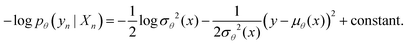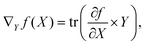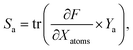Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DeepSIBA: chemical structure-based inference of biological alterations using deep learning†
C.
Fotis‡
 ,
N.
Meimetis‡
,
N.
Meimetis‡
 ,
A.
Sardis
and
L. G.
Alexopoulos
*
,
A.
Sardis
and
L. G.
Alexopoulos
*
Biomedical Systems Laboratory, National Technical University of Athens, Athens, Greece. E-mail: leo@mail.ntua.gr
First published on 9th November 2020
Abstract
Predicting whether a chemical structure leads to a desired or adverse biological effect can have a significant impact for in silico drug discovery. In this study, we developed a deep learning model where compound structures are represented as graphs and then linked to their biological footprint. To make this complex problem computationally tractable, compound differences were mapped to biological effect alterations using Siamese Graph Convolutional Neural Networks. The proposed model was able to encode molecular graph pairs and identify structurally dissimilar compounds that affect similar biological processes with high precision. Additionally, by utilizing deep ensembles to estimate uncertainty, we were able to provide reliable and accurate predictions for chemical structures that are very different from the ones used during training. Finally, we present a novel inference approach, where the trained models are used to estimate the signaling pathway signature of a compound perturbation, using only its chemical structure as input, and subsequently identify which substructures influenced the predicted pathways. As a use case, this approach was used to infer important substructures and affected signaling pathways of FDA-approved anticancer drugs.
1. Introduction
Early stage drug discovery aims to identify the right compound for the right target, for the right disease. A very important step in this process is hit identification, in which compounds that exhibit strong binding affinity to the target protein are prioritized. Traditionally, the most widely employed method for in vitro hit identification is High Throughput Screening (HTS). In vitro HTS can produce hits with strong binding affinity that may later be developed into lead compounds through lead optimization. However, due to the vast chemical space, even large scale in vitro HTS offers limited chemical coverage. On this front, the development of Computer Aided Drug Design (CADD) methods has enabled the virtual High Throughput Screening (vHTS) of vast compound libraries, thus effectively increasing the search space of hit identification. CADD methods for vHTS focus on compounds’ chemical structures and prioritize those that are likely to have activity against the target, for further experiments.1 More specifically, ligand-based approaches are based on the hypothesis that similar chemical structures will cause similar biological response, by binding to the same protein.2 However, there are many cases of compounds and drugs, which although structurally dissimilar, cause similar biological effect, either because of off-target effects or by targeting proteins in the same pathway.3 As a whole, CADD approaches focus on optimal binding affinity, by assessing a compound's structural attributes, often disregarding the effect of the perturbation on the biological system, which is closely related to clinical efficacy and toxicity.4Advances in systems-based approaches and ‘omics technologies have led to the development of systems pharmacology methods that aim to lower the attrition rates of early stage drug discovery. Systems pharmacology approaches couple ‘omics data with knowledge bases of molecular interactions and network analysis methods in order to assess compounds based on their biological effect.5 One approach that has gained considerable attraction is the use of gene expression (GEx) profiling to characterize the systematic effects of compounds. On this front, Verbist et al. showed how GEx data were able to influence decision making in eight drug discovery projects by uncovering potential adverse effects of the lead compounds.6 Additionally, Iorio et al. utilized similarities between drugs’ transcriptional responses to create a drug network and identified the mechanism of action of new drugs based on their position in the network.7 Since its release, the Connectivity Map (CMap) and the LINCS project have been a cornerstone of transcriptomic-based approaches by providing a large scale database of transcriptomic signatures from compound perturbations along with essential signature matching algorithms.8,9 CMap's approach is based on the hypothesis that compounds with similar transcriptomic signatures will cause similar physiological effects on the cell and has been widely adopted by the field of drug repurposing.10 However, signature-based approaches are not only limited in the search space of compounds with available GEx data but are also missing key structural information that is pivotal for drug design. Thus, an interdisciplinary framework that translates a compound's structural attributes to its biological effect holds promise in augmenting the application of both CADD and systems-based approaches for drug discovery. A computational approach that meets the requirements of such an interdisciplinary framework is Machine Learning (ML) and especially Deep Learning (DL).
The recent increase in available data and computing power has given rise to Deep Learning (DL) methods for various drug discovery tasks, including bioactivity and toxicity prediction as well as de novo molecular design.11–15 DL methods offer the advantage of flexible end-to-end architectures that learn task specific representations of chemical structures, without the need for precomputed features.16 One particular DL architecture that has achieved state of the art results in several drug discovery benchmark datasets is the Graph Convolutional Neural Network (GCNN).17,18 Molecular GCNNs operate on chemical structures represented as undirected graphs, with nodes being the atoms and edges the bonds between them. Kearnes et al. developed the Weave graph convolution module, which encodes both atom and bond representations and combines them using fuzzy histograms to extract meaningful molecule-level representations.19 Despite their improved performance over traditional ML methods, end-to-end models including GCNNs are still prone to generalization errors on new chemical scaffolds. This is mainly because of the limited coverage of the chemical space by the training data.20 In order to tackle this limited chemical coverage, methods like one-shot learning are promising candidates for drug discovery applications. One-shot learning techniques, such as Siamese networks, aim to learn a meaningful distance function between related inputs and have shown increased performance over traditional methods in tasks with few data points.21–24 Altae-Tran et al. implemented one-shot learning for drug discovery by combining graph convolutions and Long Short Term Memory (LSTM) networks with attention and achieved better results than traditional GCNNs.25 Furthermore, for drug discovery applications, uncertainty estimation is crucial, since incorrect predictions e.g. regarding toxicity can lead to incorrect prioritization of compounds for further experimental testing.26–29 On this front, Ryu et al. developed Bayesian GCNNs for molecular property, bioactivity and toxicity predictions and showed that quantifying predictive uncertainty can lead to more accurate virtual screening results.30 The flexibility provided by GCNN architectures along with one-shot learning and uncertainty estimation approaches can combine aspects from both systems and ligand-based methods into an interdisciplinary framework for early stage drug discovery.
In this paper, we employ deep learning to decipher the complex relationship between a compound's chemical structure and its biological effect. To make this complex problem computationally tractable, we focus on learning a combined representation and distance function that maps structural differences to biological effect alterations. For this task, we propose a deep Siamese GCNN model called deepSIBA. DeepSIBA takes as input pairs of compound structures, represented as graphs and outputs their biological effect distance, in terms of enriched biological processes (BPs) along with an estimated uncertainty. DeepSIBA is trained to minimize the loss between predicted and calculated distances of enriched BPs for compound pairs with available GEx data. In order to account for the biological factors that influence the learning task, we train cell line-specific deep ensembles only on carefully selected chemical structures, for which high quality GEx data are available. The performance of our approach was evaluated with a realistic drug discovery scenario in mind, where gene expression data are available for only one compound per pair and compared with ML methods for pairwise (dyadic) data.31,32 Finally, we present a novel inference approach, in which the trained models can be used to infer the signaling pathway signature of a target compound, without available GEx data. This inference approach is coupled with a novel method, based on graph saliency maps,33 which can identify substructures that are responsible for a compound's inferred biological footprint. As a use case, this approach was tasked to infer the signaling pathway signature and important substructures of approved anticancer drugs for which no transcriptomic signatures are available in our data sets, using only their chemical structure as input. DeepSIBA can be used in combination with existing in silico drug discovery pipelines to identify structures that not only exhibit maximal binding affinity but also cause a desired biological effect. Thus, by incorporating deepSIBA's interdisciplinary approach, the drug discovery process can produce candidates with improved clinical efficacy and toxicity.
2. Material and methods
2.1 The deepSIBA approach
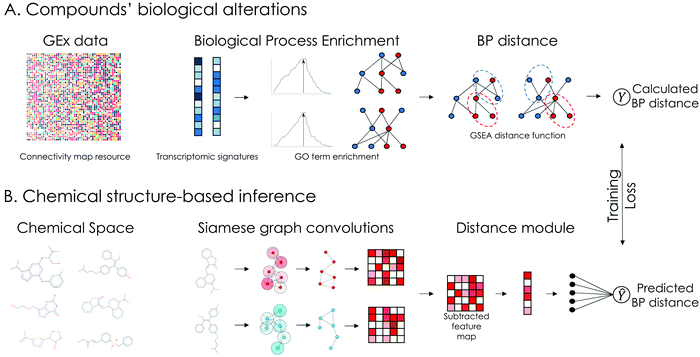
The overview of our approach is presented in Fig. 1. Transcriptomic signatures from compound perturbations along with their respective chemical structures were retrieved from the CMap dataset.9 For each compound perturbation, normalized enrichment scores (NES) of GO terms related to BPs were calculated using Gene Set Enrichment Analysis (GSEA). Afterwards, the lists of enriched BPs were ranked based on NES and a Kolmogorov–Smirnov based distance function, similar to GSEA, was used to calculate their pairwise distance (Fig. 1A). During the learning phase, the proposed model is trained to predict the pairwise distance between compounds’ affected BPs using only their chemical structure as input. The input chemical structures are represented as undirected graphs, with nodes being the atoms and edges the bonds between them and encoded using a Siamese GCNN architecture (Fig. 1B). In our approach, compounds with available GEx data, representing a small portion of the chemical space, serve as reference for the inference phase. During inference, the model is tasked to predict the biological effect distance between reference and unknown compounds (without available GEx data).2.2 Data preprocessing and quality control
Transcriptomic signatures (level 5 z-score transformed) following compound treatment were downloaded from the L1000 CMap resource.34 In this project, only the differential expression of the 978 landmark genes in the L1000 assay was considered. For each signature, a quality score was derived, based on its transcriptional activity score (TAS), the number of biological replicates and whether the signature is considered an exemplar. This quality score ranges from Q1 to Q8, with Q1 representing the highest quality. TAS is a metric that measures a signature's strength and reproducibility and is calculated as the geometric mean of the number of differentially expressed (DEx) transcripts and the 75th quantile of pairwise replicate correlations. Furthermore, exemplar signatures are specifically designated for further analysis in the CLUE platform.35 For each compound per cell line, among signatures from different dosages and time points, the signature with the highest quality was selected. An overview of the processed dataset is presented in ESI† 1.1.2.3 Biological process enrichment and pairwise distance calculation
Gene ontology (GO) terms for biological processes (BP) involving the landmark genes of the L1000 assay were retrieved using the topGO R package in Bioconductor.36 Only GO terms with at least 10 genes were considered. For each signature, GO term enrichment was calculated using the R package FGSEA in Bioconductor.37 Thus, the gene-level feature vector of each perturbation was transformed to a BP-level feature vector of Normalized Enrichment Scores (NES). Pairwise distances between BP-level feature vectors were calculated similar to Iorio et al.,7 using the R package Gene Expression Signature in Bioconductor.38 Given two feature vectors ranked by NES, A and B, GSEA is used to calculate the ES of the top and bottom GO terms of A in B and vice versa. The distance between the vectors is computed as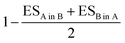 and ranges from 0 to 2. An important parameter that can introduce bias in the distance calculation is the number of top and bottom GO terms to consider during GSEA. On this front, an ensemble approach was developed, by calculating pairwise distances between BP-level feature vectors for 5 different numbers of top and bottom GO terms. The numbers we considered were selected based on the average number of significantly enriched GO terms across all perturbations in the dataset (see ESI† 1.3 for details). The distance scores were finally averaged and normalized between 0 and 1.
and ranges from 0 to 2. An important parameter that can introduce bias in the distance calculation is the number of top and bottom GO terms to consider during GSEA. On this front, an ensemble approach was developed, by calculating pairwise distances between BP-level feature vectors for 5 different numbers of top and bottom GO terms. The numbers we considered were selected based on the average number of significantly enriched GO terms across all perturbations in the dataset (see ESI† 1.3 for details). The distance scores were finally averaged and normalized between 0 and 1.
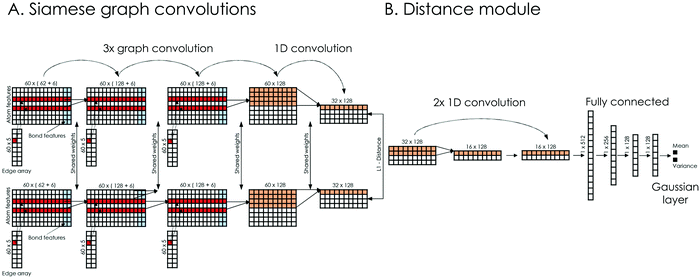
2.4 Siamese GCNN architecture
A schematic representation of our model's architecture is presented in Fig. 2. The learning model takes as input the chemical structures of compound pairs and predicts their biological distance, at the level of affected biological processes (GO terms). Regarding the input, chemical structures are represented as undirected graphs, with nodes being the atoms and edges the bonds between them. Each input is encoded using 3 matrices: the atom array, which contains atom-level features, the bond array, which contains bond-level features and the edge array, which describes the connectivity of the compound (see ESI† 2.1 for details). The learning model consists of two Siamese encoders (shared weights) that embed the input graphs into a high dimensional latent space and a trainable distance module that outputs the final distance prediction. The Siamese encoders consist of 3 graph convolutional layers that learn neighborhood-level representations, followed by a convolutional layer that extracts compound-level features (Fig. 2A). Graph convolutions were implemented similar to Duvenaud et al.17 (see ESI† 2.2 for details). The overall goal of the Siamese encoder is to learn task-specific compound representations. The feature maps of the last Siamese layer are then subtracted and their absolute difference is passed to the distance module. The distance module consists of 2 convolutional layers, which extract important features from the difference of the feature maps and 3 fully connected layers that aim to combine those features, while progressively reducing the dimensions (Fig. 2B). Finally, a Gaussian regression layer outputs a mean and variance of the biological effect distance between the compound pair. By treating the distance as a sample from a Gaussian distribution with the predicted mean and variance, the model is trained end-to-end by minimizing the negative log-likelihood criterion27 given byFor each cell line, an ensemble model combining 50 models was created. The ensemble's output is also a Gaussian, with mean and variance calculated from the uniformly weighted mixture of each model. The coefficient of variation (CV) of the Gaussian mixture is used as the model's estimate of predictive uncertainty. The model's hyperparameters, along with the equations for the Gaussian mixture's mean and variance are presented in ESI† 2.3 and 2.4.
2.5 Dataset splitting and evaluation metrics
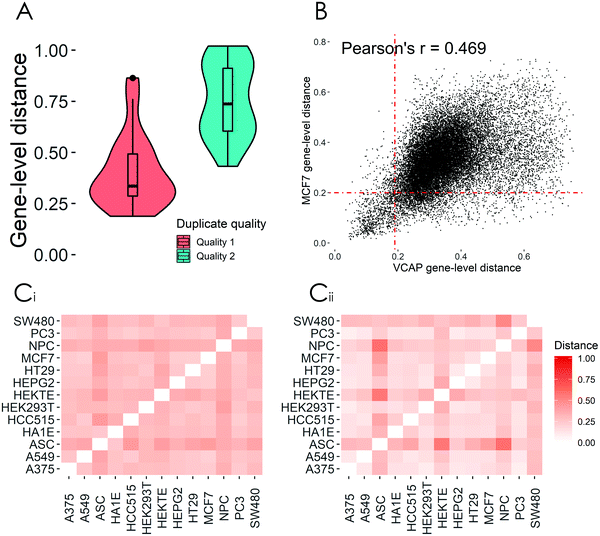
For each cell line, available compounds were split into training and test. Each cell line specific training set consists of the pairwise distances between training compounds’ affected BPs, while each test set contains distances between test and training compounds. Additionally, the Tanimoto similarity between the ECFP4 fingerprints of all training and test compounds was calculated and test compounds that exhibited a similarity higher than 0.85 to any training compound were excluded. An overview of the training and test sets is presented in ESI† 4.1. Across all test scenarios, model performance was evaluated in terms of Mean Squared Error (MSE), Pearson's r and precision. MSE and Pearson's r were calculated between the predicted and computed distance values. In order to calculate precision, the continuous distance values were transformed to binary form by comparing them with an appropriate distance threshold. Even though the learning task is a regression problem, given its nature and potential applications, high precision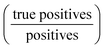 is important in order to avoid false positive hits for validation experiments. The appropriate distance threshold for precision was set at 0.2, based on the distance distribution of duplicate compound signatures, the threshold equivalent to a 90% Connectivity Score and the relationship between the threshold and the actual average number of common enriched BPs. Duplicate signatures indicate transcriptomic signatures from the same compound perturbation, cell line, dose and time point that were assayed on different L1000 plates. Thus, the distribution of distances between duplicate signatures most closely approximates the reference distribution of truly similar biological effect. A thorough investigation of the distance threshold to distinguish compounds with similar biological effect is presented in ESI† 5.1.
is important in order to avoid false positive hits for validation experiments. The appropriate distance threshold for precision was set at 0.2, based on the distance distribution of duplicate compound signatures, the threshold equivalent to a 90% Connectivity Score and the relationship between the threshold and the actual average number of common enriched BPs. Duplicate signatures indicate transcriptomic signatures from the same compound perturbation, cell line, dose and time point that were assayed on different L1000 plates. Thus, the distribution of distances between duplicate signatures most closely approximates the reference distribution of truly similar biological effect. A thorough investigation of the distance threshold to distinguish compounds with similar biological effect is presented in ESI† 5.1.
2.6 Signaling pathway inference for target structure
The predictions of a trained deepSIBA model can be used to infer a pathway signature for a target structure without the need for GEx data, in terms of the most upregulated and downregulated signaling pathways. The inference approach is similar to the k-Nearest Neighbor algorithm (KNN). Given a target structure, a trained ensemble model for the cell line of choice is used to predict all pairwise distances between target and training compounds. The predicted distance represents the difference between compounds’ enriched BPs (GO terms). Training set compounds with predicted distance less than a specified threshold dth are selected as the target's neighbors. If a target structure has more than k neighbors, a signaling pathway signature can be inferred in the following way. For each neighbor Ni, the lists of the top 10 most upregulated and most downregulated pathways, based on NES, are constructed. Pathway enrichment is calculated using FGSEA with KEGG as a knowledge base.39 KEGG signaling pathways were chosen for inference due to their interpretability. Signaling pathways that appear in the neighbors’ lists with a frequency score higher than a threshold fth are selected. Additionally, to account for signaling pathways that are frequently upregulated or downregulated in the set of training compounds, a p-value for each inferred pathway is also calculated. On this front, sets of k neighbors are randomly sampled 5000 times from the training set and a Null distribution of frequency scores for each pathway is derived. A p-value is computed as the sum of the probabilities of observing equally high or higher frequency scores. Finally, only pathways with p-value lower than a threshold pth are inferred. Thus, for each chemical structure, our approach infers two signatures of variable length (up to 10 each) of potentially downregulated and upregulated pathways respectively. For the MCF7 cell line, the aforementioned thresholds and parameters of the inference approach were selected by evaluating the results, in terms of precision and number of inferred pathways, on its respective test set (see ESI† 6.1 for details).2.7 Substructure importance using graph-based gradients
A graph-based gradient approach, similar to saliency maps, was developed to identify important substructures that influence the biological effect similarity of chemical structure pairs. First, the derivative of deepSIBA's output w.r.t the input matrices that contain the atom features of each compound, in the input pair, is calculated using Tensorflow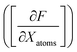 . Subsequently, for each compound atom importance is scored, using a directional derivative approach. Thus, similar to vector calculus, the directional derivative of a scalar f(X), with X being a matrix, in the direction of a matrix Y is
. Subsequently, for each compound atom importance is scored, using a directional derivative approach. Thus, similar to vector calculus, the directional derivative of a scalar f(X), with X being a matrix, in the direction of a matrix Y iswhere,
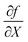 is the gradient matrix, or in our case
is the gradient matrix, or in our case 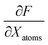 , while Y can be considered a matrix with zeros everywhere, except the row containing the specific atom's feature. Thus, an importance score for each atom of a compound can be calculated as
, while Y can be considered a matrix with zeros everywhere, except the row containing the specific atom's feature. Thus, an importance score for each atom of a compound can be calculated aswhere the only non-zero part of Ya is the one-hot encoded feature vector of atom a. For each atom the importance score Sa was transformed to a count score Ca, based on how many times each atom was in the top 20% most important atoms for each model in a deepSIBA ensemble. When scoring atom importance during the pathway inference approach, a similar score was calculated based on the times an atom was present in the top 20% for each target-reference pair. Finally, due to the GCNN core module of deepSIBA, important substructures are formed by important atoms that are neighbors in the compound's molecular graph. Atom importance is visualized using the RDKit library.40
3 Results and discussion
3.1 Biological factors influence the model's learning task
The presented model is tasked to predict the biological effect distance between compounds, using their molecular graphs as input. Considering that this distance is calculated from experimental GEx data following compound treatment, there are specific biological factors that can influence the learning task. The CMap dataset contains over 110 K transcriptomic signatures from over 20 K compounds assayed across 70 cell lines. By carefully analyzing these signatures and their pairwise distances, we were able to pinpoint the most influential factors and identify their effect on the model's target value.| Structural distance threshold | Pairs with similar chemical structure | Pairs affecting similar BPsa (%) | Pairs affecting similar genesb (%) |
|---|---|---|---|
| a BP distance threshold to consider compounds similar = 0.2. b Gene distance threshold to consider compounds similar = 0.19. | |||
| 0.10 | 91 | 76.9 | 68.1 |
| 0.15 | 114 | 75.4 | 65.7 |
| 0.20 | 200 | 74.0 | 61.0 |
| 0.25 | 316 | 69.9 | 57.6 |
| 0.30 | 494 | 65.3 | 51.0 |
Through the careful analysis of the processed data sets, we showed that raw data quality greatly affects the distribution of distance values and that lower quality transcriptomic signatures of the same compound, with the same perturbation parameters (duplicates), often exhibit large differences in terms of DEx genes (Fig. 3A). Based on these findings, we chose to develop deepSIBA using only compounds with available Q1 transcriptomic signatures. Furthermore, we showed that the transcriptomic distance of a compound pair can vary depending on the choice of cellular model (Fig. 3B). One common approach to address this issue is to aggregate either transcriptomic signatures or distance values across cell lines. While aggregating enables the training of a general model on all available compound pairs, it can often produce misleading results and cause information loss. Thus, we decided to make our approach cell line specific and develop our models for cell lines that have the highest number of Q1 transcriptomic signatures following compound treatment. Finally, we highlighted that a distance function operating on enriched BPs, rather than genes, can better capture the expected biological effect similarities of perturbations with similar structure or biological nature (Table 1 and Fig. 3C). We reason that this is the case due to the BP enrichment analysis that precedes the distance calculation, which can capture smaller changes in the expression of genes that interact with each other to form a biological process. By analyzing the relationship between the aforementioned experimental factors and our target variable, we were able to make data-driven decisions to propose a learning task that minimizes their effect. In the following sections we evaluate the ability of deepSIBA to learn the proposed task and test whether our approach can identify dissimilar structures that affect similar BPs in a meaningful way.
3.2 Performance evaluation
Model performance was evaluated on pairs of reference and test compounds. Test compounds were removed from the training sets and thus represent new chemical structures without available experimental GEx data. Additionally, the effect of the structural similarity between input compounds on performance, along with the utility of the model's estimate for uncertainty, were investigated. Finally, we evaluated the performance of our approach on test chemical structures that are very different from the ones used in training.| Cell line | Model | MSE | MSE @1% | Pearson's r | Precision (%) | Precision @1% (%) | Predicted similar pairs |
|---|---|---|---|---|---|---|---|
| A375 | DeepSIBA | 0.008 | 0.006 | 0.59 | 98.22 | 98.22 | 169 |
| ReSimNet | 0.012 | 0.022 | 0.60 | 32.23 | 56.80 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 243 243 |
|
| TwoStepRLS | 0.010 | 0.008 | 0.51 | 44.61 | 78.68 | 4024 | |
| PC3 | DeepSIBA | 0.011 | 0.007 | 0.53 | 89.29 | 89.29 | 28 |
| ReSimNet | 0.017 | 0.032 | 0.49 | 25.02 | 46.89 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 195 195 |
|
| TwoStepRLS | 0.013 | 0.041 | 0.44 | 29.98 | 38.96 | 1758 | |
| VCAP | DeepSIBA | 0.033 | 0.026 | 0.41 | 71.63 | 71.63 | 141 |
| ReSimNet | 0.039 | 0.105 | 0.38 | 32.69 | 52.97 | 9245 | |
| TwoStepRLS | 0.034 | 0.049 | 0.43 | 32.34 | 31.12 | 3120 | |
| MCF7 | DeepSIBA | 0.012 | 0.007 | 0.56 | 61.03 | 61.03 | 195 |
| ReSimNet | 0.015 | 0.029 | 0.59 | 26.93 | 51.20 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 420 420 |
|
| TwoStepRLS | 0.015 | 0.010 | 0.47 | 33.55 | 70.14 | 4322 | |
| Cell-line | MSE | MSE @1% | Pearson's r | Precision (%) |
|---|---|---|---|---|
| HT29 | 0.010 | 0.013 | 0.60 | 84.88 |
| A549 | 0.013 | 0.012 | 0.62 | 83.00 |
| HA1E | 0.015 | 0.009 | 0.58 | 100 |
| HEPG2 | 0.013 | 0.014 | 0.61 | 99.10 |
| HCC515 | 0.014 | 0.010 | 0.52 | 97.92 |
| NPC | 0.006 | 0.005 | 0.67 | 73.64 |
| Test set | Max similarity to training set | MSE | Pearson's r | Precision (%) | Predicted similar pairs |
|---|---|---|---|---|---|
| #1 | [0–0.85] | 0.0083 | 0.59 | 97.26 | 876 |
| #2 | [0.35–0.65] | 0.0092 | 0.52 | 76.48 | 330 |
| #3 | [0–0.3] | 0.0107 | 0.44 | 50.37 | 135 |
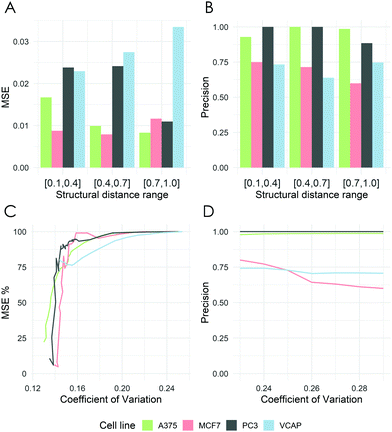
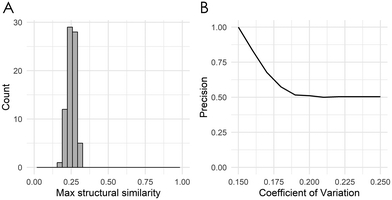
Across all examined cell lines, deepSIBA was able to identify chemical structures that affect similar BPs, outperforming, especially in terms of precision, the distance learning methods that utilize compounds’ ECFP4 fingerprints as input (Table 2). Even though the learning task is regression, we reason that precision is a crucial metric, considering the potential screening applications of deepSIBA in order to identify compounds that exhibit similar biological effect to a query. In this scenario, high precision, rather than a large number of identified hits, is required to correctly prioritize compounds for downstream experimental validation. We chose not to compare our approach with traditional machine learning methods, e.g. Random Forests and SVMs, because we argue that these are not optimal for a distance/similarity learning task. Furthermore, deepSIBA was able to maintain its high performance regardless of the structural similarity between input compounds and identify cases of structurally dissimilar compounds that affect similar BPs (Fig. 4A and B). Thus, the employed GCNN architecture shows promise towards this highly interdisciplinary task. However, there were some cases of compounds affecting similar BPs that were missed by the model. These cases, in combination with the decrease in performance as the minimum structural distance between test and training compounds increases highlight key limitations in our approach (Table 4). On this front, limited coverage of the chemical space by compounds with available GEx data is a major issue that limits our ability to model in its entirety the complex function that translates changes in chemical structure to BP alterations. Even though each training set for each cell line contains on average around 320 K samples, these are comprised from the pairing of around 800 compounds. The limitations that arise from this low coverage of the chemical space can’t be solved by changes in deep learning architecture and require more training compounds and/or extra input information. On this front, we applied a data augmentation technique, where each training set was augmented with randomly sampled pairs between Q1 and Q2 compound signatures (see ESI† 4.2). However, due to conflicting evidence between Q1 and Q2 transcriptomic signatures the performance of the models varied significantly across cell lines (see ESI† 5.3). A rather efficient workaround that we utilized in our approach is to quantify predictive uncertainty using deep ensembles. We showed that the model's performance, even when tested on compounds that are structurally different from the ones used in training, is higher for samples with lower uncertainty (Fig. 5). Thus, the model's estimate of predictive uncertainty can be used to provide more reliable and accurate results. For instance, if an application imposes a constraint on the maximum allowed error, the appropriate uncertainty threshold can be identified and only point predictions with uncertainty lower than this threshold can be considered. Finally, we showed that transfer learning is a suitable approach to expand the biological coverage of deepSIBA to additional cellular models with fewer available data points (Table 3). For example, in the NPC cell line, which has approximately 50% fewer compound signatures than A375, deepSIBA was still able to achieve reasonable performance.
3.3 Signaling pathway inference for target structure
The predictions of deepSIBA can be used to infer a signaling pathway signature, in terms of the most upregulated and downregulated pathways, for a target chemical structure without available GEx data. The inference is performed following a KNN-like approach, in which reference compounds with the smallest distance to the target, as predicted by the model, are selected as its neighbors and their pathway signatures are retrieved. Then, pathways that frequently belong in the 10 most upregulated or downregulated pathways of the neighbors are inferred as the target's signature. The performance of the approach was evaluated on the test compounds of the MCF7 model and then, as a use case, it was tasked to infer the signaling pathways affected by FDA approved anticancer drugs, for which no GEx data are available in our dataset. Additionally, the chemical substructures that mostly influence the inferred pathways were identified and visualized using a graph gradient-based approach.| Number of inferred pathways | Precision (%) | |
|---|---|---|
| Downregulated | 5 | 73.3 |
| Upregulated | 2.5 | 69.7 |
| Drug | Mechanism of action | Inferred downregulated KEGG signaling pathways | Inferred upregulated KEGG signaling pathways |
|---|---|---|---|
| Fludarabine | Nucleic acid synthesis inhibitor | Purine metabolism, pyrimidine metabolism , RNA transport, spliceosome, cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation, MicroRNAs in cancer | Leukocyte transendothelial migration, oxytocin signaling pathway, Alzheimer's disease, pertussis, rheumatoid arthritis |
| Clofarabine | Nucleic acid synthesis inhibitor | RNA transport, spliceosome, cell cycle, ubiquitin mediated proteolysis, progesterone-mediated oocyte maturation, MicroRNAs in cancer | Natural killer cell mediated cytotoxicity, leukocyte transendothelial migration, oxytocin signaling pathway, pertussis, rheumatoid arthritis |
| Pralatrexate | Inhibits dihydrofolate reductase (DHFR) and thymidylate synthase | Purine metabolism, pyrimidine metabolism, metabolic pathways, RNA transport, spliceosome | NF-kappa B signaling pathway, natural killer cell mediated cytotoxicity, TNF signaling pathway, leukocyte transendothelial migration |
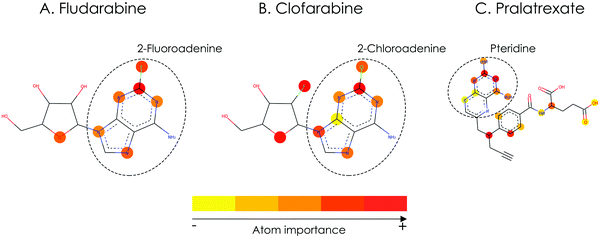 | ||
| Fig. 6 Important atoms related to the inferred biological footprint of the compounds of the use case, as identified by the deep learning model (the red color signifies the most important atoms). | ||
In the presented use case, we demonstrated that by utilizing the training compounds as reference, the inferred signaling pathway signatures for each of the anticancer drugs were found to be closely connected to their respective MoA (Table 6). Thus, our inference method has the potential to provide an early estimate regarding the pathways affected by a compound, using only its chemical structure as input. Additionally, we showed that for each compound the highlighted substructures are also directly related to their respective MoA (Fig. 6). This fact not only increases the interpretability of the model's predictions, which is a crucial topic of DL methods for drug discovery, but also shows that a GCNN model trained end-to-end on molecular graphs is able to learn meaningful structural representations that are related to compounds’ biological effects.46–48 To the best of our knowledge, this is the first time a DL model was used to identify important substructures and infer the signaling pathway signature of a target compound without available experimental GEx data. A possible limitation of our approach might be its resolution capabilities in specific use-cases of compounds with similar chemical structure but different MoA. Although the comparison of the fludarabine and clofarabine use-cases suggests that our approach might be able to identify small structural differences between drugs with similar MoA (Fig. 6), we haven’t systematically compared use-cases of structurally similar compounds that affect different BPs. From the analysis of the CMap dataset we have showed that compounds with high structural similarity tend to have similar biological effect (see ESI† Fig. S6). This lack of data regarding compounds that are derivatives but affect different BPs limits our ability to systematically perform the aforementioned comparison and pinpoint the maximum resolution of our approach. Furthermore, due to the nature of the inference method, limiting factors may also arise from the lack of diversity in affected BPs by the training compounds. This lack of diversity can influence the signaling pathway inference for an unknown target structure, when its true biological footprint is not represented in the reference compounds. In such cases, the inference of incorrect signatures can be avoided by focusing on target compounds with at least k reference neighbors (here k = 5) and only infer statistically significant pathways, using our method's calculated p-value.
4. Conclusion
In this paper, we developed a deep learning framework to match the chemical structure of compound perturbations to their biological effect on specific cellular models. We showed, that the careful formulation of the learning problem and the flexibility of the Siamese GCNN architecture enabled our models to achieve high performance across all test scenarios. Additionally, we highlighted the utility of the uncertainty estimate, provided by deep ensembles, in test cases where the unknown chemical structures are very different from the structures used to train the models. Finally, we presented a novel inference pipeline, which can infer a signaling pathway signature for a target compound and subsequently identify which substructures mostly influenced the prediction. The novelty, performance and interpretability of our methods paves the way for further investigation in order to expand their coverage and utility.Possible efforts for further investigation can be concentrated on the input representation, the biological response distance and the model's uncertainty estimate. Regarding the input, one interesting idea is to include binding information in order to capture the potential protein target of the input molecules. This extra information can be passed to the model either in the form of latent space embeddings from a trained binding affinity prediction model or in the form of predictions against a panel of protein kinases.49 Regarding the biological distance between compound perturbations, this can be augmented by calculating the compound's effect on different levels of biological hierarchy, i.e. GEx, signaling pathways, transcription factors and signaling networks.50,51 Afterwards, these distances could be combined or separate models could be trained in order to better capture the similar effect of compounds. Additionally, instead of using a distance metric between all affected BPs, specific biological processes could be selected and application specific models could be developed to identify compounds that affect these biological processes. Regarding the model's uncertainty estimate, an interesting avenue for investigation is to take into account the transcriptomic signatures of replicates from the CMap dataset and calculate distributions of pairwise distances between compounds. Then, models could be trained on these distributions to better capture the variation of the experimental ground truth. Finally, collecting more data regarding derivative compounds with different MoA is an interesting avenue for further investigation in order to identify the resolution capabilities of the substructure importance approach.
The highly interdisciplinary framework of deepSIBA combines aspects from both the CADD and ‘omics domains in order to incorporate the structural and systematic effects of small molecule perturbations, which are closely related to their efficacy and toxicity profiles. We believe that our methods have the potential to augment in silico drug discovery, either by exploring on a massive scale the biological effect of compounds/libraries without available GEx data, or by suggesting new chemical structures with desired biological effect.
Data and code availability
All analyzed data that were used to train our models and produce all tables and figures are available at https://github.com/BioSysLab/deepSIBA. Furthermore, the R source code to analyze the CMap dataset and create the training, validation and test sets is available at https://github.com/BioSysLab/deepSIBA/preprocessing. Finally, the Keras/TensorFlow implementation of our deep learning models, alongside trained ensemble models for each cell line are available at https://github.com/BioSysLab/deepSIBA/learning.Author contributions
C. F. and N. M. conceived the idea and trained the models. C. F., N. M. and A. S. preprocessed the data. L. G. A. supervised the project. All authors analyzed the results and wrote the manuscript together.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank T. Sakellaropoulos and M. Neidlin for their constructive feedback on the manuscript and N. Emmanouilidi for her help in designing the figures. Finally, we would like to acknowledge the reviewers for their valuable feedback and criticism. This research has been co-financed by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (project code: T1EDK-02829).References
- G. Sliwoski, S. Kothiwale, J. Meiler and E. W. Lowe, Pharmacol. Rev., 2014, 66, 334–395 CrossRef.
- J. Hert, P. Willett, D. J. Wilton, P. Acklin, K. Azzaoui, E. Jacoby and A. Schuffenhauer, J. Chem. Inf. Model., 2006, 46, 462–470 CrossRef CAS.
- F. Sirci, F. Napolitano, S. Pisonero-Vaquero, D. Carrella, D. L. Medina and D. di Bernardo, NPJ Syst. Biol. Appl., 2017, 3, 1–12 CrossRef.
- J. P. Bai and D. R. Abernethy, Annu. Rev. Pharmacol. Toxicol., 2013, 53, 451–473 CrossRef CAS.
- C. Fotis, A. Antoranz, D. Hatziavramidis, T. Sakellaropoulos and L. G. Alexopoulos, Drug Discovery Today, 2018, 23, 626–635 CrossRef CAS.
- B. Verbist, G. Klambauer, L. Vervoort, W. Talloen, Z. Shkedy, O. Thas, A. Bender, H. W. Göhlmann, S. Hochreiter and QSTAR Consortium, Drug Discovery Today, 2015, 20, 505–513 CrossRef CAS.
- F. Iorio, R. Bosotti, E. Scacheri, V. Belcastro, P. Mithbaokar, R. Ferriero, L. Murino, R. Tagliaferri, N. Brunetti-Pierri and A. Isacchi, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14621–14626 CrossRef CAS.
- J. Lamb, E. D. Crawford, D. Peck, J. W. Modell, I. C. Blat, M. J. Wrobel, J. Lerner, J.-P. Brunet, A. Subramanian and K. N. Ross, Science, 2006, 313, 1929–1935 CrossRef CAS.
- A. Subramanian, R. Narayan, S. M. Corsello, D. D. Peck, T. E. Natoli, X. Lu, J. Gould, J. F. Davis, A. A. Tubelli and J. K. Asiedu, Cell, 2017, 171, 1437–1452. e17 CrossRef CAS.
- S. Pushpakom, F. Iorio, P. A. Eyers, K. J. Escott, S. Hopper, A. Wells, A. Doig, T. Guilliams, J. Latimer and C. McNamee, Nat. Rev. Drug Discovery, 2019, 18, 41–58 CrossRef CAS.
- H. Chen, O. Engkvist, Y. Wang, M. Olivecrona and T. Blaschke, Drug Discovery Today, 2018, 23, 1241–1250 CrossRef.
- H. Öztürk, A. Özgür and E. Ozkirimli, Bioinformatics, 2018, 34, i821–i829 CrossRef.
- W. Jin, R. Barzilay and T. Jaakkola, arXiv preprint, 2018, arXiv:1802.04364.
- DeepChem: Deep-learning models for Drug Discovery and Quantum Chemistry.
- Z. Wu, B. Ramsundar, E. N. Feinberg, J. Gomes, C. Geniesse, A. S. Pappu, K. Leswing and V. Pande, Chem. Sci., 2018, 9, 513–530 RSC.
- Y. LeCun, Y. Bengio and G. Hinton, Nature, 2015, 521, 436–444 CrossRef CAS.
- D. K. Duvenaud, D. Maclaurin, J. Iparraguirre, R. Bombarell, T. Hirzel, A. Aspuru-Guzik and R. P. Adams, Advances in Neural Information Processing Systems 28 (NIPS 2015), 2015 Search PubMed.
- W. Torng and R. B. Altman, J. Chem. Inf. Model., 2019, 59, 4131–4149 CrossRef CAS.
- S. Kearnes, K. McCloskey, M. Berndl, V. Pande and P. Riley, J. Comput.-Aided Mol. Des., 2016, 30, 595–608 CrossRef CAS.
- I. Wallach and A. Heifets, J. Chem. Inf. Model., 2018, 58, 916–932 CrossRef CAS.
- F. Schroff, D. Kalenichenko and J. Philbin, In Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, 815–823 Search PubMed.
- L. Bertinetto, J. Valmadre, J. F. Henriques, A. Vedaldi and P. H. Torr, Computer Vision – ECCV 2016 Workshops, Springer, 2016, pp. 850–865 Search PubMed.
- O. Vinyals, C. Blundell, T. Lillicrap and D. Wierstra, Advances in Neural Information Processing Systems 29 (NIPS 2016), 2016, 3630–3638 Search PubMed.
- Y. Bai, H. Ding, Y. Sun and W. Wang, arXiv preprint, 2018, arXiv:1810.10866.
- H. Altae-Tran, B. Ramsundar, A. S. Pappu and V. Pande, ACS Cent. Sci., 2017, 3, 283–293 CrossRef CAS.
- Y. Gal and Z. Ghahramani, Proceedings of The 33rd International Conference on Machine Learning, 2016, 1050–1059 Search PubMed.
- B. Lakshminarayanan, A. Pritzel and C. Blundell, Advances in Neural Information Processing Systems 30 (NIPS 2017), 2017, 6402–6413 Search PubMed.
- S. Jain, G. Liu, J. Mueller and D. Gifford, arXiv preprint, 2019, arXiv:1906.07380.
- A. Kendall and Y. Gal, Advances in Neural Information Processing Systems 30 (NIPS 2017), 2017, 5574–5584 Search PubMed.
- S. Ryu, Y. Kwon and W. Y. Kim, Chem. Sci., 2019, 10, 8438–8446 RSC.
- M. Jeon, D. Park, J. Lee, H. Jeon, M. Ko, S. Kim, Y. Choi, A.-C. Tan and J. Kang, Bioinformatics, 2019, 35, 5249–5256 CrossRef CAS.
- T. Pahikkala and A. Airola, J. Mach. Learn. Res., 2016, 17, 7803–7807 Search PubMed.
- K. Simonyan, A. Vedaldi and A. Zisserman, 2014, arXiv:1312.6034 [cs].
- GEO GSE92742, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92742, (accessed 2020-03-20).
- CLUE platform, https://clue.io/, (accessed 2020-03-20).
- A. Alexa and J. Rahnenführer, Bioconductor Improv.
- A. Sergushichev, BioRxiv, 2016, 060012 Search PubMed.
- F. Li, Y. Cao, L. Han, X. Cui, D. Xie, S. Wang and X. Bo, OMICS: J. Integr. Biol., 2013, 17, 116–118 CrossRef CAS.
- M. Kanehisa, The KEGG Database, Wiley Online Library, 2002, pp. 91–100 Search PubMed.
- RDKit: Open-source cheminformatics.
- M. Iwata, R. Sawada, H. Iwata, M. Kotera and Y. Yamanishi, Sci. Rep., 2017, 7, 40164 CrossRef CAS.
- D. Rogers and M. Hahn, J. Chem. Inf. Model., 2010, 50, 742–754 CrossRef CAS.
- J. Sun, Q. Wei, Y. Zhou, J. Wang, Q. Liu and H. Xu, BMC Syst. Biol., 2017, 11, 87 CrossRef.
- O. Shuvalov, A. Petukhov, A. Daks, O. Fedorova, E. Vasileva and N. A. Barlev, Oncotarget, 2017, 8, 23955 CrossRef.
- D. Wu, B. Han, L. Guo and Z. Fan, J. Obstet. Gynaecol., 2016, 36, 615–621 CrossRef CAS.
- P. Pope, S. Kolouri, M. Rostrami, C. Martin and H. Hoffmann, 2019, arXiv:1812.00265 [cs, stat].
- K. Preuer, G. Klambauer, F. Rippmann, S. Hochreiter and T. Unterthiner, in Explainable AI: Interpreting, Explaining and Visualizing Deep Learning, ed. W. Samek, G. Montavon, A. Vedaldi, L. K. Hansen and K.-R. Müller, Springer International Publishing, Cham, 2019, pp. 331–345 Search PubMed.
- J. Jiménez-Luna, F. Grisoni and G. Schneider, 2020, arXiv:2007.00523 [cs, stat].
- A. Cichonska, B. Ravikumar, R. J. Allaway, S. Park, F. Wan, O. Isayev, S. Li, M. Mason, A. Lamb and Z. Tanoli.
- L. Garcia-Alonso, F. Iorio, A. Matchan, N. Fonseca, P. Jaaks, G. Peat, M. Pignatelli, F. Falcone, C. H. Benes and I. Dunham, Cancer Res., 2018, 78, 769–780 CrossRef CAS.
- A. Liu, P. Trairatphisan, E. Gjerga, A. Didangelos, J. Barratt and J. Saez-Rodriguez, NPJ Syst. Biol. Appl., 2019, 5, 1–10 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0mo00129e |
| ‡ Equal contributions. |
| This journal is © The Royal Society of Chemistry 2021 |