Open Access Article
Open Access ArticleCellular regeneration and proliferation on polymeric 3D inverse-space substrates and the effect of doxorubicin†
Chandrashekhar D.
Bobade‡
a,
Semonti
Nandi‡
 a,
Narendra R.
Kale
a,
Shashwat S.
Banerjee
a,
Narendra R.
Kale
a,
Shashwat S.
Banerjee
 b,
Yuvraj N.
Patil
b,
Yuvraj N.
Patil
 *b and
Jayant J.
Khandare
*b and
Jayant J.
Khandare
 *c
*c
aMAAER's Maharashtra Institute of Pharmacy, Kothrud, Pune 411038, India
bMaharashtra Institute of Medical Education and Research Medical College, Talegaon-Dabhade, Pune 410507, India. E-mail: yuvrajpatil@mitmimer.com
cSchool of Pharmacy, Dr Vishwanath Karad MIT World Peace University, MIT Campus, S. No. 124, Paud Road, Kothrud, Pune 411038, India. E-mail: jayant.khandare@mippune.edu.in
First published on 1st April 2020
Abstract
Spatial arrangement for cells and the opportunity thereof have implications in cell regeneration and cell proliferation. 3D inverse space (3DIS) substrates with micron-sized pores are fabricated under controlled environmental conditions from polymers such as poly(lactic-co-glycolic) acid (PLGA), poly(lactic acid) (PLA) and poly(styrene) (PS). The characterization of 3DIS substrates by optical microscopy, scanning probe microscopy (SPM), etc. shows pores within 1–18 μm diameter and prominent surface roughness extending up to 3.9 nm in height over its base. Conversely, to compare two-dimensional (2D) versus 3DIS substrates, the crucial variables of cell height, cell spreading area and cell volume are compared using lung adenocarcinoma (A549) cells. The results indicate an average cell thickness of ∼6 μm on a glass substrate whereas cells on PLGA 3DIS were ∼12 μm in height, occasionally reaching 20 μm, with a 40% decreased cell spreading area. A549 cells cultured on polymer 3DIS substrates show a cell regeneration growth pattern, dependent on the available spatial volume. Furthermore, PLGA 3DIS cell culture systems with and without graded doxorubicin (DOX) pre-treatment result in potent cell inhibition and cell proliferation, respectively. Additionally, standard DOX administration to A549 cells in the PLGA 3DIS system revealed altered drug sensitivity. 3DIS demonstrates utility in facilitating cellular regeneration and mimicking cell proliferation in defined spatial arrangements.
Introduction
Synthetic biodegradable polymers such as poly(L-lactic acid) (PLLA), poly(glycolic acid) (PGA), PLGA, poly(caprolactone) (PCL) etc. have been previously reported as scaffolding materials to exhibit cellular behavior and characteristics.1–3 Tissue regeneration and wound healing has been extensively studied in cell culture models.4–6 Comparably, the use of 3D cell culture tools in cell proliferation and drug-mediated cytotoxicity studies is limited, and is primarily studied using 2D cell cultures.7 An observable issue with such planar tools as a cell attachment/proliferation model is the morphological change induced in 2D cultured cells; the cells appear thinly spread with a predominantly flattened profile.8 While enhanced cell adhesion as a feature is desirable for cell studies, structurally altered biological features may be responsible for a varied cell response and function in such cells.8–10 Cell adhesion and interfacial interactions exert morphological changes based on attachment-substrate geometry, surface texture and stiffness among others.11,12Tissue regeneration/wound healing involves healthy cells utilizing interactive feedback such as contact inhibition, preventing healthy cells from multiplying and stacking beyond their physiological role.13,14 On the other hand, cancerous cells continue to proliferate beyond spatial contact inhibition and often grow as uncontrolled tumor masses as well as enable dissemination of cancerous cells leading to metastasis.13 Planar cell culture models with enhanced cell adhesion features significantly lack a vertical profile and may not reflect the ability of cells to simulate wound closure based on cell–cell interaction alone.2,15 Furthermore, structural components of in vivo tissues support a more spatially relaxed cell profile, compared to glass or compatible planar surfaces, where tissue sections reveal more geometrically shaped cells which can stack against each other.16,17
3DIS is a film-embedded negative or ‘inverted’ space, embodied by porous cavities. While true 3D structures have mass and distinct spatial coordinates, 3DIS presents a niche which can be exploited for cell attachment, growth and culture. The film matrix surrounding the 3DIS pore constitutes the cell scaffolding. We hypothesize that the 3DIS substrates and the corresponding cell culturing strategies may offer greater spatial opportunity compared to 2D substrates. 3DIS substrates with their predicted optimal cell attachment properties are further hypothesized to retain cell topography to mimic ex vivo cells in their natural environment. In the present study, the correlation of chemotherapy failure due to sub-lethal anticancer therapy leading to tumor cell regression and cell proliferation, thereafter using 3DIS ex vivo polymeric systems, is envisioned. Thus, the objectives of the study were (a) to design 3DIS substrates composed of PS, PLA and PLGA and characterize the same; (b) to demonstrate the utility of spatial scaffolding to allow cells to grow freely in a 3D microenvironment, thereby enabling a near-physiological outcome of DOX exposure to cancerous cells and (c) to compare the differences in 2D and 3DIS cell cultures, with regard to their effect on cell morphology and the effect of the substrate juxtaposed with DOX exposure and the fate of the cells thereof.
Experimental section
Materials
PLGA with lactide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycolide (ratio 75
glycolide (ratio 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) and a Mw ∼ 66–107 kDa and PLA with an average Mn ∼ 40 kDa were obtained from Sigma Aldrich, PS with a Mw ∼ 40–60 kDa was obtained from Analab Fine Chemicals, India. Doxorubicin hydrochloride (DOX) was obtained from Sigma Aldrich. Toluidine blue O (TBO) was obtained from SRL Pvt. Ltd, India. Methanol, chloroform, and dimethylsulfoxide (DMSO) were of analytical grade.
25) and a Mw ∼ 66–107 kDa and PLA with an average Mn ∼ 40 kDa were obtained from Sigma Aldrich, PS with a Mw ∼ 40–60 kDa was obtained from Analab Fine Chemicals, India. Doxorubicin hydrochloride (DOX) was obtained from Sigma Aldrich. Toluidine blue O (TBO) was obtained from SRL Pvt. Ltd, India. Methanol, chloroform, and dimethylsulfoxide (DMSO) were of analytical grade.
Preparation of glass, PLGA, PS and PLA substrates
Plane, unmodified microscopic cover glasses were obtained and used as substrates for cell culture post sterilization. PLGA, PS and PLA 3DIS substrates were prepared through a typical breath figure approach. First, 18 mm × 18 mm glass cover slips were washed with methanol to remove impurities. 5 mg. ml−1 polymer solution of PLGA, PS and PLA was prepared in chloroform; 50 μl of the resultant polymer solution was placed with the aid of a pipette slowly onto the glass slide under humid atmospheric conditions (∼80–90% Relative Humidity (RH) and temperature 22.5 to 23.5 °C) in a sealed acrylic chamber. The prepared 3DIS polymeric substrates were observed under brightfield microscopy. The smooth polymeric substrates (lacking inverted 3D structures) were prepared using the same method under dry conditions (40% RH and temperature 26 °C).Characterization of the polymeric scaffold
Morphological characters such as pore diameter, rim width and substrate thickness of PLGA, PS and PLA substrates were determined by calculating the average of three-point measurements. The surface area for smooth and 3DIS substrates was determined by image analysis. SPM (JSPM-5200, JEOL) analysis provided the topography data of the designed polymer substrates. Other parameters noted were polymer substrate stability in various exposure conditions such as chemical reagents, pH sensitivity, ultraviolet radiation etc.Determination of surface carboxyl (–COOH) groups using the TBO assay
The substrates were immersed in 1.5 ml of 2 mM TBO solution for 24 h at room temperature (25 °C), during which the dye bound via electrostatic interaction to the ionized acidic charges. Substrates were thoroughly rinsed with 0.015 M NaCl at pH 11.0 to wash away the unbound dye molecules. Once air dried, the substrates were placed in 1 ml of 0.2 M NaCl solution at pH 2.0 for 60 min while stirring. During this step, the TBO molecules bound to the acidic groups of the substrate were eluted from the analyzed surface and diffused into the solution, coloring it blue. The light absorbance of the solutions at 630.8 nm wavelength was measured. The blank consisted of a 0.2 M NaCl solution at pH 2.0.Measurement of wettability
The contact angle (θ) of the prepared substrates was studied and correlated with the structural geometry and wettability characteristics of the prepared substrates. A deionized water drop of 5 μl, (n = 6) was placed on dry substrates (PLGA, PS and PLA smooth and 3DIS architecture) at room temperature and images of the wetting process of the placed water drop were captured with a high speed digital camera. The captured images were processed using LBDSA Drop Shape plug-in the image analysis software ImageJ (NIH, Bethesda, MD) for θ determination.Preparation of Dox pre-treated PLGA substrates
DOX solution volumes which are mole-identical to IC50 and IC25 of free DOX were pipetted onto PLGA (smooth and 3DIS) substrates and air dried to leave a DOX coat onto the film. These prepared substrates were further used for A549 cell culturing and analyzed for morphological parameters.Cell culture
The prepared substrates were rinsed in 70% ethanol solution and kept for 30 min under UV light to sterilize before cell seeding. The substrates were immersed in Dulbecco's Modified Eagle's Media (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. A549 cells (National Center for Cell Science, Pune) were used for the cell study. After rinsing cells in the flask with phosphate buffered saline (PBS) (pH 7.4), cells were harvested with trypsin (0.5%) ethylenediaminetetraacetic acid (EDTA). A549 cells were seeded at a high density (400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml) on the substrates in 12 well plates and cultured for 3, 6, 24 and 48 h in 5% CO2 in a humidified incubator.
000 cells per ml) on the substrates in 12 well plates and cultured for 3, 6, 24 and 48 h in 5% CO2 in a humidified incubator.
Cell imaging and quantification
Cell morphology was characterized using an inverted fluorescence microscope Axio Observer A1 (Carl Zeiss, Germany). The cells were fixed with 4% paraformaldehyde for 20 min. The substrates were mounted on glass slides and observed under 20× magnification. The microscopic images of cell morphology were visualized with fluorescent dyes FITC (cytoplasm) and 4′,6-diamidino-2-phenylindole(DAPI) (nuclei) and were quantitatively analyzed using ImageJ® software (NIH, Bethesda, MD). High resolution images were obtained using a Confocal Laser Scanning Microscope (Leica Microsystems); Z-stack images for spatial data were obtained for all samples. Quantification and visual data were extracted with Fiji® software (NIH, Bethesda, MD). The volume of cells was obtained by defining specific regions of interest, followed by signal thresholding. The resulting spatial signal was compiled with the Voxel Counter plug-in in Fiji® and calculated as the volume in cubic microns. Imaging was carried out four separate times with multiple samples. The calculated data is expressed as the mean data with a standard error of mean.Biocompatibility/cell viability test
The cell viability of A549 cells on glass and PLGA polymer substrate samples, both smooth and 3DIS, was quantitatively determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Briefly, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded on each substrate in a 48-well plate and maintained for 24 h at 37 °C in 180 μl DMEM supplemented with 10% FBS, following which a stock MTT reagent, (20 μl) was added and cells were incubated for 4 h. After 4 h, the entire media was aspirated and DMSO (100 μl) was added to each well. DMSO dissolved the precipitate following which the absorbance was measured at 570 nm. Background readings (blank) were obtained from cell-free wells containing only DMSO. A549 cells grown on glass substrates were considered as the control. Percentage cell viability was calculated as
000 cells per well were seeded on each substrate in a 48-well plate and maintained for 24 h at 37 °C in 180 μl DMEM supplemented with 10% FBS, following which a stock MTT reagent, (20 μl) was added and cells were incubated for 4 h. After 4 h, the entire media was aspirated and DMSO (100 μl) was added to each well. DMSO dissolved the precipitate following which the absorbance was measured at 570 nm. Background readings (blank) were obtained from cell-free wells containing only DMSO. A549 cells grown on glass substrates were considered as the control. Percentage cell viability was calculated as| (A × 100)/C | (1) |
PLGA-3DIS DOX release study
PLGA substrates (smooth and 3DIS) were surface coated with 200 μg DOX and dried. PBS (pH 7.4) was used as the dissolution media of which 1 ml was added to the substrates and aliquots were collected at fixed time intervals (1, 3, 6, 24 and 48 h). Fresh PBS was replaced at every time point to maintain constant media volume. Fluorescence emission intensity was measured at 590 nm upon excitation at 480 nm. The DOX released was calculated as the % cumulative release against all time points. All experiments were performed in triplicate.Statistical analysis
A student t-test was performed on the data sets to determine the p-value for testing the significance of quantified data (volume, area, height, drug concentration etc.). A p-value of 0.05 was assumed as the limit of significance. Statistical processing was carried out with GraphPadPrism, GraphPad Software, San Diego, California, USA.Results
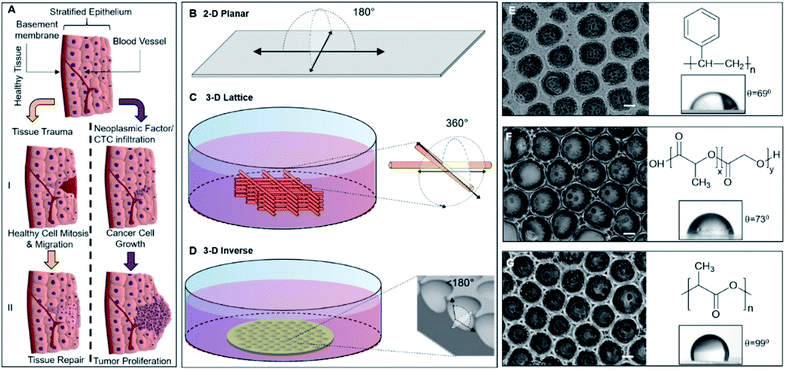
The complex model for tissue repair or regeneration utilizing multiple cell types and signaling components, as depicted in Fig. 1A, may not be easily replicated in vitro; however, the ability of epithelial cells to mimic the gap-bridging may be studied in vitro using appropriate 3D substrate architecture. Conversely, while cancer cell proliferation also presents a complex model of unregulated cell division, the changes in cell morphology are readily observed. Fig. 1A depicts the cellular fates a healthy tissue may experience upon being subjected to physical injury or cellular insult with onco-genetic potential, including infiltration of circulating or metastatic tumor cells. The 3DIS platform proposed here mimics tissue substratum offering cultured cancer cells the spatial opportunity for proliferation as well as presenting a broken surface simulating tissue trauma which in turn presents a spatial opportunity for studying tissue monolayer repair and rebuilding.Apparent degrees of freedom of cell culturing substrates
2D surfaces such as tissue culture flasks or glass offer 180° of spatial freedom for cell growth (Fig. 1B). Some 3D culture methods utilizing cell substrates as scaffolding may even approach 360° of freedom allowing cells to spread along any accessible direction (Fig. 1C). Conversely, 3DIS reduces available spatial freedom (<180°) and cells are confined to a restricted volume while allowing spatial cell adhesion opportunity (Fig. 1D), virtually absent in the above two models.Mechanism of formation of 3DIS polymer architecture
The polymer 3DIS films were generated by a method known as the ‘breath-figure’ method which exploits higher atmospheric/environmental moisture content or humidity to accelerate pore formation on the film surface during the course of film drying. When a drop of polymer solution is cast on a substrate, the volatile solvent begins to evaporate in the humid atmosphere. During evaporation, the latent heat of vaporization is absorbed due to which the temperature at the solution surface decreases to a point at which condensation begins. These condensed water droplets interact and rearrange on the solution surface to remain isolated from each other. When the temperature of the solution surface increases high enough, further condensation cannot occur. Thus, the water droplets begin to evaporate from the solution surface and the polymer precipitates around each water droplet which leaves behind cavities (pores) in the solid polymer film, after complete evaporation.18,19 The greater the humidity, the greater is the water vapor sequestration in the chloroform-polymer slurry leading to condensation of water droplets onto the drying film. Thus with a greater water content, smaller pores coalesce and form larger pores (>10 μm).Physicochemical traits of 3DIS substrates
Polymer substrates on glass cover slips were fabricated from PS, PLGA and PLA (chemical structures depicted in Fig. 1E, F, G respectively) and analyzed to verify either the smooth or 3DIS geometry of the substrates (Fig. 1E, F, G). The 3DIS substrates were distinguished as 3DIS(+) or 3DIS(−) based on their large (>12 μm) or small (<10 μm) pore sizes, respectively. Each of the substrates showed an even distribution of the 3DIS aspect with even rim-width and pore sizes (12–18 μm). The pore size of 3DIS increased (1 μm to 18 μm) with increasing polymer strength (0.3–0.7% w/v) and also with greater environmental moisture content and temperature (∼80–90% RH and 22.5 to 23.5 °C, (Fig. S1A and B†)).The polymer substrates cast on the glass surface had an average thickness of 25.4 ± 9 μm. The total surface area of 3DIS substrates was computed using the following rationale:| [πR2 + (2πr2 × n)] − πr2 × n | (2) |
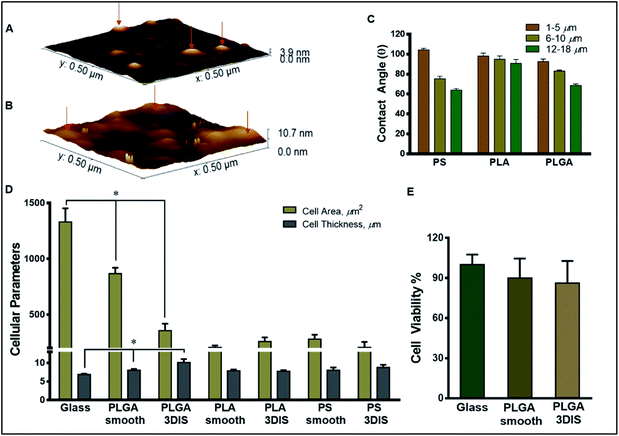
The roughness of the glass surface and PLGA substrates was evaluated with SPM. The analyzed area (0.5 μm × 0.5 μm) for PLGA revealed an intermittently textured area with prominent outgrowths not greater than 3.9 nm in height over the substrate base (Fig. 2A). Further, the calculated roughness depicted smaller features distributed about 10 nm apart. In comparison, the SPM image of glass showed significantly greater surface roughness with frequent protrusions extending up to 10 nm in height (Fig. 2B).
Further, we analyzed free active carboxyl groups using titrimetric analysis of the polymer surfaces which revealed higher surface carboxylic acid content on 3DIS substrates compared to smooth substrates (∼30% for PLA and ∼33% for PLGA). PS substrates do not carry free carboxyl groups. It was determined that the test materials, PS, PLA and PLGA were chemically and physically stable against surface sterilization techniques such as exposure to 70% ethanol/isopropyl alcohol solution and UV radiation (λ = 253.7 nm) for 30 min. Similarly, 3DIS and smooth polymer substrates immersed in cell culture media at pH 7.4 for a period of 30 days failed to demonstrate substrate fractures or physical deformation, indicating polymer resistance against mechanical degradation.
Wettability
As Fig. 2C depicts, with an increasing range of pore sizes (1 μm to 18 μm), PLGA demonstrated a decreased contact angle (θ) from 92.67 ± 2.52° to 68.67 ± 1.53°. Similarly, θ for PS at the 1–5 μm pore size was 104.33 ± 1.53° which lowered to 75.33 ± 2.52° for the 6–10 μm pore size and further decreased to 64.0 ± 1.53° for the 12–18 μm pore size range. Interestingly, the PLA substrate did not display a strong correlation between pore size and wettability and θ ranged between 100.13 ± 2.87° to 90.84 ± 3.90° for the entire pore size range (1 μm to 18 μm).PLGA DOX release study
Pretreatment of DOX on PLGA 3DIS followed by cell media immersion revealed a cumulative DOX release profile depicting a biphasic trend suggesting a more rapid drug release in the first six hours (∼36%) followed by a steady slower release up to ∼66% in 48 h (Fig. S2A†).3DIS architecture mimics in vivo cancer cell microenvironment
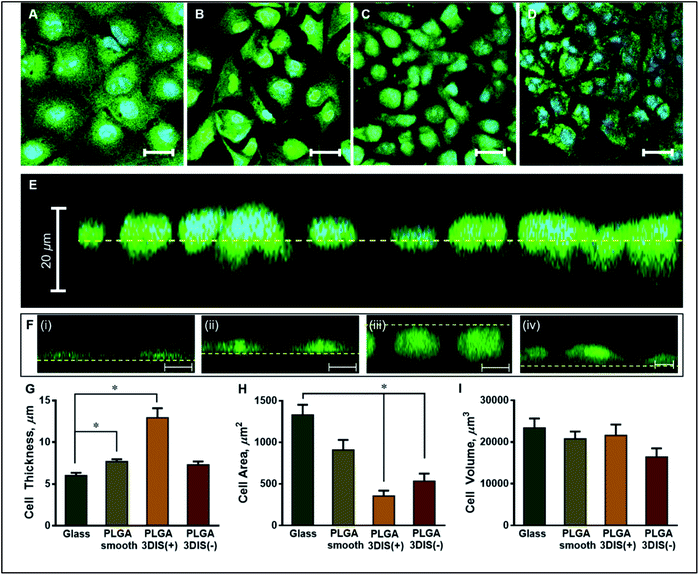
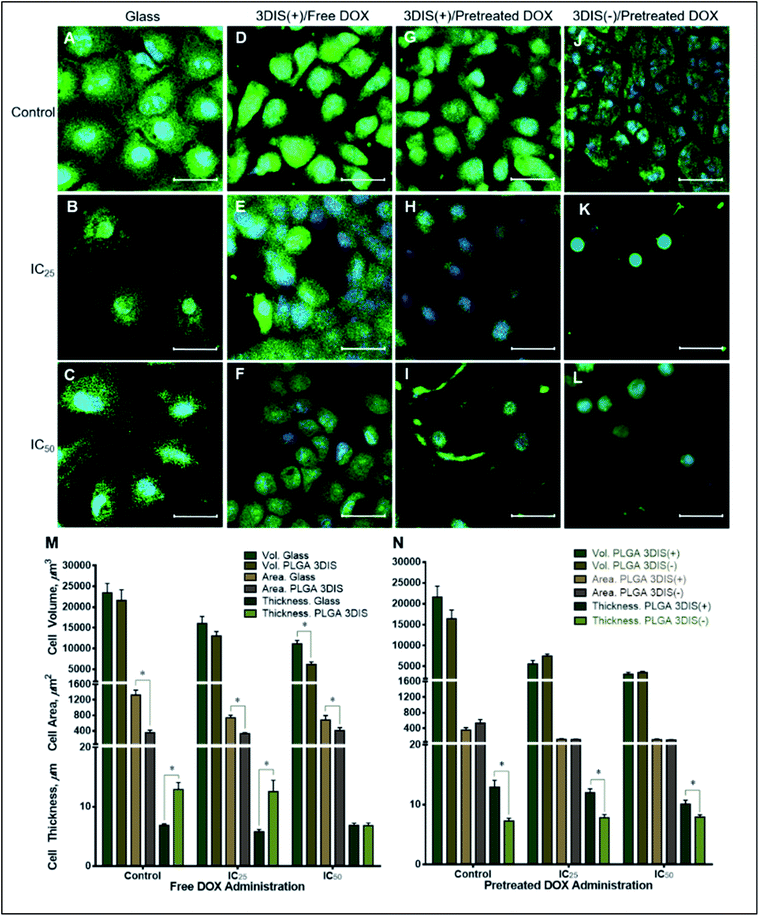
The orthogonal confocal sectioning of cells on the glass surface (ESI video 1†) highlighted a thinner spreading of the attached cells (cell height = 6.4 ± 0.35 μm), whereas the orthogonal section of PLGA smooth substrates revealed a raised cell profile with an increased cell thickness (7.7 ± 0.28 μm) (Fig. 3G). The sub-surface cytoplasmic regions appeared nestled inside the pores. The cells on PLGA 3DIS(+) displayed up to 18 μm thickness with an average cell thickness of 12.9 ± 1.15 μm (ESI video 2†). The quantification of the cellular area on the glass surface after 48 h revealed a significant cytoplasmic area (1329.68 ± 122.11 μm2) while the dorso-ventrally flattened nucleus was 241 ± 12 μm2. Phalloidin stained actin fibers spanned the volume of the cell attached on the glass cover slip, and the dense terminal protrusions of the actin fibers indicate the cell adhesion points (Fig. S2B†). However, A549 cells on PLGA smooth substrates demonstrated comparatively reduced cellular spreading area with cellular projections indicating substantial cell adhesion (Fig. 3H).
On 3DIS(−) substrates, the cells appeared to have little to no access to the depth of the pores, resulting in cells spreading over the porous structures with cytoplasmic area ∼533.6 ± 91.08 μm2 and a corresponding cell thickness of ∼7.3 ± 0.41 μm (ESI video 3†). The surface area of the cells on PLGA 3DIS(+) was sharply reduced, and compared to glass and PLGA smooth, the decrease in area was ∼74% and ∼60% respectively. The cells cultured on glass and PLGA-smooth substrates showed statistically similar cellular volume (23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424 ± 2243.40 μm3 and 20
424 ± 2243.40 μm3 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 798 ± 1729.90 μm3 respectively) (Fig. 3I). PLGA 3DIS(+) cells showed the maximum thickness which compensated for the gross decrease in cell area and consequently the cells grown on glass, PLGA smooth and PLGA 3DIS(+) surfaces were statistically comparable with cell volumes varying between 23
798 ± 1729.90 μm3 respectively) (Fig. 3I). PLGA 3DIS(+) cells showed the maximum thickness which compensated for the gross decrease in cell area and consequently the cells grown on glass, PLGA smooth and PLGA 3DIS(+) surfaces were statistically comparable with cell volumes varying between 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424 ± 2243.40 μm3 to 21
424 ± 2243.40 μm3 to 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 618 ± 2601.03 μm3.
618 ± 2601.03 μm3.
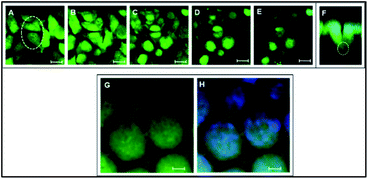
The cells showed a tendency to occupy 3DIS evenly and to form monolayers, bridging the pore gaps (ESI video 4†). As depicted in Fig. 4A–D, the ability of the 3D (spatially restored) cells in bridging small (∼15 μm) gaps was demonstrated. In Fig. 4F, as few as two cells were shown capable of bridging a micro-gap and forming cell–cell and cell–substrate adhesions.
Multiple cells are shown to fill the large (∼65 μm) pores in Fig. 4G and H effectively demonstrating the ability of PLGA 3DIS in allowing cells to grow spatially and create cell–cell adhesions as well. The 3DIS pore-rims serve as foot and hand holds for cells (Fig. 4G and H).
During the course of treatment with DOX, the cells maintained a flat profile (5.79 ± 0.37 μm at IC25 and 6.88 ± 0.34 μm at IC50) and demonstrated lateral spreading (Fig. 5M). When exposed to sub-lethal doses (IC25) of DOX, there appeared to be a mild decrease in the size of the cells (741.07 ± 61.90 μm2) with a general reduction in the number of cellular projections. A higher DOX concentration (IC50) in the cell media appeared to decrease the cell area further (679.83 ± 117.68 μm2) with a subtle shrinking effect on the nucleus.
Cell volume was reduced by about 30% in sub-lethal doses of DOX while IC50 caused a roughly 50% drop in cell volume. The cell area also significantly decreased due to DOX exposure compared to the control but not significantly between the two drug treatments (pretreated DOX, IC25 by ∼44% and IC50 by ∼48%) suggesting a near-maximal effect at IC25. Interestingly, cells on PLGA 3DIS(+) dosed with a sub-lethal (IC25) DOX concentration did not demonstrate a significant change in cell area and cell thickness compared to the control. However, upon exposure to the IC50 dose of DOX the cells demonstrated a reduction in cell size such as volume (3548.57 ± 220 μm3) and height (7.927 ± 0.37 μm). Additionally, the nuclei appeared to be proportionally shrunk.
Discussion
Enhanced adhesion along a single plane may not allow true spatial freedom for cell growth and the cell may likely compensate for the loss of 3D cell architecture by spreading laterally. On the other hand, planar cell attachment substrates consequently may not mimic the physiological responses in the cancer cell microenvironment. Thus, the mechanism by which cells conform to available spaces and geometry and the specific role of the void spaces in enabling cell attachments and proliferation needed elucidation.Spatial availability within tissues may likely result in tissue expansion via cell reorganization or multiplication; however, the availability may not be perceived in a similar fashion in conventional cell culture systems. As depicted in Fig. 1, the unrestricted space around the cells conforming to 180° for a glass surface and 360° spatial freedom for the illustrated 3D culture system respectively, appeared conducive for spatial growth; however, cells reliant on surface adhesion components were paradoxically bound to and spread along the available surface. However, restriction in 3DIS spaces with <180° of spatial freedom provided cell adhesion opportunities across the available perimeter in 3DIS. Such spatial confinements prevented planar cell adhesion localization and allowed the cells to grow in 3D spaces and have a raised profile.
In consequence, the cell and its organelles such as the nucleus, remained free of the stress fiber mediated compression which in turn restored the in vivo physiological behavior of cells. 3DIS is thus an interesting and competent model to study in vivo cell growth patterns, and in addition, it was appropriate to interpret cell morphology behavior on exposure to cytotoxic drugs.
Subsequently, PS, PLA and PLGA polymer substrates were fabricated with 3DIS architecture and subjected to numerous physicochemical characterization experiments to determine their compatibility with A549 spatial cell growth. PLGA 3DIS substrates underwent SPM analysis and the depicted texture in Fig. 2A was hypothesized to span the substrate top surface and pore surface and provide adhesion support to adherent cells. The large protuberances were an indication of a potential cell adhesion site, with an average cell area of ∼400 μm2 (on PLGA 3DIS); it followed that a cell had access to a large number of adhesion-competent sites on PLGA substrates. However, SPM analysis of the glass surface demonstrated frequent outgrowths with greater height than in the PLGA topography. Thus, the glass surface promoted higher affinity of cells with abundant cell adhesion features and significantly increased the cell surface area, owing to the presence of highly uneven surface topography.
It was noted that PLGA and PS had lower contact angles in the pore size range of 12–18 μm, which indicated higher wettability. Thus, PLGA and PS polymers were inferred to possess higher apparent affinity for cells compared to PLA which was evident for the measured cell surface area (Fig. 2). All cell studies on glass and polymer surfaces were reported after a 48 h incubation period for both morphology analysis and Dox treatment studies. An abundance of the hydrophilic surface area and compatible functional groups on the glass surface lead cells such as A549 and HeLa (data not shown) to demonstrate significant cellular spreading. Among the polymers, PS smooth substrates demonstrated cell attachment and spreading inferior to PLGA and glass surfaces.
The lower wettability of PLA likely reduced its utility in promoting cell adhesion and spreading making PLA the least favorable cell substrate among the materials under study. However, the greater hydrophilicity of PLGA, partially due to glycolic acid content (25%), resulted in the greater cell affinity of PLGA compared to PS or PLA. Owing to its superior selective cell adhesion trait, PLGA was identified in this study to further investigate 3DIS-cell behavior.
The ordering of spatial organization of the polymer substrate with regard to generation of 3DIS led to significant changes in cell morphology. In comparison to glass-bound A549 cells, cells cultured on PLGA substrates for 48 h displayed varied morphological signatures depending on the substrate geometry. Unlike glass, PLGA smooth substrates offered relatively less cell adhesive or retentive surface chemistry, leading to less dense cellular confluence. Indeed, a surface retraction of cells on PLGA smooth substrates was apparently compensated by increased cell thickness. For example, the orthogonal section in 3DIS(−) despite their restricted spatial confines allowed the cell to articulate with the adjoining pore walls and form adhesive junctions to act as anchors (Fig. 3). Conversely, since the cell adhesion features were distributed in 3D spaces within the 3DIS, there was conceivably a relaxation of the net-downward force, allowing the cell to grow while maintaining a tall profile, compared to a flattened profile seen on glass-bound cells.
Furthermore, the inter-pore substrate surface was limited in area, likely causing the cells to utilize the pores as additional cell adhesion surfaces. Specifically, as shown in Fig. 3, the cells were observed seated on the substrate surface (dotted line) while a portion of the cells appeared below the surface level. Thus in a controlled environment without drug pressure, cells demonstrate the ability to maintain a specific volume, comprising of both cytoplasmic and organelle volumes. While the volume of cells on glass, PLGA smooth and PLGA 3DIS(+) seemed invariable, cellular areas of cells on PLGA smooth and PLGA 3DIS(+) showed a marked decrease. In contrast, the cell height showed an opposing trend and showed increasing values when grown on PLGA smooth and PLGA 3DIS.
Besides allowing cells, in principle, to bear a more physiologically relevant phenotypical form, the 3DIS also generated a platform to explore cellular regeneration across simulated gaps (pores). The ability of the epithelial cells in wound closure was investigated by observing the cells adhere to the pore, spatially adapt or multiply, thereby filling the pore cavity (Fig. 4). PLGA 3DIS with tunable pore sizes presented an appropriate model of small tissue gaps or wounds which was exploited to determine the regenerative abilities of cells or co-cultures. The adhesion feature demonstrated here provided a platform upon which tumor models maybe developed as well. PLGA 3DIS demonstrated the ability to mimic cells in their near in vivo morphology; thus, further experiments involved comparison of the drug effect on cells on 3DIS and on standard 2D cultures to determine if the cells were altered with changes in their morphology and if these changes depicted in vivo outcomes.
On the other hand, we did not use collagen coating which may effectively nullify the polymeric 3DIS geometrical advantages seen in uncoated 3DIS substrates, presumably by enhanced cell attachment. Thus, we studied the interactions of A549 cells with the polymer substrate without the interfering influence of extra-cellular matrix (ECM) components. Furthermore, collagen and fibronectin may eliminate the localized surface charges of 3DIS polymer substrate structures, decrease the influence of 3D substrates and finally reduce the 3D inverse spaces milieu.
DOX was used in this study to contrast the difference in the drug effect on cells grown on different attachment substrates which manipulated cell morphology. The effect of DOX treatment on cells grown on glass and PLGA 3DIS revealed a complex interplay of the morphological features which were shown in the confocal microscopy images of cells depicted in Fig. 5; the cells cultured on glass serve as global controls. Various cell parameters such as thickness, surface area and volume were measured and the differences were depicted graphically. The parameters of cellular area (cytoplasmic area) and cell thickness (height) were considered distinct dimensions whereas cell volume was reliant on area as well as cell thickness. This phenomenon was restricted to the spatially-restored cells on 3DIS substrates and suggested a varied pharmacokinetics/pharmacodynamics balance as compared to control cells on 2D culture surfaces (glass). It may be inferred that the 3DIS(+) cells were more resistant to DOX than the results of DOX treatment on glass-bound cells suggested. The 3DIS platform exhibited cell adhesion and growth in the context of drug kinetics and activity in the cell microenvironment that was otherwise complicated to simulate in 2D cultures. The effect of altered cell morphology on the sensitivity to cytotoxic drugs was also explored; the greater surface area of cultured cancer cells on planar surfaces may likely enhance the capacity of xenobiotic uptake via multiple pathways including receptor mediated endocytosis etc.15
When considered in conjugation with upregulated drug efflux pumps in cancer cells, the 2D planar cell culture model presents a complex transport system which allows a rapid internalization and rapid efflux of the administered molecule.20,21 However such models are not expected to provide a true kinetic profile for an administered compound in the given context. Consequently, cells with lesser deviation from their in vivo cell structure were preferred with limited surface area and a more elevated 3D profile. It was likely that 3DIS(+) allowed cells to briefly adapt spatially despite drug pressure due to the close proximity of the 3DIS walls leading to cell elongation in the vertical aspect. The behavior of cells in the pre-treated DOX context was explained by the slow release of DOX from the substrate as depicted in Fig. S2A.† The rapid release of DOX in the initial six hours was expected from rim-surface and surrounding area-accumulated DOX, whereas the slower release over 48 h was likely the result of DOX slowly diffusing out of pores (from the extended surface area as described earlier by eqn (1)). The slow drug release into the cell media retains the drug in the immediate substrate vicinity leading to a pronounced cytotoxic effect.
Indeed, the utility of optimized inhibitory anticancer drug concentrations at the local site produced a far more pronounced anticancer effect than that due to drug-infused media with comparable drug content. The phenomenon depicted a failure in attachment of cancer cells to the substrate in addition to cumulative drug pressure over time. It was conceivable that such a strategy might prevent the attachment and survival of cancer cells at a given tissue site; stated differently, it might imply prevention of metastasis at secondary sites if prophylactically treated in a site-specific manner. With a reduced cell surface area, it was likely that the drug uptake mechanisms were unable to counteract the activity of drug efflux pumps which resulted in reduced overall cytotoxicity. The results implied that reduced systemic DOX content in the body would result in potential failure of anticancer activity and allow cancer growth and even metastasis. Further, low blood drug concentrations might occur due to termination of chemotherapy.
In contrast, the presence of localized content of DOX, simulating IC50 drug content in the tissue as opposed to systemic circulation may generate a potent cytotoxic environment for cancer cells. IC50 and sub-lethal IC25 DOX concentrations were applied directly to PLGA 3DIS substrates instead of cell media dispersion and the pre-treated 3DIS was used to culture A549 cells. The presence of a localized, pre-existing drug environment strongly deterred the growth and spreading of cells in both sub-lethal and IC50 drug contents as evidenced in Fig. 5. The cells appeared to be shrunk with an apparently reduced cytoplasmic compartment.
The toxicity of the treatment resulted in a very small number of viable but near-apoptotic cells. Cell volumes were drastically affected for both drug treatments with a roughly 8-fold drop determined for IC50-treated cells compared to the control. The cell height showed a modest drop for the sub-lethal dose and IC50 treatments, which resulted from the severe shrinkage of volume. It was likely that upon DOX treatment the freshly seeded cells on the PLGA 3DIS were unable to adapt to the substrate and consequently failed to adhere fully and spread. As a result, the cells retained their round shape and likely failed to deploy cytoskeletal scaffolding to attach and spread, in addition to undergoing cytotoxic damages, which were more pronounced for IC50 treated cells. It followed that localized IC50 and even the sub-lethal IC25 dose, administered as pretreatment upon the 3DIS, were far more effective in eliciting the cytotoxic activity of DOX in A549 cells than free DOX administration in the cell media. Overall, anticancer therapy often fails in achieving complete tumor regression due to sub-lethal dose concentrations and the cells that survive following the therapy continue to survive.22 Here we demonstrated the ex vivo effect using polymeric 3DIS substrates.
Conclusions
Upon comparison of PS, PLA and PLGA as 3DIS substrates, PLGA presented a viable 3DIS platform for the study of cancer cells in a near-in vivo morphological context. The ability of A549 cells to defend their cytoplasmic volume across glass and PLGA substrates strongly suggested the biocompatibility of PLGA in the composition of 3DIS, which is supported by the viability assay comparing the substrates. The superior cellular affinity, as evidenced by cell spreading, allowed PLGA to support spatial growth in the confines of 3DIS. The results presented here highlighted the behavior of A549 cells in the 3DIS culture in mimicking physiological responses. While the 3DIS architecture was central to altering cell morphology, it also presented a discontinuous surface mimicking broken tissue membranes. PLGA 3DIS served as an appropriate tissue repair model to study epithelial cell growth and gap-bridging as an index of tissue repair/regeneration. The A549 cells were shown to grow rapidly to fill a larger pore thereby forming a continuous cell monolayer bridging the gap.The evidence of morphological changes influencing cellular responses was demonstrated in the DOX study of cells grown on PLGA 3DIS. Compared to the PLGA 3DIS control, cells exposed to a sub-lethal IC25 DOX dose were undeterred. The study indicated a sustained IC50 dose strategy to elicit a noticeable anticancer (cytotoxic) effect. With its abnormal flattened morphology, cancer cells on glass lacked the physiological integrity to depict realistic in vivo responses to drugs. Similarly, the pre-treatment of DOX on the 3DIS illustrated its ability to mimic alternative drug dosing conditions which were more successful in tempering the cancerous growth of cells and in inhibiting their spread and survival altogether.
Advanced strategies can be adapted for use with 3DIS such as the use of flow-through analytical chambers with embedded PLGA 3DIS for real time monitoring of spatially restored cells, essentially mimicking entire tissues.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors would like to acknowledge the financial support of DBT-Nano-Biotechnology, DST-FIST and DST-Nano Mission, Government of India.Note and references
- P. X. Ma and J. W. Choi, Tissue Eng., 2001, 7, 23–33 CrossRef CAS PubMed.
- S. Yang, K. F. Leong, Z. Duo and C. K. Chua, Tissue Eng., 2001, 7, 679–689 CrossRef CAS PubMed.
- R. Zhang and P. X. Ma, J. Biomed. Mater. Res., 1999, 45, 285–293 CrossRef CAS PubMed.
- C. C. Liang, A. Y. Park and J. L. Guan, Nat. Protoc., 2007, 2, 329–333 CrossRef CAS PubMed.
- M. Mirbagheri, V. Adibnia, B. R. Hughes, S. D. Waldman, X. Banquy and D. K. Hwang, Mater. Horiz., 2019, 6, 45–71 RSC.
- Y. Xia, Nat. Mater., 2008, 7, 758 CrossRef CAS PubMed.
- A. S. Curtis, J. V. Forrester, C. McInnes and F. Lawrie, J. Cell Biol., 1983, 97, 1500–1506 CrossRef CAS PubMed.
- A. PrinaMello, N. Jain, B. Liu, J. I. Kilpatrick, M. A. Tutty, A. P. Bell, S. P. Jarvis, Y. Volkov and D. Movia, Tissue Cell, 2018, 50, 15–30 CrossRef CAS PubMed.
- H. J. Mulhall, M. P. Hughes, B. Kazmi, M. P. Lewis and F. H. Labeed, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 5136–5141 CrossRef CAS PubMed.
- R. Domura, R. Sasaki, Y. Ishikawa and M. Okamoto, J. Funct. Biomater., 2017, 8, 18 CrossRef PubMed.
- M. Ferrari, F. Cirisano and M. C. Morán, Colloids Interfaces, 2019, 3, 48 CrossRef CAS.
- J. Rosales-Leal, M. Rodríguez-Valverde, G. Mazzaglia, P. Ramón-Torregrosa, L. Díaz-Rodríguez, O. García-Martínez, M. Vallecillo-Capilla, C. Ruiz and M. Cabrerizo-Vílchez, Colloids Surf., A, 2010, 365, 222–229 CrossRef CAS.
- R. Fernandez-Gonzalez and J. A. Zallen, Cell, 2012, 149, 965–967 CrossRef CAS PubMed.
- K. Pietras and A. Östman, Exp. Cell Res., 2010, 316, 1324–1331 CrossRef CAS PubMed.
- I. Levinger, Y. Ventura and R. Vago, Advances in Cancer Research, ed. K. D. Tew and P. B. Fisher, Academic Press, 2014, vol. 121pp. 383–414 Search PubMed.
- B. Alberta, J. Lewis, K. Roberta, A. Johnson, M. Raff and P. Walter, Molecular Biology of the Cell, Garland Pub, USA, 2008 Search PubMed.
- B. Young, P. Woodford and G. O'Dowd, Wheater's Functional Histology E-Book: A Text and Colour Atlas, Elsevier Health Sciences, 2013 Search PubMed.
- H. Battenbo, R. Cobley and S. Wilks, Soft Matter, 2011, 7, 10864–10873 RSC.
- O. Karthaus, N. Maruyama, X. Cieren, M. Shimomura, H. Hasegawa and T. Hashimoto, Langmuir, 2000, 16(15), 6071–6076 CrossRef CAS.
- M. M. Gottesman, T. Fojo and S. E. Bates, Nat. Rev. Cancer, 2002, 2, 48–58 CrossRef CAS PubMed.
- J. P. Gillet and M. M. Gottesman, Multi-Drug Resistance in Cancer, ed. J. Zhou, Humana Press, Totowa, NJ, 2010, pp. 47–76 Search PubMed.
- S. Nandi, N. R. Kale, V. Takale, G. C. Chate, M. Bhave, S. S. Banerjee and J. J. Khandare, J. Mater. Chem. B, 2020, 8(9), 1852–1862 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1, Fig. S2, Fig. S3, video 1, video 2, video 3 and video 4. See DOI: 10.1039/d0na00075b |
| ‡ C. D. B. and S. N. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |