Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of copper-regulated proteins in an oceanic diatom, Thalassiosira oceanica 1005†
Liangliang
Kong
 ab and
Neil M.
Price
ab and
Neil M.
Price
 *a
*a
aDepartment of Biology, McGill University, Montréal, Québec H3A 1B1, Canada. E-mail: neil.price@mcgill.ca
bCollege of Marine Life Sciences, Ocean University of China, Qingdao, Shandong 266003, China
First published on 14th May 2020
Abstract
Copper (Cu) is an essential cofactor of photosynthetic and respiratory redox proteins in phytoplankton and a scarce resource in parts of the open sea. Although its importance for growth is well recognized, the molecular mechanisms by which phytoplankton respond and acclimate to Cu deficiency are not well known. In this study, we identified the dominant Cu-regulated proteins and measured key physiological traits of Thalassiosira oceanica (CCMP 1005) under Cu-limiting and sufficient conditions. Growth limitation of T. oceanica occurred at environmentally relevant Cu concentrations (1 nM) as a result of decreased photosynthetic efficiency (ΦPSII). In Cu-limited cells, levels of plastocyanin decreased by 3-fold compared to Cu-replete cells and rates of maximum photosynthetic electron transport were reduced. Proteins associated with light harvesting complexes also declined in response to Cu limitation, presumably to adjust to reduced photosynthetic electron flow and to avoid photodamage to the photosystems. Key enzymes involved in carbon and nitrogen assimilation were down-regulated in low-Cu cells, as were steady state rates of C and N uptake. Relatively fewer proteins were up-regulated by Cu limitation, but among them were two enzymes involved in fatty acid oxidation (FAO). The increase in FAO may be a sign of increased turnover of cellular lipids caused by damage from oxidative stress. A putative transcription factor containing three, repetitive methionine motifs (MpgMgggM; MpgMggM) increased significantly in Cu-limited cells. The collective results provide a general description of how plastocyanin-dependent diatoms adjust metabolism to cope with chronic Cu deficiency.
Significance to metallomicsCopper is an essential metal for phytoplankton, required in large amounts by plastocyanin-containing species. Its low environmental concentration restricts phytoplankton growth in the open sea, but the molecular mechanisms by which marine species acclimate to Cu deficiency are not well understood. This study identifies Cu-regulated proteins and measures key physiological traits of Thalassiosira oceanica (CCMP 1005) grown under Cu-limiting and sufficient conditions. Our results provide a general model of how a plastocyanin-dependent diatom adjusts cellular metabolism to cope with Cu deficiency. |
Introduction
Copper is indispensable for phytoplankton because it serves as a cofactor of redox proteins involved in electron transfer and oxygen cycling.1,2 Its requirements for growth vary among taxa and are influenced by environmental factors that modulate the activity of cellular metabolic pathways and the levels of cuproenzymes that function within them. Minimizing superfluous metabolic pools reduces Cu demand, allowing fast growth when Cu is scarce, and reallocating intracellular stores maintains levels of critical Cu-proteins and buffers short-term changes in cellular Cu requirements and Cu availability.3–5 However, under Cu-deficient conditions, effective scavenging of Cu from the environment is required to minimize growth limitation because copper use efficiency is maximized.6,7In photoautotrophs, most metabolically active Cu is contained in three metalloproteins: plastocyanin (PC), an electron carrier involved in the light reactions of photosynthesis;8,9 cytochrome c oxidase (COX), the terminal enzyme in the respiratory electron transport chain;4 and multicopper oxidase (MCO), part of the high-affinity iron assimilation pathway.4,10 Other essential cuproproteins, present in smaller amounts, include: amine oxidase, Cu–Zn superoxide dismutase, urate oxidase and tyrosinase.11 Chronic Cu limitation is expected to compromise the functions of the Cu-containing proteins and proteins that play roles in Cu homeostasis, while secondarily affecting proteins in metabolic pathways that depend on or feed into pathways requiring Cu.
Field experiments show Cu addition increases phytoplankton biomass and production rates, suggesting Cu is in short supply.12,13 These results are consistent with lab studies showing diatom growth is limited by Cu concentrations similar to the open sea.6,13,14 Copper-limited diatoms have significantly slower rates of Fe uptake and reduced photosynthetic efficiency compared to Cu-replete cells. The decrease in Fe uptake is attributed to a decrease in multicopper oxidase that provides Fe to the uptake system10,13 and slower rates of photosynthetic electron flow are thought to result from a decrease in plastocyanin.6,9 Thus, Cu deficiency affects specific physiological functions of diatoms that should be reflected in their proteomic profiles.
Identifying proteins and genes that are regulated by metal availability can reveal compensatory responses of phytoplankton to metal limitation and discover novel pathways of metal acquisition.15–20 Increases in uptake capacity and recycling efficiency and decreases in specific metalloproteins are common responses to iron deficiency,15 but the details of how diatoms respond to low concentrations of Cu are poorly known. Under Cu-limiting conditions, Thalassiosira oceanica 1005 upregulates a high affinity copper transport system, similar to that of yeast,21,22 that requires an extracellular reduction step to produce Cu(I) that is subsequently internalized.23,24 Green algae and cyanobacteria, which use plastocyanin in photosynthesis replace it with a functionally equivalent, heme-containing cytochrome c6 (CYC6), to maintain photosynthetic functions when environmental Cu concentration declines.25,26 Under these conditions, coproporphyrinogen III oxidase increases to supply additional heme for cytochrome c6 synthesis,27 and plastocyanin-bound Cu is reallocated to other metabolic functions.4 Genome-wide screening of Cu-regulated genes and proteins shows that Cu availability also affects the levels of other cuproenzymes and proteins, including Cu(II) reductases, and proteins involved in lipid metabolism and oxygen sensing.16,19,28,29
Thalassiosira oceanica 1005, a plastocyanin-containing diatom isolated from the Sargasso Sea, requires 10 times more cellular Cu to grow than a related coastal species.9 Its high cellular requirement and widespread oceanographic distribution make T. oceanica an ideal model species to study physiological and molecular responses to Cu deficiency. The genome of T. oceanica is available,30 so molecular tools, like proteomics, can be applied to elucidate Cu-regulated biochemical and molecular pathways. Here we report results of experiments to identify Cu-responsive proteins and physiological changes in T. oceanica 1005 subjected to chronic Cu deficiency.
Materials and methods
Culture and growth conditions
Thalassiosira oceanica CCMP 1005, was obtained from the National Center for Marine Algae and Microbiota (http://ncma.bigelow.org) and grown in artificial seawater medium, Aquil.31 The medium was prepared according to the recipe of Price et al.,32 but with a modified trace metal nutrient enrichment, consisting 1290 nM Fe, 125 nM Mn, 79.3 nM Zn, 100 nM Mo, 50 nM Co and 10 nM Se, buffered with 100 μM EDTA. Copper was added separately as a premixed Cu–EDTA complex in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.05 molar ratio at a total concentration of 1.0 and 21.4 nM. Semi-continuous batch cultures of Cu-limited (1.0 nM Cu) and Cu-replete (21.4 nM) T. oceanica were maintained in exponential growth for 250 generations prior to the start of the experiments. The cells were cultivated in 28 mL polycarbonate tubes at 20 °C under continuous illumination of 200 μmol photons m−2 s−1. Biomass was monitored by measuring in vivo chlorophyll a (chl a) fluorescence and specific growth rates (per day: d−1) were calculated from linear regression analysis of ln (fluorescence) versus elapsed time.13 Cell size was determined by measuring the linear dimensions of 20 randomly selected cells using a Zeiss light microscope, assuming a cylindrical shape.33 Teflon and polycarbonate bottles and tubes used in medium preparation and culturing were soaked in 10% hydrochloric acid and rinsed thoroughly with Milli-Q water (Millipore, Milli-Q® Reference).
1.05 molar ratio at a total concentration of 1.0 and 21.4 nM. Semi-continuous batch cultures of Cu-limited (1.0 nM Cu) and Cu-replete (21.4 nM) T. oceanica were maintained in exponential growth for 250 generations prior to the start of the experiments. The cells were cultivated in 28 mL polycarbonate tubes at 20 °C under continuous illumination of 200 μmol photons m−2 s−1. Biomass was monitored by measuring in vivo chlorophyll a (chl a) fluorescence and specific growth rates (per day: d−1) were calculated from linear regression analysis of ln (fluorescence) versus elapsed time.13 Cell size was determined by measuring the linear dimensions of 20 randomly selected cells using a Zeiss light microscope, assuming a cylindrical shape.33 Teflon and polycarbonate bottles and tubes used in medium preparation and culturing were soaked in 10% hydrochloric acid and rinsed thoroughly with Milli-Q water (Millipore, Milli-Q® Reference).
Cu bioassay
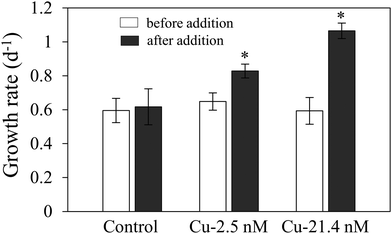
A Cu bioassay was conducted to verify that Cu was the limiting nutrient in low Cu medium. Replicate cultures of acclimated T. oceanica were grown in medium containing 1 nM Cu and enriched with 2.5 and 21.4 nM Cu (Cu–EDTA in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.05 molar ratio) in early exponential phase. Control cultures received an equivalent volume (5 μL) of Milli-Q water. Growth rates were measured before and after Cu addition.
1.05 molar ratio) in early exponential phase. Control cultures received an equivalent volume (5 μL) of Milli-Q water. Growth rates were measured before and after Cu addition.
Photosynthetic physiology
Chlorophyll a concentration was measured spectrophotometrically in mid-exponential phase cultures.34 Cells were harvested by filtration on to a glass fiber filter (25 mm diameter; GF 75; ADVANTEC; Bunkyo, Tokyo, Japan) with 1 mL of 1% magnesium carbonate added to the samples during filtration. Pigments were extracted from harvested cells (∼9 × 106 cells) overnight in 5 mL prechilled 90% acetone at 4 °C. Chlorophyll a concentration was measured by absorbance at 630, 647 and 664 nm using a CARY 1E UV/Vis Spectrophotometer (Agilent Technologies, ON, Canada) and calculated by the equation chl a = 11.85E664 − 1.54E647 − 0.08E630.34 Chlorophyll fluorescence-derived photosynthetic parameters, maximum quantum yield (Fv/Fm), effective quantum yield of photosystem II (ΦPSII) and maximum relative electron transport rate (rETRmax), were measured using a Water-PAM Fluorometer (Heinz Walz GmbH, Effeltrich, Germany) according to Kim and Price.6Protein extraction
Small aliquots of acclimated, Cu-deplete (1 nM Cu) and Cu-replete (21.4 nM Cu) cultures were inoculated into four replicate, 1 L polycarbonate bottles, and grown under identical conditions. Total cellular protein was measured in 50 mL samples harvested in mid-exponential phase by filtration onto 25 mm, 3 μm polycarbonate membrane filters and rinsed twice with fresh culture medium. The cells on the filters (∼2 × 107 cells) were extracted in 400 μL extraction buffer containing 68 mM Na2CO3, 2% sodium dodecyl sulfate and 1% protease inhibitor cocktail (Sigma-Aldrich, ON, Canada), pH 11, according to procedures described by Lommer et al.30 Cells were disrupted by sonication and then centrifuged to remove the cell debris. Total protein concentration in the supernatant was measured by a BCA Protein Assay Kit (Sigma-Aldrich) with bovine serum albumin as a standard. Protein purified for proteomic analysis was prepared from cultures harvested in mid-exponential phase by filtration onto 47 mm, 3 μm polycarbonate membrane filters. Cells on the filters (∼4 × 108 cells) were rinsed and resuspended in buffer (10 mM Tris–HCl, 250 mM sucrose, 1% protease inhibitor cocktail, pH 7.5) and then transferred to 1.7 mL micro-centrifuge tubes. Cells were concentrated by centrifugation and the cell pellets re-suspended in 600 μL pre-chilled extraction buffer (500 mM Tris–HCl, 50 mM EDTA, 700 mM sucrose, 100 mM KCl, 40 mM dithiothreitol, 0.5% Triton X-100 and 1% protease inhibitor cocktail, pH 8.0). Resuspended cells were homogenized on ice with a Branson 250 Sonifier (30% duty cycle, output 3 and 30 s–30 s on–off cycles for a total of 8 min). After sonication, protein was extracted, precipitated and washed according to Faurobert et al.35 Purified protein was dissolved in rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris) and its concentration quantified by a modified Bradford assay (Bio-Rad, ON, Canada) with bovine serum albumin as a standard.36Two-dimensional difference gel electrophoresis (2D-DiGE)
Purified proteins were labeled with CyDye DIGE Fluor minimal dyes (GE Healthcare Life Sciences, ON, Canada) following the manufacturer's instructions. Each protein sample (50 μg) was randomly labeled with either Cy3 or Cy5; the pool of the 8 algal protein samples (2 treatments × 4 replicates) was labeled with Cy2 and used as an internal standard. The combined 150 μg CyDye-labeled proteins (50 μg each for Cy2, Cy3 and Cy5-labeled proteins) for each replicate gel and 800 μg unlabeled proteins were loaded onto 24 cm, pH 4–7 (linear) immobilized pH gradient (IPG) strips (GE Healthcare Life Sciences) with 1% DTT and 1% IPG buffer (pH 4–7) in 450 μL rehydration buffer, respectively. The IPG strips were rehydrated for 16 hours with an active rehydration mode (50 V, 20 °C), followed by isoelectric focusing (IEF) in an IPGphor IEF system (GE Healthcare Life Sciences). The first dimension, IEF, was run at 500 V for 1 hour, followed sequentially by linear increases in voltage from 500 V to 1000 V for 1 hour and 1000 V to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V for 3 hours. The gels were then kept at 10
000 V for 3 hours. The gels were then kept at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V for 3.5 hours for a total of 53
000 V for 3.5 hours for a total of 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V h. The maximum current per strip was less than 40 μA. The focused IPG strips were equilibrated in 10 mL equilibration buffer (6 M urea, 100 mM Tris, 30% glycerol, 2% SDS and 0.002% bromophenol blue) containing 5 mg mL−1 DTT followed by 10 mL equilibration buffer containing 45 mg mL−1 iodoacetamide. The equilibrated IPG strips were loaded on the top of 12.5% rhinohide polyacrylamide gel (26 × 20 cm) and sealed with 1% low melting point agarose. The second dimension, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), was run in an Ettan DALTsix electrophoresis system (GE Healthcare Life Sciences) at 10 mA per gel and 30 °C in the dark for 16 hours. The 2D-DiGE technology allows analysis of multiple protein samples labeled with different fluorescent dyes on a single polyacrylamide gel, which improves the accuracy and reproducibility of identified proteins.37 Use of the internal protein standard makes comparison of the protein fold-change ratios among biological replicates statistically more reliable.
000 V h. The maximum current per strip was less than 40 μA. The focused IPG strips were equilibrated in 10 mL equilibration buffer (6 M urea, 100 mM Tris, 30% glycerol, 2% SDS and 0.002% bromophenol blue) containing 5 mg mL−1 DTT followed by 10 mL equilibration buffer containing 45 mg mL−1 iodoacetamide. The equilibrated IPG strips were loaded on the top of 12.5% rhinohide polyacrylamide gel (26 × 20 cm) and sealed with 1% low melting point agarose. The second dimension, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), was run in an Ettan DALTsix electrophoresis system (GE Healthcare Life Sciences) at 10 mA per gel and 30 °C in the dark for 16 hours. The 2D-DiGE technology allows analysis of multiple protein samples labeled with different fluorescent dyes on a single polyacrylamide gel, which improves the accuracy and reproducibility of identified proteins.37 Use of the internal protein standard makes comparison of the protein fold-change ratios among biological replicates statistically more reliable.
Gel image analysis
Each replicate gel was imaged with a Typhoon Trio plus scanner (GE Healthcare Life Sciences) immediately after SDS-PAGE. An initial scan was run at 1000 μm pixel resolution to adjust the photomultiplier (PMT) voltage to avoid reaching saturated fluorescence density. The full scan was run at 100 μm pixel resolution with the PMT voltage of 550 V. The parallel gel containing 800 μg unlabeled proteins was fixed in 40% methanol and 10% acetic acid, and then stained using colloidal Coomassie G-250 (Bio-Rad Laboratories, UK).38 Fluorescent gel images were analyzed by a DeCyder 2D software (v.7.0, GE Healthcare Life Sciences). Protein spots were identified on each gel image in the Differential In-gel Analysis module. Protein expression levels in Cu-replete and Cu-limited cells were calculated by normalizing the spot volume against the internal standard and displayed as a volume ratio. The processed 4 replicate gel images were then aligned and matched in the Biological Variation Analysis module. Average spot fold-change ratios (f) were calculated by comparing expression levels of Cu-limited proteins to Cu-replete proteins in the 4 replicates. Differentially expressed protein spots were determined and a protein pick list was generated according to the selection criteria: protein spots were identified in all 12 gel images and the average fold-change ratio f ≥ 1.5 or f ≤ −1.5; p < 0.05 (t-test).Mass spectrometry and protein annotation
Selected protein spots were isolated from the Coomassie G-250 stained gel according to the position of the differentially expressed proteins identified on the fluorescent gel images. Isolated proteins were destained, trypsin digested and analyzed on a Q-Exactive Mass Spectrometer (ThermoFisher Scientific, ON, Canada) by liquid chromatography, electrospray ionization, tandem mass spectrometry at the Institute for Research in Immunology and Cancer, University of Montreal. Details about the peptide processing and sequencing protocol are provided in the ESI.† Peptide interpretation and protein identification were performed in Mascot server (http://www.matrixscience.com) against the NCBI non-redundant protein sequences and the contaminants database. The tolerance parameters were set to 5 ppm and 0.02 Da for the precursor (peptide) and the MS/MS fragment ions, respectively. These criteria allowed for a false-positive rate of less than 1% (p < 0.01). Carbamidomethyl was selected as the fixed modification; deamidation, oxidation and phosphorylation were selected as the variable modifications. Identified T. oceanica protein sequences that were not annotated were searched in NCBI (https://www.ncbi.nlm.nih.gov/) and KEGG (http://www.genome.jp/kegg/) databases. The KEGG pathways and functional groups of identified protein spots were defined by the best Blastp hit of annotated homologous proteins with an E-value <10−10 and sequence coverage >50%.Results and discussion
Physiological responses to Cu deficiency
Growth rate of Cu-limited T. oceanica was ∼50% slower than the Cu-replete rate (Fig. 1A) and increased rapidly when Cu was added to the medium (Fig. 2). The increase in growth followed a dose response relationship with Cu addition, confirming that Cu was the limiting resource. In Cu-replete medium containing 21.4 nM Cu, cells grew at the maximum rate.23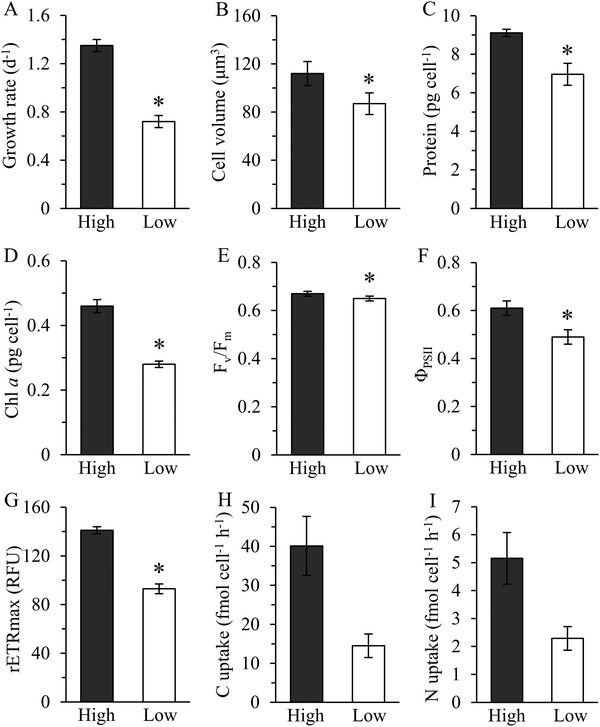 | ||
| Fig. 1 Physiological traits and composition of T. oceanica 1005 in high (Cu-replete, 21.4 nM Cu) and low (Cu-limited, 1 nM Cu) Cu medium. Growth rate (A), cell volume (B), cellular protein (C), chlorophyll a (D), maximum quantum yield of photosystem II (Fv/Fm) (E), effective quantum yield of photosystem II (ΦPSII) (F), maximum relative electron transport rate (rETRmax) (G), steady-state carbon (H) and nitrogen (I) uptake rate. Units of rETRmax are relative fluorescence units (RFU). Values reported are means ± 1 standard deviation (SD) of three to four biological replicates. An asterisk (*) indicates a significant difference between low and high Cu cultures (p < 0.01, t-test). Steady-state carbon and nitrogen uptake rates were calculated from the product of cellular quota and specific growth rate determined under the same growth conditions.6 Error bars for C and N uptake rates represent propagated errors. | ||
Copper-deplete cells contained less chlorophyll a than Cu-replete cells, but only by about 22% after accounting for differences in size (Fig. 1B and D). Similar results were obtained by Kim and Price,6 although the absolute amount of chl a they measured was lower than reported here. In addition, Kim and Price found decreases in volumetric C, N, and Cu quotas, and Cu uptake rates as a function of Cu-limited growth.6 Steady state C and N assimilation rates were roughly 60% slower in Cu-delete than in Cu-replete cells (Fig. 1H and I). Maximum quantum yield (Fv/Fm) declined minimally, as noted previously,6,9 although there was a statistically significant treatment effect (Fig. 1E). Large decreases in the maximum relative electron transport rate (rETRmax) (p < 0.01, t-test) and effective quantum yield of PSII (ΦPSII) (p < 0.01) showed photosynthetic capacity was depressed and photosynthetic electron flow inhibited (Fig. 1F and G), as reported.9 Thus, Cu limitation strongly affected photosynthetic performance of this oceanic diatom.
The physiological responses of T. oceanica 1005 reported here differ markedly from Hippmann et al.28 In that study, growth rates of the cultures were significantly different in low and high Cu medium, yet cellular carbon fixation rates were similar. Equally surprising was that effective quantum yield of PSII was unaffected by Cu limitation,28 despite the documented role of Cu in maintaining photosynthetic electron flow in this diatom.6,9 The lack of physiological changes reported by Hippmann et al. suggests to us that the cultures they examined were either not well acclimated to the treatments or were in a similar Cu-nutritional state at the time of sampling.28 This would also explain why they observed no change in the proteome of T. oceanica 1005 in high and low Cu medium. Our results show consistently that photosynthetic reactions are compromised in Cu-limited T. oceanica 1005. As described below, the decrease in photosynthetic efficiency agrees well with the decrease we observed in abundance of Cu-containing plastocyanin and other photosynthetic proteins in Cu-deplete cells.
Whole proteome analysis
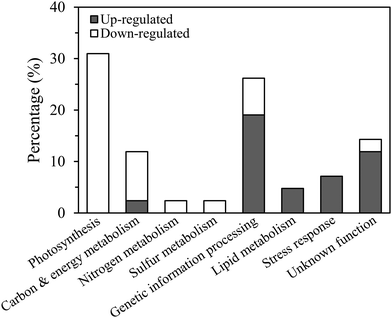
Although cellular protein content decreased by 24% in the low Cu cultures this was because the cells were smaller in size than in the high Cu cultures, as protein normalized to cell volume was identical between the treatments (p > 0.05, t-test, Fig. 1B and C). Approximately 3000 proteins were resolved by 2D-DiGE: 73 were identified as differentially expressed; 28 were up-regulated and 45 were down-regulated in Cu-limited compared to Cu-replete cells (Fig. S1, ESI†). These results were similar to other proteomic studies, which identified between 750 and 2000 proteins using non-gel based methods and reported 50 to 140 differentially expressed proteins in Fe-limited and cobalt-limited cultures compared to their respective controls.17,18,20,39Cleanly excising the proteins from the Coomassie G-250 stained gel was difficult when they were in low abundance, so only 54 highly expressed proteins were isolated and analyzed by mass spectrometry (Table S1, ESI†). Forty-two of the 54 sequenced proteins were successfully identified in the database belonging to T. oceanica (39 proteins) and T. pseudonana (3 proteins), including 23 that were down-regulated and 19 that were up-regulated (Table S2, ESI†). Functional annotation showed that most of the down-regulated proteins (83%) were associated with photosynthesis, carbon, nitrogen or sulfur metabolisms, while the majority of up-regulated proteins (68%) were involved in genetic information processing, lipid metabolism and stress response (Fig. 3).
Transcript abundance of genes encoding the identified proteins, estimated from an mRNA sequencing project conducted with T. oceanica 1005,40 showed 32 (76%) ranked within the top 10% of the most abundant mRNAs (Fig. S2, ESI†). Gene transcripts of 18 of these proteins differed significantly between the low and high Cu cells (false discovery rate (FDR) < 0.05; p < 0.01) and showed the same up- or down-regulation as the proteins (Fig. 4). There were no significant changes in transcript abundance of the remaining 24 proteins, possibly because of translational or post-translational processing.
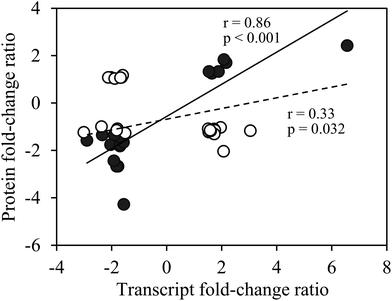 | ||
| Fig. 4 Fold-change ratios of proteins versus transcripts of differentially expressed proteins of Cu-limited T. oceanica. Protein fold-change ratios are reported in Tables 1 and 2, and transcript fold-change ratios were obtained from an RNA sequencing project on the same cultures.40 The solid circles are for transcripts that changed significantly in Cu-limited cells (false discovery rate <0.05; p < 0.01), whereas the open circles are for transcripts that showed no significant change in abundance. A significant correlation between protein and transcript fold-change ratios was observed for genes that were significantly regulated by Cu at both the protein and transcript levels (solid line, Pearson's r = 0.86, p < 0.001); a weak correlation was observed for the whole dataset (solid and open circles, dashed line, Pearson's r = 0.33, p = 0.032). | ||
Decrease in cuproenzymes
A major and immediate consequence of Cu deficiency was reduction in cellular Cu quota6 and a decrease in PC and other Cu-regulated proteins (Table 1). Plastocyanin, the most abundant Cu-containing protein in T. oceanica 1005,9 declined 3-fold when Cu was limiting (Table 1). The decrease in PC corresponded to a decrease in Cu quota of about 2 μmol Cu L−1 cell volume (from 3 in Cu-replete to 1 μmol Cu L−1 cell volume in Cu-limited cells),6 and was consistent with the observed changes in physiological traits showing restricted photosynthetic electron flow and reduced growth rate. Hippmann et al. observed a similar reduction in PC in T. oceanica 1003, but not in 1005,28 possibly for the reasons discussed above. Under Cu-limitation, about 70% of the cellular Cu was bound in PC,6 as non-essential (non-metabolic) pools were reduced.| Metabolic pathways | KEGG pathway id | Protein annotation | Spot Id. | Protein fold change |
|---|---|---|---|---|
| Photosynthetic metabolism | ||||
| Photosynthesis | ko00195 | Plastocyanin | 29 | −3.01 |
| Chlorophyll metabolism | ko00860 | Geranylgeranyl reductase | 24 | −2.03 |
| Chlorophyll metabolism | ko00860 | Geranylgeranyl reductase | 25 | −2.12 |
| Chlorophyll metabolism | ko00860 | Geranylgeranyl reductase | 36 | −1.64 |
| Photosynthesis | ko00195 | Oxygen-evolving enhancer protein 3 | 49 | −1.90 |
| Photosynthesis | ko00196 | Fucoxanthin chlorophyll a/c protein | 28 | −2.04 |
| Photosynthesis | ko00196 | Fucoxanthin chlorophyll a/c protein | 71 | −1.79 |
| Photosynthesis | ko00196 | Fucoxanthin chlorophyll a/c protein | 50 | −1.72 |
| Photosynthesis | ko00196 | Fucoxanthin chlorophyll a/c protein | 44 | −1.69 |
| Photosynthesis | ko00196 | Fucoxanthin chlorophyll a/c protein | 45 | −1.57 |
| Photosynthesis | ko00196 | Fucoxanthin chlorophyll a/c protein | 41 | −1.51 |
| Photosynthesis | Thylakoid lumenal 15 kDa protein 1 | 46 | −1.60 | |
| Photosynthesis | ko00195 | ATP synthase CF1 beta chain | 35 | −1.56 |
| Carbon & energy metabolism | ||||
| Glycolysis | ko01200 | Glyceraldehyde-3-phosphate dehydrogenase | 26 | −2.90 |
| Oxidative phosphorylation | ko00190 | Cytochrome c oxidase subunit 6b | 32 | −2.37 |
| Carbon fixation | ko00710 | RuBisCO large chain | 59 | −1.77 |
| Carbon fixation | ko00710 | RuBisCO small chain | 52 | −1.82 |
| Nitrogen metabolism | ||||
| Nitrate assimilation reduction | ko00910 | Nitrite reductase | 34 | −1.89 |
| Sulfur metabolism | ||||
| Sulfate assimilation | ko00920 | Phosphoadenosine-phosphosulphate reductase | 62 | −1.92 |
| Genetic information processing | ||||
| RNA transport | ko03013 | Small ubiquitin-like modifier | 31 | −2.34 |
| Translation factors | ko03012 | Translation factor to domain 2 | 53 | −1.82 |
| Transfer RNA biogenesis | ko03016 | Pseudouridylate synthase | 65 | −1.70 |
| Unknown metabolism | ||||
| Unknown function | Hypothetical protein THAOC_25588 | 39 | −1.68 | |
In cyanobacteria and green algae, Cu-containing PC and Fe-containing CYC6 are used interchangeably to shuttle electrons between cytochrome b6f and PSI. Restricting Cu supply causes a decrease in the amount of PC and an increase in CYC6 to maintain electron transport and photosynthesis.25,41 This Cu sparing pathway facilitates re-distribution of intracellular Cu from PC to other Cu-dependent proteins, such as cytochrome c oxidase and ferroxidase.4Thalassiosira oceanica 1005, however, appears to lack a functional CYC6 to replace PC9,42 and so, consequently, photosynthetic electron flow was impaired once levels of PC decreased. BLAST search against the T. oceanica 1005 genome identified two copies of CYC6 genes, but their expression levels were much lower than PC and unresponsive to Cu limitation in the presence of high Fe, suggesting that CYC6 genes in this diatom were non-functional (Fig. S3, ESI†).
Other important cuproenzymes were reduced at low Cu as well, including the small subunit 6b of cytochrome c oxidase (COX6B, spot 32), which declined by 2.4-fold. A ferroxidase-like MCO was also identified among down-regulated proteins (spot 63), but traces of contaminating proteins were detected in this sample which may have confounded our estimates of MCO abundance. Copper, Zn-superoxide dismutase was not encoded in the T. oceanica genome.30 The nitrite reductase identified in spot 34 was a ferredoxin-dependent enzyme. Tyrosinase and amino oxidase were not among the differentially expressed proteins we identified.
Down-regulation of photosynthesis
The Cu-induced decrease in photosynthetic electron transport rate induced compensatory adjustments to other components of the photosynthetic apparatus. Indeed, more than 65% of down-regulated proteins we identified were predicted to function in photosynthesis, including light harvesting, electron transport and carbon fixation (Table 1). Among them, a geranylgeranyl reductase (ChlP) involved in chlorophyll and tocopherol biosynthesis decreased by 1.6 to 2.0-fold (spots 24, 25, 36). Previous work on ChlP in algae and higher plants showed that reduced activity caused a decrease in cellular chlorophyll,43–45 consistent with our results for T. oceanica (Fig. 1D).Six fucoxanthin chlorophyll a/c-binding proteins (FCPs) were also down-regulated in Cu-limited cells. The FCPs are associated with PSII and PSI and involved in light harvesting and energy transfer and dissipation.46–48 In T. oceanica 1003, 17 FCPs were differentially regulated by Cu.28 Five of the 6 (spots 28, 44, 45, 50, 71) identified here belong to the Lhcf protein family that functions primarily in light harvesting.28 The remaining FCP (spot 41) was in the Lhcx family that comprises proteins involved in stress responses, like non-photochemical quenching.28,48,49 Two of the FCPs that decreased in T. oceanica 1005 under Cu-limitation (THAOC_08095 and 05777) (Table 1 and Table S1, ESI†) actually increased at low Cu in T. oceanica 1003.28 In addition, an oxygen evolving enhancer protein 3 (PsbQ) was down-regulated by 1.9-fold (spot 49). PsbQ is one of three extrinsic subunits of the PSII oxygen evolving complex that increases the binding affinity of the Cl− and Ca2+ cofactors and stabilizes the manganese cluster of PSII.50–52 Hippmann et al. observed a similar decline in PsbQ, as well as other subunits of PSII, in T. oceanica 1003 in response to low Cu.28
A chloroplastic ATP synthase CF1 beta subunit (AtpB) decreased by 1.6-fold (spot 35). AtpB is one of five subunits that constitutes the catalytic, F1 portion (CF1) of ATP synthase and contains catalytic sites that are primarily responsible for ATP synthesis.53,54 In AtpB-defective mutants of Chlamydomonas, α, γ and β subunits of CF1 detached from the thylakoid membranes, so assembly and localization of CF1 was disrupted.55 Collectively, the results show that down-regulation of PC in the chloroplast led to inhibited electron transport to PSI and eventually the supply of reducing equivalents, ferredoxin and NADPH, and ATP synthesis. Upstream processes of light energy capture and water splitting were coincidentally reduced, presumably to avoid excess electron flow that could lead to photodamage of PSII and generate reactive oxygen species.56,57
Down-regulation of carbon and nitrogen assimilation
Diminished supply of reducing power and energy caused by down-regulation of PC and AtpB in the chloroplast may subsequently affect cellular biochemical reactions that are energy and reducing power-dependent. Indeed, as illustrated in Fig. 1, C and N assimilation rates declined substantially in the low Cu cultures. These metabolic changes were consistent with decreased levels of the large and small subunits of RuBisCO, and chloroplastic ferredoxin-dependent nitrite reductase (NiR) (spot 34) (Table 1). Down-regulation of nitrate assimilation is typical in Fe-limited diatoms because nitrate and nitrite reductases contain Fe and require reducing equivalents.15,58 Under Cu-limitation, reduced NiR activity may be associated with a decreased photosynthetic rate and production of reduced ferredoxin. We note that the C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratio of T. oceanica decreased by about 20% as Cu concentration declined,6 suggesting assimilation rates of carbon may have been more compromised than N, although both rates remained relatively well balanced. The simultaneous decrease in RuBisCO and NiR expression reflects the tight coupling between C and N metabolisms and is likely a response to slower rates of photosynthetic electron transport caused by reduced PC.
N ratio of T. oceanica decreased by about 20% as Cu concentration declined,6 suggesting assimilation rates of carbon may have been more compromised than N, although both rates remained relatively well balanced. The simultaneous decrease in RuBisCO and NiR expression reflects the tight coupling between C and N metabolisms and is likely a response to slower rates of photosynthetic electron transport caused by reduced PC.
Up-regulation of fatty acid beta-oxidation
Two proteins involved in fatty acid metabolism, long-chain fatty acid CoA synthetase (LC-FACS) (spot 55) and acyl-CoA dehydrogenase (ACAD) (spot 3), were strongly up-regulated in Cu-limited cells (Table 2). These enzymes degrade free fatty acids to acyl-CoA thioester in the cytosol and catalyze the first step of fatty acid beta-oxidation (FAO) in the mitochondria.59 We hypothesize that up-regulation of FAO in Cu-deficient cells may be a result of increased oxidative stress, leading to peroxidation of lipids and damage to cell membranes.60–62 Although proteins involved in scavenging reactive oxygen species produced by oxidative stress were not detected in our analysis, they increased in Fe-deficient algae,15 primarily in response to suppressed photosynthesis and excessive light energy.63–65 We note that gene transcripts of ascorbate peroxidase, glutathione peroxidase and thioredoxin were strongly up-regulated in Cu-limited T. oceanica,40 consistent with enhanced ROS production. Copper-limited cells may thus increase fatty acid oxidation and synthesis to allow rapid turnover of lipids to maintain membrane integrity. Although remodeling of chloroplast membranes in Cu-deplete algae could account for upregulation of the FAO pathway under transient conditions,16 it would unlikely be responsible once acclimation to low Cu was achieved.| Metabolic pathways | KEGG pathway id | Protein annotation | Spot Id. | Protein fold change |
|---|---|---|---|---|
| Lipid metabolism | ||||
| Fatty acid metabolism | ko01212 | Acyl-CoA dehydrogenase | 3 | 2.16 |
| Fatty acid metabolism | ko01212 | Long-chain fatty acid CoA synthetase | 55 | 2.08 |
| Stress response | ||||
| Ca2+ dependent protein regulation | C2 domain containing protein | 2 | 6.56 | |
| Photosynthesis | ko00195 | Cytochrome c550 | 1 | 2.07 |
| mRNA biogenesis | Transcription factor BTF3-like protein | 23 | 1.53 | |
| Carbon & energy metabolism | ||||
| Oxidative phosphorylation | ko00190 | NADH dehydrogenase (ubiquinone) Fe–S protein 4 | 19 | 1.51 |
| Genetic information processing | ||||
| mRNA surveillance pathway | ko03015 | Polyadenylate binding protein | 58 | 3.04 |
| mRNA biogenesis | ko03019 | Regulator of chromosome condensation | 11 | 1.96 |
| Aminoacyl-tRNA biosynthesis | ko00970 | Threonyl-tRNA synthetase | 7 | 1.88 |
| Membrane trafficking | ko04131 | Ypt1-like rab-type small G protein | 22 | 1.72 |
| RNA transport | ko03013 | Eukaryotic translation initiation factor | 72 | 1.64 |
| Aminoacyl-tRNA biosynthesis | ko00970 | Threonyl-tRNA synthetase | 5 | 1.54 |
| Biosynthesis of amino acids | ko01230 | Arginine biosynthesis bifunctional protein | 21 | 1.64 |
| Amino acid metabolism | ko01002 | X-prolyl aminopeptidase | 54 | 1.59 |
| Unknown metabolism | ||||
| Unknown function | Hypothetical protein THAOC_29771 | 9 | 1.75 | |
| Unknown function | Hypothetical protein THAOC_03961 | 20 | 1.73 | |
| Unknown function | Hypothetical protein THAOC_33694 | 18 | 1.60 | |
| Unknown function | Hypothetical protein THAOC_29771 | 8 | 1.58 | |
| Unknown function | Hypothetical protein THAOC_29771 | 10 | 1.58 | |
FAO also provides an additional source of acetyl-CoA and reducing equivalents for the citric acid cycle and respiratory electron transport chain. These substrates are typically produced by glycolysis, in the form of pyruvate and NADH, and transported into the mitochondria as required although part of the glycolysis pathway occurs in the mitochondria in stramenophiles.66,67 In Cu-depleted cells, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was down-regulated (spot 26), suggesting glycolysis was suppressed and the supply of pyruvate and NADH limited. Up-regulation of FAO may thus partially compensate for a shortage of these metabolites.
Stress response
Cytochrome c550 (PsbV) (spot 1) was the only differentially expressed photosynthetic protein that was upregulated in response to low Cu. PsbV is an extrinsic subunit of the PSII oxygen evolving complex that plays a role in stabilizing Cl− and Ca2+ and is required for efficient water-splitting.68 The physiological role of its covalently-bound heme is not completely understood although some observations suggest it may be involved in redox reactions during anaerobic catabolism of carbohydrate during periods of reduced photosynthesis69 or possibly electron transport on the donor-side of PSII.70,71 Up-regulation of PsbV in Cu-limited T. oceanica could function as a protective strategy, reducing production of reactive oxygen species by removing excess electrons created by the bottleneck in photosynthetic electron flow.68An unidentified, C2 domain-containing protein was most highly up-regulated in Cu-limited cells (by 6.6-fold, spot 2). C2 domains are calcium dependent, lipid binding regions (CaLB) that target proteins to the cell membrane.72 They are widespread among eukaryotic proteins and have a variety of biochemical functions, mostly in signaling pathways, such as protein kinases, GTPase-activating proteins and synaptotagmin-mediated vesicular transport.73 Stress-induced CaLBs identified in plants regulate gene expression to increase tolerance to unfavourable abiotic factors.74–76 The function of CaLB identified in this study is currently unknown, but its dramatic up-regulation and Ca2+ binding motif suggests it may be involved in signal transduction pathways as a Cu sensor.
The low Cu response of T. oceanica included up-regulation of a basic transcriptional factor 3 (BTF3)-like protein (spot 23). BTF3 is known to bind to RNA polymerase II as a co-activator and is involved in RNA polymerase II mediated transcription.77,78 In yeast, homologues of BTF3 act as negative regulators of gene transcription.79 The BTF3 in T. oceanica contained a unique 30 amino acid peptide insertion at the C-terminus that was absent from human BTF3 and related phytoplankton proteins (Fig. 5). The peptide insert contained three tandem MpgMgg(g)M motifs that bear a striking resemblance to the Cu(I) binding motifs of high-affinity CTR-type Cu(I) transporters.23 Such arrangement of the methionine residues is known to selectively bind Cu(I) in a stable trigonal coordination involving three methionine residues.80,81 If ToBTF3 is a functional transcriptional factor, then its regulatory effect on gene expression could be Cu-dependent.
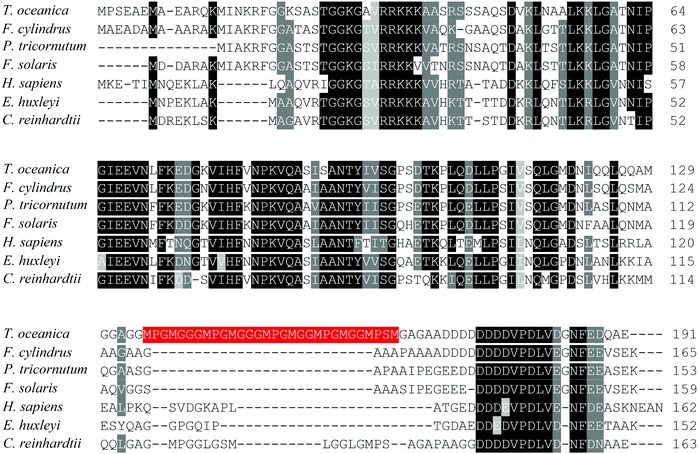 | ||
| Fig. 5 Alignment of BTF3-like transcription factors of T. oceanica and other species. The data set includes protein sequences of T. oceanica (EJK74743), Chlamydomonas reinhardtii (EDO98665), Emiliania huxleyi (EOD08611), Fistulifera solaris (GAX23608), Fragilariopsis cylindrus (OEU15333), Homo sapiens (NP_001198) and Phaeodactylum tricornutum (EEC50944) aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).82 Identical or functionally similar amino acids are shaded in black or grey using Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/color_align_cons.html).83 The 30 amino acid peptide of T. oceanica BTF3 containing tandem methionine motifs is labeled in red. | ||
Conclusions
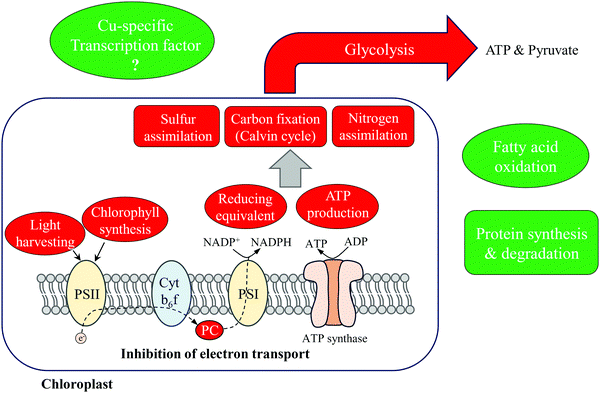
The results reported here allow us to describe the primary metabolic adjustments of T. oceanica 1005 to Cu deficiency (Fig. 6). More than 40% of the differentially expressed proteins identified were involved in photosynthesis and carbon fixation, and down-regulated under Cu limitation. A 3-fold decrease in Cu-containing PC inhibited photosynthetic electron transport and induced compensatory changes to the photosynthetic apparatus. Light harvesting capacity was reduced (down-regulation of FCPs and ChlP) to accommodate the bottleneck in electron flow and avoid photodamage to PSII. Consequently, production of ATP and reducing equivalents was inhibited. The dramatic down-regulation of photosynthetic metabolism further influenced metabolic pathways that were energy or reducing equivalent-dependent, including carbon, nitrogen and sulfur assimilation. Fatty acid oxidation was up-regulated in mitochondria, suggesting increased cycling of lipids and fatty acids, possibly in response to increased membrane damage due to oxidative stress. We also note that increased FAO would provide substrates and reducing equivalents for cellular respiration that might offset inhibited glycolysis.Two up-regulated, stress-responsive proteins were hypothesized to function in Cu sensing and signal transduction in T. oceanica. A BTF3-like transcription factor was identified containing Mets motifs that may be involved in Cu-binding. Its specific function in regulating gene expression is unknown and requires further study. About 20% of differentially expressed proteins were involved in genetic information processing pathways, such as mRNA and amino acid biosynthesis and protein processing. These up-regulated processes may be required by Cu-limited cells to accommodate faster rates of protein turnover induced by oxidative stress60,61 and required for Cu acquisition and homeostasis. Currently, we are analyzing the results of an mRNA sequencing project to obtain a more comprehensive understanding of how T. oceanica 1005 acclimates to Cu deficiency.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Stephanie Shousha for collecting the data of Fig. S3 (ESI†). Funding for this study was provided by the Natural Sciences and Engineering Research Council of Canada.References
- J. L. Burkhead, K. A. Gogolin Reynolds, S. E. Abdel-Ghany, C. M. Cohu and M. Pilon, Copper homeostasis, New Phytol., 2009, 182, 799–816 CrossRef CAS PubMed.
- P. G. Ridge, Y. Zhang and V. N. Gladyshev, Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen, PLoS One, 2008, 3, e1378, DOI:10.1371/journal.pone.0001378.
- C. Cobbett and P. Goldsbrough, Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis, Annu. Rev. Plant Biol., 2002, 53, 159–182 CrossRef CAS PubMed.
- J. Kropat, S. D. Gallaher, E. I. Urzica, S. S. Nakamoto, D. Strenkert and S. Tottey, et al., Copper economy in Chlamydomonas: Prioritized allocation and reallocation of copper to respiration vs. photosynthesis, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2644–2651 CrossRef CAS PubMed.
- T. V. O'Halloran and V. C. Culotta, Metallochaperones: an intracellular shuttle service for metal ions, J. Biol. Chem., 2000, 275, 25057–25060 CrossRef PubMed.
- J. W. Kim and N. M. Price, The influence of light on copper-limited growth of an oceanic diatom, Thalassiosira oceanica (Coscinodiscophyceae), J. Phycol., 2017, 53, 938–950 CrossRef CAS PubMed.
- J. A. Raven, The iron and molybdenum use efficiencies of plant growth with different energy, carbon and nitrogen sources, New Phytol., 1988, 109, 279–287 CrossRef CAS.
- S. S. Merchant, M. D. Allen, J. Kropat, J. L. Moseley, J. C. Long and S. Tottey, et al., Between a rock and a hard place: trace element nutrition in Chlamydomonas, Biochim. Biophys. Acta, 2006, 1763, 578–594 CrossRef CAS PubMed.
- G. Peers and N. M. Price, Copper-containing plastocyanin used for electron transport by an oceanic diatom, Nature, 2006, 441, 341–344 CrossRef CAS PubMed.
- M. T. Maldonado, A. E. Allen, J. S. Chong, K. Lin, D. Leus and N. Karpenko, et al., Copper-dependent iron transport in coastal and oceanic diatoms, Limnol. Oceanogr., 2006, 51, 1729–1743 CrossRef CAS.
- R. A. Festa and D. J. Thiele, Copper: an essential metal in biology, Curr. Biol., 2011, 21, R877–R883 CrossRef CAS PubMed.
- K. H. Coale, Effects of iron, manganese, copper, and zinc enrichments on productivity and biomass in the subarctic Pacific, Limnol. Oceanogr., 1991, 36, 1851–1864 CrossRef CAS.
- G. Peers, S. A. Quesnel and N. M. Price, Copper requirements for iron acquisition and growth of coastal and oceanic diatoms, Limnol. Oceanogr., 2005, 50, 1149–1158 CrossRef CAS.
- A. L. Annett, S. Lapi, T. J. Ruth and M. T. Maldonado, The effects of Cu and Fe availability on the growth and Cu:C ratios of marine diatoms, Limnol. Oceanogr., 2008, 53, 2451–2461 CrossRef CAS.
- A. E. Allen, J. LaRoche, U. Maheswari, M. Lommer, N. Schauer and P. J. Lopez, et al., Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10438–10443 CrossRef CAS PubMed.
- M. Castruita, D. Casero, S. J. Karpowicz, J. Kropat, A. Vieler and S. I. Hsieh, et al., Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps, Plant Cell, 2011, 23, 1273–1292 CrossRef CAS PubMed.
- N. R. Cohen, W. Gong, D. M. Moran, M. R. McIlvin, M. A. Saito and A. Marchetti, Transcriptomic and proteomic responses of the oceanic diatom Pseudo-nitzschia granii to iron limitation, Environ. Microbiol., 2018, 20, 3109–3126 CrossRef CAS PubMed.
- B. L. Nunn, J. F. Faux, A. A. Hippmann, M. T. Maldonado, H. R. Harvey and D. R. Goodlett, et al., Diatom proteomics reveals unique acclimation strategies to mitigate Fe limitation, PLoS One, 2013, 8, e75653, DOI:10.1371/journal.pone.0075653.
- I. F. Scheiber, J. Pilatova, R. Malych, E. Kotabova, M. Krijt and D. Vyoral, et al., Copper and iron metabolism in Ostreococcus tauri – the role of phytotransferrin, plastocyanin and a chloroplast copper-transporting ATPase, Metallomics, 2019, 11, 1657–1666 RSC.
- N. G. Walworth, F.-X. Fu, E. A. Webb, M. A. Saito, D. Moran and M. R. McIlvin, et al., Mechanisms of increased Trichodesmium fitness under iron and phosphorus co-limitation in the present and future ocean, Nat. Commun., 2016, 7, 12081, DOI:10.1038/ncomms12081.
- A. Dancis, D. S. Yuan, D. Haile, C. Askwith, D. Eide and C. Moehle, et al., Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport, Cell, 1994, 76, 393–402 CrossRef CAS PubMed.
- E. Georgatsou, L. A. Mavrogiannis, G. S. Fragiadakis and D. Alexandraki, The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator, J. Biol. Chem., 1997, 272, 13786–13792 CrossRef CAS PubMed.
- L. Kong and N. M. Price, Functional CTR-type Cu (I) transporters in an oceanic diatom, Environ. Microbiol., 2019, 21, 98–110 CrossRef CAS PubMed.
- L. Kong and N. M. Price, A reduction-dependent copper uptake pathway in an oceanic diatom, Limnol. Oceanogr., 2020, 65, 601–611, DOI:10.1002/lno.11329.
- S. Merchant and L. Bogorad, Metal ion regulated gene expression: use of a plastocyanin-less mutant of Chlamydomonas reinhardtii to study the Cu(II)-dependent expression of cytochrome c-552, EMBO J., 1987, 6, 2531–2535 CrossRef CAS PubMed.
- J. M. Quinn and S. Merchant, Copper-responsive gene expression during adaptation to copper deficiency, Methods Enzymol., 1998, 297, 263–279 CAS.
- J. M. Quinn, S. S. Nakamoto and S. Merchant, Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript, J. Biol. Chem., 1999, 274, 14444–14454 CrossRef CAS PubMed.
- A. A. Hippmann, N. Schuback, K.-M. Moon, J. P. McCrow, A. E. Allen and L. J. Foster, et al., Contrasting effects of copper limitation on the photosynthetic apparatus in two strains of the open ocean diatom Thalassiosira oceanica, PLoS One, 2017, 12, e0181753, DOI:10.1371/journal.pone.0181753.
- S. I. Hsieh, M. Castruita, D. Malasarn, E. Urzica, J. Erde and M. D. Page, et al., The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii, Mol. Cell. Proteomics, 2013, 12, 65–86 CrossRef PubMed.
- M. Lommer, M. Specht, A.-S. Roy, L. Kraemer, R. Andreson and M. A. Gutowska, et al., Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation, Genome Biol., 2012, 13, R66, DOI:10.1186/gb-2012-13-7-r66.
- F. M. M. Morel, J. G. Rueter, D. M. Anderson and R. R. L. Guillard, Aquil: A chemically defined phytoplankton culture medium for trace metal studies, J. Phycol., 1979, 15, 135–141 CrossRef CAS.
- N. M. Price, G. I. Harrison, J. G. Hering, R. J. Hudson, P. M. V. Nirel and B. Palenik, et al., Preparation and chemistry of artificial algal culture medium Aquil, Biol. Oceanogr., 1989, 6, 443–461 Search PubMed.
- H. Hillebrand, C.-D. Durselen, D. Kirschtel, U. Pollingher and T. Zohary, Biovolume calculation for pelagic and benthic microalgae, J. Phycol., 1999, 35, 403–424 CrossRef.
- T. R. Parsons, Y. Maita and C. M. Lalli, A Manual of Chemical and Biological Methods for Seawater Analysis, Pergamon Press, New York, 1984, pp. 101–104 Search PubMed.
- M. Faurobert, E. Pelpoir and J. Chaïb, Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues, Methods Mol. Biol., 2007, 355, 9–14 CAS.
- L. S. Ramagli and L. V. Rodriguez, Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer, Electrophoresis, 1985, 6, 559–563 CrossRef CAS.
- A. Alban, S. O. David, L. Bjorkesten, C. Andersson, E. Sloge and S. Lewis, et al., A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard, Proteomics, 2003, 3, 36–44 CrossRef CAS PubMed.
- G. Candiano, M. Bruschi, L. Musante, L. Santucci, G. M. Ghiggeri and B. Carnemolla, et al., Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis, Electrophoresis, 2004, 25, 1327–1333 CrossRef CAS PubMed.
- E. M. Bertrand, A. E. Allen, C. L. Dupont, T. M. Norden-Krichmar, J. Bai and R. E. Valas, et al., Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1762–E1771 CrossRef CAS PubMed.
- L. Kong, Molecular and physiological responses of an oceanic diatom to copper deficiency, PhD thesis, McGill University, 2019.
- G. Sandmann and P. Böger, Copper-induced exchange of plastocyanin and cytochrome c-533 in cultures of Anabaena variabilis and Plectonema boryanum, Plant Sci. Lett., 1980, 17, 417–424 CrossRef CAS.
- R. F. Strzepek and P. J. Harrison, Photosynthetic architecture differs in coastal and oceanic diatoms, Nature, 2004, 431, 689–692 CrossRef CAS PubMed.
- A. V. Shpilyov, V. V. Zinchenko, S. V. Shestakov, B. Grimm and H. Lokstein, Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803, Biochim. Biophys. Acta, Bioenerg., 2005, 1706, 195–203 CrossRef CAS PubMed.
- R. Tanaka, U. Oster, E. Kruse, W. Rüdiger and B. Grimm, Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase, Plant Physiol., 1999, 120, 695–704 CrossRef CAS PubMed.
- Y. Zhou, Z. Gong, Z. Yang, Y. Yuan, J. Zhu and M. Wang, et al., Mutation of the light-induced yellow leaf 1 gene, which encodes a geranylgeranyl reductase, affects chlorophyll biosynthesis and light sensitivity in rice, PLoS One, 2013, 8, e75299, DOI:10.1371/journal.pone.0075299.
- D. Bhaya and A. R. Grossman, Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum, Nucleic Acids Res., 1993, 21, 4458–4466 CrossRef CAS PubMed.
- I. Grouneva, A. Rokka and E.-M. Aro, The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery, J. Proteome Res., 2011, 10, 5338–5353 CrossRef CAS PubMed.
- R. Nagao, M. Yokono, A. Teshigahara, S. Akimoto and T. Tomo, Light-harvesting ability of the fucoxanthin chlorophyll a/c-binding protein associated with photosystem II from the diatom Chaetoceros gracilis as revealed by picosecond time-resolved fluorescence spectroscopy, J. Phys. Chem. B, 2014, 118, 5093–5100 CrossRef CAS PubMed.
- T. Mock, R. P. Otillar, J. Strauss, M. McMullan, P. Paajanen and J. Schmutz, et al., Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus, Nature, 2017, 541, 536–540 CrossRef CAS PubMed.
- V. Calderone, M. Trabucco, A. Vujičić, R. Battistutta, G. M. Giacometti and F. Andreucci, et al., Crystal structure of the PsbQ protein of photosystem II from higher plants, EMBO Rep., 2003, 4, 900–905 CrossRef CAS PubMed.
- A. Seidler, The extrinsic polypeptides of photosystem II, Biochim. Biophys. Acta, Bioenerg., 1996, 1277, 35–60 CrossRef.
- L. E. Thornton, H. Ohkawa, J. L. Roose, Y. Kashino, N. Keren and H. B. Pakrasi, Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803, Plant Cell, 2004, 16, 2164–2175 CrossRef CAS PubMed.
- P. D. Boyer, The ATP synthase—a splendid molecular machine, Annu. Rev. Biochem., 1997, 66, 717–749 CrossRef CAS PubMed.
- G. Groth and H. Strotmann, New results about structure, function and regulation of the chloroplast ATP synthase (CF0CF1), Physiol. Plant., 1999, 106, 142–148 CrossRef CAS.
- D. Robertson, J. P. Woessner, N. W. Gillham and J. E. Boynton, Molecular characterization of two point mutants in the chloroplast atpB gene of the green alga Chlamydomonas reinhardtii defective in assembly of the ATP synthase complex, J. Biol. Chem., 1989, 264, 2331–2337 CAS.
- J. Barber and B. Andersson, Too much of a good thing: light can be bad for photosynthesis, Trends Biochem. Sci., 1992, 17, 61–66 CrossRef CAS PubMed.
- P. Müller, X.-P. Li and K. K. Niyogi, Non-photochemical quenching. A response to excess light energy, Plant Physiol., 2001, 125, 1558–1566 CrossRef PubMed.
- N. M. Price, L. F. Andersen and F. M. M. Morel, Iron and nitrogen nutrition of equatorial Pacific plankton, Deep Sea Res., Part I, 1991, 38, 1361–1378 CrossRef.
- S. M. Houten and R. J. Wanders, A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation, J. Inherited Metab. Dis., 2010, 33, 469–477 CrossRef CAS PubMed.
- N. A. Anjum, A. Sofo, A. Scopa, A. Roychoudhury, S. S. Gill and M. Iqbal, et al., Lipids and proteins—major targets of oxidative modifications in abiotic stressed plants, Environ. Sci. Pollut. Res., 2015, 22, 4099–4121 CrossRef CAS PubMed.
- P. Sharma, A. B. Jha, R. S. Dubey and M. Pessarakli, Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions, J. Bot., 2012, 217037, DOI:10.1155/2012/217037.
- S. C. Singh, R. P. Sinha and D.-P. Hader, Role of lipids and fatty acids in stress tolerance in cyanobacteria, Acta Protozool., 2002, 41, 297–308 CAS.
- K. K. Niyogi, Photoprotection revisited: genetic and molecular approaches, Annu. Rev. Plant Biol., 1999, 50, 333–359 CrossRef CAS PubMed.
- P. Pospíšil, Production of reactive oxygen species by photosystem II, Biochim. Biophys. Acta, Bioenerg., 2009, 1787, 1151–1160 CrossRef PubMed.
- B. C. Tripathy and R. Oelmüller, Reactive oxygen species generation and signaling in plants, Plant Signaling Behav., 2012, 7, 1621–1633 CrossRef CAS PubMed.
- D. L. Nelson and M. M. Cox, Lehninger Principles of Biochemistry, 7th edn, Macmillan Publishers, New York, 2017, p. 1312 Search PubMed.
- C. Río Bártulos, M. B. Rogers, T. A. Williams, E. Gentekaki, H. Brinkmann and R. Cerff, et al., Mitochondrial glycolysis in a major lineage of eukaryotes, Genome Biol. Evol., 2018, 10, 2310–2325 CrossRef PubMed.
- J. L. Roose, K. M. Wegener and H. B. Pakrasi, The extrinsic proteins of photosystem II, Photosynth. Res., 2007, 92, 369–387 CrossRef CAS PubMed.
- D. Krogmann and S. Smith, Low potential cytochrome c 550 function in cyanobacteria and algae, in Current Research in Photosynthesis, ed. M. Baltscheffsky, Springer, Dordrecht, 1990, pp. 1647–1650 Search PubMed.
- F. Guerrero, A. Sedoud, D. Kirilovsky, A. W. Rutherford, J. M. Ortega and M. Roncel, A high redox potential form of cytochrome c550 in photosystem II from Thermosynechococcus elongatus, J. Biol. Chem., 2011, 286, 5985–5994 CrossRef CAS PubMed.
- M. Roncel, D. Kirilovsky, F. Guerrero, A. Serrano and J. M. Ortega, Photosynthetic cytochrome c550, Biochim. Biophys. Acta, Bioenerg., 2012, 1817, 1152–1163 CrossRef CAS PubMed.
- J. Rizo and T. C. Südhof, C2-domains, structure and function of a universal Ca2+-binding domain, J. Biol. Chem., 1998, 273, 15879–15882 CrossRef CAS PubMed.
- E. A. Nalefski and J. J. Falke, The C2 domain calcium-binding motif: structural and functional diversity, Protein Sci., 1996, 5, 2375–2390 CrossRef CAS PubMed.
- K. de Silva, B. Laska, C. Brown, H. W. Sederoff and M. Khodakovskaya, Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): a novel repressor of abiotic stress response, J. Exp. Bot., 2011, 62, 2679–2689 CrossRef CAS PubMed.
- A. Ouelhadj, P. Kuschk and K. Humbeck, Heavy metal stress and leaf senescence induce the barley gene HvC2d1 encoding a calcium-dependent novel C2 domain-like protein, New Phytol., 2006, 170, 261–273 CrossRef CAS PubMed.
- N. Yokotani, T. Ichikawa, Y. Kondou, S. Maeda, M. Iwabuchi and M. Mori, et al., Overexpression of a rice gene encoding a small C2 domain protein OsSMCP1 increases tolerance to abiotic and biotic stresses in transgenic Arabidopsis, Plant Mol. Biol., 2009, 71, 391, DOI:10.1007/s11103-009-9530-x.
- M. Kanno, C. Chalut and J.-M. Egly, Genomic structure of the putative BTF3 transcription factor, Gene, 1992, 117, 219–228 CrossRef CAS PubMed.
- X.-M. Zheng, V. Moncollin, J.-M. Egly and P. Chambon, A general transcription factor forms a stable complex with RNA polymerase B (II), Cell, 1987, 50, 361–368 CrossRef CAS PubMed.
- G.-Z. Hu and H. Ronne, Yeast BTF3 protein is encoded by duplicated genes and inhibits the expression of some genes in vivo, Nucleic Acids Res., 1994, 22, 2740–2743 CrossRef CAS PubMed.
- J. Jiang, I. A. Nadas, M. A. Kim and K. J. Franz, A Mets motif peptide found in copper transport proteins selectively binds Cu(I) with methionine-only coordination, Inorg. Chem., 2005, 44, 9787–9794 CrossRef CAS PubMed.
- J. T. Rubino and K. J. Franz, Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function, J. Inorg. Biochem., 2012, 107, 129–143 CrossRef CAS PubMed.
- F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus and W. Li, et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol., 2011, 7, 539, DOI:10.1038/msb.2011.75.
- P. Stothard, The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences, Biotechniques, 2000, 28, 1102–1104 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0mt00033g |
| This journal is © The Royal Society of Chemistry 2020 |