 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Developmental bioengineering: recapitulating development for repair
Eugene C.
Goldfield
 *ab and
Marc-Olivier
Coppens
*ab and
Marc-Olivier
Coppens
 cd
cd
aBoston Children's Hospital, 300 Longwood Avenue, Boston, Massachusetts 02115, USA. E-mail: Eugene.goldfield@childrens.harvard.edu
bWyss Institute for Biologically Inspired Engineering, Harvard University, USA
cDepartment of Chemical Engineering, University College London (UCL), Torrington Place, WC1E 7JE London, UK. E-mail: m.coppens@ucl.ac.uk
dEPSRC “Frontier Engineering” Centre for Nature-Inspired Engineering, UCL, UK
First published on 6th August 2020
Abstract
A nature-inspired bioengineering methodology is presented. The methodology includes (1) identifying a set of “bottom-up” normative models or fundamental mechanisms by which nature builds tissues and organs from progenitor cells during development, (2) recapitulating these bottom-up developmental processes in vitro by using stem cells to grow tissue and organ-like structures, called organoids, and (3) implementing “top-down” technologies that provide specific supportive niches for growing structures at each developmental level. Also presented, is a systematic nature-inspired solution framework that applies universal concepts in natural development to inform designs for organ repair. This organizational framework, which integrates developmental models, recapitulation of development in vitro, and engineered niches is illustrated by considering repair of two body organs: the retina and spinal cord, respectively. The former repair involves transplanting a retinal patch grown in vitro on a synthetic sheet, while the latter uses implanted stem cells to promote growth of relay circuits in the injured human spinal cord or activity-based neurotechnology.
Design, System, ApplicationCould we make the blind see and the lame walk again? Recent advances in the field of bioengineering bring such dreams closer to reality. Examples include the restoration of damaged tissues, formerly deemed irreparable, such as the retinal pigment epithelium (RPE) in age-related macular degeneration (AMD) and spinal cord injuries (SCI) leading to paralysis. This could be achieved by recapitulating developmental processes that nature herself uses to repair tissues. This perspective article first considers five prevalent, fundamental natural mechanisms in tissue development (self-organization and symmetry breaking, dynamic feedback between cells and environment, differentiation into local ensembles, assembly of the latter into larger groups, and the formation of integrated ecosystems). Then, a general nature-inspired solution framework is discussed to apply these concepts through nature-inspired designs, which are prototyped to serve a medical application. This framework is illustrated via RPE and SCI repair. In practice, top-down and bottom-up manufacturing approaches need to be combined. Furthermore, merging molecular-based and holistic perspectives requires a cross-disciplinary effort that spans (bio)materials synthesis to novel manufacturing tools, medicine, and personalized activity-based therapy. This nature-inspired solution framework for developmental bioengineering offers a systematic design strategy, which could help ultimately realize the hopes of those who seemed uncurable. |
Introduction
The development of macroscopic living systems from cellular beginnings is one of nature's marvels. A fertilized egg self-organizes into a more differentiated blastocyst, and then into a multi-layered gastrula that generates the complex and functionally interconnected organs characteristic of each animal species.1 The embryo is not alone in this developmental journey, though. Long-term time-lapse imaging of mouse embryo reveals its cell lineages as well as two extra-embryonic epithelial tissues, the precursors of the yolk sac and placenta, respectively.2 Once embedded in the uterine wall, the embryo is ready to share its journey.Compared to insects or small animals, humans may live to an age of over 100 years. At the same time, our constituent cells may have life spans of only hours, develop with errors, or become diseased or damaged. Thus, these constituent cells require intrinsic surveillance and maintenance via systems for repair and/or regeneration during homeostasis, as well as injury or disease. Repair refers to restoration of damaged tissue due to aging, disease, or injury.3 Regenerative processes continually replace blood, bone, and skin, but not entire organs.3 The focus of this article is on repair.
Nature is economical in using her resources, repurposing or recapitulating developmental processes for repair.4 Nature's recapitulation of development refers to the processes of (a) activating a cell that has remained in a quiescent state so that it now begins to undergo differentiation, or (b) switching an already functionally differentiated cell back to a growth-competent de-differentiated state, so that it can be used for repair.4 An example of the former is the pools of quiescent stem cells that may be reactivated in response to injury. For example, a cell in an active state, such as an axolotl connective tissue cell, is induced to revert to an earlier undifferentiated growth-competent state following experimental limb amputation.5 It is important to note, however, that in some cases, such as the visual system, only some of the developmental mechanisms that initially establish circuitry are available for reactivation in adulthood.6 Our goal in this article is to motivate a nature-inspired solution methodology based upon recapitulating development for repair, or developmental bioengineering.
Integrating top-down and bottom-up: a bioengineering methodology to recapitulate development for repair
Embryogenesis emerges from a continual crosstalk, or dynamic feedback, between the inseparable embryonic and post-embryonic cells.2 Historically, the field of embryology has taken a “top-down” approach to revealing dynamic feedback, such as grafting salamander tissue from the left to the right side of the body to induce (i.e., influence by close-range tissue interaction) additional supernumerary limbs.7 Here, the starting point is the whole animal, which becomes a supportive structure for the grafting of tissue. More recently, the field of developmental biology, a scion of embryology, has emphasized “bottom-up” methodologies. These begin with in vitro models in which individual building blocks (stem cells) self-organize into complex forms, and ultimately into an entire embryo.8Integration of top-down and bottom-up methods are gaining traction in synergies between biologists and engineers working on both development and repair of tissues and organs. For example, top-down technologies, such as bioprinting and microfluidics, have been used to provide micro-environments, or niches, that support self-organizing cells for bottom-up fabrication of tissue. And yet, there remains little guidance in bioengineering for selecting particular methodologies that integrate bottom-up self-organization with top-down supportive niches.
As illustrated in this perspective article, a nature-inspired approach to recapitulating development may offer a new developmental bioengineering methodology for tissue and organ repair in three ways: (1) by identifying a set of “bottom-up” normative models by which nature builds tissues and organs from progenitor cells during development, (2) by recapitulating these bottom-up developmental processes in vitro by using stem cells to grow tissue and organ-like structures, called organoids, and (3) by implementing “top-down” technologies that provide specific supportive niches for growing structures at each level of developmental complexity, from hydrogel matrices for growing personalized tissue, to embedded 3D printing of supportive scaffolds for repair at the organ level of the spinal cord. To be successful in this endeavor, bioengineers will be faced with devising top-down strategies that emulate the supportive niches nature provides at successive levels of organizational complexity of bottom-up development. Fig. 1 provides an illustration of a “top-down” supportive niche to repair the retinal pigment epithelium (RPE), discussed in a later section.
