Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer
Zhonghua
Ma
ab,
Shengying
Gu
c,
Min
Song
d,
Changsheng
Yan
e,
Bingqing
Hui
ab,
Hao
Ji
ab,
Jirong
Wang
b,
Jianping
Zhang
f,
Keming
Wang
*ab and
Qinghong
Zhao
*f
aThe Second Clinical Medical College of Nanjing Medical University, Nanjing, 210000, Jiangsu, People's Republic of China
bDepartment of Oncology, Second Affiliated Hospital, Nanjing Medical University, Nanjing, 210000, Jiangsu, People's Republic of China. E-mail: kemingwang@126.com; Fax: +86-25-58509994; Tel: +86-18951762692
cDepartment of Nursing, Second Affiliated Hospital, Nanjing Medical University, Nanjing, 210000, Jiangsu, People's Republic of China
dDepartment of Hematology, Second Affiliated Hospital, Nanjing Medical University, Nanjing, 210000, Jiangsu, People's Republic of China
eDepartment of Obstetrics and Gynecology, First Affiliated Hospital, Nanjing Medical University, Nanjing, 210000, Jiangsu, People's Republic of China
fDepartment of General Surgery, Second Affiliated Hospital, Nanjing Medical University, Nanjing, 210000, Jiangsu, People's Republic of China. E-mail: qhznanjing@sohu.com; Fax: +86-25-58509994; Tel: +86-18951762868
First published on 24th February 2020
Abstract
Correction for ‘Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer’ by Zhonghua Ma et al., Mol. BioSyst., 2017, 13, 2350–2361.
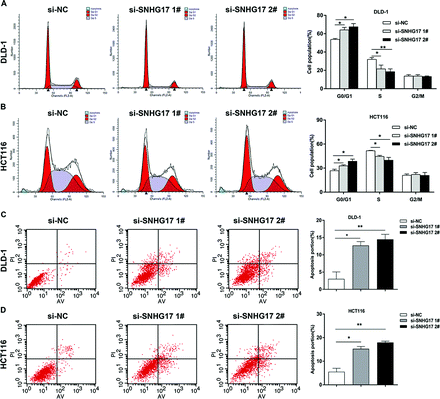
The authors regret that incorrect flow cytometry images were inadvertently used for Fig. 3 in the original article. The corrected Fig. 3 is included herein.
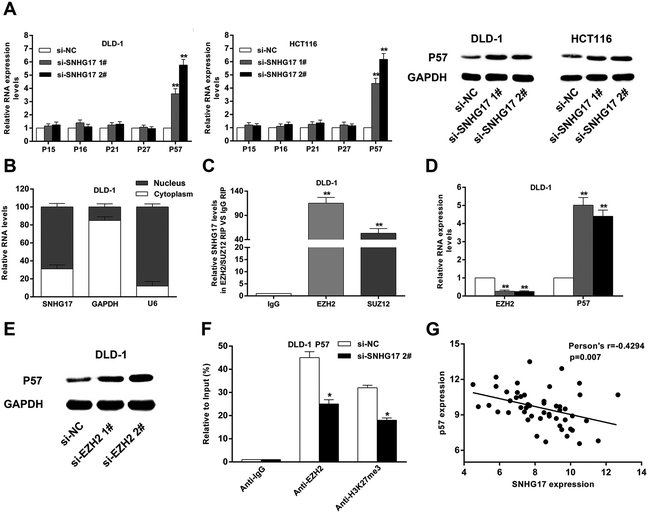
The authors regret that the siRNA labels in the two graphs in Fig. 5A of the original article are incorrect. The labels si-SNHG17 2# and si-SNHG17 3# in the original article should read si-SNHG17 1# and si-SNHG17 2#, respectively. The corrected Fig. 5 is included herein.
The authors would also like to add a clearer citation to the CRC gene expression data from the Gene Expression Omnibus (GEO) dataset GSE21510 used in the study (ref. 1). These data were cited in the original article using the link https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21510.
Finally, the authors would like to draw attention to the fact that the analysis presented in Fig. 1A in the original manuscript is the same as that presented in Fig. 1a of ref. 2. In each case the data were used to select the candidate gene to study.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
References
- S. Tsukamoto, T. Ishikawa, S. Iida, M. Ishiguro, K. Mogushi, H. Mizushima, H. Uetake, H. Tanaka and K. Sugihara, Clin. Cancer Res., 2011, 17, 2444–2450 CrossRef CAS PubMed.
- J. Ding, J. Li, H. Wang, Y. Tian, M. Xie, X. He, H. Ji, Z. Ma, B. Hui, K. Wang and G. Ji, Cell Death Dis., 2017, 8, e2997 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |