4D hydrogel for dynamic cell culture with orthogonal, wavelength-dependent mechanical and biochemical cues†
Yijun
Zheng
 *a,
Mitchell Kim
Liong Han
*a,
Mitchell Kim
Liong Han
 a,
Qiyang
Jiang
ab,
Bin
Li
a,
Qiyang
Jiang
ab,
Bin
Li
 a,
Jun
Feng
ab and
Aránzazu
del Campo
a,
Jun
Feng
ab and
Aránzazu
del Campo
 *ab
*ab
aINM-Leibniz Institute for New Materials, Campus D2 2, 66123 Saarbrücken, Germany. E-mail: Yijun.Zheng@leibniz-inm.de; aranzazu.delcampo@leibniz-inm.de
bChemistry Department, Saarland University, 66123 Saarbrücken, Germany
First published on 22nd July 2019
Abstract
Cooperative action of biochemical and biomechanical signals regulates the interactions between cells and the supporting matrix in natural tissues. Herein, we describe a hydrogel for 4D cell culture which allows user-defined stiffening of the cellular environment and presentation of bioadhesive cues in an orthogonal manner using light of different wavelengths. Stiffening of the gel is initiated by VIS light, while activation of the biochemical function is triggered by UV light. We demonstrate the versatility of this system by triggering, directing and/or hindering cell migration from spheroids based on photoactivated stiffening or integrin-binding to the hydrogels. This material allows in situ and independent manipulation of the physicochemical cues in the cellular microenvironment in vitro, and could eventually be extended to in vivo.
New conceptsLight is a biocompatible method to remotely modulate the physiochemical properties of synthetic cell culture systems. Since different chromophores show different responsiveness to light with different wavelengths, light can be used to regulate more than one parameter of physiological environments, by varying the irradiation wavelength. This concept has been applied to tune both biochemical and biophysical cues, but the examples currently available depend on the extra addition of a ligand. Herein, we propose a pre-embedded strategy to achieve bio-orthogonal control. The strategy is based on the coupling of a UV-activatable caged ligand and visible-light-triggerable polymerization of the methacrylate side chain. We demonstrate this strategy by a dual-responsive 4D cell culture hydrogel platform that allows independent physiochemical tunability and dose-dependent cellular response studies. |
4D hydrogels allow external modulation of their physicochemical properties over time and have gained attention as dynamic biomaterials to instruct and guide the responses of the embedded cells.1–5 In order to modulate the hydrogel properties, in situ activatable mechanisms compatible with the embedded living cells are to be built into the hydrogel architecture.6 Strategies currently available include enzyme-induced degradation,7 Ca2+-triggered stiffening,8 or photoregulation.1,9–11 Among them, the light-based approaches show unique advantages for regulating the hydrogel properties in terms of orthogonality, spatiotemporal control, and dosing.12,13
From a synthetic point of view, reported approaches to light-responsive 4D hydrogels rely on the photoaddition or photocleavage reactions. The former reactions include the thiol–ene addition14,15 for light-triggered incorporation of soluble ligands to the network,15,16 and the photo-triggered polymerization of the acrylate side groups for stiffening hydrogels.17,18 Photocleavage reactions have also been used to either remove the ligands from a hydrogel containing photocleavable spacers,19,20 or to soften a hydrogel network by photolysis of the photocleavable comonomers.21,22 Our group has also exploited the photolysis reactions to regulate the presentation of bioactive ligands previously incorporated into the network as latent, photoactivatable functionalities.23–27 During cell culture, or after implantation in vivo, the biochemical activity of the ligand can be restored by simply exposing to light.28 This approach has been applied to regulate the concentration of different adhesive ligands (i.e. RGDfK,29 IKVAV30 or α5β1-specific integrin ligand31) in biomaterials in vitro32 and in vivo33 leading to changes in the adhesion, migration,34,35 and differentiation36 of the embedded cells. Compared to photoaddition approaches, in situ photoactivation of the latent biofunctional groups allows higher precision in the biochemical regulation in situ, as it does not require exposing the embedded cells to the ligand in solution and subsequent washing steps. This can trigger competing interactions with the cellular receptors and would not be viable in in vivo scenarios.
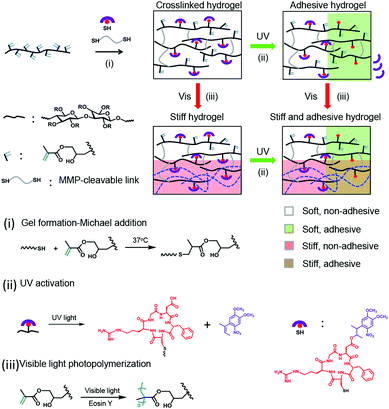
Cell-specific responses to simultaneous or sequential biochemical or mechanical matrix cues are difficult to study in natural systems and also in synthetic ones. Hydrogel designs with dual or multiple responses are needed to recreate mechanical and biochemical changes in the same hydrogel system. Two remarkable attempts have been reported for that. In a pioneering work, simultaneous photodegradation and photoaddition reactions were utilized to soften the PEG hydrogels and incorporate adhesive ligands to the network and applied to guide the migration of the embedded cells.37 More recently, photostiffening of a methacrylated hyaluronic acid (MeHA) matrix was demonstrated on a 2D surface38 on which cells were attached after the ex situ patterned photoaddition of the adhesive ligands (because of the cytotoxicity of the reaction13). Although these two systems highlight the great potential that the wavelength-selective reactions could have for developing effective 4D biomaterials, none of them realize a dual property change in the 3D cell cultures orthogonally and in the presence of cells (in situ). In this work, we report a light-controlled 4D hydrogel that allows for orthogonal control of biochemical and biomechanical cues in a 3D cellular culture. The hydrogel combines the VIS-light polymerizable methacrylate groups and UV-photoactivatable cell-adhesive peptides on a dextran backbone (Scheme 1). The methacrylate-modified dextran has been extensively used as a cell culture matrix with adjustable stiffness studies of cellular mechanosensing.39 Here, the methacrylate groups are also used to increase the stiffness of the cellular microenvironment after gelation, during cell culture, via photo-regulated crosslinking.17 The cyclo[RGD(DMNPB)fC] has been used by our group as a phototriggerable cell-adhesive peptide.19,29,33,34 Here, we demonstrate how the combination of these two systems renders cellular microenvironments with regulable mechanical and bioadhesive properties in a 4D fashion, and over a broad range of stiffness and ligand concentrations. This is proven by independent and dynamic regulation of the migratory behavior of fibroblasts from the spheroids embedded in the new material.
The 4D hydrogels were obtained by mixing dextran-MA with cyclo[RGD(DMNPB)fC] and the matrix metalloproteinase (MMP) degradable crosslinker (Pro-Leu-Gly-Leu-Trp-Ala). The thiol–ene reaction allows gelation of the dextran by reaction with the diCys-terminated crosslinker and simultaneous bioconjugation of the Cys-terminated photoactivatable adhesive peptide in one step. The dextran-MA used in this study was highly grafted with MA groups39 (methacrylate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dextran repeat unit ratio: 0.77
dextran repeat unit ratio: 0.77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; Fig. S1, ESI†). The gelation can be conducted at 37 °C in the presence of cells and results in the formation of stable hydrogels within 15 min. By tuning the concentration of crosslinker in the mixture, hydrogels with Young's modulus between 61 ± 15 Pa and 3600 ± 1900 Pa can be prepared (Fig. S2, ESI†). These gels still contained sufficient unreacted methacrylate groups in the dextran chain for bioconjugation of the photoactivatable adhesive ligand and later stiffening of the network via photo-initiated crosslinking.17
1; Fig. S1, ESI†). The gelation can be conducted at 37 °C in the presence of cells and results in the formation of stable hydrogels within 15 min. By tuning the concentration of crosslinker in the mixture, hydrogels with Young's modulus between 61 ± 15 Pa and 3600 ± 1900 Pa can be prepared (Fig. S2, ESI†). These gels still contained sufficient unreacted methacrylate groups in the dextran chain for bioconjugation of the photoactivatable adhesive ligand and later stiffening of the network via photo-initiated crosslinking.17
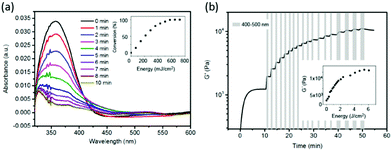
The introduction of cyclo[RGD(DMNPB)fC] units in the dextran-MA gel was corroborated by UV-spectroscopy. The thin hydrogel films supported on a quartz substrate showed an absorption maximum at 358 nm, which corresponds to λmax of the DMNPB group. The removal of the DMNPB group and consequent activation of the RGD peptide was followed by changes in the UV spectrum upon exposure to UV light and washing.40 The decay in absorbance at λmax = 358 nm indicated the loss of the DMPNB (Fig. 1a). The decay was linearly dependent on the irradiation dose over a wide range, indicating that the ligand presentation can be tailored by the exposure conditions (Fig. 1a inset).
We then tested the possibility to stiffen the gel by photopolymerization of the methacrylate side groups. In order to get orthogonal control of the biochemical and mechanical properties, the protecting group DMNPB should be stable under the irradiation conditions used for the photopolymerization. For this purpose, the irradiation wavelength of λ > 415 nm was selected, which did not lead to photolysis of DMNPB (Fig. S3, ESI†). The cytocompatible photoinitiator eosin Y41 (λmax = 520 nm) needs to be added to the polymer mixture. To study the changes in the mechanical properties of the gel during light exposure, the rheological properties of the hydrogels during irradiation were monitored in situ. A hydrogel with initial Young's modulus (E) of 3.6 kPa and containing eosin Y (10 μM) was prepared and irradiated with visible light (400–500 nm, 11 mW cm−2). The Young's modulus of the hydrogel increased up to 30.4 kPa after irradiation for 2 min (Fig. S4, ESI†). We also demonstrated that irradiation of the hydrogel at 360 nm, which is used for photocleavage of DMNPB, in the absence of photoinitiator did not lead to any stiffening (Fig. S5, ESI†). These results demonstrate effective tuning of stiffness upon light exposure. Moreover, the Young's modulus of the hydrogel was precisely regulated by simply tuning the light dose (Fig. 1b inset). Stepwise stiffening between 1 and 10 kPa was demonstrated (Fig. 1b). In conclusion, the dextran-MA hydrogels conjugated with RGD(DMNPB)fC ligand allow orthogonal regulation of RGD ligand density and stiffness using light of different wavelengths.
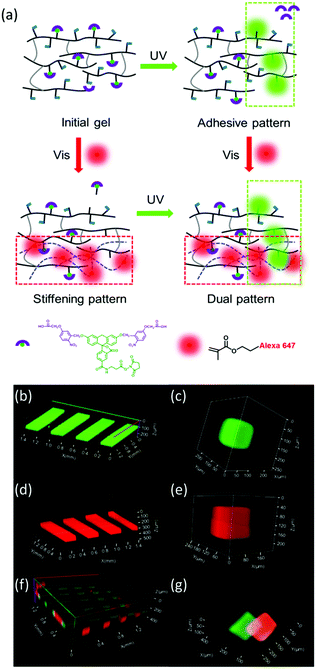
Next, we tested the possibility to spatially control the property changes in hydrogels by light exposure with a scanning laser at different wavelengths. In order to visualize the two light-triggered reactions, we added two fluorescent dyes to the hydrogel: (i) thiol-terminated photoactivatable fluorescein and (ii) methacrylated Alexa 647. The photoactivatable fluorescein bears a 5-carboxymethoxy-2-nitrobenzyl photolabile protecting group and becomes fluorescent after light exposure at exposure conditions similar to the DMNPB group.42 This allows visualization of the photolysis reaction at the regions exposed to 360 nm light within the hydrogel (Fig. 2a). The photoactivatable fluorescein was added to the hydrogel during polymerization, and is conjugated to the hydrogel through the thiol functionality. The methacrylated Alexa 647 was used to visualize the photoinitiated reaction of the methacrylic groups with visible light. The methacrylic group of the fluorophore reacted with the dextran-MA in the presence of eosin Y photoinitiator, and the fluorescence (after washing) revealed the phototriggered reaction volumes. The hydrogels were irradiated with 360 nm or 470 nm light through a chrome stripe pattern (200 μm lines spaced by 200 μm apart). The obtained fluorescence pattern revealed the exposed volumes, reflecting that photoactivation is site-selective (Fig. 2b and d). The activation patterns were also achieved by scanning with a laser at 375 or 488 nm (Fig. 2c and e). Fig. 2f and g show that the photoactivation occurs orthogonally when both fluorophores are contained in the hydrogel. A bicolor pattern with well-defined sharp regions was obtained by scanning first at 375 nm and then at 488 nm. Spatially separated or superposed activation regions are possible (Fig. 2f and g).
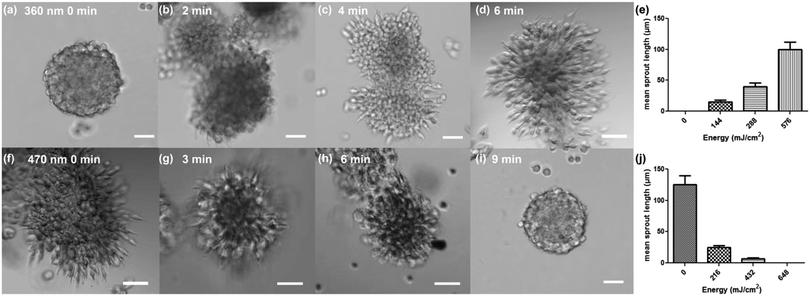
We tested the regulation of the adhesive cues of the cellular microenvironment by studying L929 fibroblast migration from the spheroids embedded in the dextran-MA hydrogel functionalized with cyclo[RGD(DMNPB)fC] with a stiffness of 3.6 kPa. The spheroid cultures were maintained for 3 days and then imaged. Fibroblasts encapsulated in the original hydrogel remained confined within the spheroid and did not migrate into the non-adhesive surrounding (Fig. 3a). In contrast, RGD activation upon UV irradiation (360 nm, 1.2 mW cm−2) allowed fibroblasts to migrate out of the primary spheroid (Fig. 3b and d). The attenuation of the UV light (360 nm) across the hydrogel thickness (800 μm) due to the absorbance of cyclo[RGD(DMNPB)fC] was measured. A drop from 1.2 to 1.15 mW cm−2 was detected, indicating that the degree of photoactivation across the z-direction should be homogeneous. Higher irradiation doses resulted in larger sprouting from the spheroid (Fig. 3e). These results confirmed that the presentation and concentration of the RGD ligand in the hydrogel was regulated by light and supported adhesion and migration of the cells through the hydrogel in a dose-dependent manner. The regulation of stiffness of the cellular microenvironment was then demonstrated. Soft dextran-MA hydrogels functionalized with RGDfC and with an initial stiffness of 3.6 kPa showed radial outgrowth of fibroblasts (Fig. 3f). Exposure at 470 nm (1.2 mW cm−2) restricted the outgrowth of cells as a consequence of the increased crosslinking degree of the hydrogel (Fig. 3g–i). The outgrowth level was dose-dependent (Fig. 3j), and full restriction of migration was observed after 9 min of irradiation, which corresponded to a Young's modulus of the hydrogel of 30 kPa. Live/dead assays on Day 3 and Day 7 demonstrated high cell viability after secondary polymerization using eosin Y as a visible photoinitiator (Fig. S6, ESI†). Moreover, taking advantage of the spatial resolution of the light beam, we were able to locally regulate the presentation of ligand or the environment stiffness to certain parts of the hydrogel and confine migration to selected spaces (Fig. S7, ESI†).
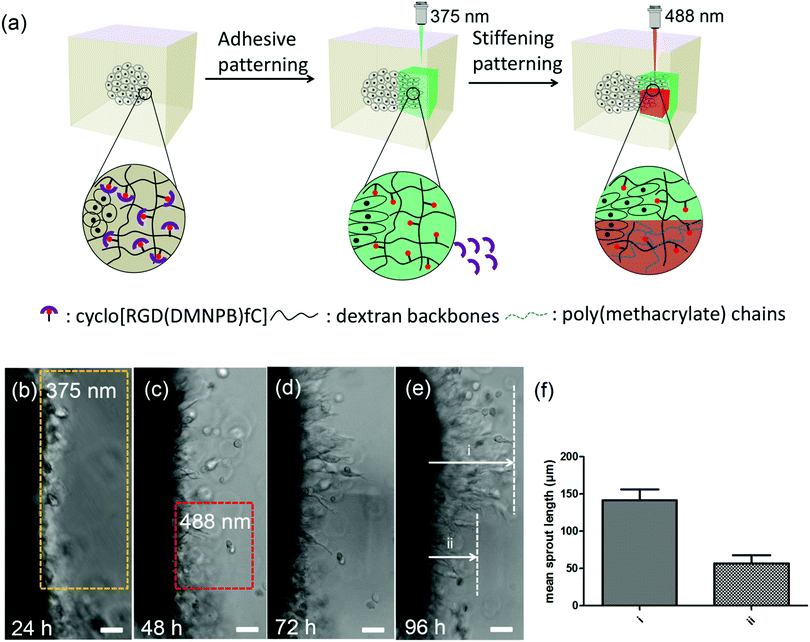
The 4D properties of the developed dextran-MA hydrogels were further demonstrated by sequential modulation of the RGD density and stiffness of the hydrogel at different days during cell culture (Fig. 4a). The L929 fibroblast spheroid was encapsulated within the hydrogel. In the absence of light, the cells remained confined in the spheroids (Fig. 4b). After 24 hours of culture, a selected volume close to the spheroid was scanned at 375 nm to activate RGD ligand (yellow marked area in Fig. 4b). In the next 24 hours, cells migrated out of the spheroid into the illuminated region (Fig. 4c). At 48 hours culture, local stiffening was induced by exposure with a 488 nm laser (red highlighted area in Fig. 4c). Imaging at 72 and 96 hours showed that cell migration was restricted within the illuminated (stiffened) region (i in Fig. 4d and e), while cells outside of the stiffened areas continued outgrowth (ii in Fig. 4d and e). The mean sprout outgrowth length within the two areas was significantly different (Fig. 4f). These results indicate that the developed hydrogel allows for on-demand control of biochemical and biophysical cues in 3D cellular cultures within ranges to which cells are responsive.
MA-grafted dextran is a widely used matrix polymer for 3D cell cultures. It allows light-regulated adjustment of the initial mechanical parameters within physiologically relevant mechanical ranges in bulk (0.45–45 kPa) and fibrillar (2.8–55 kPa) gels.39 We demonstrate that this system allows a post-polymerization dynamic stiffening of the matrix up to a ten-fold increase (3.6–30 kPa) in the Young's modulus. The required light dose for stiffening is cytocompatible within the range of stiffness changes that can trigger cell responses. This approach is not dextran-specific and can be potentially transferred to the biocompatible polymers used for 3D culture and cell therapies, e.g. gelatin,43 hyaluronic acid,17 or chitosan.44
Dual-responsive hydrogels with independent, light-modulated softening and adhesive ligand presentation have been realized by the Anseth group using derivatized PEG backbones.37 The combined effects of RGD presentation and photodegradation of a 5 kPa PEG gel were used to direct the migration of 3T3 fibroblasts into the light-mediated eroded and RGD-decorated channels within the gel. The system presented here allows the control of migration by either ligand presentation or matrix stiffening, allowing the study of cellular response to dynamic changes of individual factors. Note that the gel contains a degradable peptide which is unaffected by the stiffening reaction and whose degradation rate can be tuned by the peptide sequence.
In our experiments, L929 fibroblast sprouting speed was proportional to the adjusted local RGD concentration and inversely proportional to the light-tuned local density of non-degradable crosslinks. The similar dual-light-responsive concept was also applied by Anseth and Burdick et al.45 to enhance the diversity of change in the mechanical properties of hydrogels. In their system containing a photodegradable nitrobenzyl crosslinker and photopolymerizable methacrylate side group, on-demand softening or stiffening can be achieved via varying the irradiation wavelength. However, the system only focuses on one parameter, i.e. mechanical properties.
Methacrylated hyaluronic acid has been recently reported to enable light-based tunable stiffening (between 0.5 and 1.5 kPa) and tunable fibronectin density via thiol–maleimide reaction.38 However, this system has only been used in cell studies in 2D systems. The lack of bioorthogonality and eventual cytotoxic reactions of the thiol–maleimide chemistry13 limit the extension of this system to 3D cultures. In our study, the latent character of the cyclo[RGD(DMNPB)fC] ligand allows its direct integration into the polymer matrix before the encapsulation step and avoids post-gelation reactions to realize compositional changes.37
Conclusions
We present 4D hydrogels that allow orthogonal, wavelength-regulated stiffening and control of cell adhesiveness. With this system, real-time (in situ) and dosed manipulation of cell function within a synthetic microenvironment can be realized. The photochemical reactions and the subsequent property changes enable programmable 4D cell niches. The photochemical reactions used are not material-specific. We envision that this approach could be translated to other polymer backbone compositions or application scenarios (e.g. in vivo).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge Dr Emmanuel Terriac for helping with confocal microscopy imaging. A. del Campo acknowledges funding from the SFB1027 Collective Research Center and from the European Union's Horizon 2020 Research and Innovation Programme under the FET PROACTIVE grant agreement No. 731957 (Mechano-Control).Notes and references
- A. M. Hilderbrand, E. M. Ovadia, M. S. Rehmann, P. M. Kharkar, C. Guo and A. M. Kloxin, Curr. Opin. Colloid Interface Sci., 2016, 20, 212 CAS
.
- J. A. Burdick and W. L. Murphy, Nat. Commun., 2012, 3, 1269 CrossRef PubMed
.
- A. M. Rosales and K. S. Anseth, Nat. Rev. Mater., 2016, 1, 15012 CrossRef CAS PubMed
.
- C. Cimmino, L. Rossano, P. A. Netti and M. Ventre, Front. Bioeng. Biotechnol., 2018, 6, 2296 Search PubMed
.
- K. Vats and D. S. W. Benoit, Tissue Eng., Part B, 2013, 19, 455 CrossRef CAS PubMed
.
- P. M. Kharkar, K. L. Kiick and A. M. Kloxin, Chem. Soc. Rev., 2013, 42, 7335 RSC
.
- R. Y. Tam, L. J. Smith and M. S. Shoichet, Acc. Chem. Res., 2017, 50, 703 CrossRef CAS PubMed
.
- B. M. Gillette, J. A. Jensen, M. Wang, J. Tchao and S. K. Sia, Adv. Mater., 2010, 22, 686 CrossRef CAS PubMed
.
- Y. Zheng, A. Farrukh and A. del Campo, Langmuir, 2018, 34, 14459 CrossRef CAS
.
- R. S. Stowers, S. C. Allen and L. J. Suggs, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1953 CrossRef CAS
.
- Q. Shao and B. Xing, Chem. Soc. Rev., 2010, 39, 2835 RSC
.
- D. L. Alge and K. S. Anseth, Nat. Mater., 2013, 12, 950 CrossRef CAS
.
- E. R. Ruskowitz and C. A. DeForest, Nat. Rev. Mater., 2018, 3, 17087 CrossRef CAS
.
- L. A. Sawicki and A. M. Kloxin, Biomater. Sci., 2014, 2, 1612 RSC
.
- J. C. Grim, T. E. Brown, B. A. Aguado, D. A. Chapnick, A. L. Viert, X. Liu and K. S. Anseth, ACS Cent. Sci., 2018, 4, 909 CrossRef CAS PubMed
.
- S. Khetan, J. S. Katz and J. A. Burdick, Soft Matter, 2009, 5, 1601 RSC
.
- M. Guvendiren and J. A. Burdick, Nat. Commun., 2012, 3, 792 CrossRef
.
- S. Khetan, M. Guvendiren, W. R. Legant, D. M. Cohen, C. S. Chen and J. A. Burdick, Nat. Mater., 2013, 12, 458 CrossRef CAS
.
- M. Wirkner, J. M. Alonso, V. Maus, M. Salierno, T. T. Lee, A. J. García and A. del Campo, Adv. Mater., 2011, 23, 3907 CrossRef CAS
.
- M. A. Azagarsamy and K. S. Anseth, Angew. Chem., Int. Ed., 2013, 52, 13803 CrossRef CAS
.
- A. M. Kloxin, A. M. Kasko, C. N. Salinas and K. S. Anseth, Science, 2009, 324, 59 CrossRef CAS
.
- T. E. Brown, I. A. Marozas and K. S. Anseth, Adv. Mater., 2017, 29, 1605001 CrossRef
.
- C. Brieke, F. Rohrbach, A. Gottschalk, G. Mayer and A. Heckel, Angew. Chem., Int. Ed., 2012, 51, 8446 CrossRef CAS
.
- V. San Miguel, C. G. Bochet and A. del Campo, J. Am. Chem. Soc., 2011, 133, 5380 CrossRef CAS
.
- Y. Ohmuro-Matsuyama and Y. Tatsu, Angew. Chem., Int. Ed., 2008, 47, 7527 CrossRef CAS
.
- G. Mayer and A. Heckel, Angew. Chem., Int. Ed., 2006, 45, 4900 CrossRef CAS
.
-
J. E. T. Corrie, T. Furuta, R. Givens, A. L. Yousef and M. Goeldner, Dynamic Studies in Biology, Wiley-VCH Verlag GmbH & Co. KGaA, 2005, ch. 1, pp. 1–94, DOI:10.1002/3527605592
.
- H.-M. Lee, D. R. Larson and D. S. Lawrence, ACS Chem. Biol., 2009, 4, 409 CrossRef CAS
.
- S. Petersen, J. M. Alonso, A. Specht, P. Duodu, M. Goeldner and A. del Campo, Angew. Chem., Int. Ed., 2008, 47, 3192 CrossRef CAS
.
- A. Farrukh, F. Ortega, W. Fan, N. Marichal, J. I. Paez, B. Berninger, A. D. Campo and M. J. Salierno, Stem Cell Rep., 2017, 9, 1432 CrossRef CAS
.
- A. del Campo, R. V. Nair and A. Farrukh, ChemBioChem, 2018, 19, 1280 CrossRef
.
- M. Wirkner, S. Weis, V. San Miguel, M. Álvarez, R. A. Gropeanu, M. Salierno, A. Sartoris, R. E. Unger, C. J. Kirkpatrick and A. del Campo, ChemBioChem, 2011, 12, 2623 CrossRef CAS
.
- T. T. Lee, J. R. García, J. I. Paez, A. Singh, E. A. Phelps, S. Weis, Z. Shafiq, A. Shekaran, A. del Campo and A. J. García, Nat. Mater., 2015, 14, 352 CrossRef CAS
.
- M. J. Salierno, A. J. García and A. del Campo, Adv. Funct. Mater., 2013, 23, 5974 CrossRef CAS
.
- M. J. Salierno, L. García-Fernandez, N. Carabelos, K. Kiefer, A. J. García and A. D. Campo, Biomaterials, 2016, 82, 113 CrossRef CAS PubMed
.
- S. Weis, T. T. Lee, A. del Campo and A. J. Garcia, Acta Biomater., 2013, 9, 8059 CrossRef CAS
.
- C. A. DeForest and K. S. Anseth, Nat. Chem., 2011, 3, 925 CrossRef CAS
.
- A. D. Rape, M. Zibinsky, N. Murthy and S. Kumar, Nat. Commun., 2015, 6, 8129 CrossRef
.
- B. M. Baker, B. Trappmann, W. Y. Wang, M. S. Sakar, I. L. Kim, V. B. Shenoy, J. A. Burdick and C. S. Chen, Nat. Mater., 2015, 14, 1262 CrossRef CAS PubMed
.
- A. Farrukh, W. Fan, S. Zhao, M. Salierno, J. I. Paez and A. del Campo, ChemBioChem, 2018, 19, 1271 CrossRef CAS PubMed
.
- H. Shih and C.-C. Lin, Macromol. Rapid Commun., 2013, 34, 269 CrossRef CAS PubMed
.
- A. Farrukh, J. I. Paez and A. del Campo, Adv. Funct. Mater., 2018, 1807734 Search PubMed
.
- D. Loessner, C. Meinert, E. Kaemmerer, L. C. Martine, K. Yue, P. A. Levett, T. J. Klein, F. P. W. Melchels, A. Khademhosseini and D. W. Hutmacher, Nat. Protoc., 2016, 11, 727 CrossRef CAS PubMed
.
- M. Diolosà, I. Donati, G. Turco, M. Cadenaro, R. Di Lenarda, L. Breschi and S. Paoletti, Biomacromolecules, 2014, 15, 4606 CrossRef PubMed
.
- A. M. Rosales, S. L. Vega, F. W. DelRio, J. A. Burdick and K. S. Anseth, Angew. Chem., Int. Ed., 2017, 56, 12132 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9mh00665f |
| This journal is © The Royal Society of Chemistry 2020 |