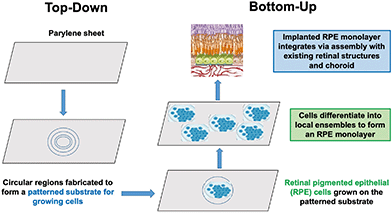 | ||
| Fig. 1 Top-down supportive niches and bottom-up self-organization are integrated in this example of retinal pigment epithelium (RPE) repair. | ||
Table 1 offers an organizational framework for bottom-up developmental models, based on a methodology for nature-inspired engineering proposed by Coppens,9 and adapted to biomedical engineering by Perera and Coppens.10 This methodology applies fundamental mechanisms underpinning desired, superior properties in the natural system to inform the design and implementation of solutions to an engineering challenge. In this design approach, the key concepts abstracted from nature typically require adaptation to acknowledge contextual differences between the source of inspiration in nature, and the engineering application, for example due to the aforementioned differences between in vivo synthesis and manufacturing. Each mechanism in Table 1—self-organization, dynamic feedback, differentiation into local ensembles, assembly of these ensembles into larger groups, and formation of integrated ecosystems—encapsulates a developmental process. Table 1 further proposes model-specific bioengineering niches that may support recapitulation of development with respect to each of the models from the bottom-up. Table 2 then provides a translational step that outlines top-down bioengineering technologies to support repair of diseased or damaged organs. This perspective article includes a case study with two illustrations of translation: (1) repairing the retina of the aging human eye following macular degeneration (Fig. 2), and (2) promoting growth of relay circuits to repair the human spinal cord following injury (Fig. 3).
| Bottom-up developmental mechanism | Nature-inspired concept | Nature-inspired design | Application: niches for in vitro recapitulation of development |
|---|---|---|---|
| Self-organization and symmetry breaking | Morphogens and mechanical forces guide stem cell differentiation and organization | Geometrically patterned substrates and signaling centers | Laboratory stem cell conversion and self-organization |
| Dynamic feedback between cells and environment | Mechanochemical transduction | Designer biomaterials with cell/niche communication | Hydrogels with transplanted stem cells as a synthetic niche for tissue, such as bone repair |
| Differentiation into local ensembles | Cell type diversity, e.g. in the retina or spinal motor neurons | Heterogeneous cell composition | Tissue for retina and spinal cord repair |
| Assembly of local ensembles into larger groups | Organogenesis: “Organizers” in organs by orchestration of interactions between tissues | Bioprinted structures using microfluidic chips, with tissue–tissue exchange | Organ chips for eye, intestine, kidney, lung, brain |
| Formation of integrated ecosystems | Holobiont: emergent behavior from dynamic, integrated signaling pathways between host and microorganisms | Transplantation of engineered bacteria in gut. EcoFABs | Microbiota-based treatments to alleviate gut infections or brain development |
| Fluidically interconnected organoids | Homo chippiens | ||
| Bottom-up developmental model | Top-down technologies for repair | |
|---|---|---|
| Mechanism | Retina | Spinal cord |
| Self-organization | In vitro self-assembly of pluripotent stem cells | Reactivate growth capacity of neural stem cells (descending propriospinal neurons) |
| Dynamic feedback between cells and environment | Use growth factor to promote axon pathfinding and target matching | Provide axon growth-supportive substrate; provide growth factors and chemo-attraction |
| Differentiation into local ensembles | Transplant sheet of differentiated retinal epithelial cells derived from stem cells into retina | Establish new relay circuits |
| Assembly of local ensembles into larger groups | Assembly occurs via new synaptic connectivity | Use activity-based therapy, pro-regenerative chemical environment and pulsed epidural stimulation to promote synaptic connectivity |
| Formation of integrated ecosystems | Integrate looking with other sensory modalities via active exploratory behavior for agent-environment ecosystem | Targeted neurotechnology: program of spatio-temporal stimulation to re-establish voluntary control of locomotion, including ecological settings |
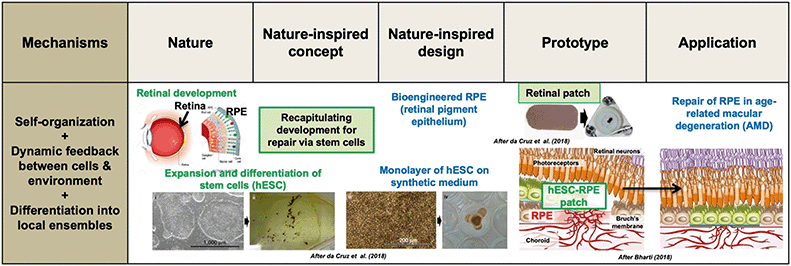 | ||
| Fig. 2 Retinal pigment epithelium (RPE) repair by retinal patch for wet, age-related macular degeneration (AMD) of the human eye. Several developmental mechanisms are utilized: self-organization, dynamic feedback between human embryonic stem cells (hESC) and the environment, and differentiation. Expanded and differentiated stem cells are introduced as a monolayer on a synthetic polymer film; this patch integrates within the RPE and closes a hole in Bruch's membrane (associated to AMD), thus preventing the choroid (vascular layer between retina and sclera) to break through to the conical photoreceptors. Thus, the nature-inspired concept of “recapitulating development for repair via stem cells” is used to inform a retinal patch design that is implanted in the eye. In early-stage human trials, patients reported a remarkable increase in reading speed. Adapted by permission from SpringerNature: Nature Biotechnology,35,85 copyright 2018. | ||
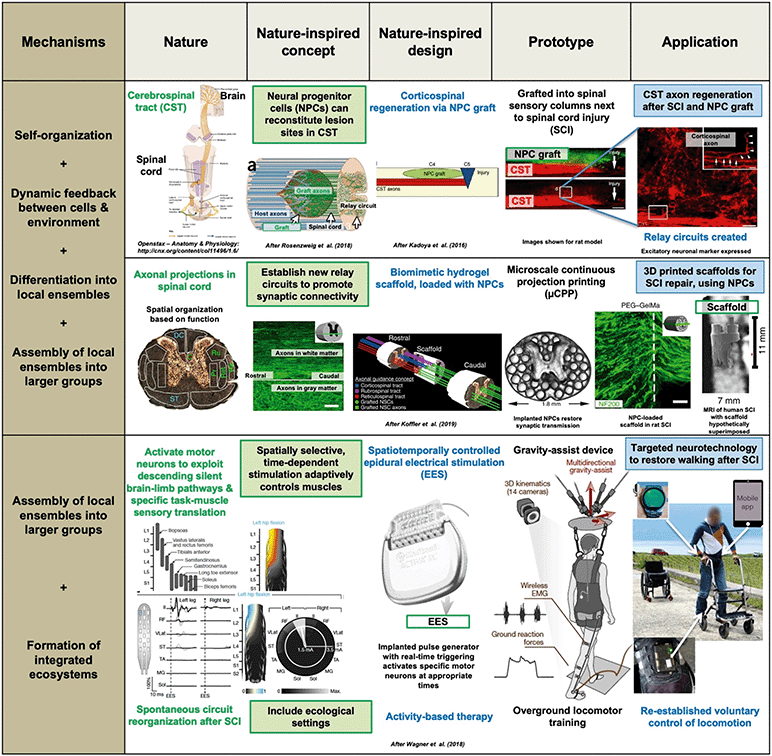 | ||
| Fig. 3 Spinal cord injury (SCI) repair by three different developmental bioengineering approaches. The top two use neural progenitor cells (NPCs): cerebrospinal tract axon regeneration is achieved via an NPC graft42,91 or a 3D printed, biomimetic hydrogel scaffold in which NPCs grow,93 using different variants of the nature-inspired concept of “establishing new relay circuits to promote synaptic connectivity”. This involves several developmental mechanisms: self-organization, dynamic feedback between NPCs and the environment, differentiation into local neuronal ensembles, and assembly of these as axons grow into the graft or through the scaffold, and synaptic transmission is restored. The bottom example94 concerns targeted neurotechnology to restore walking and cycling after SCI; it uses the nature-inspired concept of “spatially selective, time-dependent stimulation, which activates specific motor neurons and adaptively controls muscles” via an implanted pulse generator delivering real-time epidural electrical stimulation at specific locations on the spine. Crucially, this is combined with “ecological settings” via activity-based therapy, using over ground locomotor training. After some months of rehabilitation with EES, patients with paralyzed limbs were able to walk or cycle, with or even without EES, delivered via a program run through a voice-controlled watch and a mobile app. Adapted by permission from SpringerNature: Nature Medicine,42,91,93 copyright 2016, 2018 and 2019, and Nature,94 copyright 2018. | ||
To begin, let us progress through the successive developmental processes by which nature builds and repairs with special reference to mammals. Each section corresponds to a mechanism in Table 1.
How nature builds
The constructive power of self-organization has, as its partner, the process of symmetry breaking.15 Symmetry breaking occurs as an initially homogeneous signal gives rise to a signaling gradient that promotes changes in cell fate.16 An example is breaking the anterior–posterior symmetry that occurs during mammalian gastrulation, the formation of three germ layers.17 In the 1950s, the great mathematician Alan Turing turned his genius to biological processes. He gave the name “morphogens” to, then unknown, signals choreographing self-organization and symmetry breaking. We now know that morphogens are chemical informational molecules, reacting together and diffusing through a tissue, which can determine the fate of a cell by means of their concentration.18 It is noteworthy that Turing and subsequent scientists have also recognized that mechanical forces play significant roles in morphogenesis. For example, the villi in the gut form via morphogen-driven mechanical buckling,14 while intestinal smooth muscle is patterned by molecular signals, and aligned by mechanical forces.19 As we shall see, by leveraging mechano-chemical feedback loops, bioengineers have been able to induce cell and tissue morphogenesis.20
Bioengineers use both two-and three-dimensional in vitro models to emulate development. Two-dimensional in vitro systems have been used to model self-organization during embryogenesis, driving morphogens to differentiate embryonic stem cells into ectoderm, endoderm, and mesoderm layers. One illustration of how these models are used involves placing embryonic stem cells on a geometrically micro-patterned substrate. This system recapitulates in vitro how specific morphogens provide signals for guiding self-organization.16,21 For example, Deglincerti et al. have demonstrated that stem cells geometrically confined on a morphogen-laden two-dimensional disc formed distinctive concentric rings, emulating the differentiation of embryonic germ layers.22 But, as noted by Manfrin et al., differentiation into radially symmetric rings does not completely capture the symmetry breaking seen in development of natural systems.23 To make such models more physiologically plausible, Manfrin et al. have developed engineered microfluidic signaling centers, localized groups of cells that secrete morphogens, creating counteracting gradients that break radial symmetry.23 Three-dimensional in vitro models, discussed below in the section on “Assembly of Local Ensembles into Larger Groups”, capture to an even greater degree the forms built by nature.
For decades, leveraging nature's assembly process to emulate repair of human tissue required that bioengineers have available a supply of human embryonic stem cells. However, embryonic stem cells have historically been in short supply, or were unavailable due to ethical concerns. In a stunning Nobel prize-winning breakthrough in 2006, building upon earlier work by John Gurdon and colleagues,24 Yamanaka and his team discovered how to induce somatic (body) cells to return to their pluripotent state for use in in vitro stem cell technologies.25 These induced pluripotent stem cells, or iPSCs, are not derived from an embryo, but may still be used for in vitro exploration of embryonic development, tissue repair, and regeneration. With an abundant supply of iPSCs available through the use of “Yamanaka factors”, a revolution has ensued in the field of regenerative medicine. An example of the potential of iPSC for emulating nature is their direct conversion into neural progenitor cells (NPCs) that may be used to better understand brain function, development, and disease.26 Such laboratory in vitro stem cell conversion techniques require an understanding of the role of the cell environment for development in nature.
Breakthrough technologies in chemistry and materials science have advanced the laboratory formulation of “designer biomaterials” for emulating niches inspired by nature.30 These synthetic cellular niches include multiple covalently crosslinked polymer hydrogels, with specific parameters (crosslinking density, degradability, fiber architecture, and viscoelasticity). Each of these parameters emulates the dynamic mechanical properties of the ECM.30 A major advance in the engineering of ECM-based biomaterials is ECM hydrogels, containing polymer networks and water molecules.31 Experiments in the relatively new field of mechanobiology have demonstrated that it is possible to repair tissue, such as bone (e.g., the mandible), by transplanting stem cells on or within hydrogel matrices of defined stiffness and viscoelasticity.32 There are several means of fabrication with ECM hydrogels, including soft lithography, electrospinning, and extrusion-based 3D printing.29 For example, ECM scaffolds may be fabricated as patch grafts placed on damaged tissues, or may be directly injected into tissue.33 Stem cells may be cultured in matrices with specific stiffness prior to their delivery to a local site, such as skeletal muscle.32
The human retina exhibits stunning complexity, as well as specializations achieved only in primates. It is comprised of about 100 different cell types that form specific synaptic connections and reside in functionally distinct microcircuits.36 During development, axons (outputs) from retinal ganglion cells (RGCs) grow out of the eye down the optic nerve, and when they reach the optic chiasm at the base of the brain, they either cross or remain on the same side, to find their respective brain targets.37 There are also critical wiring events within the retina that determine the features in the outside world to which a given RGC and its target neuron in the brain will respond.37 RGCs lack the capacity to regenerate, and degenerative diseases that cause retinal and RGC damage result in vision loss. Repair of the retina to attempt to reverse such vision loss is discussed as a case study below.
A dramatic level of circuit complexity is evident also in the connectivity between brain, spinal cord, and muscles. Skills such as walking, or reaching and grasping with the hand, are made possible by interactions between multiple regions of the brain and spinal cord. The spinal networks that govern the precise sequencing of muscle contractions arise from the interactions among spinal motor neurons, as well as from sensory input of inhibitory interneurons and proprioceptive afferents.38,39 Spinal motor neurons differentiate into eleven distinctive populations during development.96 These cell groupings make it possible to reciprocally activate flexor and extensor muscles, and to recruit muscles in a graded fashion, crucial for engaging distinctive locomotor patterns such as walking versus swimming or climbing.40,41
There are multiple sources of input converging on the spinal cord. The cortico-spinal tract, which arises from the cerebral cortex and projects to the spinal cord, is critical for enacting a decision to perform particular functional activities, such as reaching for an object, and/or walking.42 Somatosensory feedback from proprioceptive afferents residing in the dorsal root ganglia of each spinal cord segment transmits information about the state of muscle contraction.43 Proprioceptive information reaches spinal circuits throughout the spinal cord to stabilize and refine motor output.44 Chedotal45 identifies the developmental process of molecular guidance by which sensory axons find their targets within the spinal cord, and motor neurons exit the spinal cord to reach their targets. Such knowledge is critical, because restoration of function following disease or injury may recapitulate the developmental process by which axons create new circuits by following molecular pathways. An implication of the discovery of nature's intrinsic repair process for establishing new connectivity as the basis for differentiated local assemblies is that it may be possible to (1) reactivate the chemical molecular axon guidance pathways in the injured adult spinal cord to repair lesioned circuits, or (2) activate the formation of “detour circuits” that bridge or bypass the lesion.44,46,47 Leveraging detour circuits to repair a spinal cord injury is discussed as a case study in a further section.
Body organs with diverse architectures, such as the kidney, develop higher order structures by virtue of self-organizing interactions of a variety of cells, as well as by the mechanical influences of fluid flows.53 In nature, kidney nephron progenitors form a convoluted proximal tube that resorbs nearly 100% of glucose, albumin, phosphate, amino acids, and other organic solutes.54 Lin et al. have been able to recapitulate a 3D vascularized proximal tube in vitro by means of a process called bioprinting.55 A Pluronic ink is used to 3D print the convoluted tube shape. This is then cast, the ink is evacuated, and the open lumens emulating vascular channels are seeded with proximal tube epithelial cells and vascular endothelial cells. Lin et al. have found that the bioprinted structure exhibits active reabsorption via tubular-vascular exchange of solutes, as in vivo kidney tissue does.55
In order to recapitulate the development of the glomerulus vasculature, Homan et al. cultured kidney organoids under different flow conditions within microfluidic chips.56 At a high rate of fluid flow, they found enhanced vascularization (a five-fold increase in vessel percent area compared to organoids cultured under low flow). The cells that regulate selective permeability in the glomerulus are called podocytes. Musah et al. co-cultured human iPSC-derived podocytes with a layer of human kidney glomerular endothelium as a nature-inspired recapitulation of glomerular-capillary-wall function.54 Their microfluidic device emulates the tissue–tissue interface and molecular filtration capabilities of the glomerular capillary wall.
Perhaps the greatest scientific and ethical challenge in recapitulating development is the attempt to build an entire embryo in vitro. Since publication of the 1984 Warnock report, ethicists and scientists have agreed that research on human embryos should be limited to the first 14 days of development, what is now called the 14 day rule.52,57 This is just prior to the stage of human gastrulation, and the beginnings of the nervous system.58
To address questions about how embryos develop beyond gastrulation, scientists have turned to other mammalian models, most notably mice and non-human primates. The raw materials required for developing a mouse embryo in vitro consist of three stem cell types found in the natural mammalian embryo: embryonic, trophoblast, and extra-embryonic stem cells. The latter are the precursors of the yolk sac and placenta, respectively.2 It has been possible to generate embryo-like structures in mice by culturing these three types of stem cells together, and allowing crosstalk between them.59 Thus far, in non-human primates, embryos in culture recapitulate in vivo development up to gastrulation, but only a fraction (less than 25%) develop normally beyond this stage.60 Together, then, experiments with organoid models reveal that interactions between neighboring tissues induce the formation of higher-order structures. Organs, themselves, form higher-order networks, here called integrated ecosystems.
An ecosystem model of the mammalian intestinal tract and nervous system, called the gut–brain axis, reveals the surprising ways by which bidirectional biochemical signaling between the intestinal tract and brain influence our behavior and health.65 A set of gut–brain pathways enable bottom-up modulation of brain function and behavior via neural, endocrine, and immune systems, as well as top-down influence of our nervous system and emotional states. A specific illustration is how digestion of carbohydrates and proteins may modulate brain function and behavior.
The principal metabolites produced by bacterial fermentation of dietary fibers in the gastrointestinal tract are short-chain fatty acids (SCFA), such as acetic acid, propionic acid, and butanoic acid.66 SCFAs may influence gut–brain communication and brain function directly or indirectly via immune, endocrine, vagal (i.e., vagus nerve), and other humoral pathways.66 SCFAs modulate levels of neurotrophic factors and contribute to biosynthesis of the neurotransmitter serotonin, thus directly or indirectly modulating processes associated with neural functioning, learning, memory, and mood.66 Also critical for understanding gut–brain influences is the finding that SCFAs regulate the tissue macrophages of the brain, called microglia.67 Macrophages, including microglia, are the only immune cells that appear to populate the healthy brain and spinal cord.68 Microglia play a critical role not only in the development of the brain, but also may overreact to the build-up of amyloid plaques and tau filaments, and drive neurodegenerative and neuro-inflammatory diseases of the nervous system.69
With the advent of accurate and cost-effective characterizations of the microbiome of a host and a recipient, the near future holds the significant potential for microbiota-based therapeutic interventions into gut and brain health. For example, one of the most successful procedures for treating Clostridioides difficile infections has been transplantation of fecal microbiota from a host to a recipient.70 Caveats still remain, though, due to the complexity and variability of donor stools.71 Probiotics, live micro-organisms that may confer a health benefit when consumed in adequate amounts, are now widely available to consumers. However, their effectiveness remains debatable.72 Other approaches are also on the horizon, such as treating inflammatory bowel disease by engineering bacteria that assemble in situ a scaffold for reinforcing the barrier function of the mucosal layer of the gut epithelium.73 A particularly promising microbiota-based approach may be to prevent later disease by intervening in early life to promote microbiome development.74 However, precisely how to achieve this goal remains unclear. Overall, then, the challenge for bioengineers will be to continue to develop interventions that modify a person's microbiome in order to influence brain development, as well as to alleviate gut infections and neuro-inflammatory diseases of the nervous system.65
Recognition of the pervasive communication between body organs has additionally fomented a revolution in the bioengineering of organ-scale and multi-organ in vitro systems. Some of these systems are designed specifically as physiologically-based pharmacokinetic models for evaluating human drug responses, while others serve as niches for inducing growth of multiple progenitors, in order to build synthetic organs with highly differentiated compartments, such as kidneys.56,75 Among these systems are (1) body-on-chip organ systems, fluidically-linked to emulate integrated multi-organ function,75–78 (2) body-on-chip organoid systems that consist of fluidically interconnected organoids,79 and (3) EcoFABs, specially-designed as microbial ecosystems coupled with standardized workflows, computational tools, and computational models.80
Case study: repair of retina and spinal cord
This section presents two case studies. Each illustrates the nature-inspired approach to repairing human tissue and organs: (1) surgically implanting a “patch” of cells grown on a synthetic sheet to repair the retinal pigment epithelium (RPE) of age-related macular degeneration (AMD) patients, and (2) implanting stem cells to promote growth of relay circuits in order to repair a human spinal cord injury. Together, the repair of two very different organs (eye and spinal cord, respectively), organized within the same framework, highlights the generality of the nature-inspired solution approach. Within this framework, the steps for integrating the top-down and bottom-up methods are tailored to the requirements of supporting the growth of implanted cells to replace injured tissue.A developmental bioengineering approach to restoring lost vision in AMD patients is to repair the RPE, so that it can again function to provide nutrients and remove waste via the choroid, and thereby restore photoreceptor function. Table 2 summarizes repair technologies to realize a developmentally based methodology for RPE, as illustrated by two studies that have implanted a synthetic RPE into the retina85,86 (Fig. 2).
A cell monolayer is the level of organization required to recapitulate development in order to repair the RPE in AMD patients. The procedure used by Da Cruz et al. begins as self-organizing human embryonic RPE stem cells are nurtured in vitro on a synthetic scaffold, called a “retinal patch”. In the Kashani et al. study, a 6 μm parylene sheet is fabricated with a smooth surface and circular regions to facilitate cell adherence and growth factor diffusion.86 In both procedures, there is a dynamic feedback between the cells and growth factors as the cells differentiate into local ensembles. Once implanted, there is a critical developmental step which involves the integration, via assembly of the implanted RPE monolayer with the existing retinal structures and choroid. Results are encouraging: after a period of post-operative recovery and opportunities for visual experience, vision improved in most patients.86 Bioengineering advances in other studies include the use of induced pluripotent stem cells to implement the processes of self-organization, dynamic feedback, differentiation, and integration in repairing the RPE.87
In order for our eyes to explore the environment and regulate the information flow across our retinas, our head must turn this way and that, as our legs carry us to destinations near and far; looking is integrated with other sensory modalities. This integration is achieved by the spinal cord.
Spinal cord damage causes the death of nerve cells, either through loss of blood supply or by triggering the process of apoptosis, or cell death. The leading causes of spinal cord injuries (SCI) include vehicle crashes, falls, gunshot wounds, and sports/recreation activities. There are approximately 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new SCI in the U.S. each year, and up to 500
000 new SCI in the U.S. each year, and up to 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 worldwide, with about 80% of these cases occurring in males between 29 and 42 years of age.89
000 worldwide, with about 80% of these cases occurring in males between 29 and 42 years of age.89
Fig. 3 illustrates several developmental bioengineering approaches for spinal cord injury (SCI) repair. Animal models demonstrate that following a SCI, there is some spontaneous reorganization of circuits. After incomplete SCI, intraspinal projection neurons form new “detour circuits” that relay descending supraspinal information to sites below the lesion. In turn, corticospinal and reticulospinal tract neurons sprout onto these intraspinal relay neurons that connect to regions below the lesion.46 Such spontaneous circuit reorganization has motivated repair strategies for both incomplete SCI and complete SCI. In the former, therapeutic repair is directed toward augmenting spontaneous relay circuit formation, while, in the latter, repair strategies seek to restore neural connectivity by bridging axon regrowth across the blood-starved and necrotic regions of the damaged cord, even for short distances, into functional neural tissue.46
Achieving robust regrowth of axons across lesions requires (1) activation of intrinsic regrowth programs (driving neural stem cells' capacity to self-organize), (2) making available an axon-supportive growth substrate, and (3) providing growth factors that are chemo-attractive to axons (exemplifying dynamic feedback mechanisms).90 Toward that end, synthetic biomaterials developed as niches for repair have included injectable and resorbable polypeptide hydrogels for prolonged release of growth factors that attract growing axons, and bioengineered scaffolds with linear guidance channels to direct outgrowth along linear paths. An approach in the Tuszynski laboratory has been to transplant neural progenitor cells (NPCs) into an injured spinal cord in order to form a neuronal relay circuit across the gap.91 For example, Kumamaru et al. grafted spinal cord neural stem cells into rat spinal cords.92 They found not only that the grafts extended large numbers of axons over long distances, but also that the grafts integrated into both intraspinal and supraspinal systems. Next, this group developed a microscale 3D printing method to fabricate a rodent-scaled hydrogel scaffold loaded with NPCs.93 Remarkably, host axons regenerated into the scaffold. Moreover, the implanted NPCs extended axons out of the scaffold and into the host spinal cord below the injury. Turning again to Table 2 and Fig. 3, this work illustrates how dynamic feedback within a growth-supportive niche promotes extension of axons to establish relay circuits.
A fundamental question impacting the success of axon repair strategies is an ecological one: how to use newly formed circuits to restore task-specific behaviors, such as standing, walking or climbing. This question has been pursued during decades of animal and human studies of epidural electrical stimulation (EES) during standing and walking. These studies find that EES takes advantage of spared but functionally silent descending pathways, such as the projections from the reticular nucleus, called the reticulospinal pathway, in order to produce movements in paralyzed limbs. Moreover, EES activates motor neurons by recruiting proprioceptive circuits within the posterior roots of the spinal cord.
To better leverage EES, investigators in the Courtine laboratory have developed treatment protocols for human SCI patients that surgically implant a pulse generator to deliver EES trains to lumbosacral posterior spinal roots with timing that coincides with intended movements.94 This spatiotemporally modulated EES was used during over-ground locomotor training with a gravity-assist device, an example of bio-electronic therapeutics.94 In this study, three SCI patients followed an intensive gait training program.94 The protocol included pulse generated EES for spatiotemporal neuromodulation, inertial measurement units attached to their feet to adjust spatiotemporal stimulation, and gravity assist that personalized the amount of force applied to the trunk for natural energy flow between gravitational forces and gait dynamics. After less than a month of training, all three patients were able to walk over ground during stimulation, and some recovery without EES. Establishing new connectivity patterns between descending tracts and spinal neurons illustrates the assembly of local ensembles into larger groups, and the opportunity for over ground mobility promotes the formation of integrated ecosystems (see Table 2 and Fig. 3).
Considered together, surgical repair with the retinal patch and implantation of cells for growing spinal relay circuits, used in conjunction with opportunities for functional activities, offer hope of restoring vision and mobility, respectively. The development-inspired methodologies summarized in Tables 1 and 2 may provide a launch pad for further advances that recapitulate development for repair.
Conclusions and outlook
A systematized nature-inspired solution methodology holds great promise for developmental bioengineering, extending success in other areas, from energy and environmental technology, to chemical process intensification.9 We have illustrated this new framework for discovery and innovation via two, very different cases of organ repair: the retinal pigment epithelium (RPE) and the spinal cord. Being systematic, the proposed methodology allows to move beyond ad hoc solutions, so that it can be applied more generally to other bioengineering challenges as well.The extended nature-inspired solutions framework for developmental bioengineering helps to structure an inherently extremely complex research domain. We hope that this approach may facilitate the proposal of potential solution pathways, thus spurring and accelerating innovation. Nevertheless, it must be cautioned that each implementation in practice still relies on extensive research and medical trials, requiring widely cross-disciplinary teams that combine expertise from biomaterials science, chemistry and chemical engineering, biomedical engineering and medicine, to robotics, physical therapy, psychology, and beyond.
The discussed nature-inspired, developmental bioengineering approach develops solutions by first identifying and then utilizing fundamental mechanisms underpinning natural developmental processes. These universal principles include self-organization, dynamic feedback between cells and their environment, differentiation into local ensembles, the assembly of these local ensembles into larger groups, and the formation of integrated ecosystems. Not discussed in the examples here, but relevant to other applications,9 are hierarchical transport networks (e.g., in the respiratory or the vascular network) and force balancing at multiple scales, from nano-confinement effects (e.g., in cellular membrane transport or in chaperonins) to capillary action (e.g., in blood vessels) and mechanical force balancing (e.g., in bones). In living systems, several of these principles act not individually, but in combination, and then often synergistically. This combination could be crucial to support a versatile functional response. Where this is the case, these mechanisms should preferably be applied jointly in the bioengineered solutions inspired by them as well. Such nature-inspired concepts may inform several designs that embody the key concepts, adapted to the environment in which they are used. These are implemented in prototypes, the creation of which benefits from rapid progress in biomaterials, 3D printing and various nascent manufacturing precision technologies. Both top-down and bottom-up synthesis methods can be applied. These prototypes are likely to evolve by iteration, based on medical outcomes, but also new fundamental insights, manufacturing methods, advances in diagnostics, therapies and patient response. To be successful, a holistic systems approach that appreciates the context of the application is essential, including dynamical interaction with the environment in an ecological setting, here illustrated through the activity-based therapy for spinal cord injuries.
In this perspective article, the presentation was limited to repair of tissue and organs. However, nature may also recapitulate developmental processes for regenerating tissue. For example, one of the remaining mysteries in developmental biology is why some animals, such as axolotls, but not humans, are capable of regenerating body parts.95 Will a nature-inspired methodology provide new directions for solutions to this and other developmental mysteries?
Conflicts of interest
There are no conflicts to declare.Acknowledgements
ECG received support for the writing of this article from the Wyss Institute for Biologically Inspired Engineering at Harvard. MOC gratefully acknowledges support from the EPSRC via “Frontier Engineering” and “Frontier Engineering: Progression” Awards (EP/K038656/1, EP/S03305X/1).Notes and references
- H. Shen, Embryo assembly 101, Nature, 2018, 559, 19–22 CrossRef CAS.
- M. N. Shahbazi and M. Zernicka-Goetz, Deconstructing and reconstructing the mouse and human early embryo, Nat. Cell Biol., 2018, 20, 878–887 CrossRef CAS.
- J. M. Wells and F. M. Watt, Diverse mechanisms for endogenous regeneration and repair in mammalian organs, Nature, 2018, 557(7705), 322–328 CrossRef CAS.
- M. Mahar and V. Cavalli, Intrinsic mechanisms of neuronal axon regeneration, Nat. Rev. Neurosci., 2018, 19(6), 323–337 CrossRef CAS.
- T. Gerber, P. Murawala, D. Knapp, W. Masselink, M. Schuez and S. Hermann, et al., Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration, Science, 2018, 362(6413), eaaq0681 CrossRef.
- B. Laha, B. K. Stafford and A. D. Huberman, Regenerating optic pathways from the eye to the brain, Science, 2017, 356, 1–4 CrossRef.
- E. Nacu and E. M. Tanaka, Limb regeneration: a new development?, Annu. Rev. Cell Dev. Biol., 2011, 27, 409–440 CrossRef CAS.
- M. N. Shahbazi, E. Siggia and M. Zernicka-Goetz, Self-organization of stem cells into embryos: A window on early mammalian development, Science, 2019, 364, 948–951 CrossRef CAS.
- M. O. Coppens, A nature-inspired approach to reactor and catalysis engineering, Curr. Opin. Chem. Eng., 2012, 1, 281–289 CrossRef CAS.
- A. S. Perera and M. O. Coppens, Re-designing materials for biomedical applications: from biomimicry to nature-inspired chemical engineering, Philos. Trans. R. Soc., A, 2019, 377(2138), 20180268 CrossRef CAS.
- D. Cyranoski, The cells that sparked a revolution, Nature, 2018, 555, 428–430 CrossRef CAS.
- M. Tewary, N. Shakiba and P. W. Zandstra, Stem cell bioengineering: building from stem cell biology, Nat. Rev. Genet., 2018, 19, 595–614 CrossRef CAS.
- E. Davidson, Emerging properties of animal gene regulatory networks, Nature, 2010, 468, 911–920 CrossRef CAS.
- D. Gilmour, M. Rembold and M. Leptin, From morphogen to morphogenesis and back, Nature, 2017, 541(7637), 311–320 CrossRef CAS.
- S. Wennekamp, S. Mesecke, F. Nedelec and T. Hiiragi, A self-organization framework for symmetry breaking in the mammalian embryo, Nat. Rev. Mol. Cell Biol., 2013, 14(7), 452–459 CrossRef.
- M. Simunovic and A. H. Brivanlou, Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis, Development, 2017, 144(6), 976–985 CrossRef CAS.
- M. Simunovic, J. J. Metzger, F. Etoc, A. Yoney, A. Ruzo and I. Martyn, et al., A 3D model of a human epiblast reveals BMP4-driven symmetry breaking, Nat. Cell Biol., 2019, 21(7), 900–910 CrossRef CAS.
- S. F. Gilbert and M. J. Barresi, Developmental Biology, Sunderland MA, Sinauer, 11th edn, 2016 Search PubMed.
- T. R. Huycke, B. M. Miller, H. K. Gill, N. L. Nerurkar, D. Sprinzak and L. Mahadevan, et al., Genetic and Mechanical Regulation of Intestinal Smooth Muscle Development, Cell, 2019, 179(1), 90–105 e21 CrossRef CAS.
- E. Hannezo and C. P. Heisenberg, Mechanochemical Feedback Loops in Development and Disease, Cell, 2019, 178(1), 12–25 CrossRef CAS.
- A. Warmflash, B. Sorre, F. Etoc, E. D. Siggia and A. H. Brivanlou, A method to recapitulate early embryonic spatial patterning in human embryonic stem cells, Nat. Methods, 2014, 11(8), 847–854 CrossRef CAS.
- A. Deglincerti, F. Etoc, M. C. Guerra, I. Martyn, J. Metzger and A. Ruzo, et al., Self-organization of human embryonic stem cells on micropatterns, Nat. Protoc., 2016, 11(11), 2223–2232 CrossRef CAS.
- A. Manfrin, Y. Tabata, E. R. Paquet, A. R. Vuaridel, F. R. Rivest and F. Naef, et al., Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells, Nat. Methods, 2019, 16(7), 640–648 CrossRef CAS.
- J. B. Gurdon, Nuclear reprogramming and cell replacement therapies, Nat. Rev. Mol. Cell Biol., 2016, 17(3), 137–138 CrossRef CAS.
- K. Takahashi and S. Yamanaka, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell, 2006, 126(4), 663–676 CrossRef CAS.
- J. Mertens, M. C. Marchetto, C. Bardy and F. H. Gage, Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience, Nat. Rev. Neurosci., 2016, 17(7), 424–437 CrossRef CAS.
- S. W. Lane, D. A. Williams and F. M. Watt, Modulating the stem cell niche for tissue regeneration, Nat. Biotechnol., 2014, 32(8), 795–803 CrossRef CAS.
- J. D. Humphrey, E. R. Dufresne and M. A. Schwartz, Mechanotransduction and extracellular matrix homeostasis, Nat. Rev. Mol. Cell Biol., 2014, 15(12), 802–812 CrossRef CAS.
- G. S. Hussey, J. L. Dziki and S. F. Badylak, Extracellular matrix-based materials for regenerative medicine, Nat. Rev. Mater., 2018, 3(7), 159–173 CrossRef CAS.
- L. Li, J. Eyckmans and C. S. Chen, Designer biomaterials for mechanobiology, Nat. Mater., 2017, 16, 1164–1168 CrossRef CAS.
- J. L. Drury and D. J. Mooney, Hydrogels for tissue engineering: scaffold design variables and applications, Biomaterials, 2003, 24(24), 4337–4351 CrossRef CAS.
- K. H. Vining and D. J. Mooney, Mechanical forces direct stem cell behaviour in development and regeneration, Nat. Rev. Mol. Cell Biol., 2017, 18(12), 728–742 CrossRef CAS.
- C. M. Madl, S. C. Heilshorn and H. M. Blau, Bioengineering strategies to accelerate stem cell therapeutics, Nature, 2018, 557(7705), 335–342 CrossRef CAS.
- R. H. Masland, The neuronal organization of the retina, Neuron, 2012, 76(2), 266–280 CrossRef CAS.
- K. Bharti, Patching the retina with stem cells, Nat. Biotechnol., 2018, 36(4), 311–313 CrossRef CAS.
- B. Roska and J. A. Sahel, Restoring vision, Nature, 2018, 557(7705), 359–367 CrossRef CAS.
- S. G. Varadarajan and A. D. Huberman, Assembly and repair of eye-to-brain connections, Curr. Opin. Neurobiol., 2018, 53, 198–209 CrossRef CAS.
- J. B. Bikoff, M. I. Gabitto, A. F. Rivard, E. Drobac, T. A. Machado and A. Miri, et al., Spinal Inhibitory Interneuron Diversity Delineates Variant Motor Microcircuits, Cell, 2016, 165(1), 207–219 CrossRef CAS.
- S. Arber, Motor circuits in action: specification, connectivity, and function, Neuron, 2012, 74(6), 975–989 CrossRef CAS.
- M. Goulding, Circuits controlling vertebrate locomotion: moving in a new direction, Nat. Rev. Neurosci., 2009, 10(7), 507–518 CrossRef CAS.
- J. Zhang, G. M. Lanuza, O. Britz, Z. Wang, V. C. Siembab and Y. Zhang, et al., V1 and v2b interneurons secure the alternating flexor-extensor motor activity mice require for limbed locomotion, Neuron, 2014, 82(1), 138–150 CrossRef CAS.
- K. Kadoya, P. Lu, K. Nguyen, C. Lee-Kubli, H. Kumamaru and L. Yao, et al., Spinal cord reconstitution with homologous neural grafts enables robust corticospinal regeneration, Nat. Med., 2016, 22(5), 479–487 CrossRef CAS.
- U. Windhorst, Muscle proprioceptive feedback and spinal networks, Brain Res. Bull., 2007, 73, 155–202 CrossRef CAS.
- A. Takeoka, I. Vollenweider, G. Courtine and S. Arber, Muscle spindle feedback directs locomotor recovery and circuit reorganization after spinal cord injury, Cell, 2014, 159(7), 1626–1639 CrossRef CAS.
- A. Chedotal, Roles of axon guidance molecules in neuronal wiring in the developing spinal cord, Nat. Rev. Neurosci., 2019, 20(7), 380–396 CrossRef CAS.
- G. Courtine and M. V. Sofroniew, Spinal cord repair: advances in biology and technology, Nat. Med., 2019, 25(6), 898–908 CrossRef CAS.
- H. Kumamaru, K. Kadoya, A. F. Adler, Y. Takashima, L. Graham and G. Coppola, et al., Generation and post-injury integration of human spinal cord neural stem cells, Nat. Methods, 2018, 15(9), 723–731 CrossRef CAS.
- J. Cao, M. Spielmann, X. Qiu, X. Huang, D. M. Ibrahim and A. J. Hill, et al., The single-cell transcriptional landscape of mammalian organogenesis, Nature, 2019, 566(7745), 496–502 CrossRef CAS.
- E. M. De Robertis, Spemann's organizer and self-regulation in amphibian embryos, Nat. Rev. Mol. Cell Biol., 2006, 7, 296–302 CrossRef CAS.
- E. M. De Robertis, Spemann's organizer and the self-regulation of embryonic fields, Mech. Dev., 2009, 126(11–12), 925–941 CrossRef CAS.
- M. A. Lancaster and J. A. Knoblich, Organogenesis in a dish: modeling development and disease using organoid technologies, Science, 2014, 345(6194), 1247125 CrossRef.
- A. L. Bredenoord, H. Clevers and J. A. Knoblich, Human tissues in a dish: The research and ethical implications of organoid technology, Science, 2017, 355(6322), eaaf9414 CrossRef.
- A. Ochoa-Espinosa and M. Affolter, Branching morphogenesis: from cells to organs and back, Cold Spring Harbor Perspect. Biol., 2012, 4(10), a008243 Search PubMed.
- S. Musah, N. Dimitrakakis, D. M. Camacho, G. M. Church and D. E. Ingber, Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip, Nat. Protoc., 2018, 13(7), 1662–1685 CrossRef CAS.
- N. Y. C. Lin, K. A. Homan, S. S. Robinson, D. B. Kolesky, N. Duarte and A. Moisan, et al., Renal reabsorption in 3D vascularized proximal tubule models, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(12), 5399–5404 CrossRef CAS.
- K. A. Homan, N. Gupta, K. T. Kroll, D. B. Kolesky, M. Skylar-Scott and T. Miyoshi, et al., Flow-enhanced vascularization and maturation of kidney organoids in vitro, Nat. Methods, 2019, 16, 255–262 CrossRef CAS.
- N. C. Rivron, J. Frias-Aldeguer, E. J. Vrij, J. C. Boisset, J. Korving and J. Vivie, et al., Blastocyst-like structures generated solely from stem cells, Nature, 2018, 557(7703), 106–111 CrossRef CAS.
- E. D. Siggia and A. Warmflash, Modeling Mammalian Gastrulation With Embryonic Stem Cells, Curr. Top. Dev. Biol., 2018, 129, 1–23 CAS.
- B. Sozen, G. Amadei, A. Cox, R. Wang, E. Na and S. Czukiewska, et al., Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures, Nat. Cell Biol., 2018, 20, 979–989 CrossRef CAS.
- P. L. Tam, Modeling the early development of a primate embryo, Science, 2019, 366, 798–799 CrossRef CAS.
- E. C. Goldfield, Emergent Forms: Origins and Early Development of Human Action and Perception, Oxford University Press, New York, NY, 1995, p. 384 Search PubMed.
- G. B. West, The importance of quantitative systemic thinking in medicine, Lancet, 2012, 379(9825), 1551–1559 CrossRef.
- T. C. Fung, C. A. Olson and E. Y. Hsiao, Interactions between the microbiota, immune and nervous systems in health and disease, Nat. Neurosci., 2017, 20(2), 145–155 CrossRef CAS.
- M. R. Charbonneau, L. V. Blanton, D. B. DiGiulio, D. A. Relman, C. B. Lebrilla and D. A. Mills, et al., A microbial perspective of human developmental biology, Nature, 2016, 535(7610), 48–55 CrossRef CAS.
- M. I. Butler, J. F. Cryan and T. G. Dinan, Man and the Microbiome: A New Theory of Everything?, Annu. Rev. Clin. Psychol., 2019, 15, 1–28 CrossRef.
- B. Dalile, L. Van Oudenhove, B. Vervliet and K. Verbeke, The role of short-chain fatty acids in microbiota–gut–brain communication, Nat. Rev. Gastroenterol. Hepatol., 2019, 16(8), 461–478 CrossRef.
- D. Erny, A. L. Hrabe de Angelis, D. Jaitin, P. Wieghofer, O. Staszewski and E. David, et al., Host microbiota constantly control maturation and function of microglia in the CNS, Nat. Neurosci., 2015, 18(7), 965–977 CrossRef CAS.
- M. Prinz, S. Jung and J. Priller, Microglia Biology: One Century of Evolving Concepts, Cell, 2019, 179(2), 292–311 CrossRef CAS.
- T. Masuda, R. Sankowski, O. Staszewski, C. Bottcher, L. Amann and C. Scheiwe, et al., Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution, Nature, 2019, 566(7744), 388–392 CrossRef CAS.
- S. V. Lynch, S. C. Ng, F. Shanahan and H. Tilg, Translating the gut microbiome: ready for the clinic?, Nat. Rev. Gastroenterol. Hepatol., 2019, 16(11), 656–661 CrossRef.
- E. M. Giles, G. L. D'Adamo and S. C. Forster, The future of faecal transplants, Nat. Rev. Microbiol., 2019, 17(12), 719 CrossRef CAS.
- J. Suez, N. Zmora, E. Segal and E. Elinav, The pros, cons, and many unknowns of probiotics, Nat. Med., 2019, 25(5), 716–729 CrossRef CAS.
- P. Praveschotinunt, A. M. Duraj-Thatte, I. Gelfat, F. Bahl, D. B. Chou and N. S. Joshi, Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut, Nat. Commun., 2019, 10(1), 5580 CrossRef CAS.
- K. E. Fujimura, A. R. Sitarik and S. Havstad, et al., Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation, Nat. Med., 2016, 22, 1187–1191 CrossRef CAS.
- R. Prantil-Baun, R. Novak, D. Das, M. R. Somayaji, A. Przekwas and D. E. Ingber, Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips, Annu. Rev. Pharmacol. Toxicol., 2018, 58, 37–64 CrossRef CAS.
- S. E. Park, A. Georgescu and D. Huh, Organoids-on-a-chip, Science, 2019, 364, 960–965 CrossRef CAS.
- R. Novak, M. Ingram, S. Clauson, D. Das, A. Delahanty and A. Herland, et al., A robotic platform for fluidically-linked human body-on-chips experimentation, BioRxiv, 2019, p. 569541.
- K. Ronaldson-Bouchard and G. Vunjak-Novakovic, Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development, Cell Stem Cell, 2018, 22(3), 310–324 CrossRef CAS.
- A. Sjkardal, T. Shupe and A. Atala, Body-on-a-chip: Regenerative Medicine for Personalized Medicine, in Principles of Regenerative Medicine, ed. A. Atala, R. Lanza, A. G. Mikos and R. Nerem, Elsevier, 3rd edn, 2019 Search PubMed.
- K. Zengler, K. Hofmockel, N. S. Baliga, S. W. Behie, H. C. Bernstein and J. B. Brown, et al., EcoFABs: advancing microbiome science through standardized fabricated ecosystems, Nat. Methods, 2019, 16(7), 567–571 CrossRef CAS.
- P. Mitchell, G. Liew, B. Gopinath and T. Y. Wong, Age-related macular degeneration, Lancet, 2018, 392, 1147–1159 CrossRef.
- B. D. Gelfand and J. Ambati, A Revised Hemodynamic Theory of Age-Related Macular Degeneration, Trends Mol. Med., 2016, 22(8), 656–670 CrossRef CAS.
- S. Kim, A. S. Lowe and R. Dharmat, Generation, transcription profiling, and functional validation of cone-rich human retinal organoids, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 10824–10833 CrossRef CAS.
- T. A. Seabrook, T. J. Burbridge, M. C. Crair and A. D. Huberman, Architecture, Function, and Assembly of the Mouse Visual System, Annu. Rev. Neurosci., 2017, 40, 499–538 CrossRef CAS.
- L. da Cruz, K. Fynes, O. Georgiadis, J. Kerby, Y. H. Luo and A. Ahmado, et al., Phase 1 clinical study of an embryonic stem cell-derived retinal pigment epithelium patch in age-related macular degeneration, Nat. Biotechnol., 2018, 36, 328–337 CrossRef CAS.
- A. H. Kashani, J. S. Lebkowski and F. M. Rahhal, et al., A bioengineered retinal pigment epithelial monolayer for advanced, dry age-related macular degeneration, Sci. Transl. Med., 2018, 10, 1–10, DOI:10.1126/scitranslmed.eaao4097.
- M. Mandai, A. Watanabe, Y. Kurimoto, Y. Hirami, C. Morinaga and T. Daimon, et al., Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration, N. Engl. J. Med., 2017, 376(11), 1038–1046 CrossRef CAS.
- D. Holmes and L. Reading-Ikkanda, Repairing the neural highway, Nature, 2017, 552, S50–S51 CrossRef CAS.
- National Spinal Cord Injury Statistical Center, Facts and figures at a glance, University of Alabama, Birmingham, 2016 Search PubMed.
- M. A. Anderson, T. M. O'Shea, J. E. Burda, Y. Ao, S. L. Barlatey and A. M. Bernstein, et al., Required growth facilitators propel axon regeneration across complete spinal cord injury, Nature, 2018, 561, 396–400 CrossRef CAS.
- E. S. Rosenzweig, J. H. Brock, P. Lu, H. Kumamaru, E. A. Salegio and K. Kadoya, et al., Restorative effects of human neural stem cell grafts on the primate spinal cord, Nat. Med., 2018, 24, 484–490 CrossRef CAS.
- H. Kumamaru, K. Kadoya, A. F. Adler, Y. Takashima, L. Graham and G. Coppola, et al., Generation and post-injury integration of human spinal cord neural stem cells, Nat. Methods, 2018, 15(9), 723–731 CrossRef CAS.
- J. Koffler, W. Zhu, X. Qu, O. Platoshyn, J. N. Dulin and J. Brock, et al., Biomimetic 3D-printed scaffolds for spinal cord injury repair, Nat. Med., 2019, 25, 263–269 CrossRef CAS.
- F. B. Wagner, J. B. Mignardot, C. G. Le Goff-Mignardot, R. Demesmaeker, S. Komi and M. Capogrosso, et al., Targeted neurotechnology restores walking in humans with spinal cord injury, Nature, 2018, 563(7729), 65–71 CrossRef CAS.
- E. Nacu and E. M. Tanaka, Limb regeneration: a new development?, Annu. Rev. Cell Dev. Biol., 2011, 27, 409–440 CrossRef CAS.
- M. Goulding, Circuits controlling vertebrate locomotion: moving in a new direction, Nat. Rev. Neurosci., 2009, 10, 507–518 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |
