of Salmonella Typhi and growth media were purchased from the American Type Culture Collection (ATCC, Rockville, MD). We cultured these bacteria according to the ATCC protocol, as we have reported before.18,19,24,29,30 Once the different bacteria had grown to 106 CFU mL−1, we used them for the experiment.SERS based determination of antimicrobial susceptibility
For the determination of antimicrobial susceptibility, Salmonella Typhi and Salmonella DT104 bacteria were incubated with the Augmentin antibiotic for 2 hours. During this process, we measured time dependent SERS spectra of the bacteria on the heterostructure surface for two hours of incubation.Finding the percentage of live bacteria after exposure to the Augmentin antibiotic
We used a colony-countable plate to determine the percentage of live bacteria before and after treatment. For this purpose, we transferred antimicrobial susceptible, Salmonella Typhi and Salmonella DT104 bacteria to colony-countable plates. We incubated them for 24 h at 37 °C and then the colony number was counted with a colony counter (Bantex, Model 920 A). We also used molecular probes' LIVE/DEAD® BacLightTM Bacterial Viability Kits (from Fisher Scientific) to determine the amount of antimicrobial susceptible, Salmonella Typhi and Salmonella DT104 bacteria that was dead after antibiotic treatment. This kit is well known to provide a two-color fluorescence assay, which utilizes mixtures of our SYTO® 9 green-fluorescent nucleic acid stain which can bind with live bacteria and the red-fluorescent nucleic acid stain, propidium iodide, which is known to bind with dead bacteria.ATP leakage experiment
To better understand cell wall and membrane damage during antibiotic treatment, we performed a bacterial ATP leakage experiment using the molecular probes® ATP determination kit (Thermo Fisher Scientific). For this purpose, bacteria cultures without antibiotics were used as a reference. This ATP determination kit is based on the luminescence results from the firefly luciferase enzyme.Results and discussion
Finding the Raman enhancement, stability and reproducibility
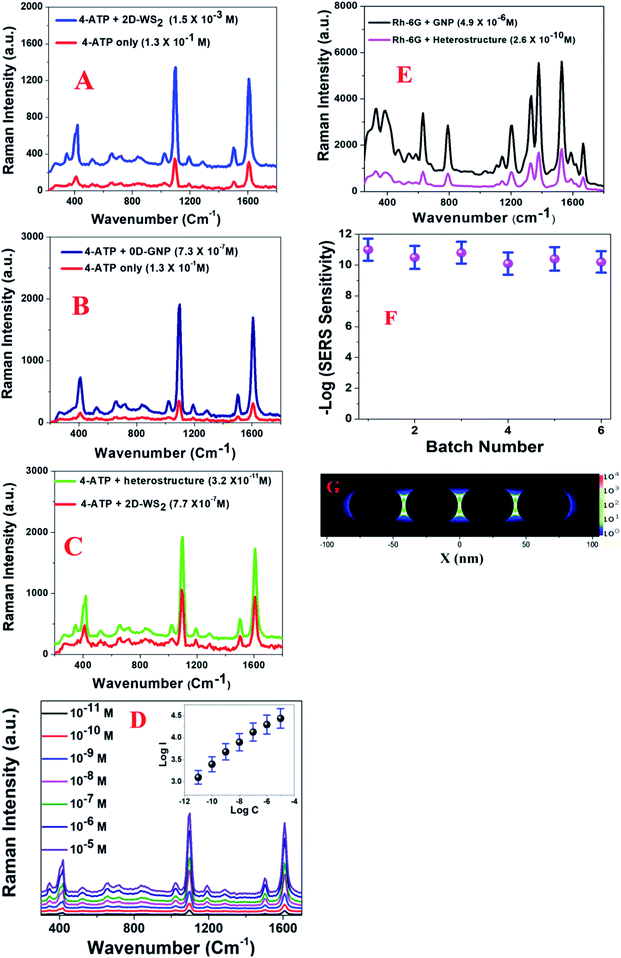
Since the enhancement factor is the most important parameter for designing a novel Raman substrate, we determined the Raman EF using 4-aminothiophenol (4-ATP) for the 2D-WS2, 0D-GNPs and heterostructure materials. To remove the Eg and A1g band contribution from WS2, we subtracted the Raman spectra of 4-ATP with the heterostructure SERS substrate from the spectrum of the heterostructure SERS substrate on its own. To find the enhancement factor for each type of material, we used the confocal Raman system with a laser excitation of 670 nm. Fig. 2A–C show the Raman spectra of 4-aminothiophenol on 2D-WS2, 0D-GNPs and the heterostructure. The reported Raman data in Fig. 2A show that significant changes in the intensities of the 4-ATP Raman peaks are observed on the 2D-WS2 substrate in comparison to the Raman spectrum of 4-ATP on its own. The observed Raman peaks are dominated by a1 vibrational modes and these are ν(C–C + NH2 bend) at ∼1590 cm−1 and ν[ν(C–C) + δ(C–H)] at ∼1078 cm−1.13–20,24 In the reported spectra, we also observed Raman peaks due to b2 modes at ∼1435 cm−1, which can be ascribed to the CC stretch in the Ph ring + NH2 rock.13–20,24 Another Raman peak due to b2 modes is observed at ∼1170 cm−1 which can be ascribed to the CH bend vibration.13–20,24 Similarly, we also observed a Raman band at 464 cm−1 due to the ν(C–N) + ν(C–S) + γ(CCC) vibrations.13–20,24As shown in the TEM and SEM images reported in Fig. 1B and C, the distribution of GNP is not uniform. As a result, the Raman intensity may vary at different spots of the heterostructure surface due to the possible aggregation of the 0D-GNPs on the 2D surface. Our experimental results indicate that Raman intensity can vary more than 10% from one spot to another spot due to the variation of GNP aggregate sizes at different spots. As a result, we measured the SERS intensity at 10 different spots and the average values are reported in this manuscript. Using the 4-ATP probe molecule, we determined that the SERS sensitivity is in the range of ∼10−3 M for 2D-WS2. The observed Raman sensitivity from 2D-WS2 can be attributed to the chemical enhancement mechanism as well as to the resonance with near- and far-field optical properties.11,16,20–24Fig. 2B shows the Raman enhancement of 4-ATP on the 0D-GNP surface. Using the reported data in Fig. 2B, we estimated that the average SERS sensitivity is in the range of ∼10−7 M for 0D-GNP. The observed high SERS sensitivity for 0D-GNP can be attributed due to the strong electromagnetism as well as the chemical enhancement effect caused by the charge-transfer process happening on the ATP-GNP surface.12–19 Similarly, using the reported data in Fig. 2C, we estimated that the average SERS sensitivity is in the range of ∼10−11 M for the heterostructure and the average SERS sensitivity is in the range of ∼10−7 M for 0D-GNP. The observed 104 higher Raman sensitivity from the heterostructure surface is due to the strong electromagnetism as well as the strong chemical enhancement capability for the heterostructure. The 4-ATP concentration dependent SERS spectra are reported in Fig. 2D, which also indicates that the SERS sensitivity is in the range of ∼10−11 M for the heterostructure.
Fig. 2E shows the SERS spectra from Rh-6G on 0D-GNPs and the heterostructure surface. Using the Rh-6G probe molecule, we determined that the SERS sensitivity is in the range of ∼10−10 M, for the heterostructure and ∼10−6 M for 0D-GNPs. The observed 104 higher Raman sensitivity from the heterostructure surface is due to the strong electromagnetism as well as the strong chemical enhancement capability of the heterostructure. We performed the finite-difference time-domain (FDTD) simulation13,18,19,24 to better understand the near-field distribution around the 0D-GNPs. The calculation details have been reported before by our group.13,18,19,24Fig. 2G shows that the square of the field enhancement (|E|2) for the 0D-GNP “hot spots” can be around two orders of magnitude higher than that of an individual nanoparticle. It is now well documented13–19,24 that the Raman EF α|E|4. As a result, we expect to increase the Raman enhancement to around 4 orders of magnitude for the heterostructure, which is only due to the formation of aggregates by 0D-GNPs on the surface of 2D-WS2. To understand the reproducibility, we developed a heterostructure in five different batches using 0D-GNP and 2D-WS2. After that, we monitored the average Raman sensitivity using 4-ATP in 10 different spots for each batch. As reported in Fig. 2F, our experimental data show that the relative standard deviation is around ∼10%.
Use of heterostructure-based SERS for the label-free determination of antimicrobial susceptibility
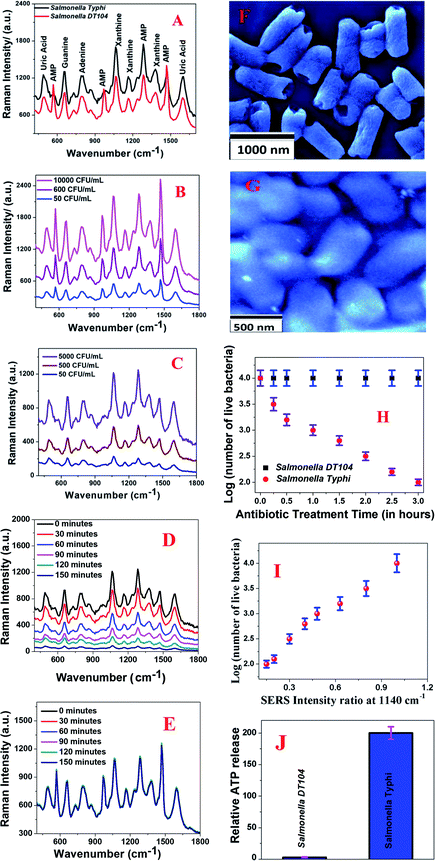
Next, to understand whether 0D–2D heterostructure-based SERS can be used for rapid and accurate determination of antimicrobial susceptibility, we used multidrug resistant-Salmonella DT104 and a normal strain of Salmonella Typhi. In our approach, we used 0D–2D heterostructure-based SERS for monitoring structural changes in the pathogen's cell wall during the Augmentin antibiotic treatment. To remove the Eg and A1g band contribution from WS2, we subtracted the Raman spectra of pathogens with the heterostructure SERS substrate from the spectrum for the heterostructure SERS substrate on its own. As reported in Fig. 3A–E, we observed very strong Raman peaks from the multidrug resistant Salmonella DT104 and the normal strain of Salmonella Typhi in the presence of the 0D–2D heterostructure. As shown in Fig. 3A, the observed vibrational Raman bands from multidrug resistant Salmonella DT104 and the normal strain of Salmonella Typhi can be assigned to the spectral contributions of different purines such as adenine, xanthine, guanine, uric acid, and adenosine monophosphate (AMP) as reported by Premasiri and Ziegler et al.41,42 By comparing the SERS data reported in Fig. 3A, we can conclude that most of the Raman peaks are very similar for multidrug resistant Salmonella DT104 and the normal strain of Salmonella Typhi, except the AMP Raman bands at ∼570 cm−1, ∼968 cm−1 and ∼1386 cm−1, which are much stronger and unique for multidrug resistant Salmonella DT104. As shown in Fig. 3B and C, concentration dependent SERS data clearly indicate that the heterostructure based SERS platform can be used for the identification of multidrug resistant Salmonella DT104 and the normal strain of Salmonella Typhi even at a concentration of 50 CFU mL−1.Our SERS based determination of antimicrobial susceptibility is based on the fact that the Augmentin antibiotic will alter the biochemical components of the normal strain of Salmonella Typhi bacterial cell wall and it will induce variation in the SERS spectra. On the other hand, since multidrug resistant Salmonella DT104 is resistant to the Augmentin antibiotic, it will not able to alter the biochemical components of the Salmonella DT104 cell wall and as a result, there will be no variation in the SERS spectra. For the determination of antimicrobial susceptibility, Salmonella Typhi and Salmonella DT104 bacteria cultures without an antibiotic were used as a reference. During the incubation of Salmonella Typhi and Salmonella DT104 bacteria cultures with the Augmentin antibiotic, we measured the time dependent SERS spectra of bacteria on the heterostructure surface for two hours of incubation.
We used a bacterial viability assay kit to determine the amount of dead bacteria after antibiotic treatment. Fig. 3D shows how the characteristic Raman peaks from the Salmonella Typhi bacteria vary during Augmentin antibiotic exposure. We used 1000 CFU mL−1 of Salmonella Typhi bacteria for this experiment. The reported data clearly show that the Raman bands from the Salmonella Typhi bacteria decreased significantly during antibacterial treatment, which indicates that the observed Raman modes originated from the bacterial cell wall. To understand better, we also performed the SEM image experiment, as reported in Fig. 3F. Reported SEM data clearly shows that the bacterial cell walls are damaged during Augmentin antibiotic treatment for the Salmonella Typhi bacteria. On the other hand, Fig. 3E shows how the characteristic Raman peaks from the multidrug resistant Salmonella DT104 bacteria varies during Augmentin antibiotic exposure. We used 1000 CFU mL−1 of Salmonella DT104 bacteria for this experiment.
The reported data clearly shows that the Raman bands remain unaltered during the antibacterial treatment, which indicates that there may be no damage to the bacterial cell wall during antibiotic treatment for Salmonella DT104. Fig. 3G shows the SEM image of Salmonella DT104 after antibiotic treatment for two hours, which also indicates no damage on the bacterial cell wall during antibiotic treatment. The loss of the SERS signal from Salmonella Typhi bacteria during Augmentin antibiotic treatment for the Salmonella Typhi bacteria is due to the decrease in the number of “hot spots” on the bacteria surface due to cell wall damage. Since the SERS intensity is only an indication of the number of bacteria experiencing EF, during antibiotic treatment, the number of unaffected bacteria on the hotspots will decrease with time and as a result, the SERS intensity from the Salmonella Typhi bacteria decreases.
To better understand cell wall and membrane damage during antibiotic treatment, we performed a bacterial ATP leakage experiment. Experimental details have been reported before.29,30 The ATP leakage experiment data as reported in Fig. 3J clearly shows high leakage of cellular ATP during Augmentin antibiotic treatment for the Salmonella Typhi bacteria. On the other hand, as reported in Fig. 3J, experimental data also indicate minimum leakage of cellular ATP during Augmentin antibiotic treatment for the Salmonella DT104 bacteria. All the above data clearly shows that cell wall and membrane damage during antibiotic treatment is high for Salmonella Typhi and it is minimal for Salmonella DT104.
Cell viability data as measured by the bacterial viability assay kit and as reported in Fig. 3H also indicate that multidrug resistant Salmonella DT104 bacteria remain alive during Augmentin antibiotic treatment for three hours. On the other hand, more than 90% of the Salmonella Typhi bacteria died during Augmentin antibiotic treatment for three hours. All the above reported experimental data clearly shows that the 2D–0D heterostructure based SERS platform has the capability for separation of the Salmonella DT104 drug resistant strain and the Salmonella Typhi normal strain via the antibiotic susceptibility test. Fig. 3I shows how the SERS intensity at 1140 cm−1 varies as the number of live Salmonella Typhi bacteria decreases during treatment with the Augmentin antibiotic. Since among all the observed Raman bands from the bacteria, the Raman mode at 1140 cm−1 is one of the strongest band for Salmonella Typhi and it is also a common band for both the bacteria we studied here, we used this band for plotting against the number of live bacteria. We measured the time dependent SERS intensity during treatment and reported the ratio of SERS intensity at 1140 cm−1 before and after treatment. Our data indicate that SERS intensity at 1140 cm−1 decreases abruptly as the number of live Salmonella Typhi bacteria decreases during treatment with the Augmentin antibiotic. From the experimental data we determined that even at ∼102 CFU mL−1 concentration, a distinguishable SERS spectrum change can be obtained during antibiotic treatment.
Conclusions
In the current manuscript, we reported the design of a novel heterostructure material using 2D-WS2 and 0D-GNPs, which can be used to determine the antimicrobial susceptibility via surface enhanced Raman spectroscopy (SERS). Experimental data indicate that in the heterostructure EM and CM enhancement factors have been combined and a very high SERS sensitivity of ∼10−10 to 10−11 M was observed, which is several orders of magnitude higher than 2D-TMD (∼10−3) and 0D-GNPs (∼10−6 to 10−7) individually. Reported FDTD simulation data show that due to the formation of “hot spots” by 0D-GNPs in the heterostructure material, the SERS enhancement can be four orders of magnitude higher than that of an individual nanoparticle.Experimental data demonstrate that 0D-2D heterostructure based SERS has the capability for the rapid determination of the antimicrobial susceptibility of multidrug resistant Salmonella DT104 and the normal strain Salmonella Typhi. Reported data show that since cell wall and membrane damage during antibiotic treatment is high for Salmonella Typhi, Raman bands from Salmonella Typhi bacteria decreased significantly during antibacterial treatment. Experimental data also indicate that since cell wall and membrane damage during antibiotic treatment are minimal for Salmonella DT104, the Raman bands remain unaltered during antibacterial treatment. Reported data demonstrate that the 2D-0D heterostructure-based SERS platform has the capability for the rapid identification of the Salmonella DT104 drug resistant strain and separation from the Salmonella Typhi normal strain via an antibiotic susceptibility test, even at a concentration of 100 CFU mL−1.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Dr Ray thanks NSF-PREM grant # DMR-1826886 and NSF CREST grant # 1547754 for their generous funding.Notes and references
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef CAS PubMed.
- A. K. Geim and I. V. Grigorieva, Nature, 2013, 499, 419–425 CrossRef CAS PubMed.
- Q. H. Wang, K. Kalantar-Zadeh, A. Kis, J. N. Coleman and M. S. Strano, Nat. Nanotechnol., 2012, 7, 699–712 CrossRef CAS PubMed.
- K. Novoselov, A. Mishchenko, A. Carvalho and A. C. Neto, Science, 2016, 353, aac9439 CrossRef CAS PubMed.
- D. Jariwala, T. J. Marks and M. C. Hersam, Nat. Mater., 2016, 16, 170–181 CrossRef PubMed.
- Z. Meng, R. M. Stolz, L. Mendecki and K. A. Mirica, Chem. Rev., 2019, 119, 478–598 CrossRef CAS PubMed.
- H. Sun, M. Yao, Y. Song, L. Zhu, J. Dong, R. Liu, P. Li, B. Zhao and B. Liu, Nanoscale, 2019, 11, 21493–21501 RSC.
- S. Huang, L. Liang, X. Ling, A. A. Puretzky, D. B. Geohegan, B. G. Sumpter, J. Kong, V. Meunier and M. S. Dresselhaus, Nano Lett., 2016, 16, 1435–1444 CrossRef CAS PubMed.
- Z. Zhang, P. Lin, Q. Liao, Z. Kang, H. Si and Y. Zhang, Adv. Mater., 2019, 31, 1806411 CrossRef PubMed.
- X. Xing, L. Zhao, W. Zhang, Z. Wang, H. Su, H. Chen, G. Ma, J. Dai and W. Zhang, Nanoscale, 2020, 12, 2498–2506 RSC.
- G. Kang, M. Yang, M. S. Mattei, G. C. Schatz and R. P. Van Duyne, Nano Lett., 2019, 19, 2106–2113 CrossRef CAS PubMed.
- S. Y. Ding, E. M. You, Z. Q. Tian and M. Moskovits, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- S. S. Sinha, S. Jones, A. Pramanik and P. C. Ray, Acc. Chem. Res., 2016, 49, 2725–2735 CrossRef CAS PubMed.
- M. F. Cardinal, E. V. Ende, R. A. Hackler, M. O. McAnally, P. C. Stair, G. C. Schatz and R. P. Van Duyne, Chem. Soc. Rev., 2017, 46, 3886–3903 RSC.
- D. Cialla-May, X. S. Zheng, K. Weber and I. Popp, Chem. Soc. Rev., 2017, 46, 3945–3961 RSC.
- L. P. McDonnell, C. C. Huang, Q. Cui, D. W. Hewak and D. C. Smith, Nano Lett., 2018, 18, 1428–1434 CrossRef CAS PubMed.
- C. Matricardi, C. Hanske, J. L. Garcia-Pomar, J. Langer, A. Mihi and L. M. Liz-Marzan, ACS Nano, 2018, 12, 8531–8539 CrossRef CAS PubMed.
- S. Jones, S. S. Sinha, A. Pramanik and P. C. Ray, Nanoscale, 2016, 8, 18301–18308 RSC.
- A. M. Paul, Z. Fan, S. S. Sinha, Y. Shi, L. Le, F. Bai and P. C. Ray, J. Phys. Chem. C, 2015, 119, 23669–23675 CrossRef CAS PubMed.
- S. A. Ghopry, M. A. Alamri, R. Goul, R. Sakidja and J. Z. Wu, Adv. Opt. Mater., 2019, 7, 1801249 CrossRef.
- Y. Tan, L. Ma, Z. Gao, M. Chen and F. Chen, Nano Lett., 2017, 17, 2621–2626 CrossRef CAS PubMed.
- M. Li, X. Fan, Y. Gao and T. Qiu, J. Phys. Chem. Lett., 2019, 10, 4038–4044 CrossRef CAS PubMed.
- C. Kuttner, R. P. M. Höller, M. Quintanilla, M. J. Schnepf, M. Dulle, A. Fery and L. M. Liz-Marzán, Nanoscale, 2019, 11, 17655–17663 RSC.
- A. Pramanik, Y. Gao, K. Gates, S. Begum and P. C. Ray, ACS Omega, 2019, 4, 11112–11118 CrossRef CAS PubMed.
- CDC 2018: Drug-resistant ‘nightmare bacteria’ pose growing threat, https://nypost.com/2018/04/03/drug-resistant-nightmare-bacteria-pose-growing-threat-cdc, accessed, August 12, 2018 Search PubMed.
- C. A. D. Burnham, J. Leeds, P. Nordmann, J. O'Grady and J. Patel, Nat. Rev. Microbiol., 2017, 15, 697 CrossRef CAS PubMed.
- X. Xiong, Y. Huang, C. Lin, X. Yang Liu and Y. Lin, Nanoscale, 2019, 11, 22206–22221 RSC.
- A. Vangara, A. Pramanik, Y. Gao, K. Gates, A. Begum and P. C. Ray, ACS Appl. Bio Mater., 2018, 1, 298–309 CrossRef CAS.
- S. Befum, A. Pramanik, K. Gates, Y. Gao and P. C. Ray, ACS Appl. Bio Mater., 2019, 2, 769–776 CrossRef.
- K. Syal, R. Iriya, Y. Z. Yang, H. Yu, S. P. Wang, S. E. Haydel, H. Y. Chen and N. J. Tao, ACS Nano, 2016, 10, 845–852 CrossRef CAS PubMed.
- P. J. Weldrick, S. Iveson, M. J. Hardman and V. N. Paunov, Nanoscale, 2019, 11, 10472–10485 RSC.
- N. G. Schoepp, T. S. Schlappi, M. S. Curtis, S. S. Butkovich, S. Miller, R. M. Humphries and R. F. Ismagilov, Sci. Transl. Med., 2017, 9, eaal3693 CrossRef PubMed.
- W. Liao, Q. Lin, Y. Xu, E. Yang and Y. Duan, Nanoscale, 2019, 11, 5346–5354 RSC.
- K. Y. Chang, H. W. Cheng, J. Shiue, J. K. Wang, Y. L. Wang and N. T. Huang, Anal. Chem., 2019, 91, 10988–10995 CrossRef CAS PubMed.
- J. Kirchhoff, U. Glaser, J. A. Bohnert, M. W. Pletz, J. Popp and U. Neugebauer, Anal. Chem., 2018, 90, 1811–1818 CrossRef CAS PubMed.
- A. Novelli-Rousseau, I. Espagnon, D. Filiputti, O. Gal, A. Douet, F. Mallard and Q. Josso, Sci. Rep., 2018, 8, 3957 CrossRef CAS PubMed.
- K. Syal, S. M. Shen, Y. Z. Yang, S. P. Wang, S. E. Haydel and N. J. Tao, ACS Sens., 2017, 2, 1231–1239 CrossRef CAS PubMed.
- D. D. Galvan and Q. Yu, Adv. Healthcare Mater., 2018, 13, e1701335 CrossRef PubMed.
- R. Gao, Z. Lv, Y. Mao, L. Yu, X. Bi, S. X. Cui and Y. Wu, ACS Sens., 2019, 4, 938–943 CrossRef CAS PubMed.
- W. R. Premasiri, J. C. Le and A. Sauer-Budge, Anal. Bioanal. Chem., 2016, 408, 4631–4647 CrossRef CAS PubMed.
- W. R. Premasiri, J. C. Lee, A. Sauer-Budge, R. Théberge, C. E. Costello and L. D. Ziegler, Anal. Bioanal. Chem., 2017, 409, 3043–3054 CrossRef CAS PubMed.
- S. Begum, A. Pramanik, D. Davis, S. Patibandla, K. Gates, Y. Gao and P. C. Ray, ACS Omega, 2020, 5, 3116–3130 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |