Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Investigation of the impact of magnesium versus titanium implants on protein composition in osteoblast by label free quantification†
M.
Omidi
a,
N.
Ahmad Agha
b,
A.
Müller
b,
F.
Feyerabend
 b,
H.
Helmholz
b,
H.
Helmholz
 b,
R.
Willumeit-Römer
b,
H.
Schlüter
a and
B. J. C.
Luthringer-Feyerabend
b,
R.
Willumeit-Römer
b,
H.
Schlüter
a and
B. J. C.
Luthringer-Feyerabend
 *b
*b
aInstitute of Clinical Chemistry and Laboratory Medicine, University Medical Centre Hamburg-Eppendorf, 20246 Hamburg, Germany
bInstitute of Materials Research, Division for Metallic Biomaterials, Helmholtz-Zentrum Geesthacht (HZG), 21502 Geesthacht, Germany. E-mail: berengere.luthringer@hzg.de; Fax: +49 4152 87 2595; Tel: +49 4152 87 1292
First published on 30th April 2020
Abstract
Metallic implant biomaterials predominate in orthopaedic surgery. Compared to titanium-based permanent implants, magnesium-based ones offer new possibilities as they possess mechanical properties closer to the ones of bones and they are biodegradable. Furthermore, magnesium is more and more considered to be “bioactive” i.e., able to elicit a specific tissue response or to strengthen the intimate contact between the implant and the osseous tissue. Indeed, several studies demonstrated the overall beneficial effect of magnesium-based materials on bone tissue (in vivo and in vitro). Here, the direct effects of titanium and magnesium on osteoblasts were measured on proteomes levels in order to highlight metal-specific and relevant proteins. Out of 2100 identified proteins, only 10 and 81 differentially regulated proteins, compare to the control, were isolated for titanium and magnesium samples, respectively. Selected ones according to their relationship to bone tissue were further discussed. Most of them were involved in extracellular matrix maturation and remodelling (two having a negative effect on mineralisation). A fine-tuned balanced between osteoblast maturation, differentiation and viability was observed.
Significance to metallomicsFor the first time, effects of Ti and Mg biomaterials on human primary osteoblasts were studied via comparative analyses of the variation of the proteome. No adverse effect was noticed on phenotypic observations of osteoblast cells cultured on Mg or Ti samples. Out of 2100 identified proteins, only 10 and 81 differentially metal-specific regulated proteins (compare to the control) were isolated for Ti and Mg, respectively. Overall impact of degrading Mg on osteoblast is significant larger and multifaceted compared to Ti metal discs, highlighting its bioactivity. A fine-tuned balanced between osteoblast maturation, differentiation and viability was observed for Mg. |
1. Introduction
The growing interest in magnesium (Mg) and its alloys in the field of implantology is justified by their unique properties combining appropriate mechanical properties, biocompatibility and biodegradability. Previous concerns about gas production due to a too fast degradation is now controlled by combinable factors such as Mg purity, choice of alloying elements, metal microstructure, surface treatment and material processing routes. Mg-Based materials is a promising alternative to titanium (Ti)-based ones, as a removal surgery is unnecessary, reducing not only costs but also potential infection and increasing patient wellness.The majority of hitherto existing in vivo studies deal primarily with the description of degradation processes in living organisms and proving the biocompatibility. An analysis of the impact of Mg-implants in vivo in mice has been performed recently by Jähn et al.,1 showing “a significant increase in callus size due to an augmented bone formation rate and a reduced bone resorption in fractures”. Nevertheless, although the observed biological activity of Mg is intriguing, it remains largely unexplored. In 2016, Zhang et al.2 proposed that the osteogenic effects of Mg-based implant could be mediated via a mechanism involving neuronal calcitonin gene-related polypeptide-a (CGRP). Nonetheless, due to the numerous and ubiquitous roles of Mg, Mg-related osteogenesis regulation probably involve the integration of several mechanisms which are difficult to assess in vivo. Proteomics approach can be a method to discover potentially involved factors to be further studied. Mg as essential element in most living organisms is known to maintain a multitude of basic cellular functions, mainly as cofactor of metal-containing enzymes. Various pathological in vivo effects of Mg deficiency have been observed supporting the relevance of a balanced physiological Mg concentration. Mg is especially important for bone regeneration processes. It was reported that Mg deficiency causes (e.g.) osteopenia, a condition in which bone mineral density is lower than normal owing to inhibition of the differentiation and activity of osteoblasts (OB).3 The number of OB decreased in Mg-deficient rodents,4 but increased in presence of Mg-based extracts.5 On the other hand, differentiation of OB is affected by titanium (Ti)-disc surfaces in vitro and a thin layer of bone tissue, which completely covers the discs can be formed in vivo.6 Furthermore, both Mg and Ti ions have a significant role in bone formation.7
A successful design of Mg-based implants requires the understanding of the response of the bone cells at the interface of implant and bone tissue. Due to their role in bone formation and mineralisation, osteoblasts were selected to compare the osteoinductive effect of Mg and Ti. In previous studies an osteoinductive effect of Mg-implant degradation products, termed Mg-extracts, on osteoblasts was demonstrated by several biological methods and functional assays.5,8 Various osteosarcoma derived cell lines may also serve as model system to investigate the biocompatibility and impacts of implant materials. Mouse osteosarcoma cells (MC3T3-E1) showed an overexpression of proteins related with the mineralisation process as response to a magnesium-based alloy extract (AZ31 fluoride surface-modified).9 Nevertheless, human primary osteoblasts remain the most relevant OB model to study Mg-based materials for orthopaedic application.5
To contribute to the development of biodegradable metallic implants and to discover potential new effector molecules, quantitative differential proteomics was chosen to characterise and compare the response of cultured osteoblasts on Mg- and Ti-implant materials.
2. Experimental
2.1. Samples preparation
Pure Ti samples, polished non-porous (10 mm diameter, 1 mm thickness) were supply by DOT GmbH, Rostock, Germany. Prior to the biological tests, Ti culture substrates were cleaned by immersing them in 2% Hellmanex solution (Hellma, Mühlheim, Germany) and ultrasonication at room temperature for 20 min. These steps were repeated by replacing the Hellmanex solution with chloroform (Sigma-Aldrich Chemie GmbH, Munich, Germany), ethanol (Sigma-Aldrich Chemie GmbH, Munich, Germany), chloroform/methanol (80/20), and deionized water. The samples were then sterilised (autoclaved for 20 min at 121 °C).High-purity Mg (99.95%) was purchased from Magnesium Elektron (Manchester, United Kingdom – UK). Pure Mg was cast using a permanent mould direct chill cast. The cast billet was indirect extruded (Strangpreßzentrum Berlin, Berlin, Germany). Discs (diameter of 10 mm and 1.5 mm thickness) were machined from the extruded bars (Henschel KG, Munich, Germany), and the samples were sterilized using gamma radiation at a dose of 29.2 kGy (BBF GmbH, Stuttgart, Germany). In order to assure suitability of the Mg material for biological tests, samples degradation rate was characterised via weight loss over 7 days as described here.10 A value of 0.21 ± 0.08 mm per year was measured, appropriate for cell seeding.10 As for degradation or cell tests, medium is exchanged every 2 to 3 days, a maximal (calculated) concentration of 7.3 × 10−3 mol L−1 may be reached (medium Mg concentration about 0.93 × 10−3 mol L−1). However, as cells are directly cultured on the material, higher local concentrations might be attained.
2.2. Osteoblast immunophenotyping
All experiments involving human cells were performed in accordance with relevant German guidelines and regulations. Human hip joints were obtained from patients undergoing hip arthroplasty (at Hospital Eilbek, Hamburg, Germany; written consent documented) with the approval of the local ethics committee, but without passing patient data (ethics committee of the Hamburg medical association; WF-057/11). Cell isolation was prepared as described previously.11 Expression of the cell surface markers of primary OB were characterised by flow cytometry (S3e cell sorter; BioRad, Munich, Germany). OB were harvested (0.05% trypsin EDTA) and washed twice in phosphate buffer saline (PBS). For each measurements, aliquots of 100 μL of OB suspension (1.6 × 106 cells mL−1) were washed in ice cold 1% bovine serum albumin (BSA; fraction V; Carl Roth, Karlsruhe, Germany) in PBS (w/v) and then incubated with appropriate antibody (see Table 1). After 50 min incubation, cells were washed twice with ice cold 1% BSA in PBS. Nonspecific fluorescence was determined by cells stained with conjugated antibodies raised against antihuman IgG1, IgG2a and IgG2b (all purchased from BD Bioscience, Heidelberg, Germany). At least 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were acquired for each sample and cell flow cytometry data were further analysed using Prosort software version 1.5 (BioRad, Munich, Germany).
000 events were acquired for each sample and cell flow cytometry data were further analysed using Prosort software version 1.5 (BioRad, Munich, Germany).
| Label | Name – symbol | Supplier |
|---|---|---|
| Allophycocyanin (APC) | 5′-Nucleotidase (5′-NT) cluster of differentiation (CD) 73 | Invitrogen Molecular Probes, Karlsruhe, Germany |
| Fluorescein isothiocyanate (FITC) | Endoglin CD105 | Invitrogen Molecular Probes, Karlsruhe, Germany |
| Thy-1 membrane glycoprotein CD90 | Invitrogen Molecular Probes, Karlsruhe, Germany | |
| Leukocyte common antigen CD45 | Invitrogen Molecular Probes, Karlsruhe, Germany | |
| Human leukocyte antigen (HLA) class II | BD Bioscience, Heidelberg, Germany | |
| Phycoerythrin (PE) | Platelet endothelial cell adhesion molecule (PECAM-1) CD31 | Thermo Scientific – Fisher Scientific GmbH, Schwerte, Germany |
| Platelet glycoprotein 4, fatty acid translocase (FAT) CD36 | Invitrogen Molecular Probes, Karlsruhe, Germany | |
| Alkaline phosphatase (ALP) | R&D Systems, Wiesbaden, Germany | |
| PE coupled to cyan dye (PE-Cy5) | Indian blood group CD44 | Invitrogen Molecular Probes, Karlsruhe, Germany |
| HLA class I | BD Bioscience, Heidelberg, Germany |
2.3. Biological test preparation
OB were used up to the 3rd passage. Cells were cultured with DMEM-GlutaMAX (Life Technologies, Darmstadt, Germany) containing 10% (v/v) foetal bovine serum (FBS; PAA Cell Culture Company, Linz, Austria). To allow the formation of a protective degradation layer while avoiding the initial degradation burst of Mg samples, Mg-discs were preincubated overnight in DMEM-GlutaMAX I + 10% FCS (v/v) under cell culture conditions (i.e., 37 °C, 5% CO2, 20% O2, 95% relative humidity). While degrading, an alkalinisation at the material surface happened, leading to a solubility decrease of compounds that will form the degradation layer (e.g., Mg(OH)2, MgCO3, CaCO3, Mg3(PO4)2 or Ca5(PO4)3(OH)).10 Primary human osteoblasts from a single donor (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells in 50 μL) were let to adhere for 30 min on the Ti and Mg samples’ surfaces before adding fresh supplemented media. Cells were also directly cultured on tissue culture plastic (culture condition further referred as “control”). Cells were cultured for 7 days with medium change every 2 to 3 d. This set-up was selected to perform phenotypic examinations (see Section 2.3) as well as proteomics experiments (n = 4; see Section 2.4). For phenotypic examinations, several technical and biological replicates with different donors were performed. Representative pictures were then selected.
000 cells in 50 μL) were let to adhere for 30 min on the Ti and Mg samples’ surfaces before adding fresh supplemented media. Cells were also directly cultured on tissue culture plastic (culture condition further referred as “control”). Cells were cultured for 7 days with medium change every 2 to 3 d. This set-up was selected to perform phenotypic examinations (see Section 2.3) as well as proteomics experiments (n = 4; see Section 2.4). For phenotypic examinations, several technical and biological replicates with different donors were performed. Representative pictures were then selected.
2.4. Phenotypic examination
In order to appreciate general OB phenotype cultured on Mg and Ti discs, two different tests were performed: live/dead cell viability assay and actin cytoskeleton and focal adhesion staining.2.5. Cell preparation for proteomics
After 7 days, cells were harvested with 0.05% trypsin/EDTA (Life Technologies, Darmstadt, Germany) and washed in EDTA-free protease inhibitor buffer (Complete ULTRA Tablets; EDTA-free; Roche, Mannheim, Germany). The remaining cell pellet was snap-frozen in liquid nitrogen. The cells were stored at −80 °C until further processing.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min. The supernatants were transferred into 10 kDa molecular weight exclusion centrifugal filter units (Merck Millipore, Darmstadt, Germany) and tryptic in-solution digestion was performed. 500 μL 6 M urea was added. For reduction and alkylation, 1.3 μL of a 10 mM dithiothreitol solution (DTT dissolved in 100 mM NH4CO3) and 300 mM iodoacetamide (IAA dissolved in 100 mM NH4CO3) were added and incubated for 10 min at 56 °C or for 20 min in the dark at room temperature. Then, 425 μL NH4CO3 (100 mM, dissolved in HPLC-H2O) was added. For tryptic digestion 8 μL of a 0.25 μg μL−1 trypsin solution (Promega GmbH, Mannheim, Germany) was added. The samples were incubated overnight at 37 °C. Then, formic acid (FA) was added to the samples until a final concentration of 0.2% and evaporated.
000g for 30 min. The supernatants were transferred into 10 kDa molecular weight exclusion centrifugal filter units (Merck Millipore, Darmstadt, Germany) and tryptic in-solution digestion was performed. 500 μL 6 M urea was added. For reduction and alkylation, 1.3 μL of a 10 mM dithiothreitol solution (DTT dissolved in 100 mM NH4CO3) and 300 mM iodoacetamide (IAA dissolved in 100 mM NH4CO3) were added and incubated for 10 min at 56 °C or for 20 min in the dark at room temperature. Then, 425 μL NH4CO3 (100 mM, dissolved in HPLC-H2O) was added. For tryptic digestion 8 μL of a 0.25 μg μL−1 trypsin solution (Promega GmbH, Mannheim, Germany) was added. The samples were incubated overnight at 37 °C. Then, formic acid (FA) was added to the samples until a final concentration of 0.2% and evaporated.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and the supernatants were transferred into new vials for liquid-chromatography tandem mass spectrometry (LC-MSMS) analysis.
000 rpm for 10 min and the supernatants were transferred into new vials for liquid-chromatography tandem mass spectrometry (LC-MSMS) analysis.
3. Results & discussion
3.1. OB immunophenotyping
OB immunophenotyping via flow cytometry indicated the absence of isolation contaminations by endothelial cells, adipocytes and hematopoietic cells. More than 90% of the cells were CD31, CD36, CD45, and HLA class II negative. Due to the observed pattern of cell's surface markers, the mesenchymal stem cell character of the OB was proven. Cells of the first and second passage were CD44 (97%), CD73 (95%), CD90 (90%). CD105 (92%) and HLA class I (93%). Furthermore, a positive staining of 19% (P1) and 29% (P2) for ALP an osteogenic activity could be measured.3.2. Phenotypic examination
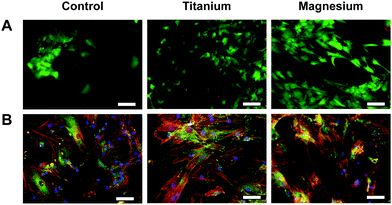
Cytocompatibility and (early) adhesion are two prerequisites and factors influencing further cell fate, (such as migration, proliferation, differentiation, appropriate functioning or long-term viability) as well as implant lifespan. Thus phenotypic examinations of osteoblasts cultured on either Mg or Ti carriers, LIVE/DEAD cell viability and actin cytoskeleton and focal adhesion stainings, were performed to assess these factors. As presented in Fig. 1A, no adverse effect of the materials on cells was observed. Indeed, at day 7, mostly alive cells (green) and only few dead cells (red) could be seen. Similarly, for both materials, cells showed a well-defined cytoskeletal structure and ovoid nuclei implying no impaired cell adhesion (Fig. 1B).On the three substrates, OB retained their fibroblastic shape. OB differentiation is generally accompanied with modification of the actin cytoskeleton (e.g., parallel arrangement and orientation of the actin filaments and stress fibre). However, on the phenotypic level, no obvious integrin structure difference could be observed depending of the substrate. Focal adhesion spots (vinculin) may be smaller and more numerous on Ti surface especially compared to Mg one (larger). With these observations, it may be assumed that Mg could promote osteogenesis.
3.3. Proteomics analyses
Proteomes of osteoblasts directly cultured on substrates made of implant material (Ti–6Al–4V and Mg) were analysed to reflect their responses towards these materials.In total, approximately 2100 proteins were identified by the LC-MSMS analysis including more than 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 peptides, and 357
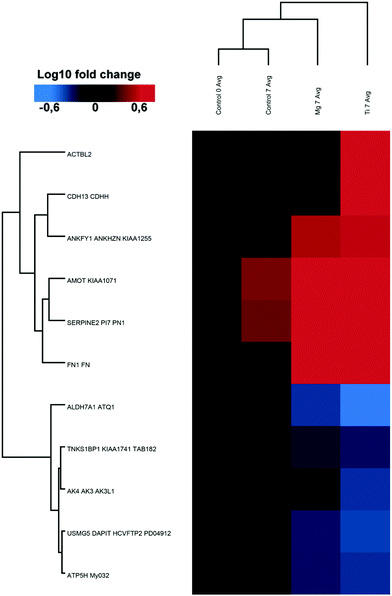
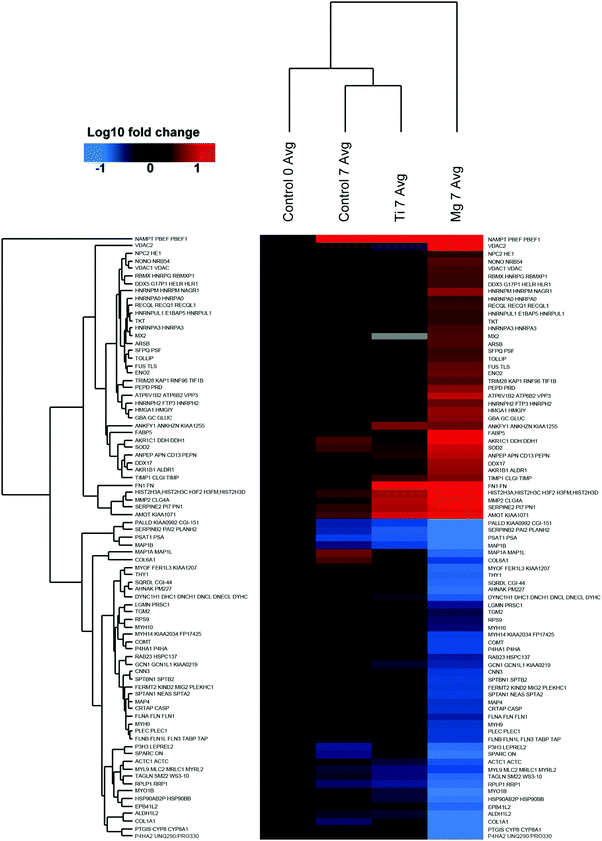
000 peptides, and 357![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 spectra were recorded. Among them, 91 (0.2%) of the proteins of osteoblasts were significantly increased or decreased by a factor of at least two or more in the presence or absence (control) of metal-discs that were listed in Tables 2 and 3. The heat-maps (Fig. 2 and 3) give an overview about the up- and downregulation of proteins. There was only a minimal difference in the proteome of the osteoblast control group and the osteoblasts cultured on Ti-discs (only 10 proteins – Fig. 2). In contrast, the protein pattern of the osteoblasts grown on Mg-discs varies remarkably indicating a clear effect of Mg-discs, with 81 proteins significantly regulated (Fig. 3). 38 proteins were considerably increased by the presence of Mg-discs while only 6 proteins were upregulated in case of Ti-discs. On the other hand, the concentration of 43 proteins was significantly decreased in presence of Mg-discs, and 7 unique proteins in presence of Ti-disc.
102 spectra were recorded. Among them, 91 (0.2%) of the proteins of osteoblasts were significantly increased or decreased by a factor of at least two or more in the presence or absence (control) of metal-discs that were listed in Tables 2 and 3. The heat-maps (Fig. 2 and 3) give an overview about the up- and downregulation of proteins. There was only a minimal difference in the proteome of the osteoblast control group and the osteoblasts cultured on Ti-discs (only 10 proteins – Fig. 2). In contrast, the protein pattern of the osteoblasts grown on Mg-discs varies remarkably indicating a clear effect of Mg-discs, with 81 proteins significantly regulated (Fig. 3). 38 proteins were considerably increased by the presence of Mg-discs while only 6 proteins were upregulated in case of Ti-discs. On the other hand, the concentration of 43 proteins was significantly decreased in presence of Mg-discs, and 7 unique proteins in presence of Ti-disc.
| Name | Symbol | Fold change | |
|---|---|---|---|
| Downregulation | Up-regulated during skeletal muscle growth 5 homolog (mouse) | USMG5 | 0.28 |
| bA792D24.4, DAPIT, DKFZp566D211, HCVFTP2, HCV F-transactivated protein 2, MGC14697, PD04912, Up-regulated during skeletal muscle growth protein 5 | |||
| Aldehyde dehydrogenase 7 family, member A1 | ALDH7A1 | 0.29 | |
| Aldehyde dehydrogenase family 7 member A1, Alpha-AASA dehydrogenase, alpha-aminoadipic semialdehyde dehydrogenase, antiquitin-1, ATQ1, betaine aldehyde dehydrogenase, delta1-piperideine-6-carboxylate dehydrogenase, EPD, FLJ11738, FLJ92814, P6c dehydrogenase, PDE | |||
| ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d | ATP5H | 0.37 | |
| ATP5JD, ATPase subunit d, ATPQ, ATP synthase subunit d, mitochondrial, My032 | |||
| Adenylate kinase 4 | AK4 | 0.45 | |
| Adenylate kinase 3-like, adenylate kinase isoenzyme 4, mitochondrial, AK3, AK3L1, AK3L2, ATP-AMP transphosphorylase, MGC166959 | |||
| Upregulation | Serpin peptidase inhibitor, clade E member 2 | SERPINE2 | 2.00 |
| DKFZp686A13110, GDN, glia-derived nexin, nexin, peptidase inhibitor 7, PI7, PI-7, PN1, PN-1, PNI, protease nexin 1, protease nexin I, serpin E2, serine (or cysteine) proteinase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | |||
| Angiomotin | AMOT | 2.35 | |
| KIAA1071 | |||
| Ankyrin repeat and FYVE domain containing 1 | ANKFY1 | 3.72 | |
| ANKHZN, ankyrin repeat and FYVE domain-containing protein 1, ankyrin repeats hooked to a zinc finger motif, DKFZp686M19106, KIAA1255, ZFYVE14 | |||
| Cadherin 13 | CDH13 | 9.75 | |
| Cadherin-13, CDHH, H-cadherin, heart cadherin, P105, T-cad, T-cadherin, truncated cadherin | |||
| Fibronectin 1 | FN1 | 14.59 | |
| CIG, cold-insoluble globulin, DKFZp686F10164, DKFZp686H0342, DKFZp686I1370, DKFZp686O13149, ED-B, fibronectin, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | |||
| Actin, beta-like 2 | ACTBL2 | 53.88 | |
| ACT, beta-actin-like protein 2, DKFZp686D0972, kappa-actin | |||
| Name | Symbol | Fold change | |
|---|---|---|---|
| Downregulation | Up-regulated during skeletal muscle growth 5 homolog (mouse) | MAP1A | 0.05 |
| FLJ77111, MAP-1A, MAP1L, microtubule-associated protein 1A, MTAP1A, proliferation-related protein p80 | |||
| Prostaglandin I2 (prostacyclin) synthase | PTGIS | 0.07 | |
| CYP8, CYP8A1, MGC126858, MGC126860, PGIS, prostacyclin synthase, prostaglandin I2 synthase, PTGI | |||
| Collagen, type VI, alpha 1 | COL6A1 | 0.09 | |
| Collagen alpha-1(VI) chain | |||
| Prolyl 4-hydroxylase, alpha polypeptide II | P4HA2 | 0.10 | |
| 4-PH alpha-2, C-P4Halpha(II), procollagen-proline, 2-oxoglutarate-4-dioxygenase subunit alpha-2, prolyl 4-hydroxylase subunit alpha-2, UNQ290/PRO330 | |||
| Myosin IB | MYO1B | 0.10 | |
| MMIa, MMI-alpha, MYH-1c, myosin I alpha, myosin-Ib, myr1 | |||
| Aldehyde dehydrogenase 1 family, member L2 | ALDH1L2 | 0.11 | |
| Aldehyde dehydrogenase family 1 member L2, DKFZp686A16126, DKFZp686M064, DKFZp686P14145, FLJ36769, FLJ38508, MGC119536, MGC119537, mtFDH, mitochondrial 10-formyltetrahydrofolate dehydrogenase | |||
| Myoferlin | MYOF | 0.11 | |
| FER1L3, Fer-1-like protein 3, FLJ36571, FLJ90777, KIAA1207, myoferlin | |||
| Microtubule-associated protein 1B | MAP1B | 0.11 | |
| DKFZp686E1099, DKFZp686F1345, FLJ38954, FUTSCH, MAP-1B, MAP5, microtubule-associated protein 1B | |||
| AHNAK nucleoprotein | AHNAK | 0.12 | |
| AHNAKRS, desmoyokin, MGC5395, neuroblast differentiation-associated protein AHNAK, PM227 | |||
| Sulfide quinone reductase-like (yeast) | SQRDL | 0.13 | |
| CGI-44, sulfide:quinone oxidoreductase, mitochondrial | |||
| Collagen, type I, alpha 1 | COL1A1 | 0.13 | |
| Alpha-1 type I collagen, collagen alpha-1(I) chain, OI4 | |||
| Thy-1 cell surface antigen | THY1 | 0.15 | |
| CD90, CDw90, FLJ33325, Thy-1 antigen, Thy-1 membrane glycoprotein | |||
| Phosphoserine aminotransferase 1 | PSAT1 | 0.17 | |
| EPIP, MGC1460, phosphohydroxythreonine aminotransferase, phosphoserine aminotransferase, PSA, PSAT | |||
| Dynein, cytoplasmic 1, heavy chain 1 | DYNC1H1 | 0.17 | |
| Cytoplasmic dynein 1 heavy chain 1, cytoplasmic dynein heavy chain 1, DHC1, DHC1a, DKFZp686P2245, DNCH1, Dnchc1, DNCL, DNECL, DYHC, dynein heavy chain, cytosolic, HL-3, KIAA0325, p22 | |||
| Heat shock protein 90 alpha family class B member 2, pseudogene | HSP90AB2P | 0.18 | |
| Heat shock protein 90Bb, heat shock protein 90-beta b, heat shock protein 90 kDa alpha (cytosolic), class B member 2 (pseudogene), HSP90BB | |||
| Catechol-O-methyltransferase | COMT | 0.19 | |
| Myosin, heavy chain 14 | MYH14 | 0.20 | |
| DFNA4, DKFZp667A1311, FLJ13881, FLJ43092, FP17425, KIAA2034, MHC16, MYH17, myosin, myosin-14, myosin heavy chain, non-muscle IIc, myosin heavy chain 14, NMHC II-C, NMHC-II-C, non-muscle myosin heavy chain IIc | |||
| Erythrocyte membrane protein band 4.1-like 2 | EPB41L2 | 0.20 | |
| 4.1G, 4.1-G, band 4.1-like protein 2, DKFZp781D1972, DKFZp781H1755, generally expressed protein 4.1 | |||
| Prolyl 4-hydroxylase subunit alpha 1 | P4HA1 | 0.23 | |
| Prolyl 4-hydroxylase, alpha polypeptide I, 4-PH alpha-1, C-P4Halpha(I), P4HA, procollagen-proline, 2-oxoglutarate-4-dioxygenase subunit alpha-1 | |||
| Serpin family B member 2 | SERPINB2 | 0.25 | |
| Serpin peptidase inhibitor, clade B (ovalbumin), member 2, HsT1201, monocyte Arg-serpin, PAI, PAI2, PAI-2, placental plasminogen activator inhibitor, PLANH2, plasminogen activator inhibitor 2, serpin B2, urokinase inhibitor | |||
| Plectin | PLEC | 0.26 | |
| EBS1, EBSO, HD1, hemidesmosomal protein 1, LGMD2Q, PCN, PLEC1, PLEC1b, plectin, plectin-1, PLTN | |||
| Legumain | LGMN | 0.26 | |
| AEP, asparaginyl endopeptidase, legumain, LGMN1, protease, cysteine 1, PRSC1 | |||
| GCN1, eIF2 alpha kinase activator homolog | GCN1 | 0.26 | |
| GCN1 general control of amino-acid synthesis 1-like 1 (yeast), GCN1, GCN1L, GCN1-like protein 1, HsGCN1, KIAA0219, PRIC295, translational activator GCN1 | |||
| Filamin B | FLNB | 0.26 | |
| ABP-278, ABP-280, ABP-280 homolog, actin-binding-like protein, AOI, beta-filamin, DKFZp686A1668, DKFZp686O033, Fh1, FH1, filamin-3, filamin-B, filamin homolog 1, FLN1L, FLN3, FLN-B, LRS1, SCT, TABP, TAP, thyroid autoantigen, truncated ABP, truncated actin-binding protein | |||
| RAB23, member RAS oncogene family | RAB23 | 0.27 | |
| DKFZp781H0695, HSPC137, MGC8900, Ras-related protein Rab-23 | |||
| Palladin, cytoskeletal associated protein | PALLD | 0.29 | |
| CGI151, CGI-151, FLJ22190, FLJ38193, FLJ39139, FLJ61376, KIAA0992, MYN, palladin, PNCA1, sarcoma antigen NY-SAR-77, SIH002 | |||
| Myosin heavy chain 9 | MYH9 | 0.30 | |
| Cellular myosin heavy chain, type A, DFNA17, EPSTS, FTNS, MGC104539, MHA, myosin-9, myosin heavy chain, non-muscle IIa, myosin heavy chain 9, NMHC-II-A, NMMHCA, NMMHC-A, NMMHC II-a, NMMHC-IIA, non-muscle myosin heavy chain A, non-muscle myosin heavy chain IIa | |||
| Transgelin | TAGLN | 0.30 | |
| 22 kDa actin-binding protein, DKFZp686B01212, DKFZp686P11128, protein WS3-10, SM22, SM22-alpha, SMCC, smooth muscle protein 22-alpha, TAGLN1, transgelin, WS3-10 | |||
| Microtubule-associated protein 4 | MAP4 | 0.32 | |
| DKFZp779A1753, MAP-4, MGC8617, microtubule-associated protein 4 | |||
| Secreted protein, acidic, cysteine-rich | SPARC | 0.32 | |
| Basement-membrane protein 40, BM-40, ON, osteonectin, secreted protein acidic and rich in cysteine | |||
| Fermitin family member 2 | FERMT2 | 0.33 | |
| DKFZp686G11125, fermitin family homolog 2, FLJ34213, FLJ44462, KIND2, kindlin-2, MIG2, mig-2, MIG-2, mitogen-inducible gene 2 protein, PH domain-containing family C member 1, pleckstrin homology domain-containing family C member 1, PLEKHC1, UNC112, UNC112B | |||
| Spectrin, alpha, non-erythrocytic 1 | SPTAN1 | 0.33 | |
| Alpha-II spectrin, EIEE5, FLJ17738, FLJ44613, fodrin alpha chain, NEAS, spectrin, non-erythroid alpha chain, spectrin alpha chain, brain, SPTA2 | |||
| Myosin, heavy chain 10 | MYH10 | 0.33 | |
| Cellular myosin heavy chain, type B, MGC134913, MGC134914, myosin-10, myosin heavy chain, non-muscle IIb, myosin heavy chain 10, NMMHCB, NMMHC-B, NMMHC II-b, NMMHC-IIB, non-muscle myosin heavy chain B, non-muscle myosin heavy chain IIb | |||
| Cartilage associated protein | CRTAP | 0.33 | |
| CASP, LEPREL3 | |||
| Ribosomal protein lateral stalk subunit P1 | RPLP1 | 0.33 | |
| 60S acidic ribosomal protein P1, FLJ27448, LP1, MGC5215, P1, RPP1, RRP1, LP1 | |||
| Filamin A | FLNA | 0.34 | |
| ABP-280, ABPX, actin-binding protein 280, alpha-filamin, CVD1, DKFZp434P031, endothelial actin-binding protein, filamin-1, filamin-A, FLN, FLN1, FLN-A, FMD, MNS, NHBP, non-muscle filamin, OPD, OPD1, OPD2, XLVD, XMVD | |||
| Actin, alpha, cardiac muscle 1 | ACTC1 | 0.35 | |
| ACTC, actin, alpha cardiac muscle 1, alpha-cardiac actin, ASD5, CMD1R, CMH11, LVNC4 | |||
| Spectrin, beta, non-erythrocytic 1 | SPTBN1 | 0.37 | |
| Beta-II spectrin, betaSpII, ELF, fodrin beta chain, spectrin, non-erythroid beta chain 1, spectrin beta chain, brain 1, SPTB2 | |||
| Transglutaminase 2 | TGM2 | 0.38 | |
| G-ALPHA-h, GNAH, protein-glutamine gamma-glutamyltransferase 2, TG(C), TG2, TGase-2, TGase C, TGase H, TGC, tissue transglutaminase, transglutaminase-2, transglutaminase C, transglutaminase H | |||
| Calponin 3 | CNN3 | 0.38 | |
| Calponin, acidic isoform, calponin-3 | |||
| Ribosomal protein S9 | RPS9 | 0.42 | |
| 40S ribosomal protein S9 | |||
| Prolyl 3-hydroxylase 3 | P3H3 | 0.44 | |
| Leprecan-like 2, LEPREL2, procollagen-proline 3-dioxygenase, GRCB, HSU47926, leprecan-like protein 2, P3H3, prolyl 3-hydroxylase 3, protein B | |||
| Myosin light chain 9 | MYL9 | 0.47 | |
| 20 kDa myosin light chain, LC20, MGC3505, MLC2, MLC-2C, MRLC1, myosin regulatory light chain 2, smooth muscle isoform, myosin regulatory light chain 9, myosin regulatory light chain MRLC1, myosin regulatory light polypeptide 9, Myosin RLC, MYRL2 | |||
| Upregulation | Heterogeneous nuclear ribonucleoprotein U-like 1 | HNRNPUL1 | 2.02 |
| Adenovirus early region 1B-associated protein 5, E1B-55 kDa-associated protein 5, E1BAP5, E1B-AP5, FLJ12944, heterogeneous nuclear ribonucleoprotein U-like protein 1, HNRPUL1 | |||
| Tripartite motif containing 28 | TRIM28 | 2.08 | |
| FLJ29029, KAP1, KAP-1, KRAB-associated protein 1, KRAB-interacting protein 1, KRIP-1, nuclear corepressor KAP-1, RING finger protein 96, RNF96, TF1B, TIF1B, TIF1-beta, transcription intermediary factor 1-beta, tripartite motif-containing protein 28 | |||
| Aldo-keto reductase family 1 member B | AKR1B1 | 2.08 | |
| ADR, aldehyde reductase, aldo-keto reductase family 1 member B1, aldose reductase, ALDR1, ALR2, AR, MGC1804 | |||
| Transketolase | TKT | 2.09 | |
| FLJ34765, TK, TKT1, transketolase | |||
| Heterogeneous nuclear ribonucleoprotein A0 | HNRNPA0 | 2.09 | |
| hnRNPA0, hnRNP A0, HNRPA0 | |||
| TIMP metallopeptidase inhibitor 1 | TIMP1 | 2.23 | |
| CLGI, collagenase inhibitor, EPA, EPO, erythroid-potentiating activity, fibroblast collagenase inhibitor, FLJ90373, HCI, metalloproteinase inhibitor 1, TIMP, TIMP-1, tissue inhibitor of metalloproteinases 1 | |||
| MX dynamin like GTPase 2 | MX2 | 2.29 | |
| Myxovirus (influenza virus) resistance 2 (mouse), interferon-induced GTP-binding protein Mx2, interferon-regulated resistance GTP-binding protein MxB, MXB, myxovirus resistance protein 2, p78-related protein | |||
| Superoxide dismutase 2, mitochondrial | SOD2 | 2.30 | |
| IPOB, MNSOD, MVCD6 | |||
| RecQ like helicase | RECQL | 2.33 | |
| RecQ protein-like (DNA helicase Q1-like), ATP-dependent DNA helicase Q1, DNA-dependent ATPase Q1, DNA helicase, RecQ-like type 1, RecQ1, RECQ1, RecQL1, RECQL1, RecQ protein-like 1 | |||
| Heterogeneous nuclear ribonucleoprotein A3 | HNRNPA3 | 2.36 | |
| 2610510D13Rik, D10S102, FBRNP, heterogeneous nuclear ribonucleoprotein A3, hnRNP A3, HNRPA3, MGC138232, MGC142030 | |||
| Nicotinamide phosphoribosyltransferase | NAMPT | 2.40 | |
| 1110035O14Rik, DKFZp666B131, MGC117256, NAmPRTase, NAMPT, nicotinamide phosphoribosyltransferase, PBEF, PBEF1, pre-B-cell colony-enhancing factor 1, pre-B cell-enhancing factor, VF, visfatin, VISFATIN | |||
| NPC intracellular cholesterol transporter 2 | NPC2 | 2.46 | |
| EDDM1, epididymal secretory protein E1, He1, HE1, human epididymis-specific protein 1, MGC1333, Niemann-pick disease type C2 protein, NP-C2 | |||
| Splicing factor proline and glutamine rich | SFPQ | 2.51 | |
| 100 kDa DNA-pairing protein, DNA-binding p52/p100 complex, 100 kDa subunit, hPOMp100, polypyrimidine tract-binding protein-associated-splicing factor, POMP100, PSF, PTB-associated-splicing factor, splicing factor, proline- and glutamine-rich | |||
| Heterogeneous nuclear ribonucleoprotein H2 | HNRNPH2 | 2.52 | |
| FTP3, FTP-3, heterogeneous nuclear ribonucleoprotein H′, heterogeneous nuclear ribonucleoprotein H2, hnRNPH′, hnRNP H′, hnRNP H2, HNRPH′, HNRPH2 | |||
| DEAD-box helicase 17 | DDX17 | 2.54 | |
| DEAD (Asp–Glu–Ala–Asp) box polypeptide 17, dead box protein 17, dead box protein p72, DKFZp761H2016, P72, RH70, RNA-dependent helicase p7 | |||
| Toll interacting protein | TOLLIP | 2.57 | |
| FLJ33531, IL-1RAcPIP, Toll-interacting protein | |||
| Aldo-keto reductase family 1 member C1 | AKR1C1 | 2.64 | |
| Aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase), 20-alpha-HSD, 20-ALPHA-HSD, 20-alpha-hydroxysteroid dehydrogenase, 2-ALPHA-HSD, aldo-keto reductase family 1 member C1, C9, chlordecone reductase homolog HAKRC, DD1, DD1/DD2, DDH, DDH1, dihydrodiol dehydrogenase 1/2, H-37, HAKRC, HBAB, high-affinity hepatic bile acid-binding protein, indanol dehydrogenase, MBAB, MGC8954, trans-1,2-dihydrobenzene-1,2-diol dehydrogenase | |||
| High mobility group AT-hook 1 | HMGA1 | 2.66 | |
| High mobility group AT-hook protein 1, high mobility group protein A1, high mobility group protein HMG-I/HMG-Y, high mobility group protein R, HMGA1A, HMG-I(Y), HMGIY, HMG-R, MGC12816, MGC4242, MGC4854 | |||
| Serpin family B member 2 | SERPINE2 | 2.70 | |
| Serpin peptidase inhibitor, clade B (ovalbumin), member 2, HsT1201, monocyte Arg-serpin, PAI, PAI2, PAI-2, placental plasminogen activator inhibitor, PLANH2, plasminogen activator inhibitor 2, serpin B2, urokinase inhibitor | |||
| Glucosylceramidase beta | GBA | 2.73 | |
| Acid beta-glucosidase, alglucerase, beta-glucocerebrosidase, D-glucosyl-N-acylsphingosine glucohydrolase, GBA1, GC, GCB, GLUC, glucosylceramidase, imiglucerase | |||
| Alanyl aminopeptidase, membrane | ANPEP | 2.79 | |
| Alanyl (membrane) aminopeptidase, alanyl aminopeptidase, aminopeptidase M, aminopeptidase N, AP-M, APN, AP-N, CD13, gp150, GP150, hAPN, LAP1, microsomal aminopeptidase, myeloid plasma membrane glycoprotein CD13, p150, P150, PEPN | |||
| Arylsulfatase B | ARSB | 2.81 | |
| Arylsulfatase B, ASB, G4S, MPS6, N-acetylgalactosamine-4-sulfatase | |||
| FUS RNA binding protein | FUS | 2.98 | |
| Fused in sarcoma, 75 kDa DNA-pairing protein, ALS6, FUS1, HNRNPP2, hnRNP-P2, oncogene FUS, oncogene TLS, POMp75, POMP75, RNA-binding protein FUS, TLS, translocated in liposarcoma protein | |||
| Enolase 2 | ENO2 | 3.09 | |
| 2-Phospho-D-glycerate hydro-lyase, enolase 2, gamma-enolase, neural enolase, neuron-specific enolase, NSE | |||
| ATPase H+ transporting V1 subunit B2 | ATP6V1B2 | 3.16 | |
| ATP6B1B2, ATP6B2, endomembrane proton pump 58 kDa subunit, HO57, vacuolar proton pump subunit B 2, VATB, V-ATPase subunit B 2, Vma2, VPP3, V-type proton ATPase subunit B, brain isoform | |||
| Peptidase D | PEPD | 3.21 | |
| Imidodipeptidase, MGC10905, peptidase D, PRD, prolidase, PROLIDASE, proline dipeptidase, Xaa-Pro dipeptidase, X-Pro dipeptidase | |||
| Non-POU domain containing octamer binding | NONO | 3.22 | |
| 54 kDa nuclear RNA- and DNA-binding protein, 55 kDa nuclear protein, DNA-binding p52/p100 complex, 52 kDa subunit, NMT55, nono protein, non-POU domain-containing octamer-binding protein, NRB54, P54, p54(nrb), p54nrb, P54NRB | |||
| RNA binding motif protein, X-linked | RBMX | 3.29 | |
| Glycoprotein p43, heterogeneous nuclear ribonucleoprotein G, hnRNP G, hnRNP-G, HNRPG, RBMXP1, RBMXRT, RNA-binding motif protein, X chromosome, RNMX | |||
| Voltage-dependent anion channel 1 | VDAC1 | 3.30 | |
| hVDAC1, MGC111064, outer mitochondrial membrane protein porin 1, plasmalemmal porin, PORIN, Porin 31HL, Porin 31HM, VDAC, VDAC-1, voltage-dependent anion-selective channel protein 1 | |||
| Ankyrin repeat and FYVE domain containing 1 | ANKFY1 | 3.39 | |
| ANKHZN, ankyrin repeat and FYVE domain-containing protein 1, ankyrin repeats hooked to a zinc finger motif, DKFZp686M19106, KIAA1255, ZFYVE14 | |||
| Histone cluster 2 H3 family member a | HIST2H3A | 3.61 | |
| H3/n, H3/o, HIST2H3C, HIST2H3D | |||
| Heterogeneous nuclear ribonucleoprotein M | HNRNPM | 4.25 | |
| CEAR, DKFZp547H118, heterogeneous nuclear ribonucleoprotein M, hnRNP M, HNRNPM4, HNRPM, HNRPM4, HTGR1, NAGR1 | |||
| DEAD-box helicase 5 | DDX5 | 4.28 | |
| DEAD (Asp–Glu–Ala–Asp) box polypeptide 5, DKFZp434E109, DKFZp686J01190, G17P1, HELR, HLR1, HUMP68, p68, RNA helicase p68 | |||
| Matrix metallopeptidase 2 | MMP2 | 4.47 | |
| 72 kDa gelatinase, 72 kDa type IV collagenase, CLG4, CLG4A, gelatinase A, matrix metalloproteinase-2, MMP-2, MMP-II, MONA, TBE-1 | |||
| Angiomotin | AMOT | 6.93 | |
| KIAA1071 | |||
| Fatty acid binding protein 5 | FABP5 | 7.34 | |
| EFABP, E-FABP, epidermal-type fatty acid-binding protein, fatty acid-binding protein, epidermal, fatty acid-binding protein 5, PAFABP, PA-FABP, psoriasis-associated fatty acid-binding protein homolog | |||
| Fibronectin 1 | FN1 | 8.88 | |
| CIG, cold-insoluble globulin, DKFZp686F10164, DKFZp686H0342, DKFZp686I1370, DKFZp686O13149, ED-B, fibronectin, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | |||
| Voltage-dependent anion channel 2 | VDAC2 | 10.54 | |
| FLJ23841, hVDAC2, outer mitochondrial membrane protein porin 2, POR, VDAC-2, voltage-dependent anion-selective channel protein 2 | |||
According to the Gene Ontology (GO) annotation given in UniProt, providing a general and basic overview about the protein functions and their involvement in diverse biological processes, a major part of the regulated proteins belongs to extracellular or membrane proteins. To gain understanding of involved proteins functions, a deeper bibliographic research was performed on possible important regulated proteins with a special focus on proteins involved in bone remodelling, energy metabolism, apoptosis and oxidative stress.
Upregulated proteins.
Fibronectin 1 (FN1). The level of FN1, a collagen-binding high molecular weight dimeric glycoprotein, was increased in osteoblasts cultured on both metal discs (8.88 fold in the presence of Mg and 14.59 fold in the presence of Ti) (Fig. 2, 3 and Tables 2, 3). Fibronectin is one of the major ECM components, interacting with cell surface receptors like integrins, which regulate cell attachment, migration, and growth. The adsorption of FN1 to solid substrates (here Ti) was figured out to improve osseointegration.18 This protein is involved in early stage of bone tissue formation by regulating the attachment of osteoblasts to ECM components18,19 and is essential for matrix mineralisation.19 Hence, it would be expected to have more bone formation by increasing the amount of fibronectin in presence of metal implants in vivo.
Angiomotin (AMOT). AMOT was one of the significantly upregulated proteins in the presence of both metal discs (Mg and Ti) compared to control cells (Fig. 2, 3 and Tables 2, 3). AMOT belongs to the motin family and has a role in tight junction maintenance, cell polarity, migration and Hippo pathway. Ortiz et al. demonstrated that AMOT is necessary for cadherin 11 (CDH11) mediated migration of prostate cancer cells.20 CDH11, also known as osteoblast cadherin, not expressed in normal prostate epithelia, is upregulated during prostate cancer progression. Furthermore, involvement of AMOT in psoriasis due to its role in promoting angiogenesis and vasodilatation was proposed by Niu et al.21 The strong upregulation of AMOT may be associated with the increased bone formation induced by Mg-implants, which was reported by several groups.1,22,23
Matrix metalloproteinase-2 (MMP2). The level of MMP2 was significantly increased in osteoblasts after incubation with Mg-discs compared to Ti-disc and control cells (Fig. 2 and Table 2). MMP2, a member of the zinc-dependent enzyme family, is capable of remodelling the extracellular matrix component by degrading bioactive molecules, leading to the improvement, and healing of the skeleton. Although MMPs actively participate in ECM remodelling, a balance is required as cleavage of cell surface receptors affect signalling and cell functions.24 MMP2 has role in mediating several cellular processes like embryonic (foetal) development, tissue repair, and tumorigenesis via degrading the components of the extracellular matrix.24 Mutation in human MMP2 gene causes multicentric osteolysis (an uncommon genetic syndrome) which provokes bone resorption. Furthermore, previous studies showed that MMP2 and MMP9 have a direct effect on the processing of transforming growth factor, beta 1 (TGFB1) into an active ligand by its cleavage. Since MMP2−/− deficient mice develop skeletal disease, it is convincing that MMP2 has a crucial role during fracture healing processes.24 Therefore, it can be assumed that upregulation of MMP2 in osteoblasts affected by Mg-implants would improves bone formation.
Dead box protein 5 (DDX5). The multifunctional DDX5 (or p68), affects ribonucleic acid (RNA) expression like transcription and splicing and interacts with many transcriptional factors. DDX5 is a known coactivator of runt-related transcription factor 2 (RUNX2), which is essential for the maturation of osteoblasts.25 However, Jensen et al.25 showed that osteoblast maturation was accelerated in DDX5-knockdown cells. Moreover, RUNX2 was found to suppress DDX5 expression.26 These data indicate that although DDX5 coactivates RUNX2 activity, it inhibits osteoblast differentiation suggesting an osteoblast differentiation and maturation control at multiple levels. This implies that upregulation of DDX5 (Fig. 3 and Table 3) in the osteoblasts induced by the presence of Mg-implants may either help osteoblast differentiation via interacting with RUNX2 or delay their maturation in order to maintain their differentiation potential. Nevertheless, both possibilities will lead to increasing bone formation and should be further studied.
RNA binding motif protein, X-linked (RBMX). RBMX, also known as heterogeneous nuclear ribonucleoprotein G (hnRNPG), was upregulated in the presence of Mg-discs compared to osteoblast control cells (Fig. 3 and Table 3). RBMX belongs to the heterogeneous family of RNA binding proteins and has several roles in the regulation of pre- and posttranscriptional processes but the full range of roles is still not well known. Nevertheless, upregulation of RBMX during osteoblast differentiation has been reported.27
Non-POU domain containing octamer binding (NONO). The amount of NONO, a 54 kDa nuclear binding protein, was considerably increased in the presence of Mg-discs compared to Ti-discs and osteoblast control cells (Fig. 2 and Table 2). During chondrogenesis, NONO was found to directly interact with SRY (sex determining region Y)-box 9 (Sox9) transcription factor, promoting its transcriptional activity (especially on collagen, type II, alpha 1 (Col2a1) promoter activity).28 Its mutation also led to delayed enchondral ossification in the transgenic mice.28 Thus, Mg-implants may improve bone repair/formation indirectly by upregulation of NONO and subsequently ECM formation.
Peptidase D (PEPD). PEPD, a cytosolic exopeptidase able to hydrolyses imidodi- and imidotripeptides c-terminal to proline and hydroxyproline, was significantly upregulated in the presence of Mg-discs (Fig. 3 and Table 3). PEPD has a key role in the metabolism of collagen due to the enrichment of iminopeptides in both synthesis and degradation of collagen, type I.29 Reduction of bone strength and bone mass was observed in PEPD deficiency.29 Therefore, Mg-implants may support bone healing also by upregulation of PEPD, which is highly required for collagen synthesis and degradation.
Enolase2 (ENO2). One of significantly upregulated protein in presence of Mg-discs, compared to Ti-disc and control cells, is ENO2. ENO2 controls DeltaFosB which is an activator protein (AP)-1 transcription factor. An increase of DeltaFosB leads to an increase in bone mass and a decrease in adipose mass.30
Arylsulfatase B (ARSB). ARSB expression was increased in osteoblasts cultured on Mg-discs (Fig. 3 and Table 3). In contrast, there was no significant change in presence of Ti-discs. Vairo et al. showed that mutation of ARBS causes mucopolysaccharidosis VI, which is a seldom-genetic disease related to bone dysplasia. This mutation prevents this lysosomal enzyme to break down of glycosaminoglycans (GAG). In another study, Bhattacharyya et al.31 demonstrated that ARSB is involved in chondroitin 4-sulfate degradation (C4S; removing the 4-sulfate group) and biosynthesis via bone morphogenetic protein 4 (BMP4). Decreased in ARSB was leading to an increased C4S sulfation and a BMP4 sequestration. BMP are involved in bone and cartilage development and fracture repair. Hintze et al. demonstrated that GAG interact with BMP2 and thereby alter its receptor interaction in a sulfation-dependent manner.32 Therefore, it can be assumed that upregulation of ARSB in the presence of Mg-implants could induce an increase ECM remodelling/development. However, its effect on osteoblastogenesis may be dual: increased long-distance BMP signalling while decreasing the local effect(s).
Alanyl aminopeptidase, membrane (ANPEP). In the culture with Mg-discs, ANPEP, or Aminopeptidase N – CD13 expression was increased (Fig. 3 and Table 3). ANPEP is one of the surface marker proteins highly synthesised in adipose-derived mesenchymal stem cells (AMSC) and deciduous tooth stem cells (DTSCs). During the differentiation of these cells into osteoblasts, they synthesise ANPEP, which can therefore serve as osteogenic factor. Furthermore, ANPEP was also found as surface marker on bone derived mesenchymal stem cells. Fiddler et al. showed that the effect of tumour necrosis factor-alpha-induced apoptosis in neutrophils is regulated by ANPEP, though no other function for ANPEP has been ascribed in this cell.33
Glucosylceramidase beta (GBA). Another significantly upregulated protein in osteoblasts in presence of Mg-discs is the enzyme GBA (Fig. 3 and Table 3). Deficiency of GBA leads to Gaucher's disease (lysosomal storage disorder due to a deficiency of glucocerebrosidase) leading to skeletal complications such as remodelling failure, osteopenia, osteoporosis, and osteolysis. However, Gaucher's disease can be alleviated by GBA enzyme replacement therapy. Since Mg-discs stimulate the formation of GBA in osteoblasts in vitro, Mg-implants may have a positive effect on bone formation in vivo.
Nicotinamide phosphoribosyltransferase (NAMPT). NAMPT is involved in the initial steps of NAD+-biosynthetic pathway. In presence of Mg-discs NAMPT is upregulated in osteoblasts (Fig. 3 and Table 3). Li et al.34 found that age-related NAMPT-deficiency in mouse affects NAD resynthesis from nicotinamide, leads to low NAD levels. Consequently the NAD depended enzyme sirtuin-1, an inducer of adipogenesis, is decreased. This deacetylase deactivates also proteins involved in the cellular stress response therefore a multitude of potential effects on cell metabolism can be impacted via this pathway.
TIMP metallopeptidase inhibitor 1 (TIMP1). TIMP1 (also known as fibroblast collagenase inhibitor or collagenase inhibitor) was also significantly upregulated with Mg-discs (Fig. 3 and Table 3). TIMP1 belongs to the natural tissue inhibitors of matrix metalloproteinase (MMP), which mostly degrade the components of ECM. Bord et al.35 observed that low levels of TIMP1 in presence of MMPs caused extreme degradation of extracellular matrix in pathological bone and resulted in a decrease in heterotopic and osteophytic bone formation. They suggested that TIMP1 might act in the regulation of bone remodelling.35 Thus, Mg may exert, via an upregulation of TIMP1, a decreased MMPs driven ECM degradation.
Downregulated proteins.
Prostaglandin I2 (prostacyclin) synthase (PTGIS). Prostacyclin synthase (PTGIS) is one of most downregulated proteins in osteoblasts in presence of Mg-discs (Fig. 3 and Table 3). PTGIS, which belongs to prostaglandin family, regulates prostacyclin production. Addition of prostacyclin or its analogue to osteoblasts increases β-catenin, which enhances matrix mineralisation and differentiation. Tuncbilek et al.36 found that bone formation was stimulated by injection of a stable analogue of PTGIS. Furthermore, Nakalekha et al.37 observed a biphasic/age-dependent relationship between PTGIS and bone remodelling: in PTGIS-knockout mice, a decrease and enhanced bone mass and mineral density were measured in young mice (5 weeks old) and in adult mice (34 weeks old), respectively.36,37 Because of these contradictory observations, further in vivo experiments are important for clarifying if Mg-implants may have negative impacts on the bone healing in children. Inflammation may be increased.
Filamin A (FLNA). The downregulated FLNA promotes cytoskeleton network by binding and anchoring various transmembrane proteins to actin. Thus, FLNA is involved in various function like cell adhesion, migration, survival and intracellular trafficking. FLN can bind several type of integrins at their cytoplasmic tail thus competing with talin, thereby potentially inhibiting integrin and cell adhesion. Hong et al.38 and Alves et al.39 found an upregulation of FLNA in differentiating osteoblast. Mezawa et al.40 observed a higher density of but less organised collagen fibres (compared to wild type mice) after 2 weeks in filamin-knock down mice. In another study, Leung et al. found FLNA41 to have a critical role in actin-dependent osteoclastogenesis. Therefore, reduced FLNA in response to the presence of Mg-implants may delay osteoblast differentiation and should have a positive effect on bone formation since osteoclastogenesis might be downregulated in vivo during Mg-implant degradation.
Secreted protein, acidic, cysteine-rich (SPARC). SPARC or osteonectin is one of the most abundant protein found in the ECM, recognised by several integrins and probably modulates osteoblast function and/or differentiation as SPARC is believed to favoured hydroxyapatite crystal nucleation. Furthermore, SPARC has a crucial role in bone remodelling by stimulating plasminogen (a serine protease) activation. Plasminogen binds to collagen in the presence of SPARC leading to ECM proteolysis. Daci et al.42 demonstrated that plasminogen activator is vital for the degradation of non-collagenous bone matrix components. In their scenario where plasminogen activator was deficient, enhanced bone formation was observed. In conclusion, downregulation of SPARC in the presence of Mg-implants should have a negative impact on bone SPARC-dependent binding of plasminogen to collagen, thereby inhibiting the degradation of noncollagenous proteins present in the non-mineralised bone.
Palladin, cytoskeletal associated protein (PALLD). PALLD, an actin-associated protein, which has a significant role in the organisation of cytoskeleton and focal adhesion,43 was significantly downregulated in the osteoblast cell culture in presence of Mg-discs (Fig. 3 and Table 3). Wall et al.43 reported the upregulation of PALLD during osteogenic differentiation in adipose-derived adult stem cells. Their results showed the reduction of actin stress fibres when the PALLD gene was silenced but also a non-affected osteogenesis.
Transgelin (TAGLN). The 22 kDa actin binding protein, TAGLN, a TGFB-inducible gene, was downregulated in the osteoblasts during incubation with Mg-discs (Fig. 3 and Table 3). Transgelin suppresses the synthesis of matrix metalloproteinase 9 (MMP9 or collagenase, type IV) a. Nair et al. showed silencing the TAGLN gene in mice causes upregulation of MMP-9 compared to wild type mice.44 Furthermore, Esafadi et al.45 demonstrate that TAGLN overexpression (reciprocally for deficiency) decreases BMSC proliferation, but enhanced cell migration, osteoblastic and adipocytic differentiation, and in vivo bone formation.
Transglutaminase 2 (TGM2). TGM2 (downregulated in presence of Mg – Fig. 3 and Table 3) is an intra/extracellular protein-cross-linking enzyme, which functions in cell attachment and probably in the development of bone matrix. Osteopontin (a main non-collagenous protein in ECM) is one substrate for this enzyme. TGM2 catalyses inter- (e.g., with collagen) and intramolecular calcium-dependant cross-linking. Polymeric osteopontin has been shown to increase integrin-mediated cell adhesion, spreading, focal contact formation, and migration.
Prolyl 4-hydroxylase, alpha polypeptide II (P4HA2). A further significantly downregulated protein in osteoblasts in presence of Mg-discs is P4HA2 (Fig. 3 and Table 3), which plays a crucial role in collagen biosynthesis & triple helix stabilisation as lack of hydroxylation would lead to collagen intracellular degradation. This isoform predominates in chondrocytes and capillary endothelial cells. Furthermore, an increase of P4HA2 was measured in cartilage from patients with necrosis of the femoral head compared to normal hip cartilage.46 Thus, P4HA2 could have a role in ECM stability and turnover but it has to be noticed that expression of COL1A1 was also downregulated (see below).
Legumain (LGMN). LGMN, also known as osteoclast inhibitory peptide 2 (OIP2), inhibits osteoclast formation and bone resorption via its asparaginyl endopeptidase activity. Choi et al.47 found that osteoclast formation was promoted by treating bone marrow culture with the antibody against legumain. Furthermore, in vivo & in vitro, degradation of Mg-based implants has been associated with decreased osteoclast number and activity.1,8 However, osteoclastogenesis is needed to allow proper bone remodelling, thus, downregulation of LGMN in presence of Mg may counteract the global osteoclastogenesis inhibition.
Prolyl 3-hydroxylase 3 (P3H3) and cartilage associated protein (CRTAP). P3H3 and CRTAP are members of the leprecan family. Together in a complex with peptidylprolyl isomerase B (PPIB cyclophilin B), they process (hydroxylation) certain forms of collagen. Mutation of CRTAP and P3H3 genes causes recessive form of osteogenesis imperfecta. Furthermore, Wilson et al. measured a decreased expression of P3H3 and CRTAP during cartilage maturation.48 Thus, Mg may promotes endochondral ossification during tissue repair.
Calponin 3 (CNN3). CNN3 from the actin-binding family of proteins is decreased in osteoblasts in the presence of Mg-discs (Fig. 3 and Table 3). CNN3 functions in neuronal development, preventing cell fusion in trophoblasts and myoblasts and negatively regulates BMP-dependent signalling pathway in chondrocytes. Haag et al.49 saw a downregulation of calponin-3 in osteoarthritic joints where the BMP expressions is generally also reduced. Moreover, an increased expression of CNN3 was measured in human MSC driven toward myogenic differentiation. Thus, Mg-based implants may preferentially drive MSC into the chondrogenesis/osteogenesis pathway.
Collagens. The collagens COL1A1 and COL6A1 were decreased in the presence of Mg-discs. Collagens are the most abundant proteins in mammals and are mostly present extracellularly in skin, bones and the other tissues. They comprise 28 members, which play various roles in differentiation, proliferation and cell migration with diverse types serving as osteogenic markers. The COL1A1 gene is one of those involved in the enhancement of bone mass through the linkage of vitamin D receptor (VDR). The lack of either COL1A1 or COL1A2 is one of the causes of osteogenesis imperfecta, characterised by bone fragility. COL6A1 is found in the “pericellular matrix” (PCM) and encapsulates chondrocytes.50 A reduction of the mechanical properties of bone in COL6A1-knockdown mice was observed in the study of Alexopoulos et al.50 In another study, Ishibashi et al.51 found that bone mineralisation was improved when COL6A1 and COL6A2 accumulation was induced by addition of interleukin-4 in osteoblasts in vitro51 but the exact role of COL6A1 remains undefined. In the study of Egusa et al., undifferentiated bone marrow stromal cells (BMSCs) and adipose tissue-derived stromal cells (ASCs) were cultured in an osteogenic media and surprisingly, they observed a decrease in many osteogenic markers including different collagen types, especially the one of our interest here (i.e., COL1A1 and COL6A1) in BMSC. Accordingly, the downregulation of collagens observed here may indicate that either Mg-implants could affect bone stability or this is a normal situation during BMSC differentiation.
Mg is necessary for proper ATP synthesis and enzymatic reactions and has thus a central role in metabolism and cell growth.52 Furthermore, increased extracellular Mg promotes cell proliferation to some extent.5 Thus, high levels of Mg may increase production of ATP.
Regulated proteins involved in energy metabolism (Tables 2 and 3) are discussed below.
Adenosine triphosphate (ATP)ase H+ transporting V1 subunit B2 (ATP6V1B2). The multi-subunit complex protein ATP6V1B2, also called proton-pumping vacuolar-type ATPase, was also upregulated during the incubation of osteoblasts with Mg-discs (Fig. 3 and Table 3). V-ATPase is involved in several biological processes such membrane trafficking, receptor-mediated endocytosis and extracellular and intracellular (organelles) acidification. It may thus be involved in either regulation of intra- extracellular pH or material degradation. Furthermore, ATP6V1B2 plays a vital role in amelogenesis in enamel development. Sarkar et al. showed ATP6V1B2 is one of the four of V-type ATPase that is significantly upregulated via maturation stage of amelogenesis in development of tooth enamel.53 Hence, upregulation of ATP6V1B2 in the presence of Mg-implants may improve bone maturation.
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d (ATP5H). ATP5H or ATP synthase subunit d was strongly downregulated in presence of Ti-discs (Fig. 2 and Table 2). Hong et al.54 found in their study that ATP5 was downregulated in the osteoblastic cell line MC3T3-E1 under dexamethasone treatment and that osteoblast differentiation and proliferation was inhibited compared to control cells. However, this protein was not regulated in osteoblasts incubated with Mg-discs. Comparing the effect of both metals indicates that the Mg-implants do not affect osteoblastic differentiation through ATP5 unlike Ti.
Fatty acid binding protein 5 (FABP5). FABP5 was significantly upregulated in the presence of Mg (Fig. 3 and Table 3). FABP5 is a low molecular weight protein responsible for transferring intracellular fatty acids with a high affinity to n − 3 polyunsaturated fatty acids. Deficiency of polyunsaturated fatty acid causes a decrease in bone matrix formation, and an increase in calcium ion release from bones, which leads to osteoporosis.
Aldo-keto reductase family 1 member B (AKR1B1). AKR1B1 is one of the significantly upregulated proteins in osteoblasts with Mg-discs (Fig. 3 and Table 3). AKR1B1 is an aldo-keto-reductase and plays an important role in the polyol pathway of glucose metabolism. In cell culture, glucose is an essential factor regulating cell proliferation. Thus, it is logical that glucose metabolism is increased. Aldose reductase does not only reduce glucose to sorbitol but also releases various aldehydes and their glutathione-conjugates. Furthermore, AKR1B1 has a detoxifying role and it mediates oxidant stress in the cells during lipid peroxidation. This protein has a vital role in different inflammation diseases such as asthma and cancer. Consequently, upregulation of aldose reductase during the incubation of osteoblasts with Mg-implants might protect cells from oxidative stress, which may occur from corrosion.
Voltage-dependent anion channel 1 (VDAC1) and channel 2 (VDAC2). VDAC1 and VDAC2 are major proteins in mitochondrial outer membrane and control the mitochondrial membrane potential. VADC1 is involved in metabolism, regulates the passage of molecules and apoptosis. Brahimi-Horn et al. observed earlier mouse cell proliferation in Vdac1−/− knockout compared to wild type mouse. This implies that VDAC1 inhibits cell proliferation56 thus, its upregulation should be advantageous to the bone-forming osteoblast. VDAC2 interacts with the multidomain proapoptotic molecules (BCL2 antagonist/killer 1 or BAK1) and thus inhibits or modulates apoptosis. Morrisson et al. found a decrease in apoptosis in the presence of VDAC2 in tet-WT1A cells in vitro.57 Thus in the presence of Mg-implants a higher viability of osteoblasts is expected (Table 3).
Tripartite motif containing 28 (TRIM28) transcription intermediary factor 1-beta. TRIM28 (also known as transcription intermediary factor 1-beta or TIF1-beta) was also upregulated in the presence of Mg-discs (Fig. 3 and Table 2). This protein is a transcriptional corepressor, which binds to DNA in association with Kruppel-associated box domain-zinc finger proteins. TRIM28 prevents the development of auto-inflammatory T-cell in mammalian cells. In TRIM28-knock-out mice, apoptosis of hematopoietic stem cells (HSCs) is stimulated and exiting of HSCs from the bone marrow is promoted which leads to the rapid reduction of hematopoietic stem cells. Thus, according to the study of Miyagi et al.58 the upregulation of TIF1b in osteoblasts in presence of Mg-implants may increase cell viability.
Heterogeneous nuclear ribonucleoprotein U-like 1 (HNRNPUL1). HNRNPUL1 has a significant role in cellular response to deoxyribonucleic acid (DNA) double-strand breaks signalling, which is crucial for cell viability. Due to the upregulation of this protein in osteoblasts incubated with Mg-discs, we can conclude that Mg-implants may have a positive effect on cell viability.
Cadherin 13 (CDH13). CDH13 was upregulated in cells cultured on Ti-substrate (Fig. 2 and Table 2). The non-canonical CDH13 belongs to the cadherin superfamily but unlike the other members, this cadherin lacks a transmembrane domain or cytoplasmic region and is thus surface-anchored. On its extracellular domain, several amino acids responsible for homodimerisation and adhesion are also missing. CDH13 is expressed in several tissues from heart, skeletal muscle, brain, to bone (osteoblast). CDH13 has a dual role in cancer. On the one hand, CDH13 downregulation will increase numerous cancer cells type proliferation and invasion. On the other hand, CDH13 expression will be upregulated under hypoxia supporting angiogenesis and dissemination. Nevertheless, Zhang et al. measured an upregulation of CDH13 in enhanced tibial bone formation after ankle loading.59 Thus, upregulation of CDH13 in presence of Ti may reflect a promoted bone formation.
Superoxide dismutase 2, mitochondrial (SOD2). SOD2, which catalyses the dismutation of O2− into either O2 or H2O2, was upregulated in the presence of Mg-discs (Fig. 3 and Table 3). H2O2 has however been illustrated to be a main cause of bone resorption, favouring both osteoclastogenesis and the activity of mature osteoclast. This is also quite important in bone remodelling, as osteoclasts are essential for resorbing calcified tissue and promoting remodelling. Hence pathological conditions like osteoporosis start when cells are subject to oxidative stress with a weak antioxidant defence system thereby shifting the balance between bone formation and bone loss. It was also noticed that none of these proteins responsible for oxidative stress defence was downregulated in the presence of Mg-discs.
Aldehyde dehydrogenase 7 family, member A1 (ALDH7A1). ALDH7A1, also known as antiquitin, was significantly decreased in response to Ti-discs (Fig. 2 and Table 2). ALDH7A1 was first renowned for its involvement in lysine catabolism. However, its role for protecting cells against hyper-osmotic stress-induced apoptosis via generating osmolytes and detoxification of toxic aldehydes throughout oxidative stress was later recognised. Thus, ALDH7A1 is involved in several metabolic functions: detoxification, role in intermediary metabolism, protection from osmotic stress and generation of nicotinamide adenine dinucleotide phosphate (NADPH). Furthermore, Guo et al. demonstrated that ALDH7A1 was strongly associated with osteoporotic fractures.60 They further stated, “ALDH7A1 degrades and detoxifies acetaldehyde, which inhibits osteoblast proliferation and results in decreased bone formation”.
Taking the increased protein expressions together.
4. Conclusions
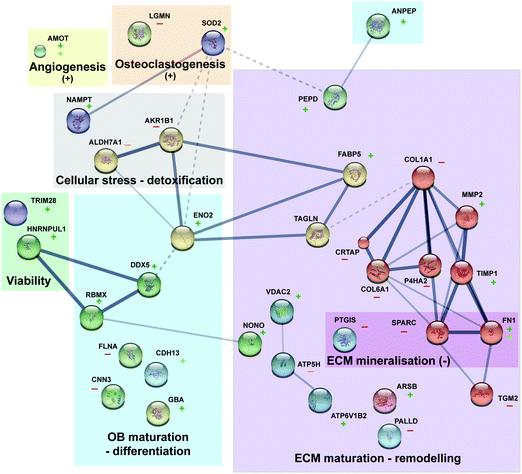
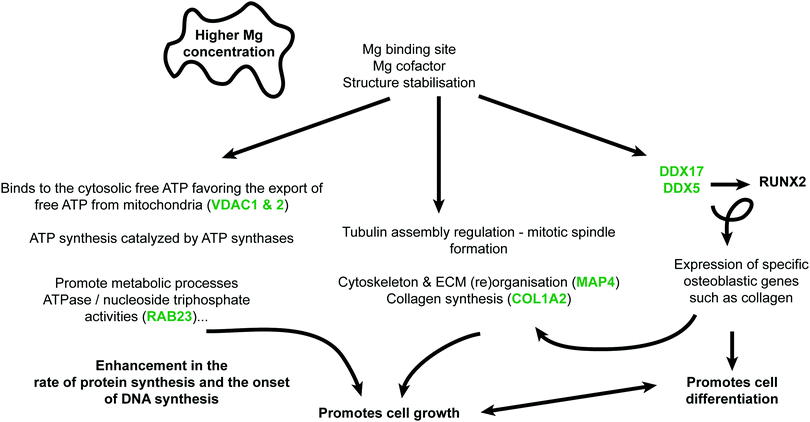
Primordially, no adverse effect was noticed on phenotypic observations of osteoblast cells cultured on Mg or Ti samples. The results of this study confirm that Mg alloys as well as Ti alloys have individual effects on osteoblasts, which are mirrored by changes in the protein patterns. The overall impact of degrading Mg on osteoblast is significant larger and multifaceted compared to Ti metal discs. In accordance with the postulated osteoprotective effect of Mg, some osteogenesis-related proteins were regulated more significantly in the presence of Mg-discs compared to Ti-disc. All discussed proteins and their functions are presented in Fig. 4 Furthermore, an important effect of Mg on protein involved in extracellular matrix (re)modelling was observed (e.g., few proteins related to collagen synthesis were downregulated). A general balance between osteoblast proliferation (TAGLN), maturation (DDX5), differentiation (RBMX) and survival (TRIM28 and HRNPUL1) was highlighted here with several acting and nuancing proteins. As already presented in an osteoblast/osteoclast coculture model cultured with various concentrations of Mg-extract,8 Mg is favouring osteoblastogenesis but do not completely abolish osteoclastogenesis, a prerequisite for suitable bone fracture healing and remodelling. Indeed, (e.g.) downregulation of LGMN and upregulation of SOD2 may favour osteoclastogenesis. This study is the first step toward deeper mechanistic studies. However, after these proteomics analyses, preliminary mechanisms can be assumed (Fig. 5). The Mg-regulated proteins can be classified into “biological processes” and “molecular functions” (analyses done via STRING website, https://string-db.org, version 11.0, see Table S1, ESI†). Knowledge about Mg actions in biology and analyses of the found biological processes and molecular functions, led to the assembly of Fig. 5. Indeed three main blocks explaining here the Mg effect can be found. First, Mg is necessary for proper ATP synthesis and enzymatic reactions involving ATP require Mg.Generally, Mg2+ is rather found in mitochondrial matrix than in the cytosol, mediating ADP/ATP exchange between the cytosol and matrix (i.e., import of free ADP in mitochondria).52 However, higher Mg cytosolic concentration favours the export of free ATP from mitochondria (e.g., upregulation of VDAC1, protein controlling ions, ATP, and other small metabolites across the mitochondrial membrane). This rise of available ATP can explain the overall increase of metabolic processes or ATase activities (e.g., GO:0009987 cellular process, GO:0071840 cellular component organization or biogenesis and GO:0008152 metabolic process). Furthermore, increased cytosolic and extracellular Mg would lead to augmented interaction with cytoskeleton or ECM proteins (e.g., GO:0007010 cytoskeleton organization). Together with the increased available ATP, this may, favour lead to a better spindle organisation, collagen synthesis or cell differentiation. Finally, increase expression of specific proteins such as DDX5 and DDX17 are directly linked to osteoblastogenesis via their effect on RUNX2 (e.g., triggering the expression of major bone matrix genes-collagens). These three axes will be further studied in vitro. Furthermore, supplementary in vivo studies will be performed in the future in order to clarify whether these effects are also induced by the presence of metal implants in the healing bone. Similar analyses for Ti were not possible with so few regulated proteins. However, FN1, CDH13, ACTBL2 are the most regulated ones which may reflect a specific sample surface response due to (e.g.,) mechanical mismatching rather than a triggered “active biological answer” as observed for Mg.
Conflicts of interest
The authors confirm having neither competing interests nor conflict of interest.Acknowledgements
Gabriele Salamon is acknowledged for expert technical assistance with cell culture. Schön-Klinik Eilbek, Hamburg is acknowledged for the supply of the primary cells.References
- K. Jahn, H. Saito, H. Taipaleenmaki, A. Gasser, N. Hort, F. Feyerabend, H. Schluter, J. M. Rueger, W. Lehmann, R. Willumeit-Romer and E. Hesse, Intramedullary Mg2Ag nails augment callus formation during fracture healing in mice, Acta Biomater., 2016, 36, 350–360 CrossRef PubMed.
- Y. Zhang, J. Xu, Y. C. Ruan, M. K. Yu, M. O'Laughlin, H. Wise, D. Chen, L. Tian, D. Shi, J. Wang, S. Chen, J. Q. Feng, D. H. Chow, X. Xie, L. Zheng, L. Huang, S. Huang, K. Leung, N. Lu, L. Zhao, H. Li, D. Zhao, X. Guo, K. Chan, F. Witte, H. C. Chan, Y. Zheng and L. Qin, Implant-derived magnesium induces local neuronal production of CGRP to improve bone-fracture healing in rats, Nat. Med., 2016, 22, 1160–1169 CrossRef CAS PubMed.
- R. K. Rude and H. E. Gruber, Magnesium deficiency and osteoporosis: animal and human observations, J. Nutr. Biochem., 2004, 15, 710–716 CrossRef CAS PubMed.
- R. K. Rude, H. E. Gruber, L. Y. Wei, A. Frausto and B. G. Mills, Magnesium deficiency: effect on bone and mineral metabolism in the mouse, Calcif. Tissue Int., 2003, 72, 32–41 CrossRef CAS PubMed.
- A. Burmester, R. Willumeit-Romer and F. Feyerabend, Behavior of bone cells in contact with magnesium implant material, J. Biomed. Mater. Res., Part B, 2017, 105, 165–179 CrossRef CAS PubMed.
- M. Haga, N. Fujii, K. Nozawa-Inoue, S. Nomura, K. Oda, K. Uoshima and T. Maeda, Detailed process of bone remodeling after achievement of osseointegration in a rat implantation model, Anat. Rec., 2009, 292, 38–47 CrossRef PubMed.
- N. J. Lakhkar, I. H. Lee, H. W. Kim, V. Salih, I. B. Wall and J. C. Knowles, Bone formation controlled by biologically relevant inorganic ions: role and controlled delivery from phosphate-based glasses, Adv. Drug Delivery Rev., 2013, 65, 405–420 CrossRef CAS PubMed.
- L. Wu, F. Feyerabend, A. F. Schilling, R. Willumeit-Romer and B. J. C. Luthringer, Effects of extracellular magnesium extract on the proliferation and differentiation of human osteoblasts and osteoclasts in coculture, Acta Biomater., 2015, 27, 294–304 CrossRef CAS PubMed.
- R. M. Lozano, B. T. Perez-Maceda, M. Carboneras, E. Onofre-Bustamante, M. C. Garcia-Alonso and M. L. Escudero, Response of MC3T3-E1 osteoblasts, L929 fibroblasts, and J774 macrophages to fluoride surface-modified AZ31 magnesium alloy, J. Biomed. Mater. Res., Part A, 2013, 101, 2753–2762 CrossRef PubMed.
- J. Gonzalez, R. Q. Hou, E. P. S. Nidadavolu, R. Willumeit-Romer and F. Feyerabend, Magnesium degradation under physiological conditions – Best practice, Bioact. Mater., 2018, 3, 174–185 CrossRef PubMed.
- J. Fischer, D. Pröfrock, N. Hort, R. Willumeit and F. Feyerabend, Improved cytotoxicity testing of magnesium materials, Mater. Sci. Eng., B, 2011, 176, 830–834 CrossRef CAS.
- M. Kwiatkowski, M. Wurlitzer, M. Omidi, L. Ren, S. Kruber, R. Nimer, W. D. Robertson, A. Horst, R. J. Miller and H. Schluter, Ultrafast extraction of proteins from tissues using desorption by impulsive vibrational excitation, Angew. Chem., Int. Ed., 2015, 54, 285–288 CrossRef CAS PubMed.
- M. Kwiatkowski, M. Wurlitzer, A. Krutilin, P. Kiani, R. Nimer, M. Omidi, A. Mannaa, T. Bussmann, K. Bartkowiak, S. Kruber, S. Uschold, P. Steffen, J. Lubberstedt, N. Kupker, H. Petersen, R. Knecht, N. O. Hansen, A. Zarrine-Afsar, W. D. Robertson, R. J. Miller and H. Schluter, Homogenization of tissues via picosecond-infrared laser (PIRL) ablation: giving a closer view on the in-vivo composition of protein species as compared to mechanical homogenization, J. Proteomics, 2016, 134, 193–202 CrossRef CAS PubMed.
- J. Cox, M. Y. Hein, C. A. Luber, I. Paron, N. Nagaraj and M. Mann, Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ, Mol. Cell. Proteomics, 2014, 13, 2513–2526 CrossRef CAS PubMed.
- C. UniProt, The Universal Protein Resource (UniProt), Nucleic Acids Res., 2010, 38, D142–D148 CrossRef PubMed.
- J. Cox and M. Mann, 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data, BMC Bioinf., 2012, 13(suppl. 16), S12 CrossRef CAS PubMed.
- J. H. Lee and J. Y. Cho, Proteomics approaches for the studies of bone metabolism, BMB Rep., 2014, 47, 141–148 CrossRef PubMed.
- S. Kim, W. C. Myung, J. S. Lee, J. K. Cha, U. W. Jung, H. C. Yang, I. S. Lee and S. H. Choi, The effect of fibronectin-coated implant on canine osseointegration, J. Periodontal Implant Sci., 2011, 41, 242–247 CrossRef CAS PubMed.
- W. B. Zhang and L. Wang, Label-free quantitative proteome analysis of skeletal tissues under mechanical load, J. Cell. Biochem., 2009, 108, 600–611 CrossRef CAS PubMed.
- A. Ortiz, Y. C. Lee, G. Yu, H. C. Liu, S. C. Lin, M. A. Bilen, H. Cho, L. Y. Yu-Lee and S. H. Lin, Angiomotin is a novel component of cadherin-11/beta-catenin/p120 complex and is critical for cadherin-11-mediated cell migration, FASEB J., 2015, 29, 1080–1091 CrossRef CAS PubMed.
- X. Niu, W. Chang, R. Liu, R. Hou, J. Li, C. Wang, X. Li and K. Zhang, mRNA and protein expression of the angiogenesis-related genes EDIL3, AMOT and ECM1 in mesenchymal stem cells in psoriatic dermis, Clin. Exp. Dermatol., 2016, 41, 533–540 CrossRef CAS PubMed.
- A. Chaya, S. Yoshizawa, K. Verdelis, N. Myers, B. J. Costello, D. T. Chou, S. Pal, S. Maiti, P. N. Kumta and C. Sfeir, In vivo study of magnesium plate and screw degradation and bone fracture healing, Acta Biomater., 2015, 18, 262–269 CrossRef CAS PubMed.
- J. Zhuang, Y. Jing, Y. Wang, J. Zhang, H. Xie and J. Yan, Degraded and osteogenic properties of coated magnesium alloy AZ31; an experimental study, J. Orthop. Surg. Res., 2016, 11, 30 CrossRef PubMed.
- M. Egeblad and Z. Werb, New functions for the matrix metalloproteinases in cancer progression, Nat. Rev. Cancer, 2002, 2, 161–174 CrossRef CAS PubMed.
- E. D. Jensen, L. Niu, G. Caretti, S. M. Nicol, N. Teplyuk, G. S. Stein, V. Sartorelli, A. J. van Wijnen, F. V. Fuller-Pace and J. J. Westendorf, p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation, J. Cell. Biochem., 2008, 103, 1438–1451 CrossRef CAS PubMed.
- F. V. Fuller-Pace and S. Ali, The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators, Biochem. Soc. Trans., 2008, 36, 609–612 CrossRef CAS PubMed.
- L. J. Foster, P. A. Zeemann, C. Li, M. Mann, O. N. Jensen and M. Kassem, Differential expression profiling of membrane proteins by quantitative proteomics in a human mesenchymal stem cell line undergoing osteoblast differentiation, Stem Cells, 2005, 23, 1367–1377 CrossRef CAS PubMed.
- K. Hata, R. Nishimura, S. Muramatsu, A. Matsuda, T. Matsubara, K. Amano, F. Ikeda, V. R. Harley and T. Yoneda, Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice, J. Clin. Invest., 2008, 118, 3098–3108 CrossRef CAS PubMed.
- R. Besio, S. Maruelli, R. Gioia, I. Villa, P. Grabowski, O. Gallagher, N. J. Bishop, S. Foster, E. De Lorenzi, R. Colombo, J. L. Diaz, H. Moore-Barton, C. Deshpande, H. I. Aydin, A. Tokatli, B. Kwiek, C. S. Kasapkara, E. O. Adisen, M. A. Gurer, M. Di Rocco, J. M. Phang, T. M. Gunn, R. Tenni, A. Rossi and A. Forlino, Lack of prolidase causes a bone phenotype both in human and in mouse, Bone, 2015, 72, 53–64 CrossRef CAS PubMed.
- G. Sabatakos, N. A. Sims, J. Chen, K. Aoki, M. B. Kelz, M. Amling, Y. Bouali, K. Mukhopadhyay, K. Ford, E. J. Nestler and R. Baron, Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis, Nat. Med., 2000, 6, 985–990 CrossRef CAS PubMed.
- S. Bhattacharyya, L. Feferman and J. K. Tobacman, Regulation of chondroitin-4-sulfotransferase (CHST11) expression by opposing effects of arylsulfatase B on BMP4 and Wnt9A, Biochim. Biophys. Acta, 2015, 1849, 342–352 CrossRef CAS PubMed.
- V. Hintze, S. A. Samsonov, M. Anselmi, S. Moeller, J. Becher, M. Schnabelrauch, D. Scharnweber and M. T. Pisabarro, Sulfated glycosaminoglycans exploit the conformational plasticity of bone morphogenetic protein-2 (BMP-2) and alter the interaction profile with its receptor, Biomacromolecules, 2014, 15, 3083–3092 CrossRef CAS PubMed.
- C. A. Fiddler, H. Parfrey, A. S. Cowburn, D. Luo, G. B. Nash, G. Murphy and E. R. Chilvers, The Aminopeptidase CD13 Induces Homotypic Aggregation in Neutrophils and Impairs Collagen Invasion, PLoS One, 2016, 11, e0160108 CrossRef PubMed.
- Y. Li, X. He, Y. Li, J. He, B. Anderstam, G. Andersson and U. Lindgren, Nicotinamide phosphoribosyltransferase (Nampt) affects the lineage fate determination of mesenchymal stem cells: a possible cause for reduced osteogenesis and increased adipogenesis in older individuals, J. Bone Miner. Res., 2011, 26, 2656–2664 CrossRef CAS PubMed.
- S. Bord, A. Horner, C. A. Beeton, R. M. Hembry and J. E. Compston, Tissue inhibitor of matrix metalloproteinase-1 (TIMP-1) distribution in normal and pathological human bone, Bone, 1999, 24, 229–235 CrossRef CAS PubMed.
- G. Tuncbilek, P. Korkusuz and F. Ozgur, Effects of iloprost on calvarial sutures, J. Craniofac. Surg., 2008, 19, 1472–1480 CrossRef PubMed.
- C. Nakalekha, C. Yokoyama, H. Miura, N. Alles, K. Aoki, K. Ohya and I. Morita, Increased bone mass in adult prostacyclin-deficient mice, J. Endocrinol., 2010, 204, 125–133 CAS.
- D. Hong, H. X. Chen, H. Q. Yu, Y. Liang, C. Wang, Q. Q. Lian, H. T. Deng and R. S. Ge, Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells, Exp. Cell Res., 2010, 316, 2291–2300 CrossRef CAS PubMed.
- R. D. Alves, M. Eijken, S. Swagemakers, H. Chiba, M. K. Titulaer, P. C. Burgers, T. M. Luider and J. P. van Leeuwen, Proteomic analysis of human osteoblastic cells: relevant proteins and functional categories for differentiation, J. Proteome Res., 2010, 9, 4688–4700 CrossRef CAS PubMed.
- M. Mezawa, V. I. Pinto, M. P. Kazembe, W. S. Lee and C. A. McCulloch, Filamin A regulates the organization and remodeling of the pericellular collagen matrix, FASEB J., 2016, 30, 3613–3627 CrossRef CAS PubMed.
- R. Leung, Y. Wang, K. Cuddy, C. Sun, J. Magalhaes, M. Grynpas and M. Glogauer, Filamin A regulates monocyte migration through Rho small GTPases during osteoclastogenesis, J. Bone Miner. Res., 2010, 25, 1077–1091 CAS.
- E. Daci, V. Everts, S. Torrekens, E. Van Herck, W. Tigchelaar-Gutterr, R. Bouillon and G. Carmeliet, Increased bone formation in mice lacking plasminogen activators, J. Bone Miner. Res., 2003, 18, 1167–1176 CrossRef CAS PubMed.
- M. E. Wall, A. Rachlin, C. A. Otey and E. G. Loboa, Human adipose-derived adult stem cells upregulate palladin during osteogenesis and in response to cyclic tensile strain, Am. J. Physiol., 2007, 293, C1532–C1538 CrossRef CAS PubMed.
- R. R. Nair, J. Solway and D. D. Boyd, Expression cloning identifies transgelin (SM22) as a novel repressor of 92 kDa type IV collagenase (MMP-9) expression, J. Biol. Chem., 2006, 281, 26424–26436 CrossRef CAS PubMed.
- M. Elsafadi, M. Manikandan, R. A. Dawud, N. M. Alajez, R. Hamam, M. Alfayez, M. Kassem, A. Aldahmash and A. Mahmood, Transgelin is a TGFbeta-inducible gene that regulates osteoblastic and adipogenic differentiation of human skeletal stem cells through actin cytoskeleston organization, Cell Death Discovery, 2016, 7, e2321 CrossRef CAS PubMed.
- R. Liu, Q. Liu, K. Wang, X. Dang and F. Zhang, Comparative analysis of gene expression profiles in normal hip human cartilage and cartilage from patients with necrosis of the femoral head, Arthritis Res. Ther., 2016, 18, 98 CrossRef PubMed.
- S. J. Choi, S. V. Reddy, R. D. Devlin, C. Menaa, H. Chung, B. F. Boyce and G. D. Roodman, Identification of human asparaginyl endopeptidase (legumain) as an inhibitor of osteoclast formation and bone resorption, J. Biol. Chem., 1999, 274, 27747–27753 CrossRef CAS PubMed.
- R. Wilson, E. L. Norris, B. Brachvogel, C. Angelucci, S. Zivkovic, L. Gordon, B. C. Bernardo, J. Stermann, K. Sekiguchi, J. J. Gorman and J. F. Bateman, Changes in the chondrocyte and extracellular matrix proteome during post-natal mouse cartilage development, Mol. Cell. Proteomics, 2012, 11, M111.014159 CrossRef PubMed.
- J. Haag and T. Aigner, Identification of calponin 3 as a novel Smad-binding modulator of BMP signaling expressed in cartilage, Exp. Cell Res., 2007, 313, 3386–3394 CrossRef CAS PubMed.
- L. G. Alexopoulos, I. Youn, P. Bonaldo and F. Guilak, Developmental and osteoarthritic changes in Col6a1-knockout mice: biomechanics of type VI collagen in the cartilage pericellular matrix, Arthritis Rheum., 2009, 60, 771–779 CrossRef CAS PubMed.
- H. Ishibashi, S. Harumiya and Y. Koshihara, Involvement of type VI collagen in interleukin-4-induced mineralization by human osteoblast-like cells in vitro, Biochim. Biophys. Acta, 1999, 1472, 153–164 CrossRef CAS.
- E. Gout, F. Rebeille, R. Douce and R. Bligny, Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: unravelling the role of Mg2+ in cell respiration, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E4560–E4567 CrossRef CAS PubMed.
- J. Sarkar, X. Wen, E. J. Simanian and M. L. Paine, V-type ATPase proton pump expression during enamel formation, Matrix Biol., 2016, 52–54, 234–245 CrossRef CAS PubMed.
- D. Hong, H. X. Chen, H. Q. Yu, C. Wang, H. T. Deng, Q. Q. Lian and R. S. Ge, Quantitative proteomic analysis of dexamethasone-induced effects on osteoblast differentiation, proliferation, and apoptosis in MC3T3-E1 cells using SILAC, Osteoporosis Int., 2011, 22, 2175–2186 CrossRef CAS PubMed.
- T. Moriishi, R. Fukuyama, T. Miyazaki, T. Furuichi, M. Ito and T. Komori, Overexpression of BCLXL in Osteoblasts Inhibits Osteoblast Apoptosis and Increases Bone Volume and Strength, J. Bone Miner. Res., 2016, 31, 1366–1380 CrossRef CAS PubMed.
- M. C. Brahimi-Horn, S. Giuliano, E. Saland, S. Lacas-Gervais, T. Sheiko, J. Pelletier, I. Bourget, F. Bost, C. Feral, E. Boulter, M. Tauc, M. Ivan, B. Garmy-Susini, A. Popa, B. Mari, J. E. Sarry, W. J. Craigen, J. Pouyssegur and N. M. Mazure, Knockout of Vdac1 activates hypoxia-inducible factor through reactive oxygen species generation and induces tumor growth by promoting metabolic reprogramming and inflammation, Cancer Metabol., 2015, 3, 8 CrossRef PubMed.
- D. J. Morrison, M. A. English and J. D. Licht, WT1 induces apoptosis through transcriptional regulation of the proapoptotic Bcl-2 family member Bak, Cancer Res., 2005, 65, 8174–8182 CrossRef CAS PubMed.
- S. Miyagi, S. Koide, A. Saraya, G. R. Wendt, M. Oshima, T. Konuma, S. Yamazaki, M. Mochizuki-Kashio, Y. Nakajima-Takagi, C. Wang, T. Chiba, I. Kitabayashi, H. Nakauchi and A. Iwama, The TIF1beta-HP1 system maintains transcriptional integrity of hematopoietic stem cells, Stem Cell Rep., 2014, 2, 145–152 CrossRef CAS PubMed.
- P. Zhang, C. H. Turner and H. Yokota, Joint loading-driven bone formation and signaling pathways predicted from genome-wide expression profiles, Bone, 2009, 44, 989–998 CrossRef CAS PubMed.
- Y. Guo, L. J. Tan, S. F. Lei, T. L. Yang, X. D. Chen, F. Zhang, Y. Chen, F. Pan, H. Yan, X. Liu, Q. Tian, Z. X. Zhang, Q. Zhou, C. Qiu, S. S. Dong, X. H. Xu, Y. F. Guo, X. Z. Zhu, S. L. Liu, X. L. Wang, X. Li, Y. Luo, L. S. Zhang, M. Li, J. T. Wang, T. Wen, B. Drees, J. Hamilton, C. J. Papasian, R. R. Recker, X. P. Song, J. Cheng and H. W. Deng, Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis, PLoS Genet., 2010, 6, e1000806 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0mt00028k |
| This journal is © The Royal Society of Chemistry 2020 |