Open Access Article
Open Access ArticleEnhanced avidin binding to lipid bilayers using PDP-PE lipids with PEG-biotin linkers†
Holly L.
Birchenough
a,
Marcus J.
Swann
b,
Egor
Zindy
a,
Anthony J.
Day
a and
Thomas A.
Jowitt
 *a
*a
aWellcome Trust Centre for Cell-Matrix Research, UK. E-mail: t.jowitt@manchester.ac.uk
bSwann Scientific Consulting Ltd, 110 Sandy Lane, Lymm WA13 9HR, UK
First published on 10th March 2020
Abstract
Two of the most important aspects of lipid bilayers that have increased their popularity in the field of nanotechnology and biosensors are their fluid nature, which is highly beneficial in ensuring the spatial organization of attached molecules, and the relative ease in which they can be manipulated to change the surface chemistry. Here we have used two different types of functionalized lipids to study the interaction of avidin, which is a common approach to attach further ligands for study. We have tested the commonly used Biotinyl-Cap-PE lipids at different molar percentages and reveal that avidin is not evenly distributed, but forms what looks like clusters even at low percentage occupancy which hampers the level of avidin that can be associated with the surface. We have then successfully employed the novel strategy of using PDP-PE lipids which contain a reducible disulphide to which we added maleamide-PEG-biotin spacers of different lengths. There is a more even distribution of avidin on these layers and thereby increasing the amount and efficiency of avidin association. The reduced levels of avidin that was being associated with the Biotinyl-Cap-PE layers as compared to the PDP-PE lipids could be analysed with QCM-D and interferometry approaches, but it was only with SEEC microscopy that the reason for the reduced occupancy was resolved.
Introduction
The utilization of lipid supports on sensor surfaces has increased dramatically over the last few years. One reason for this is the relative ease in which one can make a well-ordered layer that can be tailored to suit the needs of the experiment. These can be bilayers,1–5 self-assembled monolayers6,7 or supported/tethered bilayers, using the integration of other molecules as supports distancing the layer from the surface.8,9 The adaptability of the approach has led to exciting new developments in bio-nanotechnology10,11 and in the design of novel characterization methods for investigating membrane protein–lipid interactions and antimicrobial peptides.3,12,13 Biomimetic lipid layers, as well as being adaptable, have the potential to deliver a surface that is less chemically reactive and with native biological properties thereby allowing researchers to study more complex systems.Another reason for the rapid advance of lipid biosensor-based applications is the introduction of instrumentation capable of monitoring both the formation of the layers and subsequent interactions in real time; notable among these are quartz-crystal microbalances (QCM), ellipsometers and interferometers such as the Farfield dual polarisation interferometer (DPI). The deposition of simple lipid bilayers onto planar surfaces using these technologies is in the most part by vesicle fusion of small unilamellar vesicles onto silica surfaces with a percentage of the lipids substituted for ones that are suitable for the attachment of proteins or other macromolecules.1,4,14–20 The majority of these approaches use a molar percentage of between 5 and 10% functionalized lipids, utilizing predominantly either biotin- or NTA-moieties. On non-lipid based sensor surfaces the most widely used methods of attachment to proteins are through alkanethiol self-assembled monolayers or carboxyl-modified sugars, which can both subsequently be modified with the molecule of interest via carbodiimide chemistry and reactive NHS-esters. It is interesting that these chemistries are not often implemented on lipid layers, perhaps due to the inherent difficulty of forming bilayers with the necessary carboxyl head-groups.18 Avidin is a tetravalent biotin binding protein, with the affinity for biotin being one of the tightest known natural non-covalent molecular associations. Most of the family of related avidin molecules from different species, such as streptavidin have very similar affinity and they provide a convenient way of attaching molecules to surfaces. Being multi-valent, avidin molecules can attach to a biotin-modified surface and have 1–2 binding sites still available for attaching further biotin-modified molecules.1 New ways of attaching biotin to proteins such as with the AviTag™ or by using the twin Strep-Tag system21 and novel avidin molecules such as Captavidin™,22 which allows efficient removal of the molecules from the surface for regeneration purposes, opens up more possibilities for biotin coated sensor surfaces. Exploitation of lipid bilayer systems for biosensor applications relies on the membrane remaining fluid and fully formed without any ruptures or perturbations and without overcrowding the surface, which will restrict lateral diffusion. Most research groups use an arbitrary value of functionalized lipids between 5 and 10% (molar concentration) because of the lack of information on the levels needed for optimal protein coverage and there is a need to understand whether the most common biotin functionalized lipid, Biotinyl-Cap PE, is in fact appropriate. Here we have investigated the use of two types of functionalized lipids with three different lengths of surface linkers and have used QCM-D, dual polarization interferometry and surface enhanced ellipsometric contrast (SEEC) microscopy to provide detailed information on the optimal concentrations of functional group required to either saturate a surface or to provide a surface that has a controlled and uniform layer. We show that the traditional Biotinyl-Cap PE (BC-PE) lipids are not optimal for binding avidin moieties to the surface of the bilayers and that these form lipid clusters not detectable using non-visual techniques. Instead we identify a thiol-based alternative PDP-PE, which can be modified with PEG-biotin linkers that has a greater avidin-binding capacity and do not form these apparent clusters on the surface. These results have important implications for future studies and biomimetic bilayer design.
Results
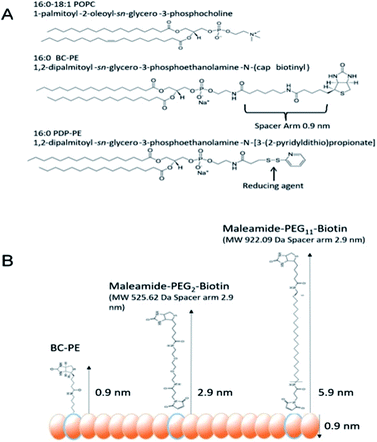
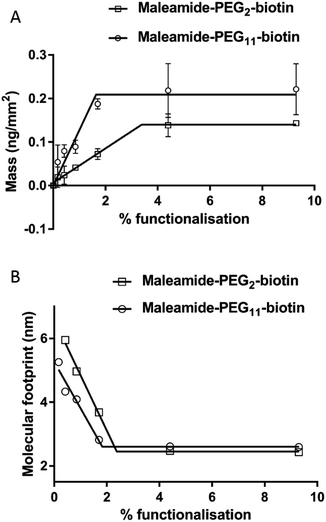
In order for us to investigate the effect of protein addition to the layers, and to study the effect of increasing the linker length between the protein and bilayer surface, we introduced varying percentages of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Cap Biotinyl) herein called BC-PE and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(3-pyridinyldithio-propionate) referred to here as PDP-PE (Fig. 1A), lipids to the POPC liposomes. The BC-PE lipids contain a 0.9 nm spacer between the phosphoethanolamine and terminal biotin, whilst the PDP-PE has a reducible disulphide bond to which, following reduction, we were able to link maleamide-PEG-biotin linkers with either 2 or 11 polyethylene glycol (PEG) units creating 2.9 or 5.9 nm spacers (Fig. 1B). These lipids were incorporated at percentages of between 0.2–10% (molar concentration) of the total POPC lipid within the bilayer and it is assumed that there is an equal distribution of lipids throughout the two leaflets. Using DPI we analysed the change in birefringence of the layer after incorporation of the different head-groups. This showed a general decrease in lipid ordering with birefringence measurements dropping from 0.0210 ± 0.0003 to 0.0150 ± 0.0011. These small differences, whilst certainly detectable are within the range of birefringence measurements seen previously for full lipid bilayers23 and indicate a small but detectable change in lipid anisotropy after incorporation of the functional lipid groups.Following PDP-PE bilayer formation and reduction using TCEP, maleimide-PEG2-biotin and maleimide-PEG11-biotin linkers were incorporated. Mass addition of these molecules (526.62 Da and 922.09 Da, respectively) onto the surface was measured for each of the two linkers (at varying percentages of functionalised lipid relative to POPC) using DPI (Fig. 2). Total linker mass at equilibrium for each of the percentage points fits to a segmented linear approximation where the X0 value of the upper plateau represents the level of layer saturation, and the intercept of the linear segments is the percentage at which total saturation is achieved. Not surprisingly larger mass addition was seen for maleimide-PEG11-biotin at ∼0.22 ng mm−2 compared to ∼0.15 ng mm−2 from maleimide-PEG2-biotin (Fig. 2A). The PEG11 layer saturated at ∼2.1% functionalisation whereas the PEG2 layers saturated at ∼3.9%. Using the average mass for each linker the number of molecules per mm2 of the lipid surface was calculated as 1.5 and 1.6 × 1011 linker molecules per mm2 at saturation for PEG11 and PEG2 respectively, indicating that there is little difference in the ability of each linker to immobilise to the functionalised lipid surface. At linker surface saturation, there is on average one linker present every 2.2 nm (PEG2) and 2.3 nm (PEG11) which is termed the molecular footprint (Fig. 2B). This also suggests that at saturation of the linkers i.e. ≥∼2.1% functionalisation, there is molecular redundancy of binding sites. This means that there is no positive benefit in using higher percentages of functionalised lipid.
Avidin addition to the layers
In order to assess the ability of the layers to immobilise avidin, we first calculated the theoretical amounts of avidin that could be coupled in an idealised system using eqn (1):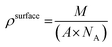 | (1) |
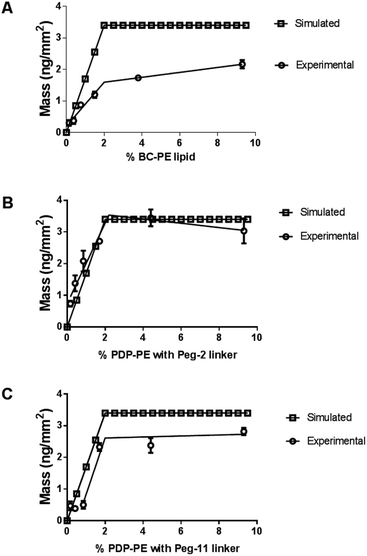
We compared the simulated avidin absorption for each of the linkers with the data recorded using DPI. Fig. 3 shows that all linkers followed the segmented linear approximations of the simulated data with X0 values (the level of the upper plateau) corresponding to ∼2% functionalisation at saturation. However, there are significant differences in the maximum level of avidin that can be bound to the surface. The BC-PE lipids (Fig. 3A) produced the lowest amount of protein mass addition of 1.60 ± 0.08 ng mm−2 at ∼2% and a maximal amount of 2.10 ng mm−2 at 10%. PEG2-biotin layers (Fig. 3B) exhibited the greatest mass addition at 3.20 ± 0.10 ng mm−2 followed by PEG11-biotin layers saturating at approximately 2.70 ± 0.15 ng mm−2 (Fig. 3C). Surprisingly, we also observed the PEG11 linkers at percentages below 1% do not follow a linear response but rather the shape of the curve is sigmoidal suggesting that at low percentages there are problems associated with biotin availability. Interestingly based on QCM-D measurements, quantitation of mass using the Sauerbrey equation estimated maximal bound mass of ∼8 ng mm−2 for PEG2 and BC-PE and 6 ng mm−2 for the PEG11-linkers (ESI Fig. 1†). These higher total mass additions compared to DPI values are due to coupled water.25,26 So it is apparent that the level of coupled water is greater for the BC-PE linker, at ∼6.4 ng mm−2 water compared to values of 5.9 and 3.3 ng mm−2 for PEG2 and PEG11 respectively, and is inversely related to linker length. This highlights that when avidin is close to the surface, i.e. within 1–3 nm, the level of water associated with the layer is significantly higher. QCM-D measurements clearly distort the apparent amount of bound protein depending upon the length of the linker used. The shorter linkers trapping more bound water than the longer more flexible ones.
At low percentage functionalization there is a strong relationship between association rate and linker length (ESI Fig. 2†). BC-PE (0.9 nm spacer arm) has the slowest association rate at 1.1 ± 0.3 × 106 M−1 min−1 at 1% functionalization. The PEG11-biotin surfaces (5.9 nm spacer arm) demonstrated an order of magnitude quicker on-rate (1.1 ± 0.2 × 107 M−1 min−1) at low functionalization (0.2%). It can be concluded that at the lower percentage-functionalization the biotin is more readily available for binding when immobilised at greater distances from the lipid surface. Above ∼2% functionalization, i.e. the level at which avidin saturation occurs, the association of avidin with both the PEG linkers is diffusion limited and restricted to ∼5 × 106 M−1 min−1 but the BC-PE linkers fail to reach the association rates exhibited by the PEG linkers until much higher percentages.
Understanding the sigmoidal behaviour of avidin association to PEG11-biotin linkers
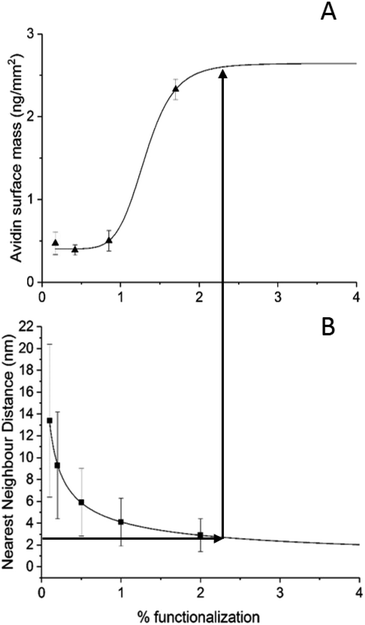
The average surface mass of immobilised avidin molecules is calculated to be ∼3.2 ng mm−2 at saturation. This is consistent with an almost complete monolayer of closely packed avidin on the lipid surface. We have shown (Fig. 3) that both BC-PE and PEG11 saturate at levels lower than PEG2 and therefore we can assume that these immobilisation methods produce less densely packed surfaces and are unable to form complete monolayers. At low percentage functionalization (i.e. below 2%) the layers for BC-PE and PEG2-biotin exhibit linear mass/percentage responses that fit to the expected segmented linear approximation for molecular association. However, as shown in Fig. 3C and 4A, the PEG11-biotin layer exhibits very low avidin binding at low percentages. The sigmoidal binding curve for PEG11-biotin, which is most pronounced using DPI suggests that at low functionality, the biotin attached to the longer linker is potentially unavailable for binding. There is no evidence from the birefringence that the PEG11 linker is inserted into the bilayer (data not shown) but the lack of avidin binding would suggest that the linkers may be associated closely to the lipid surface and unavailable, or more than one linker is attached to one avidin molecule.To understand this phenomenon we simulated the nearest neighbour spatial distribution of functionalized lipids within the bilayer for each percentage of linker addition using the method of Clark and Evans, which has been adapted for use within the ImageJ software by Ciaran Beggan and Christopher Hamilton.27 The simulation represents labelled/functionalised lipids within a planar bilayer in a random distribution within a known area (ESI Fig. 3†). Functionalized percentages of between 0.1% and 5% show that the neighbouring labelled lipids are on average 12.4 nm apart for 0.1% lipid layers, to 1.8 nm apart for 5% functionalised lipid layers. Based on the molecular footprint of avidin, interference by neighbouring attachment sites would start at levels above 1% functionalization. This is also the point at which we could start to see multi-valency. However below this, the distance between linkers is too great for multi-valency to be a significant problem assuming a random distribution of linkers. The interfering effects of neighbouring linkers, which for the PEG11-biotin have a footprint of 2.6 nm (Fig. 2B), becomes evident at >2% functionalisation. It seems therefore that the sigmoidal pattern observed for PEG11-biotin is caused by molecular crowding of the linkers. At concentrations higher than 2% functionalisation, linker availability is increased, likely due to self-assembly on the surface. At lower concentrations the lack of self-assembly leads to a decrease in biotin availability perhaps due to steric effects of the lipid surface. Interestingly, given the fast association of the avidin to the PEG11-biotin linkers (ESI Fig. 2†), those biotin moieties that are available become quickly bound to the avidin; however, the total mass of avidin on the surface is reduced compared to PEG2-biotin, most likely due to multiple occupancy of biotin binding sites.1
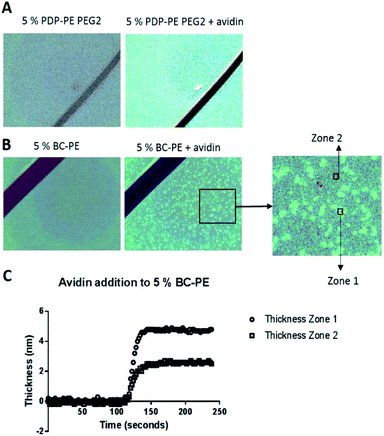
Using surface enhanced ellipsometric imaging to understand the lower avidin binding efficiency of the BC-PE lipid
In order to understand why we are seeing consistently lower avidin association to the BC-PE lipid we used a quantitative technique called time resolved Surface Enhanced Ellipsometric Contrast (SEEC) imaging. This technique, described by Frederik Hook's group in 2012 (ref. 28) and first developed by Ausserre and Valignat in 2007,29 provides high-resolution imaging and thickness measurements in real-time on thin films. The SEEC sensors are used in the place of conventional microscope slides on a widefield microscope with a ×25 objective and are composed of multiple layers of organic oxides that produce a non-reflective surface under canonical illumination.30 When material is deposited upon this surface, in our case using a microfluidics channel to achieve time-resolved measurements, the contrast enhancement is ∼100-fold compared to that produced by non-enhanced widefield microscopy.30 In addition to the enhancement in contrast, the thickness of the film can be accurately measured using the Fizeau fringes created by optical interference in the colorimetric range of the reflected light. Each pixel is converted into an optical thickness which is accurate over the range of approximately 0.1 nm to 200 nm based on the colour changes of a thickness standard.30,31 We deposited POPC bilayers functionalised with PDP-PE and BC-PE lipids onto the surface of the SEEC sensors and captured image frames every 5 seconds. Using the Sarusoft software we were able to pick one or multiple areas for the analysis of thickness change throughout sequential frames as illustrated in Fig. 5. Instantaneous rupture of the lipids occurred in all cases and there was no evidence of membrane perturbations or gaps. The thickness of the layers was ∼4.25 nm at saturation for all the lipids tested. Avidin was then added to the BC-PE (Fig. 5B) and PDP-PE lipids (following linker addition) (Fig. 5B), which showed remarkably different results. The BC-PE lipids exhibited the formation of areas of higher thickness that were not present on the PDP-PE lipids. These were evident from 0.2–10% functionalization (ESI Fig. 4†). Analysis of the thickness of the different zones showed that these zones were areas where avidin was more concentrated, presumably because the BC-PE was more localized and evidenced by the ∼5 nm change in thickness (Fig. 5C); in between the spots there was a contribution of avidin binding but at a lower level evidenced by the lower level of ∼2.5 nm at 5% functionalisation. This highlights that the BC-PE lipids are forming larger clusters whereas the PDP-PE lipids do not, which therefore explains the reduction in avidin binding efficiency observed for BC-PE lipids.Discussion
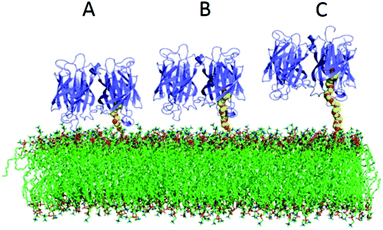
We have compared two different attachment strategies for the association of avidin molecules onto lipid bilayers. The first using biotin functionalized lipids (BC-PE), and the second using 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)-propionate] (PDP-PE) which is a novel approach to the attachment of molecules to layers using the reduced thiol. Adding a known percentage of biotinylated lipids and capturing avidin is the most common method for coupling proteins to lipid surfaces and has been used in many applications.1,7,17,32–35 The use of PDP-PE is a method used here to link avidin to lipid bilayers via spacer intermediates. By using this approach we are able to modulate the length of the spacer and therefore the distance of the attached target molecule (in our case avidin) from the surface. We have shown that this enhances the coupling of avidin molecules to the surface in both quantity and efficiency compared to the standard BC-PE method. Saturation occurred extremely rapidly for PEG11-linkers, slightly slower for the PEG2 but was an order of magnitude slower for BC-PE. Ouberai et al.26 showed that lysozyme covered a bare SiO2 surface with a density of 2.87 ng mm−2 and BSA, which has a similar size to avidin, with 1.83 ng mm−2 according to dry mass calculations using DPI measurements of proteins absorbed onto the surface. Interestingly when we use DPI to study the maximum level of addition of avidin onto the PDP-PE layers using the PEG2-biotin linker (corresponding to a 2.9 nm spacer; Fig. 1B), we obtained a surface coverage of 3.2 ± 0.1 ng mm−2 which is consistent with a complete monomeric layer. This suggests that the PEG2-biotin spacers are capable of forming a layer that has a significantly higher level of saturation compared to physisorption and is the same as the theoretical diffusion-limited maximal level of binding. This greater binding efficiency is likely due to the diffusional capability of the lipids.BC-PE lipids, which have the shortest spacer arm, are the most common form of functionalized lipid used in the literature, but we have shown these to be far from ideal. This type of linker when bound to avidin couples large amounts of water to the lipid surface. Molecular modelling of avidin bound to the lipids (PDB files extracted from Dr Peter Tieleman's website, University of Calgary) by all three linkers (Fig. 6) indicates that the BC-PE linkers could create a substantial interaction between the avidin and the lipid surface creating a super-hydrated and rigidly coupled layer which fits with our data comparing surface coverage from the DPI and QCM-D, which suggests that there is far greater coupled water for the BC-PE lipids than the other linkers. The molecular model in Fig. 6 also indicate that avidin bound to BC-PE lipids interacts with the phosphatidylcholine head-group of the lipid surface. This could have the causative effect of altering lipid surface dynamics. The PEG11-biotin and PEG2-biotin linkers in contrast have much faster kinetics for their association to avidin presumably due to higher availability of the biotin. We have shown that the longer PEG11 linkers are not very good at binding avidin below concentrations of 2% but above this, the long linkers have increased availability. Our data from the simulation of nearest neighbour distributions suggests that this could be due to the longer PEG chains arranging in a self-assembled monolayer but we have no direct proof of this. Below 2% these longer linkers are not optimal for binding proteins to the surface. However for both the PEG2-biotin and PEG11-biotin linkers there is little difference in surface immobilized mass of avidin binding at percentage functionalization above 5%.
Most importantly we have found that BC-PE lipids produce what looks like surface clusters. These areas of higher avidin concentration can be clearly viewed using SEEC microscopy and the contrast interferometric responses for avidin binding within and outside of these clusters (Fig. 5B) indicates a clear difference in the levels of binding. This phenomenon explains the overall lower dry mass response to avidin association recorded using DPI compared to that of the PDP-PE lipids and the PEG2-biotin linkers. The reason that avidin is binding in areas of higher density is unclear. This could be due to clustering of the BC-PE lipids where the difference in charges of the BC-PE lipids cause local non-random associations and like-molecules may separate into these domains. It is also possible that this is due to membrane curvature,36 but significant curvature was not observed in the membrane thickness measurements. However lateral pressure caused by changes in the packing of the BC-PE chains could cause differential pressure distributions37 which causes membrane curvature that is not detected as changes in overall thickness but is sufficient to cause clustering of like-molecules. This could also be occurring in the PDP-PE but with much smaller clusters which do not adversely affect the levels of avidin that can be associated with the surface.
Conclusion
These results show that using BC-PE lipids will result in clustering of avidin molecules on the surface of the lipid layer and this can be resolved using PDP-PE lipids, to which one can add maleamide-PEG-biotin linkers. We also show that PEG2-biotin linkers bind the most avidin, have fast kinetics and have no deleterious effects on the layer itself. We have also shown that there is no effective need to use greater than 2.5% functionalised lipid. Higher percentages than this result in molecular redundancy of the linkers.Experimental
Liposome preparation
Lyophilised 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was dissolved in chloroform, with or without head group-functionalized lipid (i.e. 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Cap biotinyl) (referred to here as BC-PE) or 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)propionate] (termed PDP-PE)) (Avanti Polar Lipids, USA), within a glass tube. Chloroform was removed under a continuous nitrogen stream to produce a thin lipid film. Tubes were placed under vacuum for a minimum of 2 h to ensure all chloroform had been removed. Lipid films were rehydrated in 20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH 7.4 at 1 mg ml−1. Lipid solutions were extruded through a 50 nm pore size filter composed of polycarbonate membranes (Whatman®, GE Healthcare Life Sciences), using a mini-extruder set (Avanti Polar-Lipids, USA). Dynamic Light Scattering (DLS) using a Zetasizer nano-S instrument (Malvern Instruments Ltd, UK) was used to check the size of the unilamellar vesicles. Only vesicles producing single peaks with a z-average of approximately 100 nm and below were used.QCM-D and DPI chip surface preparation
Q-sense (SiO2) and Analight unmodified (SiOxNy) chips were treated to a surface cleaning process prior use, however Analight chips were cleaned approximately 12 h before use and Q-sense chips immediately before use. Chips were sonicated in 0.2% (w/v) SDS for 15 min and then rinsed extensively in ddH2O. Surfaces were then rinsed in ethanol and dried under nitrogen. Chips were finally placed under UV Ozone (ProCleaner™ Plus, (Bioforce Nanosciences)) for 30 min. Q-sense chips were used immediately and Analight chips were stored dry until use.Quartz crystal microbalance with dissipation monitoring (QCM-D)
QCM-D measurements were performed on a Q-sense E1 or a Q-sense Omega with standard flow module. All sample injections were undertaken in flow mode (continuous solution delivery to the measurement chamber) injecting 200 μl at 25 μl min−1. Changes in dissipation (D) and normalised frequency (f) were measured in all overtones but are only displayed for the fifth overtone (25 MHz). For lipid bilayer production small unilamellar vesicles (SUVs) (100 μg ml−1) were injected at 25 μl min−1 over pre-equilibrated SiO2 Q-sense chips in filtered and degassed 20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH 7.4 at 21 °C. Following planar bilayer production, running buffer was changed to filtered and degassed 20 mM HEPES, 150 mM NaCl, pH 7.4 (RB). BSA (100 μg ml−1 in RB) was injected over formed bilayers at 50 μl min−1 to ensure continuous coverage. For experiments in which linkers were used, disulphide capped lipids (PDP-PE) were reduced with 10 mM tris(2-carboxyethyl)phosphine (TCEP) in RB injected over the lipid surface at a flow rate of 50 μl min−1 for a minimum of 10 minutes or until the response had plateaued. Linkers (EZ-Link® maleimide-PEG2-biotin or EZ-link® maleimide-PEG11-biotin (Thermo)) were re-suspended in RB at a concentration of 1 mM and flowed over the reduced lipid surface at a flow rate of 20 μl min−1 for a ∼10 minutes or until the response had again plateaued. For the subsequent production of an avidin layer, 10 μg ml−1 of this protein was flowed over the bilayer surface at 50 μl min−1 in RB until the surface was saturated. Analysis of layers was performed using the Qtools software (Q-sense®, Sweden).Dual polarization interferometry (DPI)
An Analight® dual polarization interferometer (DPI) (Analight® 4D, Farfield-Biolin Scientific AB, Sweden) was used to measure lipid bilayer production and protein addition. The instrument was set to 20 °C throughout all experiments. Cleaned chips were equilibrated in filtered and degassed 20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH 7.4 (RB) within the flow module for a minimum of 30 min. System calibrations were undertaken prior to each experiment which consisted of an injection of filtered and degassed 80% (w/w) ethanol for chip calibration followed by an injection of filtered and degassed ddH2O for bulk calibrations. Only experimental set-ups that passed the calibration criteria were used. Lipid bilayer production was carried out as described for QCM-D experiments (see above) before buffer was changed for filtered and degassed RB. Avidin injections were performed as described for QCM-D with a 200 μl injection loop. Initial layer analysis was undertaken using the Analight® 4D Resolver software (Farfield-Biolin Scientific AB, Sweden). Lipid refractive index (RI) was set to 1.47 and the dn/dc to 0.135 (ref. 23) with thickness, mass and birefringence measurements unfixed. The dn/dc for protein analysis was set to a standard value of 0.182 and all remaining measurements were left unfixed unless otherwise stated within the Results section.Surface enhanced interferometric microscopy
The N-Lab Station (Nanolane, France) was used to measure the real-time binding of avidin-containing bilayers to SEEC sensors. Lipids were produced in the same manner as for QCM-D and DPI. Silicon-dioxide wet Sarfus slides were used without any prior cleaning. Sarfus slides were inserted into the instrument and scratched to provide a focal point before the 1-channel fluidics system was assembled on top. Buffer (20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH 7.4) was flowed over at ∼100 μl min−1 and the microscope was focused on the scratch using a 50× objective. Lipid deposition, reduction of PDP-PE, linker addition and avidin immobilisation were performed in the same manner as for DPI and QCM-D. Images were recorded every second at each phase of the experiment for example, lipid layer addition, reduction, linker addition and avidin deposition.Dynamic light scattering
DLS measurements were performed with Zetasizer Nano-S (Malvern, Germany). The measurements of both lipid vesicles and avidin were taken in a 1 cm 45 μl quartz cuvette at a temperature of 20 °C. Avidin or vesicles were analysed at a concentration of 1 mg ml−1 taking three measurements of 13 averages each.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the Biotechnology and Biological Sciences Research Council (BBSRC), Biolin Scientific and the Wellcome Trust for funding this project.References
- G. V. Dubacheva, C. Araya-Callis, A. Geert Volbeda, M. Fairhead, J. Codee and M. Howarth, et al., Controlling Multivalent Binding through Surface Chemistry: Model Study on Streptavidin, J. Am. Chem. Soc., 2017, 139(11), 4157–4167 CrossRef CAS PubMed.
- A. Sumino, T. Dewa, T. Takeuchi, R. Sugiura, N. Sasaki and N. Misawa, et al., Construction and structural analysis of tethered lipid bilayer containing photosynthetic antenna proteins for functional analysis, Biomacromolecules, 2011, 12(7), 2850–2858 CrossRef CAS PubMed.
- S. Forbes, A. J. McBain, S. Felton-Smith, T. A. Jowitt, H. L. Birchenough and C. B. Dobson, Comparative surface antimicrobial properties of synthetic biocides and novel human apolipoprotein E derived antimicrobial peptides, Biomaterials, 2013, 34(22), 5453–5464 CrossRef CAS PubMed.
- O. Beutel, F. Roder, O. Birkholz, C. Rickert, H. J. Steinhoff and M. Grzybek, et al., Two-Dimensional Trap for Ultrasensitive Quantification of Transient Protein Interactions, ACS Nano, 2015, 9(10), 9783–9791 CrossRef CAS PubMed.
- D. Melendrez, T. Jowitt, M. Iliut, A. F. Verre, S. Goodwin and A. Vijayaraghavan, Adsorption and binding dynamics of graphene-supported phospholipid membranes using the QCM-D technique, Nanoscale, 2018, 10(5), 2555–2567 RSC.
- J. N. Barbosa, M. C. Martins, S. C. Freitas, I. C. Goncalves, A. P. Aguas and M. A. Barbosa, Adhesion of human leukocytes on mixtures of hydroxyl- and methyl-terminated self-assembled monolayers: effect of blood protein adsorption, J. Biomed. Mater. Res., Part A, 2010, 93(1), 12–19 Search PubMed.
- N. Ballav, A. Terfort and M. Zharnikov, Fabrication of mixed self-assembled monolayers designed for avidin immobilization by irradiation promoted exchange reaction, Langmuir, 2009, 25(16), 9189–9196 CrossRef CAS PubMed.
- R. Budvytyte, M. Mickevicius, D. J. Vanderah, F. Heinrich and G. Valincius, Modification of tethered bilayers by phospholipid exchange with vesicles, Langmuir, 2013, 29(13), 4320–4327 CrossRef CAS PubMed.
- R. Budvytyte, G. Valincius, G. Niaura, V. Voiciuk, M. Mickevicius and H. Chapman, et al., Structure and properties of tethered bilayer lipid membranes with unsaturated anchor molecules, Langmuir, 2013, 29(27), 8645–8656 CrossRef CAS PubMed.
- T. Albrecht, Nanobiotechnology: a new look for nanopore sensing, Nat. Nanotechnol., 2011, 6(4), 195–196 CrossRef CAS PubMed.
- E. C. Yusko, J. M. Johnson, S. Majd, P. Prangkio, R. C. Rollings and J. Li, et al., Controlling protein translocation through nanopores with bio-inspired fluid walls, Nat. Nanotechnol., 2011, 6(4), 253–260 CrossRef CAS PubMed.
- K. Hall, T. H. Lee, A. I. Mechler, M. J. Swann and M. I. Aguilar, Real-time measurement of membrane conformational states induced by antimicrobial peptides: balance between recovery and lysis, Sci. Rep., 2014, 4, 5479 CrossRef CAS PubMed.
- D. J. Hirst, T. H. Lee, M. J. Swann and M. I. Aguilar, Combined mass and structural kinetic analysis of multistate antimicrobial peptide-membrane interactions, Anal. Chem., 2013, 85(19), 9296–9304 CrossRef CAS PubMed.
- I. Reviakine, D. Johannsmann and R. P. Richter, Hearing what you cannot see and visualizing what you hear: interpreting quartz crystal microbalance data from solvated interfaces, Anal. Chem., 2011, 83(23), 8838–8848 CrossRef CAS PubMed.
- G. R. Heath, M. Li, I. L. Polignano, J. L. Richens, G. Catucci and P. O'Shea, et al., Layer-by-Layer Assembly of Supported Lipid Bilayer Poly-L-Lysine Multilayers, Biomacromolecules, 2016, 17(1), 324–335 CrossRef CAS PubMed.
- S. Shao, J. Geng, H. Ah Yi, S. Gogia, S. Neelamegham and A. Jacobs, et al., Functionalization of cobalt porphyrin-phospholipid bilayers with his-tagged ligands and antigens, Nat. Chem., 2015, 7(5), 438–446 CrossRef CAS PubMed.
- T. T. Nguyen, K. L. Sly and J. C. Conboy, Comparison of the energetics of avidin, streptavidin, neutrAvidin, and anti-biotin antibody binding to biotinylated lipid bilayer examined by second-harmonic generation, Anal. Chem., 2012, 84(1), 201–208 CrossRef CAS PubMed.
- S. Vafaei, S. R. Tabaei, K. H. Biswas, J. T. Groves and N. J. Cho, Dynamic Cellular Interactions with Extracellular Matrix Triggered by Biomechanical Tuning of Low-Rigidity, Supported Lipid Membranes, Adv. Healthcare Mater., 2017, 6(10), 1700243 CrossRef PubMed.
- M. R. Horton, C. Reich, A. P. Gast, J. O. Radler and B. Nickel, Structure and dynamics of crystalline protein layers bound to supported lipid bilayers, Langmuir, 2007, 23(11), 6263–6269 CrossRef CAS PubMed.
- N. B. Eisele, F. I. Andersson, S. Frey and R. P. Richter, Viscoelasticity of thin biomolecular films: a case study on nucleoporin phenylalanine-glycine repeats grafted to a histidine-tag capturing QCM-D sensor, Biomacromolecules, 2012, 13(8), 2322–2332 CrossRef CAS PubMed.
- T. G. Schmidt, L. Batz, L. Bonet, U. Carl, G. Holzapfel and K. Kiem, et al., Development of the Twin-Strep-tag(R) and its application for purification of recombinant proteins from cell culture supernatants, Protein Expression Purif., 2013, 92(1), 54–61 CrossRef CAS PubMed.
- C. Garcia-Aljaro, F. X. Munoz and E. Baldrich, Captavidin: a new regenerable biocomponent for biosensing?, Analyst, 2009, 134(11), 2338–2343 RSC.
- A. Mashaghi, M. Swann, J. Popplewell, M. Textor and E. Reimhult, Optical anisotropy of supported lipid structures probed by waveguide spectroscopy and its application to study of supported lipid bilayer formation kinetics, Anal. Chem., 2008, 80(10), 3666–3676 CrossRef CAS PubMed.
- C. Song, P. Wang and H. A. Makse, A phase diagram for jammed matter, Nature, 2008, 453(7195), 629–632 CrossRef CAS PubMed.
- C. Larsson, M. Rodahl and F. Hook, Characterization of DNA immobilization and subsequent hybridization on a 2D arrangement of streptavidin on a biotin-modified lipid bilayer supported on SiO2, Anal. Chem., 2003, 75(19), 5080–5087 CrossRef CAS PubMed.
- M. M. Ouberai, K. Xu and M. E. Welland, Effect of the interplay between protein and surface on the properties of adsorbed protein layers, Biomaterials, 2014, 35(24), 6157–6163 CrossRef CAS PubMed.
- C. Beggan and C. W. Hamilton, New image processing software for analyzing object size-frequency distributions, geometry, orientation, and spatial distribution, Comput. Geosci., 2010, 36(4), 539–549 CrossRef.
- A. Gunnarsson, M. Bally, P. Jonsson, N. Medard and F. Hook, Time-resolved surface-enhanced ellipsometric contrast imaging for label-free analysis of biomolecular recognition reactions on glycolipid domains, Anal. Chem., 2012, 84(15), 6538–6545 CrossRef CAS PubMed.
- D. Ausserre and M. P. Valignat, Surface enhanced ellipsometric contrast (SEEC) basic theory and lambda/4 multilayered solutions, Opt. Express, 2007, 15(13), 8329–8339 CrossRef CAS PubMed.
- A. Ducret, M. P. Valignat, F. Mouhamar, T. Mignot and O. Theodoly, Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(25), 10036–10041 CrossRef CAS PubMed.
- A. M. Egea, E. Trevisiol and C. Vieu, Direct patterning of probe proteins on an antifouling PLL-g-dextran coating for reducing the background signal of fluorescent immunoassays, Biointerphases, 2013, 8(1), 37 CrossRef PubMed.
- S. Mun and S. J. Choi, Optimization of the hybrid bilayer membrane method for immobilization of avidin on quartz crystal microbalance, Biosens. Bioelectron., 2009, 24(8), 2522–2527 CrossRef CAS PubMed.
- E. Nileback, L. Feuz, H. Uddenberg, R. Valiokas and S. Svedhem, Characterization and application of a surface modification designed for QCM-D studies of biotinylated biomolecules, Biosens. Bioelectron., 2011, 28(1), 407–413 CrossRef PubMed.
- N. S. Baranova, E. Nileback, F. M. Haller, D. C. Briggs, S. Svedhem and A. J. Day, et al., The inflammation-associated protein TSG-6 cross-links hyaluronan via hyaluronan-induced TSG-6 oligomers, J. Biol. Chem., 2011, 286(29), 25675–25686 CrossRef CAS PubMed.
- M. Edvardsson, S. Svedhem, G. Wang, R. Richter, M. Rodahl and B. Kasemo, QCM-D and reflectometry instrument: applications to supported lipid structures and their biomolecular interactions, Anal. Chem., 2009, 81(1), 349–361 CrossRef CAS PubMed.
- H. Koldso, D. Shorthouse, J. Helie and M. S. Sansom, Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers, PLoS Comput. Biol., 2014, 10(10), e1003911 CrossRef PubMed.
- H. Zhan and T. Lazaridis, Inclusion of lateral pressure/curvature stress effects in implicit membrane models, Biophys. J., 2013, 104(3), 643–654 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00060d |
| This journal is © The Royal Society of Chemistry 2020 |