Open Access Article
Open Access ArticleRemodeling of Zn2+ homeostasis upon differentiation of mammary epithelial cells†
Yu
Han
ab,
Lynn
Sanford
ab,
David M.
Simpson
ab,
Robin D.
Dowell
bc and
Amy E.
Palmer
 *ab
*ab
aDepartment of Biochemistry, University of Colorado Boulder, 3415 Colorado Ave., Boulder, CO 80303, USA. E-mail: Amy.palmer@colorado.edu; Tel: +1-303-492-1945
bBioFrontiers Institute, University of Colorado Boulder, Boulder, CO 80303, USA
cDepartment of Molecular, Cellular and Developmental Biology, University of Colorado Boulder, Boulder, CO 80309, USA
First published on 10th January 2020
Abstract
Zinc is the second most abundant transition metal in humans and an essential nutrient required for growth and development of newborns. During lactation, mammary epithelial cells differentiate into a secretory phenotype, uptake zinc from blood circulation, and export it into mother's milk. At the cellular level, many zinc-dependent cellular processes, such as transcription, metabolism of nutrients, and proliferation are involved in the differentiation of mammary epithelial cells. Using mouse mammary epithelial cells as a model system, we investigated the remodeling of zinc homeostasis during differentiation induced by treatment with the lactogenic hormones cortisol and prolactin. RNA-Seq at different stages of differentiation revealed changes in global gene expression, including genes encoding zinc-dependent proteins and regulators of zinc homeostasis. Increases in mRNA levels of three zinc homeostasis genes, Slc39a14 (ZIP14) and metallothioneins (MTs) I and II were induced by cortisol but not by prolactin. The cortisol-induced increase was partially mediated by the nuclear glucocorticoid receptor signaling pathway. An increase in the cytosolic labile Zn2+ pool was also detected in lactating mammary cells, consistent with upregulation of MTs. We found that the zinc transporter ZIP14 was important for the expression of a major milk protein, whey acid protein (WAP), as knockdown of ZIP14 dramatically decreased WAP mRNA levels. In summary, our study demonstrated remodeling of zinc homeostasis upon differentiation of mammary epithelial cells resulting in changes in cytosolic Zn2+ and differential expression of zinc homeostasis genes, and these changes are important for establishing the lactation phenotype.
Significance to metallomicsIn this manuscript we take a global approach to examine how transformation from a proliferative to a differentiated secretory state alters zinc levels, zinc regulatory genes, and zinc dependent pathways in mammary epithelial cells. This work provides insight into how epithelial cells remodel zinc homeostasis during lactation while also shedding light on the regulation of zinc transporters themselves. |
Introduction
Zinc (Zn2+) is an essential trace element in humans and its importance is underscored by the discovery that up to 10% of the proteins encoded by the human genome are predicted to contain at least one zinc binding site.1 Zinc-dependent proteins play critical roles in DNA synthesis, transcriptional regulation, and metabolic processes, and hence are important for many cellular functions.1,2 Zinc homeostasis refers to the process by which Zn2+ is acquired, distributed and maintained in tissues and cells, and homeostasis is dynamically regulated to meet the specific need for Zn2+ by proteins under different biological conditions. Many of the key players in mammalian Zn2+ homeostasis have been identified. These players include transporters (genes Slc39a1-14 encode ZIP proteins and genes Slc30a1-10 encode ZnT proteins) that are responsible for the import and export of Zn2+ across the plasma membrane and intracellular organelle membranes, buffers such as metallothioneins (MTs) that have been suggested to regulate labile Zn2+ levels in cells, and transcription factors, such as MTF-1, that regulate expression of MTs and Zn2+ exporters.2,3 While we know many of the proteins involved in mammalian Zn2+ homeostasis, we still lack a comprehensive picture of how cells sense and regulate Zn2+ availability and how the cooperative action of Zn2+ transporters, buffers and transcriptional regulators is coordinated. For example, while MTF-1 regulates expression of MTs, ZnT1, and ZnT2, how the expression of other zinc homeostasis proteins is controlled in order for cells to respond to shifting Zn2+ needs is unclear.Mammary epithelial cells (MECs) are the basic units of milk production and undergo differentiation from a proliferative to a secretory phenotype during lactation in order to secrete large amounts of zinc (1–3 mg per day) as well as other nutrients into mother's milk.4 Many of the changes in cell physiology that accompany differentiation are zinc-dependent. For example, transcriptional remodeling requires zinc-dependent transcription factors, proliferation is a zinc-dependent process,5 and matrix metalloproteinases (MMPs) are zinc-dependent enzymes that are secreted by MECs to remodel the architecture of the mammary gland and whose expression levels are regulated in a temporal fashion throughout lactation.6 Previous work has revealed that the level of total Zn2+ and the expression level of many Zn2+ transporters is altered in the mouse lactating mammary gland during lactation.7,8 However, a comprehensive global analysis of the changes in Zn2+ homeostasis and Zn2+-dependent molecular processes over the course of differentiation has not been carried out.
In this study, we used HC11 cells as a model system to define changes in Zn2+ homeostasis in lactating MECs. HC11 mouse mammary epithelial cells can be differentiated into a secretory phenotype upon treatment with lactogenic hormones (prolactin and cortisol) and have been widely used to recapitulate molecular and cellular processes in lactating MECs.9,10 In order to provide a comprehensive picture of how Zn2+ homeostasis and Zn2+-dependent processes change over the course of differentiation, we performed next-generation RNA sequencing of HC11 cells to examine global changes in gene expression. Our analysis identified that the majority of zinc homeostasis genes were differentially regulated at a late stage of cell differentiation. Secondly, we quantified the labile cytosolic Zn2+ pool over the course of HC11 differentiation using genetically-encoded Zn2+ FRET sensors and discovered a 50% increase in cytosolic Zn2+ in differentiated cells. Lastly, we identified that a cortisol-induced transporter, ZIP14, contributes to the increase of cytosolic Zn2+ level and is important for the mRNA expression of whey acid protein, a major milk protein, suggesting an important association between zinc homeostasis and milk production.
Methods
Chemicals and reagents
Tris(2-pyridylmethyl)amine (TPA) was purchased from Sigma-Aldrich (catalog number 723134) and diluted with dimethyl sulfoxide (DMSO) to prepare 20 mM stock solutions. Stock solutions (5 mM) of 2-mercaptopyridine N-oxide (pyrithione, Sigma-Aldrich, catalog number 188549) were prepared in DMSO. Nuclear staining was achieved with NucBlue Live ReadyProbes Reagent (Thermo Fisher, catalog number R37605) for live cells or Hoechst 33258 (Sigma-Aldrich 861405) for fixed cells. An aqueous ZnCl2 solution (1 mM) was diluted in phosphate-free HEPES-buffered Hanks’ balanced salt solution (HHBSS, 1.26 mM CaCl2, 1.1 mM MgCl2, 5.36 mM KCl, 137 mM NaCl, 16.65 mM D-glucose, and 30 mM HEPES, pH 7.4) to make a 200 μM ZnCl2 stock solution. An insulin stock solution (4 mg mL−1) was purchased from Life Technologies (catalog number 12585014). Human recombinant epidermal growth factor (EGF, VWR 47743-566) was prepared as a 10 μg mL−1 stock solution in water. Prolactin (Sigma-Aldrich L6520) was prepared as a 5 mg mL−1 stock solution in water. A stock solution (138 μM) of hydrocortisone (Sigma-Aldrich H0888) in absolute ethanol was also prepared. Trace-metal grade nitric acid (65–70%) was purchased from FLUKA (02650). For ICP-MS experiments, Y standard solution (10 ppm) was purchased from Inorganic Ventures (IV-stock-53-125 mL), Ga standard solution (1000 ppm) was purchased from Inorganic Ventures (ICP – CGGA1-125ML) and Zn standard solution (1000 ppm) was purchased from VWR (Cat. No. RCMSZN1KN-100).Cell culture
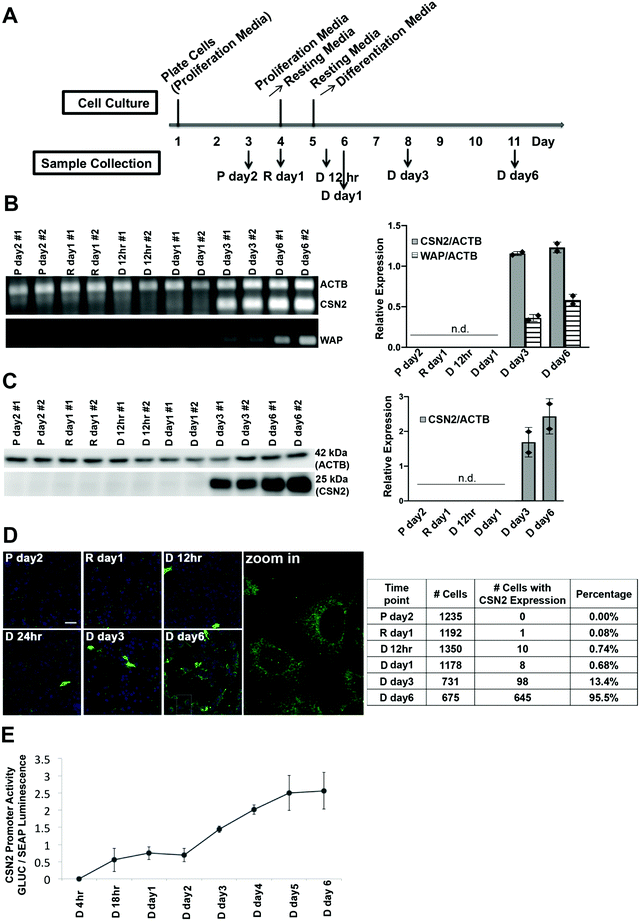
HC11 cells were grown and maintained in proliferation (P) medium (RPMI supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) penicillin/streptomycin (pen/strep), 10 ng mL−1 EGF and 5 μg mL−1 insulin). To induce cell differentiation, 4 × 105 cells were plated in a 35 mm dish (or 1 × 106 cells in a 10 cm dish) in proliferation medium. Cells reached confluency after 3 days. Cells were maintained in resting (R) medium containing RPMI, 2% FBS (v/v), 1% pen/strep (v/v), 5 μg mL−1 insulin for one additional day, then treated with differentiation (D) medium containing 2% FBS, 1% pen/strep, 5 μg mL−1 insulin, 5 μg mL−1 prolactin, and 1 μM hydrocortisone for up to 6 days. Differentiation medium was changed every 2 days. In P media the cells doubled every 36 h. Proliferation was generally inhibited when the media was switched to R. The differentiated cells exhibited different morphology compared to cells treated with R for the same time period. Differentiated cells were clumpier and more clustered (Fig. S1, ESI†), as observed in previous studies.11,12 Samples were collected at the following time points for downstream analysis: P day 2 (cells growing in proliferation medium for 2 days since plated), R day 1 or day 6 (cells growing in resting medium for 24 h or 7 days), D 12 h, 24 h, day 3 and day 6 (cells incubated in differentiation medium for the corresponding time).Lentiviral transduction
HEK293T cells were maintained in RPMI medium supplemented with 10% FBS and transfected with lentiviral packaging plasmids and viral expression plasmids (which encode the promoter reporter constructs or ZIP14 shRNAs) using the TransIT-LT1 (Mirus Bio) reagent following the manufacturer's instruction. The packaging plasmids were a gift from Dr Hubert Yin (University of Colorado, Boulder), containing pRev, which encodes the reverse transcriptase protein (Rev), pMDL, which encodes the Gag and Pol proteins, and pVSV-G, which encodes the Env protein. Media was changed at 24 h after transfection. By day 3 after transfection, media was filtered using a 0.22 μm filter and the virus-containing media supplemented with 8 μg mL−1 of polybrene was added to HC11 cells. After 3 days, puromycin (3 μg mL−1) was added to HC11 cells to select for cells with stable expression of exogenous genes. Cells that survived puromycin selection were recovered in proliferation medium and stored in liquid nitrogen for future use.shRNA knockdown (KD) of ZIP14
Four individual ZIP14-targeted shRNA constructs were stably expressed in HC11 cells using lentiviral transduction (Genecopoeia, Cat. No. LVRU6MP-MSH038111-31,32,33,34). Transduced cells were selected by puromycin and expression of a red fluorescent protein (mCherry) marker using flow cytometry. The efficiency of ZIP14 KD was examined by RT-qPCR.RT-PCR and RT-qPCR
HC11 cells were trypsinized and harvested. Cell pellets were stored at −80 °C after flash freezing in liquid nitrogen until use. RNA purification, DNA removal and reverse transcription were performed following manufacturer's protocols. Specifically, RNA was purified with the RNeasy Mini Kit (QIAGEN, Cat. No. 74106); DNA was removed by DNaseI treatment (Thermo Scientific Cat. No. EN0525); RNA was reverse transcribed to cDNA using oligodT primers (Invitrogen, Cat. No. 18418012) and the Omniscript RT Kit (QIAGEN, Cat. No. 205113). RNase inhibitor (RNaseOUT Recombinant Ribonuclease Inhibitor, Invitrogen, Cat. No. 10777019) was supplemented in all reaction mixtures to inhibit RNA degradation. The synthesized first-strand cDNAs were then used as templates in the RT-PCR or RT-qPCR reactions. RT-PCR and RT-qPCR reactions were performed using Phusion DNA polymerase (NEB, Cat. No. M0530S) and SYBRTM Green PCR Master Mix (Applied Biosystems, Cat. No. 4367659), respectively, following manufacturer's protocols. Quantitative analysis of relative expression of target genes normalized to reference genes using qPCR data was performed with the Pfaffl method as previously described.13 Detailed information including RT-qPCR protocols, primer sequences and quantification method (standard curve, efficiency) are provided in ESI.†Western blotting
HC11 cell pellets were resuspended in cold RIPA buffer supplemented with 1 mM dithiothreitol (DTT) and a protease inhibitor cocktail (one tablet per 10 mL, cOmplete, Roche, Cat. No. 04693159001). The mixture was incubated at 4 °C for 30 minutes on a rotator, followed by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 30 min. The pellet containing cell debris and nuclei was discarded. The protein concentration in the supernatant fraction was measured by the BCA assay (PierceTM BCA Protein Assay Kit, Thermo ScientificTM, Cat. No. 23225). 15 μg of protein was mixed with 5× SDS-loading dye and denatured at 95 °C for 5 min (CSN2 immunoblot). The protein mixture was separated in 10% (w/v) acrylamide gels for SDS-PAGE and then transferred to PVDF membranes at 80 V for 1 h. The membrane was blocked in 5% (w/v) non-fat milk in 1× TBS-T (1× TBS with 0.1% (v/v) Tween20, pH 7.4) at room temperature for 1 h. The membrane was incubated with the primary antibody in blocking buffer overnight at 4 °C. The next day, the membrane was washed with 1× TBS-T for 30 min (10 min × 3 washes) and incubated in secondary antibody in blocking buffer at room temperature for 1 h. Membrane was washed with 1× TBS-T for 30 min (10 min × 3 washes) again. Blots were developed with the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Cat. No. RNP2232) and imaged on an ImageQuant LAS4000 imaging system (GE Healthcare Life Sciences). The antibodies used in immunoblotting were as follows: Beta-CSN2: the primary antibody was the goat anti-β-casein polyclonal antibody (Santa Cruz Biotechnology, SC-17971), 1
000 rpm at 4 °C for 30 min. The pellet containing cell debris and nuclei was discarded. The protein concentration in the supernatant fraction was measured by the BCA assay (PierceTM BCA Protein Assay Kit, Thermo ScientificTM, Cat. No. 23225). 15 μg of protein was mixed with 5× SDS-loading dye and denatured at 95 °C for 5 min (CSN2 immunoblot). The protein mixture was separated in 10% (w/v) acrylamide gels for SDS-PAGE and then transferred to PVDF membranes at 80 V for 1 h. The membrane was blocked in 5% (w/v) non-fat milk in 1× TBS-T (1× TBS with 0.1% (v/v) Tween20, pH 7.4) at room temperature for 1 h. The membrane was incubated with the primary antibody in blocking buffer overnight at 4 °C. The next day, the membrane was washed with 1× TBS-T for 30 min (10 min × 3 washes) and incubated in secondary antibody in blocking buffer at room temperature for 1 h. Membrane was washed with 1× TBS-T for 30 min (10 min × 3 washes) again. Blots were developed with the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Cat. No. RNP2232) and imaged on an ImageQuant LAS4000 imaging system (GE Healthcare Life Sciences). The antibodies used in immunoblotting were as follows: Beta-CSN2: the primary antibody was the goat anti-β-casein polyclonal antibody (Santa Cruz Biotechnology, SC-17971), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000 dilution; secondary antibody was donkey anti-goat IgG (H + L) secondary antibody (HRP) (Novus Biologicals, Cat. No. NB7357), 1
4000 dilution; secondary antibody was donkey anti-goat IgG (H + L) secondary antibody (HRP) (Novus Biologicals, Cat. No. NB7357), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution. ACTB: the primary antibody was mouse anti-β-actin antibody (Sigma, A2228), 1
000 dilution. ACTB: the primary antibody was mouse anti-β-actin antibody (Sigma, A2228), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution; secondary antibody was goat anti-mouse IgG (H + L) secondary antibody (HRP) (Novus Biologicals, Cat. No. NB7539), 1
000 dilution; secondary antibody was goat anti-mouse IgG (H + L) secondary antibody (HRP) (Novus Biologicals, Cat. No. NB7539), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution.
000 dilution.
Immunofluorescence
For immunofluorescence detection of CSN2, HC11 cells were grown on glass coverslips and induced to differentiate. Samples were collected at the following time points: P day 2, R day 1, D 12 h, 24 h, day 3 and day 6. Cells were washed with phosphate-buffered saline (PBS) three times and then fixed in 4% paraformaldehyde (PFA) in PBS for 45 min in the dark at room temperature. Cells were washed with PBS three times, followed by incubation with 20 mM NH4Cl in PBS for 5 min at room temperature. Samples were stored in PBS at 4 °C until use. For permeabilization of the cell membrane, cells were incubated with 0.2% Triton X-100 in PBS solution for 15 min at room temperature, followed by washing 3× with PBS and blocking in 5% bovine serum albumin (BSA) for 45 min. Cells were washed with PBS three times and then incubated with primary antibody (goat anti-β-casein polyclonal antibody, Santa Cruz Biotechnology, SC-17971, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution) at room temperature for one h. Cells were washed with PBS three times and then incubated with secondary antibody (donkey anti-goat IgG-FITC, Santa Cruz Biotechnology, SC-2024, 1
10 dilution) at room temperature for one h. Cells were washed with PBS three times and then incubated with secondary antibody (donkey anti-goat IgG-FITC, Santa Cruz Biotechnology, SC-2024, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 dilution) at room temperature for one h. Cells were then washed with PBS three times. To stain nuclei, cells were incubated with 1 mg mL−1 Hoechst 33258 in H2O for 30 seconds at room temperature and then washed with PBS three times prior to mounting on glass slides.
20 dilution) at room temperature for one h. Cells were then washed with PBS three times. To stain nuclei, cells were incubated with 1 mg mL−1 Hoechst 33258 in H2O for 30 seconds at room temperature and then washed with PBS three times prior to mounting on glass slides.
CSN2 immunofluorescence images were acquired on a Nikon A1R laser scanning confocal microscope equipped with the Nikon Elements software platform, Ti-E Perfect Focus system with a Ti Z drive, using a 100× oil objective (NA 1.45) and the following channels: CSN2 (green) (488 nm laser line, PMT gain: 45 or 110, pinhole size: 1 or 4 AU, emission filter: 525/50 nm); Hoechst 33258 (blue) (405 nm laser line, PMT gain: 80 or 95, pinhole size: 1 AU, emission filter: 450/50 nm).
RNA-Seq
To prepare samples for RNA-Seq, three biological replicates of HC11 cells were trypsinized and harvested. Cell pellets were stored at −80 °C after flash freezing until use. RNA was extracted with the RNeasy Mini Kit (QIAGEN, Cat. No. 74106). DNA was removed by DNaseI treatment (Thermo Scientific Cat. No. EN0525). RNA samples were then submitted to the BioFrontiers Next-Gen Sequencing Core Facility at University of Colorado for library construction and sequencing. Specifically, libraries were prepared using the NEXTflex Rapid Illumina Directional RNA-Seq Library Prep Kit with polyA enrichment (Bioo Scientific). Sequencing was performed on the NextSeq platform using paired-end reads (High Output, 2 × 75). After sequencing, read quality was assessed using FastQC (version 0.11.2). Illumina adapters were trimmed using Trimmomatic 0.32 in paired-end mode (HEADCROP:10). Reads were then mapped to the mm10 genome (downloaded from NCBI) using Tophat 2.0.6 with the options {b2-very-sensitive -p 12 -r 240 --mate-std-dev 105 –library-type fr-secondstrand}.14 Mapped reads were counted using HTSeq (version 0.6.1)15 with the options {-f -r -s -m intersection-strict}. Differential expression of genes was analyzed using the R(version 3.4.1) package DESeq2 (version 1.16.1).16 The expression levels of Zn2+ homeostasis genes were represented as transcripts per million mapped reads (TPM) using the following equation: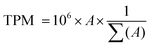 , A = (total reads mapped)/(gene length in kilo bp). Gene ontology (GO)-derived functional annotations were analyzed using DAVID 6.8 with GOTERM_BP_DIRECT (BP: biological processes), GOTERM_MF_DIRECT (MF: Molecular Function) and KEGG Pathways. Enrichment of biological processes and pathways was ranked based on the adjusted p-value (Benjamini score) associated with each annotation term, with a lower score indicating more significant enrichment.
, A = (total reads mapped)/(gene length in kilo bp). Gene ontology (GO)-derived functional annotations were analyzed using DAVID 6.8 with GOTERM_BP_DIRECT (BP: biological processes), GOTERM_MF_DIRECT (MF: Molecular Function) and KEGG Pathways. Enrichment of biological processes and pathways was ranked based on the adjusted p-value (Benjamini score) associated with each annotation term, with a lower score indicating more significant enrichment.
All raw next-generation sequencing data files and processed data files used to draw conclusions are available at the Gene Expression Omnibus.
Inductively coupled plasma mass spectrometry (ICP-MS)
All ICP-MS samples were spiked with known amounts of two internal standard elements, yttrium (Y) and gallium (Ga), to correct technical or human errors. The ion counts of zinc (Zn) in each sample measured by ICP-MS were corrected by the ratio of measured Y or Ga (ppb) to the known amount of Y or Ga (ppb). The Zn ion counts were then converted to parts per billion (ppb) using a Zn standard curve. The Zn ppb signal was normalized to total cell number to compare Zn in different samples. To prepare the HC11 cells for ICP-MS, 10 × 106 cells were trypsinized and harvested in 15 mL metal-free conical tubes (VWR, 89049-172) and then cell pellets were dried at 50 °C in a fume hood overnight. The next day, 200 μL of Trace-metal grade 65% nitric acid (FLUKA, 02650) was added to cell pellets and the mixture was heated in boiling water for 30 minutes. After the samples cooled down to room temperature, the nitric acid concentration in samples was corrected to 2% using chelex-treated Milli-Q water. 5 ppb of Y and 5 ppb of Ga were added to each sample as internal standard elements. Samples were submitted for ICP-MS analysis to the LEGS Lab at CU Boulder. Zn (ppb) was converted to the concentration ([Zn]) of Zn per cell. Detailed protocols and data analysis are presented in ESI.†Measurement of labile cytosolic Zn2+ using NES-ZapCV2
An HC11 cell line stably expressing the cytosolic Zn2+ sensor NES-ZapCV217 was generated using the PiggyBac transposase system (System Biosciences) following the manufacturer's instruction. Fluorescence imaging was performed on a Nikon Ti-E wide-field fluorescence microscope equipped with Nikon elements software, an iXon3 EMCCD camera (Andor), mercury arc lamp, and FRET (434/16 excitation, 458 dichroic, 535/20 emission), CFP (434/16 excitation, 458 dichroic, 470/24 emission), and YFP (495/10 excitation, 515 dichroic, 535/20 emission) filter sets. To perform a titration experiment, fluorescence intensities of FRET, CFP and YFP channels of a randomly selected cytoplasmic region (cytosol) and a region without cells (background) were recorded every 40 seconds for ∼3 min in the resting state. The FRET ratio was calculated as (FRETcytosol − FRETbackground)/(CFPcytosol − CFPbackground) and all ratios were then averaged to acquire the resting ratio (R). Cells were then treated with 150 μM TPA (Zn2+ chelator) for an extended time (∼20 min) to ensure that FRET R decreased and stabilized. The minimum ratio, Rmin, was acquired by averaging ratios of the last 3 time points of TPA treatment. After cells were washed twice with phosphate-free HHBSS buffer, a mixture of pyrithione (a Zn2+ ionophore, final concentration 0.75 μM) and ZnCl2 (final concentration 20 μM) was added to cells until the FRET ratio increased and reached a plateau. The maximum ratio, Rmax, was calculated by taking the mean of all ratios measured in the plateau phase. The labile Zn2+ concentration was calculated using this equation: [Zn2+] = Kd × [(R − Rmin)/(Rmax − R)]1/n. For the NES-ZapCV2 cytosolic sensor, the Kd = 2.3 nM and the Hill coefficient (n) = 0.532.17 Alternatively, the labile Zn2+ concentration can be represented as the fractional saturation (FS) of the sensor, which was defined as FS = (R − Rmin)/(Rmax − Rmin). Cells with a dynamic range (Rmax/Rmin) in the range of 1.5–2.5 were selected for analysis.Promoter reporter assay
The promoter reporter assay designed to test promoter activity in response to hormone treatment was performed using the Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia) following the manufacturer's instructions. The CSN2 promoter was amplified from the HC11 cell genome using the NucleoSpin BloodXL kit (MACHEREY-NAGEL, ref. 740950.50) and inserted into multiple cloning site 1 (MCS1) of the pEZX-LvGA01 vector (GeneCopoeiaTM, catalog No. ZX107) 5′ upstream of the Gaussia Luciferase (GLuc) ORF. The 983 bp promoter sequence includes a ∼900 bp fragment upstream of the transcription start site (TSS) and a ∼100 bp fragment downstream of the TSS. The TSS position was acquired from the Eukaryotic Promoter Database (EPD). Primers sequences are: Csn_Fwd 5′ TCTGATGAATTCTCTGTAATTACTATTATTAAAAAGATTAATGTTTGTTAAG 3′; Csn_Rev 5′ ATCAGAACCGGTCTAGTATGCATTTGAAATAATGAAAATGATATTTC. This vector also constitutively expresses an internal control reporter, SEAP (secreted alkaline phosphatase). Both GLuc and SEAP can be secreted into cell media upon being synthesized. The promoter activity is reported using the ratio of GLuc signal to the SEAP signal. HC11 cell lines stably expressing promoter constructs were generated using lentiviral transduction and selected for successful transduction by puromycin. To determine the promoter activity in response to lactogenic hormone treatment, 4 × 105 HC11 stable cells were plated in a 35 mm dish in the proliferation media (2 mL) for 3 days and then cultured in the resting media (2 mL) for one more day. Cells were then treated with the lactogenic hormones for 6 days with fresh media replaced every 2 days. For luciferase analysis, 200 μL of media was collected at 4, 18, 24 h, day 2, 3, 4, 5 and 6 post hormone treatment. 200 μL of fresh media was added back to cell culture to compensate for the volume change after media collection. All samples were frozen at −20 °C. The GLuc and SEAP catalyzed luminescence reactions were performed using the Secrete-Pair™ Dual Luminescence Assay Kit (GeneCopoeia) following the manufacturer's instructions. Luminescence was recorded using a BioTek Synergy H1 hybrid plate reader (gain: 200; integration time: 3 s). 2 biological replicates were utilized at each time point and 3 technical replicates were measured for each biological replicate.Results
Characterization of HC11 cell differentiation
The murine mammary epithelial cell line HC11 has been widely used as an in vitro model system to investigate mammary cell differentiation.18–20 The cell line was isolated from the COMMA-1D mammary epithelial cell line9 which was established from the mammary tissue of BALB/c mice in the middle of pregnancy,21 and the expression of several milk genes can be induced by a combination of lactogenic hormones (prolactin and cortisol) without the requirement of any matrix protein.9,10 HC11 cells were differentiated using a previously established protocol9,22 (Fig. 1A). The progression of differentiation was characterized by measuring the expression of two lactation markers casein (CSN2) and whey acid protein (WAP) in samples from the proliferation state (P day 2), resting state (R day 1) and upon treatment with prolactin and cortisol, representing the differentiation state (D 12 h, day 1, day 3 and day 6). Csn2 and Wap mRNA were not detected until 3 days after hormone treatment and the mRNA levels of both genes further increased at day 6 after treatment (Fig. 1B). CSN2 protein levels, as detected by Western blotting, appear at 3 days after treatment and increase further 6 days after treatment, paralleling the mRNA signature (Fig. 1C). To examine differentiation at the single cell level, immunofluorescence of CSN2 protein expression was performed over the course of differentiation. The total number of cells and number of cells that stained positively for CSN2 expression were manually counted and the percentage of cells with detectable CSN2 expression was quantified. CSN2 expression was essentially undetectable in P day 2 (0.00%) and R day 1 samples (0.08%) (Fig. 1D). After hormone treatment, the percentage of cells with detectable CSN2 was as follows: D 12 h (0.74%), day 1 (0.68%) and day 3 (13.4%) samples. By day 6 after hormone treatment, CSN2 expression was detected in almost every cell (95.5%). Although it took 3 days of hormone treatment to detect the steady-state mRNA and protein level of lactation markers, using a luciferase promoter assay, gene transcription from the CSN2 promoter was initiated by 18 h of hormone treatment and transcriptional activity continued to increase from day 1 to day 6 post lactogenic hormone treatment (Fig. 1E).Changes in global gene expression at 24 h and 6 days post hormone treatment
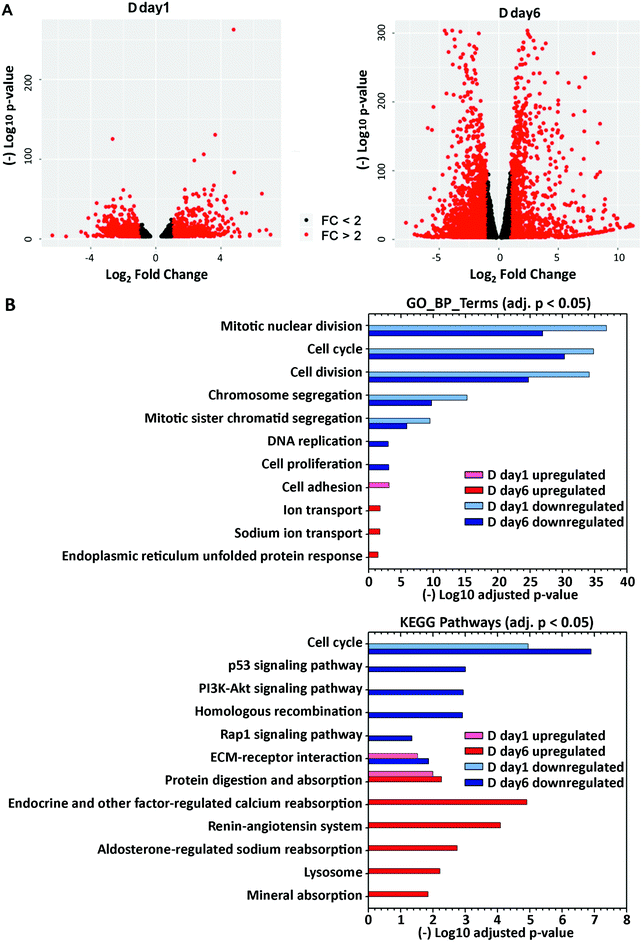
To define how treatment with lactogenic hormones remodeled global gene expression of mammary epithelial cells, next-generation RNA sequencing (RNA-Seq) was performed on R day 1 (control), D day 1 (early stage differentiation) and D day 6 (later stage differentiation) samples, with 3 biological replicates for each condition. The expression of genes in D day 1 and D day 6 samples relative to R day 1 samples was analyzed pairwise using DESeq2, revealing that 1909 and 8161 genes were differentially expressed at D day 1 and D day 6 compared to R day 1, respectively (padj < 0.001). Fig. 2A shows volcano plots depicting statistical significance vs. fold change of the differentially expressed genes. The genes that were differentially expressed by more than 2-fold are highlighted in red, with 1132 genes at day 1 and 2943 genes at day 6 post hormone treatment, which account for 4.5% or 11.7% of 25059 total mouse genes.To gain insight into the biological functions and pathways that were altered upon differentiation, gene ontology (GO)-derived functional annotations of the downregulated and upregulated genes at day 1 and day 6 (fold change >2, padj < 0.001) post-hormone treatment were analyzed using DAVID 6.8 with GOTERM_BP_DIRECT (BP: biological processes) and KEGG pathways (Fig. 2B, Supp Info RNAseq). Cell cycle and cell division processes were significantly downregulated at day 1, and further downregulated at day 6 post-hormone treatment, consistent with the shift away from a proliferation phenotype. The major biological processes and pathways that were upregulated in the early stage of differentiation D day 1 were cell adhesion, ECM–receptor interaction and protein digestion and absorption. By D day 6, pathways involved in cell metabolism and secretion were upregulated, such as protein digestion and absorption, ion transport and absorption, and lysosome. Overall, RNA-Seq revealed that lactogenic hormone treatment temporally altered the activities of different biological processes in mammary epithelial cells. A full list of regulated GO and KEGG terms is included in the Supporting Information RNAseq file.
Zinc-dependent genes are differentially expressed upon differentiation
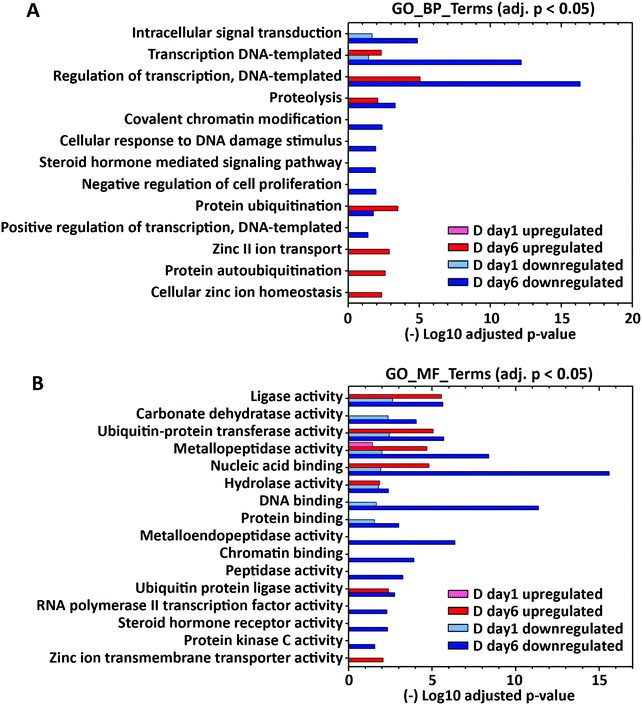
To explore how the expression of zinc-regulatory and zinc-dependent genes were modulated by lactogenic hormone treatment, the differentially expressed genes at day 1 and day 6 post hormone treatment (fold change >1.5, padj < 0.001) were analyzed using DAVID 6.8 with UniProt (UP)_Keywords. According to the definition of the keyword “Ligand” (KW9993) in the Uniprot database, the UP_Keyword “Zinc” (KW0862, which is in the “Ligand” category) was assigned to genes when the translated gene products (i.e. proteins) bind, are associated with, or have activity that is dependent on zinc. In D day 1 samples, 96 out of 942 (10.2%) downregulated genes and 46 out of 761 (6.0%) upregulated genes (fold change >1.5) were annotated with the keyword “Zinc”. In D day 6 samples, 269 out of 2572 (10.4%) downregulated genes and 198 out of 2254 (8.8%) upregulated genes (fold change >1.5) were annotated with “Zinc” (Fig. 3A). The differentially expressed “Zinc”-annotated genes at D day 6 were then sorted based on fold change or padj. Interestingly, two zinc transporters, Slc39a8 (ZIP8) and Slc39a14 (ZIP14), were reported among the top 10 genes with highest fold change or lowest padj respectively (Table S6, ESI†).To further examine functional categories of differentially expressed zinc-dependent genes, the functional annotations of downregulated and upregulated genes in D day 1 and D day 6 samples (fold change >1.5, p < 0.001) were analyzed using GOTERM_BP and GOTERM_MF_DIRECT (MF: Molecular Function), as described previously (Fig. 3, Supporting Information_RNAseq). GO_MF terms “metal ion binding” and “zinc ion binding” are excluded, as all of the zinc-dependent genes bind zinc/metal and therefore those terms are enriched in each comparison. Upon hormone treatment, 15 zinc-dependent biological and molecular processes are enriched at D day 1, including intracellular signal transduction, DNA/protein binding, regulation of gene transcription, and metabolic enzyme activities (Fig. 3A and B). At D day 6, 55 zinc-dependent processes are enriched, including cell signal transduction, transcription, protein ubiquitination and metabolic enzyme activities. Interestingly, GO enrichment analysis demonstrated that “zinc II ion transport” (GO_BP), “cellular zinc ion homeostasis” (GO_BP) and “zinc ion transmembrane transporter activity” (GO_MF) are upregulated at day 6 post hormone treatment, indicating that zinc transport and distribution may also be altered upon hormone treatment. A full list of regulated GO and KEGG terms is included in the Supporting Information RNAseq file.
Zinc homeostasis genes are differentially expressed upon differentiation
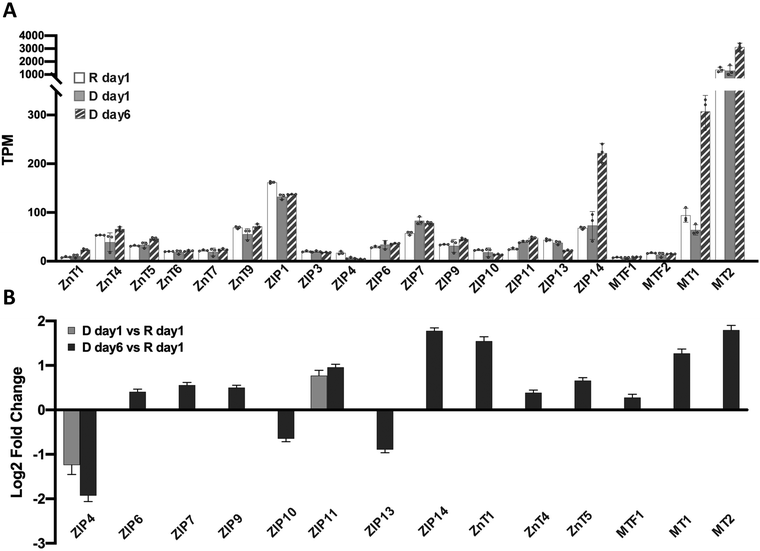
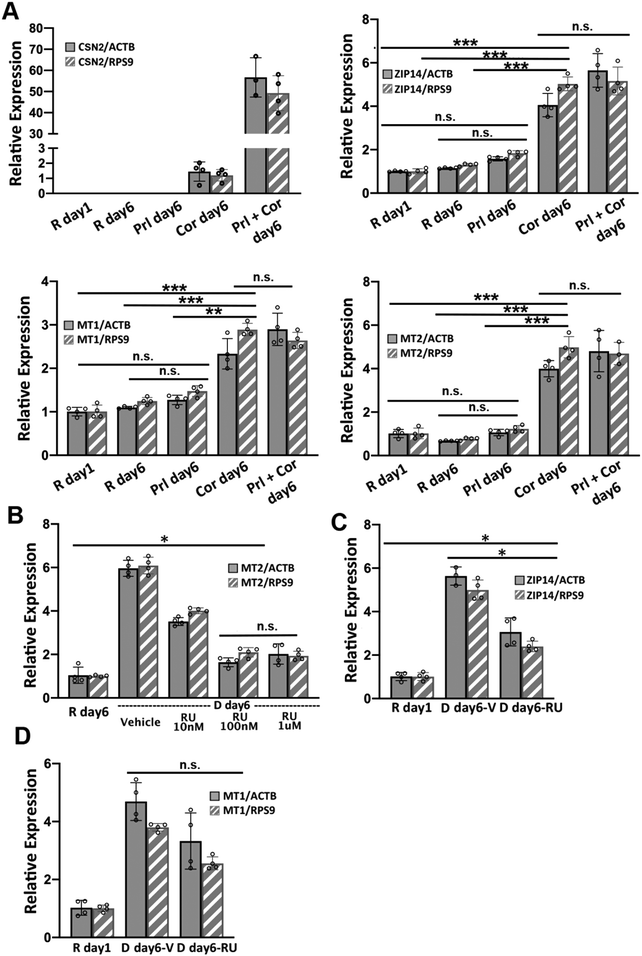
Zn2+ homeostasis is regulated by Zn2+ transporters, Zn2+ buffering protein metallothioneins (MTs) and the metal-responsive transcription factor 1 (MTF1).23 To understand how cell differentiation regulates zinc homeostasis, we used our RNA-Seq data to analyze how known zinc regulatory genes change upon differentiation. Because many Zn2+ transporters are expressed at low levels, we first examined the average transcripts per million mapped reads (TPM) for each Zn2+ regulatory gene (Fig. 4A). Genes with TPM < 1 were not included in differential expression analysis because the expression level was deemed too low to be reliable. Twenty Zn2+ homeostasis genes with TPM > 1 were detected in all conditions and analyzed for differential expression using DESeq2. Two genes, ZIP4 and ZIP11, were differentially regulated at D day 1 (Fig. 4B). After 6 days of hormone treatment, 14 genes were differentially expressed. Among these, 3 genes encoding ZIP transporters were downregulated, while 11 genes encoding 5 ZIPs, 3 ZnTs, 2 MTs and MTF1 were upregulated. These results demonstrate that expression of Zn2+ homeostasis genes is remodeled upon treatment with lactogenic hormones, consistent with previous studies using RT-qPCR,8 and reveal that Zn2+ homeostasis is more dramatically altered at a late stage of cell differentiation. For downstream analysis of how hormones affect zinc homeostasis genes, ZIP14, MT1 and MT2 were chosen as targets because of their relatively high expression level and fold change in D day 6 samples compared to that observed in resting cells.Cortisol increases the steady-state level of ZIP14, MT1 and MT2 mRNAs
Two lactogenic hormones, prolactin and cortisol, were used to initiate HC11 cell differentiation. These two hormones function through different signaling pathways and we wanted to determine which was most influential in remodeling Zn2+ homeostasis. To examine the role of each hormone in regulating the mRNA levels of Zn2+ homeostasis genes, HC11 cells were cultured in proliferation media or resting media as in the differentiation protocol and then treated with prolactin (Prl), cortisol (Cor), or both hormones (Prl + Cor) for 6 days. Cells were collected at day 1 post treatment with resting media (R day 1), and day 6 post treatment with hormone(s) (Prl day 6, Cor day 6 and Prl + Cor day 6). To eliminate the possibility that the higher levels of mRNA for Zn2+ homeostasis genes resulted from extended culturing, we also collected cells that continued to grow in the resting media for another 6 days (R day 6). Using RT-qPCR, expression of ZIP14, MT1 or MT2 was analyzed and normalized to two reference genes: beta actin (ACTB) and ribosomal protein S9 (RPS9). CSN2 was used as a positive control because its mRNA expression was strongly activated by the treatment of prolactin and cortisol, as shown previously.9 Cortisol was the primary factor that increased the steady-state level of ZIP14, MT1 and MT2 mRNAs (Fig. 5A). Prolactin treatment alone had no effect on the mRNA levels of ZIP14, MT1 and MT2. On the other hand, cortisol treatment alone (Cor day 6) significantly increased the expression of ZIP14, MT1, and MT2 compared to their expression level in R day 1, R day 6 or P day 6 samples. Finally, there was no significant difference in the mRNA levels of ZIP14, MT1 and MT2 between the cells treated with cortisol alone (Cor day 6) and the cells treated with both hormones (Prl + Cor day 6).To examine whether cortisol affects the mRNA level of ZIP14, MT1 and MT2 via the canonical glucocorticoid receptor (GR) signaling pathway, HC11 cells were treated with a GR antagonist RU486 to competitively inhibit receptor binding with cortisol.24 The optimal dosage of RU486 was determined by supplementing differentiation media with different amounts of RU486 and quantifying the mRNA of MT2 by RT-qPCR under each condition. MT2 was used as a positive control for cortisol dependence because previous work showed cortisol increased MT2 mRNA levels and RU486 abolished the effect.24 Treatment with 100 nM RU486 significantly lowered the MT2 mRNA level compared to vehicle and 10 nM RU486 treatment (p < 0.0001), while there was no difference between 100 nM and 1 μM (Fig. 5B). It was noteworthy that 100 nM RU486 did not reduce the mRNA level of MT2 to the basal level observed in the R day 1 samples. To assess the effect of RU486 on the mRNA level of MT1 and ZIP14, HC11 cells were treated with the differentiation media supplemented with 100 nM RU486 for 6 days and the relative expression of ZIP14 and MT1 was determined using RT-qPCR. The steady-state ZIP14 mRNA level was lowered by RU486 treatment compared to the vehicle group, but it was still higher than that of basal level in the R day 1 samples (Fig. 5C). The effect of RU486 on MT1 mRNA level was inconclusive due to the variance of the results (Fig. 5D). When normalized to RPS9, the MT1 mRNA level in the vehicle-treated and RU486-treated samples was significantly different. However, when normalized to ACTB, the difference in MT1 mRNA level between the vehicle-treated and the RU486-treated groups was not statistically significant (p = 0.0499). Overall, these data show that a competitive GR inhibitor significantly abrogated the cortisol-induced increase in expression of ZIP14 and MT1, indicating that cortisol induces these genes at least partially via the nuclear GR signaling pathway.
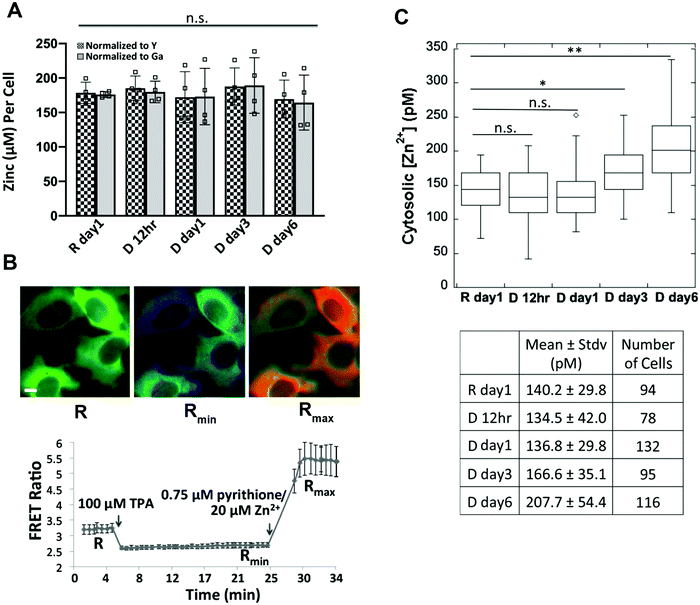
Total Zn2+ doesn’t change, but labile cytosolic Zn2+ increases over the progression of cell differentiation
Our observation that MTF1, MTs, and Zn2+ transporters are differentially expressed at a later stage of cell differentiation (day 6 after hormone treatment) led us to hypothesize that total or cytosolic labile Zn2+ might be altered. To determine total cellular Zn2+ over the course of differentiation, HC11 cells were treated with lactogenic hormones and Zn2+ was measured at different time points using ICP-MS, an elemental analysis technique that measures total metal in a bulk sample. We observed that total Zn2+ did not change over the progression of cell differentiation (Fig. 6A). Labile Zn2+ in the cytosol was measured using NES-ZapCV2, a genetically-encoded Zn2+ FRET-based ratiometric sensor that localizes to the cytosol.17 To quantify cytosolic Zn2+, an in situ calibration was performed to acquire the resting FRET ratio (R), the minimum ratio (Rmin) after treatment with the Zn2+ chelator TPA, and the maximum ratio (Rmax) after treatment with the ionophore pyrithione and Zn2+ (Fig. 6B). Statistical analysis revealed that cytosolic Zn2+ did not change at 12 or 24 h post hormone treatment, but an increase in Zn2+ was detected in the D day 3 samples compared to the resting state R day 1 (R day 1, 140 ± 30 pM; D day 3, 170 ± 35 pM, p < 0.0005), with a further increase at day 6 post hormone treatment (D day 6, 210 ± 60 pM, p < 0.0001 vs. R day 1 or D day 3). Combined with the result that the mRNA and protein expression of two differentiation markers (CSN2 and WAP) increased on day 3 of hormone treatment (Fig. 1), our data suggest that higher cytosolic Zn2+ is positively associated with the progression of cell differentiation.ZIP14 contributes to increased cytosolic Zn2+ and upregulation of WAP at day 6 post hormone treatment
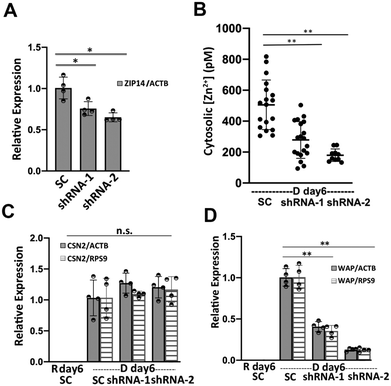
Cytosolic labile Zn2+ is balanced between influx (regulated by ZIPs), efflux (regulated by ZnTs), and buffering (regulated largely by MTs) to stay at an optimal level. What defines the set-point of labile Zn2+ in cells is not well understood, but Zn2+ transporters have been shown to alter Zn2+ in different cellular compartments.24–27 We wanted to perturb the changes in Zn2+ homeostasis upon differentiation in order to examine how Zn2+ might impact the differentiation phenotype. While there were changes in multiple transporters (both increases and decreases) upon differentiation, the 5-fold increase in ZIP14 expression was intriguing because of the overall high expression level of this trasporter. To explore how ZIP14 expression affected cytosolic Zn2+ levels, ZIP14 was knocked down using two shRNAs targeting different regions of ZIP14 mRNA and cytosolic Zn2+ was examined in D day 6 cells using the NES-ZapCV2 sensor. HC11 cells stably expressing ZIP14 shRNA-1, ZIP14 shRNA-2 or scrambled control (SC) shRNA were treated with lactogenic hormones for 6 days and ZIP14 mRNA expression was quantified by RT-qPCR (Fig. 7A). ShRNA-1 and -2 lowered the ZIP14 mRNA expression by 27% and 34%, respectively (ZIP14/ACTB in SC cells: 1.00 ± 0.11; shRNA-1 cells: 0.73 ± 0.07; shRNA-2 cells: 0.66 ± 0.07). As shown in Fig. 7B, the cytosolic Zn2+ concentration was significantly lower in ZIP14 KD cells compared to control cells. The lower concentration in shRNA-2 cells is consistent with the fact that shRNA-2 lowered ZIP14 mRNA level more than shRNA-1. Importantly, treatment with shRNA essentially abolished the increase in Zn2+ upon differentiation, providing a window to explore how Zn2+ and a key Zn2+ transporter could influence the differentiation phenotype.To investigate the role of ZIP14 in mammary cell differentiation and lactation, the mRNA expression of differentiation markers CSN2 and WAP was measured in ZIP14-KD cells using RT-qPCR. HC11 cells stably expressing ZIP14 shRNA-1, ZIP14 shRNA-2 or scrambled control (SC) shRNA were treated with lactogenic hormones for 6 days. ZIP14-KD did not change the steady state CSN2 mRNA level (Fig. 7C). However, WAP mRNA expression decreased significantly in ZIP14-KD cells, with a greater effect by shRNA-2 (Fig. 7D). With WAP relative expression in SC cells normalized to 1.00, the WAP expression level was 0.41 ± 0.06 (WAP/ACTB) and 0.35 ± 0.07 (WAP/RPS9) in ZIP14 shRNA-1 KD cells, 0.13 ± 0.02 (WAP/ACTB) and 0.12 ± 0.02 (WAP/RPS9) in ZIP14 shRNA-4 KD cells. This result suggests that ZIP14 specifically, and Zn2+ homeostasis in general, plays an important role in the induction of WAP mRNA but not CSN2 mRNA in differentiated mammary cells. This finding is surprising because WAP and CSN2 are induced by the cooperative and synergistic action of prolactin-activated JAK-STAT signaling and cortisol-activated glucocorticoid receptor (GR) signaling. Despite the similarities, other researchers have reported differences in the underlying mechanism of transcription regulation of the two genes. For example, cortisol alone is able to effectively induce WAP transcription but has little effect on CSN2 expression, whereas prolactin alone was sufficient to induce CSN2 but not WAP transcription.28,29 Additionally, a transcriptional factor, nuclear factor 1 (NF1) was found to be essential in the transcriptional activation of WAP in cooperation with STAT5 and GR.30 An intriguing study revealed that NF1 binds and activates MT1 in a Zn2+-inducible and MTF-1 dependent manner, suggesting an important link between NF1 and zinc homeostasis.31 How Zn2+ homeostasis affects WAP but not CSN2 transcription is unknown and requires further investigation.
Discussion
The HC11 cell line is a well-established model system to recapitulate systematic changes in molecular and cellular processes of mammary epithelial cells during lactation. Previous transcriptomic studies using microarray32,33 and next generation RNA sequencing34 demonstrated global changes in gene expression in HC11 cells upon lactogenic hormone treatment. However, these studies only examined HC11 cell differentiation at day 3 after lactogenic hormone (prolactin and cortisol) treatment and therefore were unable to distinguish the cellular and molecular changes occurring at an early versus late stage of cell differentiation. Our RNA-Seq analysis revealed that 1132 genes (∼4.5% of the mouse genome) were differentially regulated at 24 h after treatment, consistent with a global change at an early stage of cell differentiation. GO_BP enrichment and KEGG pathway analysis of these 1132 genes suggested two major suppressed cellular functions are mitosis/cell division and cell cycle. For example, Cdk1 (Cyclin-dependent kinase 1) is a central player in driving mammalian cell cycle through G2/M phase35 and its expression was decreased by more than 3 fold at 24 h (p < 0.0001). The major functional clusters that are regulated by differentiation hormones include cell adhesion, ECM–receptor interaction, cell cycle and division, which could play important regulatory roles in the preparation for cell differentiation.Not surprisingly, by D day 6, a higher number of biological processes/pathways were affected, including downregulation of pathways such as cell division, cell replication and a few signaling pathways (e.g. p53 and PI3K-Akt signaling pathways), as well as upregulation of ion transport and absorption, lysosome and metabolism (protein digestion and absorption). The majority of the above processes/pathways are strongly overlapping with published transcriptomic studies where cells underwent 3-day treatment.32–34 Nonetheless, our analysis uniquely identified enrichment of Endoplasmic Reticulum (ER) unfolded protein response (UPR) pathway in upregulated genes at D day 6. During lactation, mammary gland epithelial cells synthesize a large quantity of proteins, which leads to the accumulation of unfolded proteins in the ER. A few key UPR factors (e.g. XBP1 and ATF4) have been demonstrated to be upregulated in the lactating mammary gland.36,37 Furthermore, knockdown of ATF4 or XBP1 in HC11 cells suppressed the gene expression of beta-casein and the lactogenic hormone receptor,38 suggesting an essential role of the UPR in HC11 cell differentiation. In our study, enrichment of the UPR pathway and higher expression of essential UPR factors (33-fold increase of XBP1, 6.7-fold increase of ATF4, 2.7-fold increase of ATF6 Supporting Information_RNAseq) are consistent with previous studies and suggests that the UPR is upregulated during the differentiation of HC11 cells.
Using RNA-Seq, we also discovered that the majority of zinc homeostasis genes (14 out of 20 detected) and 467 zinc-dependent genes were differentially regulated by D day 6. A number of studies have interrogated the regulation of expression and functional roles of select zinc transporters (i.e. ZnT2, ZnT4 and ZIP3) in differentiated HC11 cells.39–42 For example, the expression of ZnT2 was shown to be activated by prolactin, and ZnT2 was proposed to import Zn2+ into the secretory pathway in differentiated HC11 cells, suggesting a role for ZnT2 in Zn2+ secretion into the milk lumen.39,40 Additionally, ZnT4 and ZnT5 have been shown to be functionally related with milk production. A ZnT4 nonsense-mutation in lethal milk mice resulted in low zinc concentration in milk, and pups exclusively fed with the lethal milk die even before weaning.43 In addition, reduced mRNA expression of ZnT5 was identified in breastfeeding women producing zinc-deficient milk.44 The most comprehensive study examined the mRNA and protein levels of all Zn2+ transporters in lactating versus non-lactating mouse mammary glands, revealing a number of changes in Zn2+ homeostasis upon lactation.8 However, this study profiled changes in the entire mammary gland, which contains multiple cell types,45 and it focused on a single time point, as opposed to monitoring changes over the course of differentiation. Still, these prior studies provide an important foundation for our study, by demonstrating significant changes in Zn2+ transporter expression during differentiation.
In addition to changes in Zn2+ transporters, changes in labile Zn2+ levels have been previously detected upon differentiation. Specifically, an accumulation of Zn2+ in lysosomes and vesicular bodies was detected using small-molecule fluorescent Zn2+ dyes in HC11 cells at 16–24 h post hormone treatment,7,46 suggesting a redistribution of intracellular Zn2+ among organelles during lactation. Here, we found two Zn2+ homeostasis genes and 142 Zn2+-dependent genes with altered expression at day 1, and 11 Zn2+ homeostasis genes and 467 Zn2+-dependent genes by day 6, suggesting that Zn2+ homeostasis starts to be remodeled at day 1, but is not complete until later stages of differentiation.
In exploring what causes the expression changes in key zinc regulatory genes, we demonstrated that cortisol, but not prolactin, increased the mRNA levels of ZIP14, MT1 and MT2. At first glance this was surprising given that Zn2+ can interact with prolactin and induce aggregation.47,48 However, careful consideration of the concentrations of Zn2+ and prolactin in our media conditions reveals that the concentration of prolactin in our media is very low (0.2 μM) and most of the Zn2+ in the media is complexed with serum albumin,49 which binds Zn2+ more tightly than prolactin.50,51 Therefore, we speculate that the Zn2+–prolactin interaction isn’t relevant in our model system. The induction of ZIP14 and MT2 was at least partially mediated by the classical nuclear glucocorticoid receptor (GR) signaling pathway. However, treatment with GR-specific antagonist RU486 did not completely abolished the mRNA increase of ZIP14 and MT2 in differentiated cells compared to resting cells, suggesting that another signaling pathway may also play a role in induction of these genes. In addition to the nuclear GR signaling pathway, recent studies have shown that cortisol can also activate cell surface receptors52–55 or crosstalk with other signaling pathways (e.g. JAK-STAT)24,56 which may contribute to the induction of ZIP14 and MT2. Additionally, it has been reported that ER stress and UPR responses induce ZIP14 expression.57,58 UPR factors ATF4 and ATF6 directly bind the ZIP14 promoter and induce ZIP14 transcription in human and mouse.57,59 Therefore, it is plausible that ATF4 and ATF6 directly enhance the transcription of ZIP14 in mammary cells, though how cortisol and UPR are correlated is unknown.
Zn2+ is an important structural and/or functional cofactor for ∼10% of human proteins.1 Therefore, it is reasonable that cytosolic labile Zn2+ would increase to meet the structural or functional needs of the upregulated cytosol-localized zinc-dependent proteins during mammary cell differentiation. Among the list of upregulated zinc-dependent genes at D day 6, we found genes which reportedly encode cytosolic proteins that play important or essential roles in cell differentiation or lactation, including the vitamin D receptor (VDR),60 Peptidylglycine alpha-amidating monooxygenase (PAM)61 and tristetraprolin (TTP),62–64 a zinc-finger mRNA-binding protein encoded by ZIP36. TTP is highly expressed in differentiated mammary cells and the lactating mammary gland.62,63 The importance of TTP in mammary cell function is underscored by the observation that a knockout of TTP in mice led to mammary cell death and underweight pups.62 The concurrent increase of cytosolic Zn2+ and expression of the cytosol-localized zinc-dependent proteins exemplifies a mechanism of how cells temporally remodel zinc homeostasis to regulate protein function under different biological processes.
Conclusion
In conclusion, we comprehensively examined zinc homeostasis regulation at multiple levels over the course of HC11 mammary epithelial cell differentiation. We detected differential expression of a majority of zinc transporters and an increase in cytosolic Zn2+. We discovered that ZIP14 played a significant role in increasing cytosolic Zn2+ in differentiated cells, and that Zip14 expression was important for WAP expression and may play important biological roles in mammary epithelial cells differentiation or lactation.Author contributions
Y. H. and A. P. designed the research and analyzed the results. Y. H. carried out the majority of the research. L. S. helped with RNA-Seq and enrichment analysis. D. S. carried out cloning for the promoter assay. R. D. assisted with RNA-Seq analysis and provided critical feedback. Y. H. and A. P. wrote the paper with edits from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge the following sources of financial support: NIH R01 GM105997 and Director's Pioneer Award DP1 GM114863 (to A. E. P.), a Signaling and Cell Cycle Training Grant to L. S. (T32 GM008759), and a traineeship in the IQ Biology program of the BioFrontiers Institute to L. S. (NSF IGERT 1144807). We would like to acknowledge the University of Colorado BioFrontiers Institute Next-Gen Sequencing Core Facility, which performed the Illumina sequencing and library construction, the BioFrontiers Computing Core and BioFrontiers IT for providing High Performance Computing resources, and the University of Colorado Biochemistry Cell Culture Core Facility for providing resources for cell culturing. We also would like to thank Dr Mary Allen (University of Colorado Boulder, BioFrontiers Institute) for helpful discussions and training in RNA-Seq analysis.References
- C. Andreini, L. Banci, I. Bertini and A. Rosato, Counting the Zinc-Proteins Encoded in the Human Genome, J. Proteome Res., 2006, 5, 196–201 CrossRef CAS PubMed.
- T. Kambe, T. Tsuji, A. Hashimoto and N. Itsumura, The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism, Physiol. Rev., 2015, 95, 749–784 CrossRef CAS PubMed.
- R. A. Colvin, W. R. Holmes, C. P. Fontaine and W. Maret, Cytosolic Zinc Buffering and Muffling: Their Role in Intracellular Zinc Homeostasis, Metallomics, 2010, 2, 306–317 RSC.
- M. F. Picciano and H. A. Guthrie, Copper, Iron, and Zinc Contents of Mature Human Milk, Am. J. Clin. Nutr., 1976, 29, 242–254 CrossRef CAS PubMed.
- D. Beyersmann and H. Haase, Functions of Zinc in Signaling, Proliferation and Differentiation of Mammalian Cells, Biometals, 2001, 14, 331–341 CrossRef CAS PubMed.
- S. L. Kelleher, N. H. McCormick, V. Velasquez and V. Lopez, Zinc in Specialized Secretory Tissues: Roles in the Pancreas, Prostate, and Mammary Gland, Adv. Nutr., 2011, 2, 101–111 CrossRef CAS PubMed.
- N. McCormick, V. Velasquez, L. Finney, S. Vogt and S. L. Kelleher, X-Ray Fluorescence Microscopy Reveals Accumulation and Secretion of Discrete Intracellular Zinc Pools in the Lactating Mouse Mammary Gland, PLoS One, 2010, 5, e11078 CrossRef PubMed.
- S. L. Kelleher, V. Velasquez, T. P. Croxford, N. H. McCormick, V. Lopez and J. MacDavid, Mapping the Zinc-Transporting System in Mammary Cells: Molecular Analysis Reveals a Phenotype-Dependent Zinc-Transporting Network during Lactation, J. Cell. Physiol., 2012, 227, 1761–1770 CrossRef CAS PubMed.
- R. K. Ball, R. R. Friis, C. A. Schoenenberger, W. Doppler and B. Groner, Prolactin Regulation of Beta-Casein Gene Expression and of a Cytosolic 120-Kd Protein in a Cloned Mouse Mammary Epithelial Cell Line, EMBO J., 1988, 7, 2089–2095 CrossRef CAS.
- G. R. Merlo, D. Graus-Porta, N. Cella, B. M. Marte, D. Taverna and N. E. Hynes, Growth, Differentiation and Survival of HC11 Mammary Epithelial Cells: Diverse Effects of Receptor Tyrosine Kinase-Activating Peptide Growth Factors, Eur. J. Cell Biol., 1996, 70, 97–105 CAS.
- S. Desrivières, T. Prinz, N. Castro-Palomino Laria, M. Meyer, G. Boehm, U. Bauer, J. Schäfer, T. Neumann, C. Shemanko and B. Groner, Comparative proteomic analysis of proliferating and functionally differentiated mammary epithelial cells, Mol. Cell. Proteomics, 2003, 2, 1039–1054 CrossRef PubMed.
- B. Morrison and M. L. Cutler, Mouse Mammary Epithelial Cells form Mammospheres During Lactogenic Differentiation, J. Visualized Exp., 2009, 32, 1265 Search PubMed.
- M. W. Pfaffl, A New Mathematical Model for Relative Quantification in Real-Time RT–PCR, Nucleic Acids Res., 2001, 29, e45 CrossRef CAS PubMed.
- D. Kim, G. Pertea, C. Trapnell, H. Pimentel, R. Kelley and S. L. Salzberg, TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions, Genome Biol., 2013, 14, R36 CrossRef.
- S. Anders, P. T. Pyl and W. Huber, HTSeq—a Python Framework to Work with High-Throughput Sequencing Data, Bioinformatics, 2015, 31, 166–169 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- B. L. Fiedler, S. Van Buskirk, K. P. Carter, Y. Qin, M. C. Carpenter, A. E. Palmer and R. Jimenez, Droplet Microfluidic Flow Cytometer For Sorting On Transient Cellular Responses Of Genetically-Encoded Sensors, Anal. Chem., 2017, 89, 711–719 CrossRef CAS PubMed.
- N. Cella, R. R. Cornejo-Uribe, G. S. Montes, N. E. Hynes and R. Chammas, The Lysosomal-Associated Membrane Protein LAMP-1 Is a Novel Differentiation Marker for HC11 Mouse Mammary Epithelial Cells, Differentiation, 1996, 61, 113–120 CrossRef CAS PubMed.
- R. Chammas, D. Taverna, N. Cella, C. Santos and N. E. Hynes, Laminin and Tenascin Assembly and Expression Regulate HC11 Mouse Mammary Cell Differentiation, J. Cell Sci., 1994, 107, 1031–1040 CAS.
- D. Taverna, B. Groner and N. E. Hynes, Epidermal Growth Factor Receptor, Platelet-Derived Growth Factor Receptor, and c-ErbB-2 Receptor Activation All Promote Growth but Have Distinctive Effects upon Mouse Mammary Epithelial Cell Differentiation, Cell Growth Differ., 1991, 2, 145–154 CAS.
- K. G. Danielson, C. J. Oborn, E. M. Durban, J. S. Butel and D. Medina, Epithelial Mouse Mammary Cell Line Exhibiting Normal Morphogenesis in Vivo and Functional Differentiation in Vitro, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 3756–3760 CrossRef CAS PubMed.
- N. Cella, B. Groner and N. E. Hynes, Characterization of Stat5a and Stat5b Homodimers and Heterodimers and Their Association with the Glucocortiocoid Receptor in Mammary Cells, Mol. Cell. Biol., 1998, 18, 1783–1792 CrossRef CAS PubMed.
- W. Maret, Zinc in Cellular Regulation: The Nature and Significance of “Zinc Signals”, Int. J. Mol. Sci., 2017, 18(11), 2285 CrossRef PubMed.
- L. Guo, L. A. Lichten, M.-S. Ryu, J. P. Liuzzi, F. Wang and R. J. Cousins, STAT5-Glucocorticoid Receptor Interaction and MTF-1 Regulate the Expression of ZnT2 (Slc30a2) in Pancreatic Acinar Cells, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2818–2823 CrossRef CAS PubMed.
- J. Jeong, J. M. Walker, F. Wang, J. G. Park, A. E. Palmer, C. Giunta, M. Rohrbach, B. Steinmann and D. J. Eide, Promotion of Vesicular Zinc Efflux by ZIP13 and Its Implications for Spondylocheiro Dysplastic Ehlers–Danlos Syndrome, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E3530–E3538 CrossRef CAS PubMed.
- J. P. Liuzzi, L. A. Lichten, S. Rivera, R. K. Blanchard, T. B. Aydemir, M. D. Knutson, T. Ganz and R. J. Cousins, Interleukin-6 Regulates the Zinc Transporter Zip14 in Liver and Contributes to the Hypozincemia of the Acute-Phase Response, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 6843–6848 CrossRef CAS PubMed.
- J.-H. Kim, J. Jeon, M. Shin, Y. Won, M. Lee, J.-S. Kwak, G. Lee, J. Rhee, J.-H. Ryu, C.-H. Chun and J.-S. Chun, Regulation of the Catabolic Cascade in Osteoarthritis by the Zinc-ZIP8-MTF1 Axis, Cell, 2014, 156, 730–743 CrossRef CAS PubMed.
- W. Doppler, A. Villunger, P. Jennewein, K. Brduscha, B. Groner and R. K. Ball, Lactogenic hormone and cell type-specific control of the whey acidic protein gene promoter in transfected mouse cells, Mol. Endocrinol., 1991, 5, 1624–1632 CrossRef CAS PubMed.
- J. Lechner, T. Welte, J. K. Tomasi, P. Bruno, C. Cairns, J. Gustafsson and W. Doppler, Promoter-dependent synergy between glucocorticoid receptor and Stat5 in the activation of beta-casein gene transcription, J. Biol. Chem., 1997, 272, 20954–20960 CrossRef CAS PubMed.
- S. S. Mukhopadhyay, S. L. Wyszomierski, R. M. Gronostajski and J. M. Rosen, Differential interactions of specific nuclear factor I isoforms with the glucocorticoid receptor and STAT5 in the cooperative regulation of WAP gene transcription, Mol. Cell. Biol., 2001, 21, 6859–6869 CrossRef CAS PubMed.
- O. LaRochelle, S. Labbé, J.-F. Harrisson, C. Simard, V. Tremblay, G. St-Gelais, M. V. Govindan and C. Séguin, Nuclear factor-1 and metal transcription factor-1 synergistically activate the mouse metallothionein-1 gene in response to metal ions, J. Biol. Chem., 2008, 283, 8190–8201 CrossRef CAS PubMed.
- C. Williams, L. Helguero, K. Edvardsson, L.-A. Haldosén and J.-Å. Gustafsson, Gene Expression in Murine Mammary Epithelial Stem Cell-like Cells Shows Similarities to Human Breast Cancer Gene Expression, Breast Cancer Res., 2009, 11, R26 CrossRef PubMed.
- W. Wang, C. Jose, N. Kenney, B. Morrison and M. L. Cutler, Global Expression Profiling Reveals Regulation of CTGF/CCN2 during Lactogenic Differentiation, J. Cell Commun. Signal., 2009, 3, 43–55 CrossRef PubMed.
- T. R. Sornapudi, R. Nayak, P. K. Guthikonda, A. K. Pasupulati, S. Kethavath, V. Uppada, S. Mondal, S. Yellaboina and S. Kurukuti, Comprehensive Profiling of Transcriptional Networks Specific for Lactogenic Differentiation of HC11 Mammary Epithelial Stem-like Cells, Sci. Rep., 2018, 8, 11777 CrossRef PubMed.
- M. K. Diril, C. K. Ratnacaram, V. C. Padmakumar, T. Du, M. Wasser, V. Coppola, L. Tessarollo and P. Kaldis, Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 3826–3831 CrossRef CAS PubMed.
- S. Yonekura, M. Tsuchiya, Y. Tokutake, M. Mizusawa, M. Nakano, M. Miyaji, H. Ishizaki and S. Haga, The Unfolded Protein Response Is Involved in Both Differentiation and Apoptosis of Bovine Mammary Epithelial Cells, J. Dairy Sci., 2018, 101, 3568–3578 CrossRef CAS PubMed.
- G. Invernizzi, A. Naeem and J. J. Loor, Short Communication: Endoplasmic Reticulum Stress Gene Network Expression in Bovine Mammary Tissue during the Lactation Cycle1, J. Dairy Sci., 2012, 95, 2562–2566 CrossRef CAS PubMed.
- M. Tsuchiya, Y. Koizumi, S. Hayashi, M. Hanaoka, Y. Tokutake and S. Yonekura, The Role of Unfolded Protein Response in Differentiation of Mammary Epithelial Cells, Biochem. Biophys. Res. Commun., 2017, 484, 903–908 CrossRef CAS PubMed.
- L. Qian, V. Lopez, Y. A. Seo and S. L. Kelleher, Prolactin Regulates ZNT2 Expression through the JAK2/STAT5 Signaling Pathway in Mammary Cells, Am. J. Physiol.: Cell Physiol., 2009, 297, C369–C377 CrossRef CAS PubMed.
- V. Lopez and S. L. Kelleher, Zinc Transporter-2 (ZnT2) Variants Are Localized to Distinct Subcellular Compartments and Functionally Transport Zinc, Biochem. J., 2009, 422, 43–52 CrossRef CAS PubMed.
- N. H. McCormick and S. L. Kelleher, ZnT4 Provides Zinc to Zinc-Dependent Proteins in the Trans-Golgi Network Critical for Cell Function and Zn Export in Mammary Epithelial Cells, Am. J. Physiol.: Cell Physiol., 2012, 303, C291–C297 CrossRef CAS PubMed.
- S. L. Kelleher and B. Lönnerdal, Zip3 Plays a Major Role in Zinc Uptake into Mammary Epithelial Cells and Is Regulated by Prolactin, Am. J. Physiol.: Cell Physiol., 2005, 288, C1042–C1047 CrossRef CAS PubMed.
- L. Huang and J. Gitschier, A novel gene involved in zinc transport is deficient in the lethal milk mouse, Nat. Genet., 1997, 17, 292–297 CrossRef CAS PubMed.
- M. L. Ackland and A. Michalczyk, Zinc deficiency and its inherited disorders -a review, Genes Nutr., 2006, 1, 41–49 CrossRef CAS PubMed.
- S. Cristea and K. Polyak, Dissecting the Mammary Gland One Cell at a Time, Nat. Commun., 2018, 9, 1–3 CrossRef CAS PubMed.
- Y. Han, J. M. Goldberg, S. J. Lippard and A. E. Palmer, Superiority of SpiroZin2 Versus FluoZin-3 for Monitoring Vesicular Zn2+ Allows Tracking of Lysosomal Zn2+ Pools, Sci. Rep., 2018, 8, 1–15 CrossRef PubMed.
- L. F. B. Christensen, K. G. Malmos, G. Christiansen and D. E. Otzen, A Complex Dance: The Importance of Glycosaminoglycans and Zinc in the Aggregation of Human Prolactin, Biochemistry, 2016, 55, 3674–3684 CrossRef CAS PubMed.
- B.-J. Sankoorikal, Y. L. Zhu, M. E. Hodsdon, E. Lolis and P. S. Dannies, Aggregation of human wild-type and H27A-prolactin in cells and in solution: roles of Zn(2 +), Cu(2 +), and pH, Endocrinology, 2002, 143, 1302–1309 CrossRef CAS PubMed.
- J. Lu, A. J. Stewart, P. J. Sadler, T. J. T. Pinheiro and C. A. Blindauer, Albumin as a zinc carrier: properties of its high-affinity zinc-binding site, Biochem. Soc. Trans., 2008, 36, 1317–1321 CrossRef CAS PubMed.
- J. Masuoka, J. Hegenauer, B. R. Van Dyke and P. Saltman, Intrinsic stoichiometric equilibrium constants for the binding of zinc(II) and copper(II) to the high affinity site of serum albumin, J. Biol. Chem., 1993, 268, 21533–21537 CAS.
- E. A. Permyakov, D. B. Veprintsev, G. Y. Deikus, S. E. Permyakov, L. P. Kalinichenko, V. M. Grishchenko and C. L. Brooks, pH-induced transition and Zn2+-binding properties of bovine prolactin, FEBS Lett., 1997, 405, 273–276 CrossRef CAS PubMed.
- C. Wang, Y. Liu and J.-M. Cao, Protein-Coupled Receptors: Extranuclear Mediators for the Non-Genomic Actions of Steroids, Int. J. Mol. Sci., 2014, 15, 15412–15425 CrossRef PubMed.
- C. Stahn and F. Buttgereit, Genomic and Nongenomic Effects of Glucocorticoids, Nature Reviews, Rheumatology, 2008, 4, 525–533 CAS.
- J. G. Tasker, S. Di and R. Malcher-Lopes, Minireview: Rapid Glucocorticoid Signaling via Membrane-Associated Receptors, Endocrinology, 2006, 147, 5549–5556 CrossRef CAS PubMed.
- L. Dindia, E. Faught, Z. Leonenko, R. Thomas and M. M. Vijayan, Rapid Cortisol Signaling in Response to Acute Stress Involves Changes in Plasma Membrane Order in Rainbow Trout Live, Am. J. Physiol.: Endocrinol. Metab., 2013, 304, E1157–E1166 CrossRef CAS PubMed.
- N. Gupta and D. Mayer, Interaction of JAK with steroid receptor function, Jakstat, 2013, 2, e24911 Search PubMed.
- M.-H. Kim, T. B. Aydemir, J. Kim and R. J. Cousins, Hepatic ZIP14-Mediated Zinc Transport Is Required for Adaptation to Endoplasmic Reticulum Stress, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5805–E5814 CrossRef CAS PubMed.
- M.-H. Kim, T. B. Aydemir and R. J. Cousins, Zinc and ZIP14 (Slc39a14) Are Required for Adaptation to ER Stress in Mouse Liver, FASEB J., 2016, 30, 148.2 Search PubMed.
- K. Homma, T. Fujisawa, N. Tsuburaya, N. Yamaguchi, H. Kadowaki, K. Takeda, H. Nishitoh, A. Matsuzawa, I. Naguro and H. Ichijo, SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency, Mol. Cell, 2013, 52, 75–86 CrossRef CAS PubMed.
- K. W. Colston, U. Berger, P. Wilson, L. Hadcocks, I. Naeem, H. M. Earl and R. C. Coombes, Mammary Gland 1, 25-Dihydroxyvitamin D3 Receptor Content during Pregnancy and Lactation, Mol. Cell. Endocrinol., 1988, 60, 15–22 CrossRef CAS PubMed.
- F. de, O. Andrade, S. de Assis, L. Jin, C. C. Fontelles, L. F. Barbisan, E. Purgatto, L. Hilakivi-Clarke and T. P. Ong, Lipidomic Fatty Acid Profile and Global Gene Expression Pattern in Mammary Gland of Rats That Were Exposed to Lard-Based High Fat Diet during Fetal and Lactation Periods Associated to Breast Cancer Risk in Adulthood, Chem.-Biol. Interact., 2015, 239, 118–128 CrossRef PubMed.
- M. V. Goddio, A. Gattelli, J. M. Tocci, L. P. Cuervo, M. Stedile, D. J. Stumpo, N. E. Hynes, P. J. Blackshear, R. P. Meiss and E. C. Kordon, Expression of the MRNA Stability Regulator Tristetraprolin Is Required for Lactation Maintenance in the Mouse Mammary Gland, Oncotarget, 2018, 9, 8278–8289 CrossRef PubMed.
- M. V. Goddio, A. Gattelli, V. Slomiansky, E. Lacunza, T. Gingerich, J. M. Tocci, M. M. Facchinetti, A. C. Curino, J. LaMarre, M. C. Abba and E. C. Kordon, Mammary Differentiation Induces Expression of Tristetraprolin, a Tumor Suppressor AU-Rich MRNA-Binding Protein, Breast Cancer Res. Treat., 2012, 135, 749–758 CrossRef CAS PubMed.
- A.-M. Fairhurst, J. E. Connolly, K. A. Hintz, N. J. Goulding, A. J. Rassias, M. P. Yeager, W. Rigby and P. K. Wallace, Regulation and Localization of Endogenous Human Tristetraprolin, Arthritis Res. Ther., 2003, 5, R214–R225 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9mt00301k |
| This journal is © The Royal Society of Chemistry 2020 |