Bioinspired carbon dots (biodots): emerging fluorophores with tailored multiple functionalities for biomedical, agricultural and environmental applications
Yoonah
Choi†
c,
Xin Ting
Zheng†
 b and
Yen Nee
Tan
b and
Yen Nee
Tan
 *abc
*abc
aFaculty of Science, Agriculture & Engineering, Newcastle University, Newcastle Upon Tyne NE1 7RU, UK. E-mail: yennee.tan@newcastle.ac.uk
bInstitute of Materials Research and Engineering (IMRE), Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, Singapore 138634. E-mail: tanyn@imre.a-star.edu.sg
cDepartment of Chemistry, National University of Singapore, 3 Science Drive, Singapore 117543
First published on 17th October 2019
Abstract
Nature provides abundant resources to support life on our planet. In particular, biological systems possess unique properties and functions such as bio-recognition and self-assembly, which makes them an attractive inspiration for mankind to design smart materials at the molecular level. This review reports an emerging class of bioinspired fluorescent nanodots (biodots) derived from natural resources ranging from small biomolecules, biopolymers to biomass for a wide spectrum of technological applications. These “biodots” are carbonaceous in nature with inherent biocompatibility, which can be molecularly engineered to obtain both biological and physiochemical properties in one entity similar to the molecular machines in cells. Importantly, the surface properties and functionalities of biodots can be fine-tuned depending on the types of precursors used in their synthesis. This mini-review summarises the recent development of fluorescent biodots with intrinsic biological and/or biomimetic functions, with an aim to examine their formation mechanisms and understand how different types of precursors affect the overall properties of biodots. This is followed by a thorough discussion on the successful utilizations of these biodots for biomedical (e.g., theranostic agents), agricultural (e.g., fertilizers for plant growth) and environmental (e.g., pollutant detection) applications, in order to gain critical insights into the molecular design of multifunctional biodots tailored for different applications. Lastly, we highlight the promising directions and future outlook of biodots with the aid of artificial intelligence, which will potentially accelerate the development of this promising field.
Design, System, ApplicationBioinspired carbon dots, also known as “biodots”, represent an emerging class of fluorophores derived from natural resources ranging from small biomolecules and biopolymers to renewable biomass like plant and food waste. The abundance of carbon precursors in nature allows molecular engineering of biodots through different formation mechanisms, which subsequently leads to remarkable controllability of their physicochemical properties. Biodots are typically highly fluorescent with excellent photostability, biocompatibility and aqueous solubility. Additionally, biodots often inherit the intrinsic properties of their natural building blocks, e.g., bio-recognition, therapeutic or even biomimetic functions, thereby endowing them with unique attributes from Nature. Today, these multifunctional biodots have been successfully applied in biomedical, agricultural and environmental fields as theranostic agents for bioimaging and therapy, fertilizers for enhancing plant growth as well as nanosensors for on-site pollutant detection. We envision that the advancement in artificial intelligence and better understanding of bioinspired systems will accelerate the molecular design of biodots with fine-tuned properties for a broader range of cutting edge applications in the near future. |
1. Introduction
With surging interest in carbon dots in the last 15 years, extensive research has been carried out to explore synthesis of carbon dots with tunable properties.1,2 Conventionally, carbon dot synthesis can be broadly categorized into top-down and bottom-up approaches. Top-down approaches involve deconstructing large carbon materials such as graphite, activated carbon, and carbon nanotubes through methods such as laser ablation, arc discharge and electrochemical oxidation.3,4 Bottom-up approaches, on the other hand, involve constructing carbon dots from small molecular precursors through hydrothermal, solvothermal, microwave, or pyrolysis methods.3,4 The sizes and photoluminescence properties of carbon dots depend on the synthesis conditions such as temperature, reaction time, concentration and composition of reactants.5,6 The main drawbacks of the conventional methods are the frequent use of toxic precursors or harsh synthesis routes involving strong acids/alkalis that are not environment-friendly.In recent years, the emergence of bioinspired approaches for the synthesis of multifunctional nanomaterials has attracted much attention from fundamental research to various technological applications.7–10 In particular, researchers are actively searching for greener and renewable carbon sources to produce biocompatible carbonaceous nanodots.11 Such efforts led to the exploration of biomolecules or other natural resources with good biocompatibility and inherent biological functions as precursors to synthesize bioinspired carbon dots, an emerging class of fluorophores with integrated biological and/or chemical properties, which is known as “biodots”.7,8 A wide spectrum of natural resources ranging from small molecules (e.g., glucose), biopolymers (e.g., proteins or nucleic acids) to complex matrices such as plant- and/or animal-derived materials (e.g. leaves or eggs) has been successfully employed as natural carbon sources to synthesize biodots with intrinsic biological or biomimetic functions. In this context, the synthesis routes of biodots are categorized based on the types of precursors used. As shown in Fig. 1, the “top-down” approach involves the use of natural products from large biomass as precursors, while the “bottom-up” approach utilizes small biological building blocks such as amino acids for the synthesis of biodots.
Today, there are three widely-accepted explanations for the optical properties of carbon dots, namely (1) graphitic core driven mechanism, (2) surface mediated mechanism and (3) molecular photoluminescence.12 The graphitic core driven mechanism proposes that exciton confinement in the π-conjugated graphitic core (also known as quantum confinement) is responsible for the optical properties of biodots. As a result, photoluminescence of carbon dots can be controlled by modifying their size. Meanwhile, the surface mediated mechanism suggests that the optical properties of carbon dots can be attributed to the surface defect states or the combined effects of surface defects and quantum confinement. For the third explanation, molecular photoluminescence attributes the optical properties of carbon dots to the π-conjugated islands within their nanostructures. However, due to the large variety of natural precursors and synthesis routes, the photoluminescence origin of biodots is yet to be fully understood. Thus, it is essential to review currently available approaches for the synthesis of biodots in order to gain better understanding of their molecular design in improving the photoluminescence properties, including fluorescence quantum yield, emission color and photostability, which are delicately controlled by a combination of the above mechanisms.12
Apart from their unique photoluminescence properties, biodots can be engineered to integrate the intrinsic functions such as therapeutic and bio-recognition from their natural precursors (e.g., herbs and DNA). Furthermore, natural resources are especially appealing due to the abundance and variety of heteroatoms and functional groups which can be used to decorate or passivate the biodot surfaces, and consequently enable fine-tuning of their biological (e.g., biocompatibility), chemical (e.g., stability) and optical (e.g., emission colour) properties.13 Since molecular structures and inherent functionalities of natural precursors could eventually affect the biological and/or physiochemical properties of the resultant biodots, close examination of different types of precursors in biodot synthesis is essential to better understand their molecular design strategies. This mini-review aims to look at different natural resources that have been utilized for biodot synthesis, and compare and summarize the different synthesis strategies that lead to the formation of biodots with varying physiochemical properties and biofunctionalities, as well as discuss their prominent applications in bioimaging, nanotherapeutics, and enhanced plant growth in agriculture and environmental sensing (Fig. 1). Lastly, we will highlight the promising directions and future development of these emerging materials with the aid of artificial intelligence.
2. Natural materials as renewable carbon sources for biodot synthesis
As explained in the introduction, biodots synthesized from biomass (e.g., fruits, eggs, etc.) can be obtained through the “top-down” approaches, while those derived from biomolecules or biopolymers are obtained through the “bottom-up” means (Fig. 1). To date, various types of natural materials from small biomolecules to more complex matrices such as plant extracts have been used for the synthesis of biodots. In the following sections, we will discuss the different types of natural precursors for biodot synthesis and their respective formation mechanisms leading to the tunable structures and properties of the resultant biodots.2.1. Small biomolecules
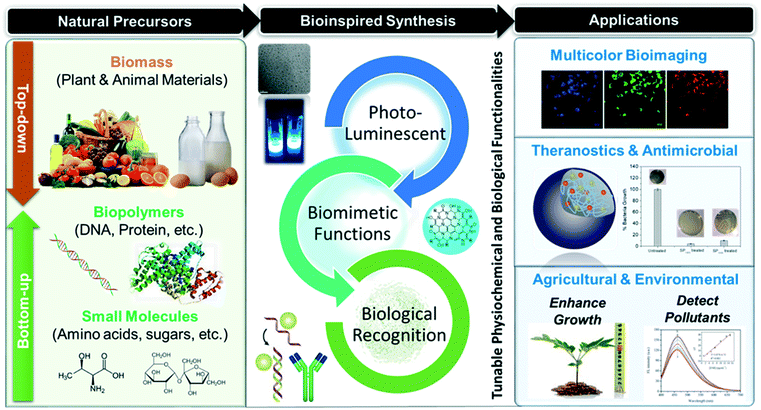
As biomolecules are good sources of heteroatoms, the as-synthesized biodots are often doped with nitrogen, sulfur or phosphorus. Doping of the dots with nitrogen and sulfur causes changes in molecular structures resulting in different surface electronic states.14 This could affect the properties of the resultant biodots, thus enabling molecular engineering from bottom-up approaches.15 Specifically, nitrogen doping has been found to greatly enhance the photoluminescence of biodots.15,16 In addition, various functional groups such as the hydroxyl groups found in these small biomolecules not only contribute significantly to fine-tuning of the fluorescence properties of biodots (including photostability, emission color, and fluorescence quantum yield),13 but also endow them with unique (bio)functionalities, such as antimicrobial,17,18 tumour inhibition19,20 and singlet oxygen generation.21 As compared to biodots derived from top-down approaches using complex natural matrices (e.g., plant leaves) as precursors, small molecule-derived biodots usually exhibit better reproducibility, because significant batch-to-batch variations may be present in biomass due to different contents of the nutrients. | ||
| Fig. 2 a) Molecular structure and photoluminescence properties of biodots synthesized from 20 natural amino acids (AA-dots) via hydrothermal synthesis. (b) Fluorescence quantum yield (%), (c) formation mechanism, and (d) summary of important functional groups (+) within the AA-dots responsible for high brightness. Reproduced, adapted and reprinted with permission from ref. 22. | ||
Combination of an amino acid with another small molecule as biodot precursors has also been investigated. For example, Nawaz et al. produced blue emissive biodots using L-lysine and ascorbic acid in hydrothermal synthesis.23 When L-lysine was replaced with phenol, the emission color was not affected, but the PL intensity was significantly reduced. This result highlighted the importance of nitrogen-containing precursors in enhancing the fluorescence QY of the as-synthesized biodots.23 In another example, Zeng et al. showed that uniform biodots (2.3 nm) with orange emission could be produced by co-doping with nitrogen (N) and sulfur (S), using a mixture of L-cysteine (sulfur source) and L-serine (nitrogen source) as precursors.14 When only L-cysteine was used, biodots with irregular shape and low fluorescence QY were formed, while nanorod-shape biodots with blue-emission were produced by L-serine as the sole precursor. In addition, the type of nitrogen-containing precursor affected the size and maximum emission wavelengths of the biodots formed. For instance, when one of the amino acids (i.e., L-serine) was replaced with L-proline, L-alanine or L-asparagine, biodots with different sizes and blue-shifted emission wavelengths were formed.14
Besides hydrothermal synthesis, microwave-assisted methods have also been applied to produce amino acid-derived nanodots. For example, Jiang et al. synthesized highly fluorescent biodots from histidine via microwave irradiation in a 700 W domestic microwave for 160 seconds, using ortho-phosphoric acid as the catalyst.24 The as-formed biodots have an average size of (2.0 ± 0.4) nm, and a high fluorescence QY of 44.9%. In the absence of an acid or base catalyst, no biodots were formed. Using the same catalytic method, hydrophobic amino acids such as leucine, isoleucine and valine led to the formation of weakly fluorescent biodots.24 On the other hand, Choi et al. succeeded in synthesizing highly fluorescent biodots (QY = 23.3%) without the use of an acid or base catalyst via facile microwave pyrolysis in a 750 W household microwave using L-lysine as a precursor. It should be noted that the amino acid precursor used in such a synthesis should be a branched, AB2 type polyamidation monomer (where A and B represent different functional groups), suggesting the importance of functional groups in biodot synthesis.25
Different isomers of amino acids have been explored as well. The chirality of the precursors may play an important role in the function of the biodots produced. Zhang et al. used citric acid in combination with L-cysteine, D-cysteine or DL-cysteine to produce chiral (L and D) and achiral biodots.26 It was found that both the chiral and achiral dots were similar in molecular structure, but different in efficacy to promote the growth of mung bean plants.26 This study shows the potential of using simple natural precursors such as amino acid isomers to tailor the functionalities of biodots (i.e., chirality) in achieving better performance for certain applications such as in agricultural and pharmacological studies for drug design.
Simple heating or ultrasonication is an alternative method for biodot synthesis. Ma et al. reported nitrogen-doped biodots using glucose as a carbon precursor and ammonia as a nitrogen source through hydrogen peroxide-assisted ultrasonic treatment.28 The resultant biodots contained 7.45 wt% nitrogen, indicating successful incorporation of nitrogen into the carbon framework.28 Chen and co-workers developed a simple synthesis method to form biodots of 2–3 nm in diameter from lactose by heating in an alkali medium under low pressure conditions.29 On the other hand, Xiao's group reported a greener and faster synthesis method to obtain ultrasmall biodots with an average diameter of 1.6 nm by heating sucrose, urea and edible oil at ambient pressure for 5 min.30 Recently, Naik et al. reported a new approach to synthesize sulfur-doped biodots from sucrose using concentrated sulfuric acid as a dehydrating agent and source of sulfur atoms.31 This method does not even require an external heat source as the heat released during the dehydration of sucrose was found to be sufficient for the reaction. However, the usage of harsh reagents such as concentrated acid makes the synthesis route less environmentally friendly, which requires a stringent post-treatment process to purify the resultant biodots.
More recently, we have conducted a systematic study to investigate the structure–property relationship of nucleotide dots (N-dots)21 synthesized hydrothermally from the four basic nucleotides, namely 2′-deoxyadenosine 5′-monophosphate (dAMP), 2′-deoxyguanosine 5′-monophosphate (dGMP), 2′-deoxythymidine 5′-monophosphate (dTMP) and 2′-deoxycytidine 5′-monophosphate (dCMP). It was found that purine bases could produce N-dots with higher PL intensity as compared to pyrimidine precursors, among which dAMP dots exhibited the highest fluorescence QY of 12.4%. Besides different types of nucleobases, the effects of the other two sub-units of nucleotide on N-dot synthesis were also examined. Results revealed that apart from the adenine base, the presence of a sugar moiety also enhances the PL intensity. Additionally, the phosphate group was found to enhance the photostability of the resulting N-dots. For example, ADP-dots exhibited the highest photostability, maintaining 97.6% of their PL intensity under continuous UV irradiation (30 minutes). Interestingly, the N-dots also possess unique single oxygen generation properties not found in their original nucleotide precursors. This study provides the first rudimentary design rules to tune the photochemical properties of biodots at the nucleotide level.21
| Precursor | Passivating agent | Sizea (nm) | QYb (%) | Treatment method | Application | λ ex/λem (nm) | Ref. | |
|---|---|---|---|---|---|---|---|---|
| a In cases where the average size of the biodot is not reported, and the range of sizes is reported. b QY is reported in reference to quinine sulfate, unless otherwise stated. c Rhodamine B in ethanol used as the QY standard. | ||||||||
| Amino acids | L-Serine | PEI | 3.83 ± 0.73 | 11.4 | Hydrothermal | Therapeutic | 360/450 | 17 |
| L-Serine with | — | 5 by 5 (film-structure) | 18.25 (61.12 in acid) | Hydrothermal | Detection of H+ and Fe3+ | 380/448 | 39 | |
| L-Tryptophan | ||||||||
| L-Lysine | — | 5 to 10 | 23.3 | Microwave pyrolysis | Bioimaging | 360/432 | 25 | |
| Asparagine | — | 2.9 ± 0.2 | NA | — | Rutin detection | 348/441 | 40 | |
| Histidine | — | 2 ± 0.4 | 44.9 | Microwave pyrolysis | Enhancement of chemiluminescence | 360/430 | 24 | |
| 20 natural amino acids | 2.62–27.66 | Up to 30.44 | Hydrothermal | Bioimaging | 360/450 | 22 | ||
| L-Lysine with ascorbic acid | — | 10–12 | NA | Hydrothermal | — | 360/440 | 23 | |
| L- or D-cysteine with citric acid | — | 4–8 | 31 (L-Cys) | Ultrasonication & hydrothermal | Agricultural | 360/442 | 26 | |
| 33 (D-Cys) | ||||||||
|
L-Cysteine with:L-Serine L-Proline L-Alanine L-Asparagine |
— | 2.66.2 8.1 13.0 |
7.0c8.4c 7.1c 12.3c |
Hydrothermal | Bioimaging | 540/610 | 14 | |
| Tryptophan with formic acid | — | 1.7 | 58.4 | Hydrothermal | One- and two-photon bioimaging | 360/442 (one-photon), 450 (two-photon) | 41 | |
| Mono-/di-saccharides | Glucose | Ammonium hydroxide | 9.21 ± 1 | 6.65 | Ultrasonification | Photocatalyst | 350/435 | 28 |
| Lactose | — | 2–3 | — | Heating with NaOH | Detection of folic acid | 308/420 | 29 | |
| Sucrose | Urea | 1.6 | 1.57–11.03 | Heating in edible oil | pH sensing | 380/440 | 30 | |
| Sucrose | H2SO4 (doping agent) | 2.3 | 5.77 | Acid carbonization | Fe3+ detection | 360/∼440 | 31 | |
| Nucleotides | AMP | — | 2.5 | 33.81 | Hydrothermal | Picric acid detection | 350/421 | 32 |
| The four nucleotides (A, T, G, C) | — | 7 ± 1 (dAMP) | 13.9 | Hydrothermal | PDT | 365/450 (dGMP at 430) | 21 | |
| Other small molecules | Ascorbic acid (vitamin C) | — | 3.1 ± 0.3 | 15 | Hydrothermal & microwave | Bioimaging | 330/405 | 37 |
| Vitamin B1 | Blue: sodium phosphate dodecahydrate | 3.7 ± 0.8 | 75.9 | Carbonization | Bioimaging | Blue: 391/441 | 38 | |
| Green: sodium tripolyphosphate | 5.9 ± 1.8 | 8.5 | Green: 400/470 | |||||
| L-Glutathione | EDA | 2.54 | 40 | Hydrothermal | L/D arginine detection | 320/390 | 33 | |
| Folic acid | Ethylene glycol | 4.5 ± 1 | 15.7 | Hydrothermal | Hg2+ detection | 390/470 | 34 | |
| Citric acid | Beta-alanine | 3.09 ± 0.51 | 21.9 | Microwave pyrolysis | Therapeutic | 335/418 | 42 | |
| Citric acid | EDA | 2–6 | 80 | Hydrothermal | Fe3+ detection | 360/443 | 36 | |
| Uric acid | — | 3.6 | 52.06 | Pyrolysis & ultrasonic | Ag+ detection | 350/402 | 35 | |
In terms of multicolour emission, Bhunia and co-workers have prepared blue and green fluorescent biodots from vitamin B1 in the presence of phosphate salt.38 The emission colours of the two biodots were tuned by varying the type of phosphate salt, reaction temperature, and solvent used in the synthesis. For instance, reacting vitamin B1 with sodium tripolyphosphate in ethylene glycol at 130 °C produced green emissive biodots while the reaction of vitamin B1 with sodium phosphate dodecahydrate in water at 90 °C produced blue emissive biodots. In the case of blue emissive biodots, the fluorescence QY was exceptionally high at 76%. There are three main reasons as proposed by the authors: firstly, vitamin B1 contains many heteroatoms and five unsaturated bonds, which enables carbonization to occur easily, even at lower temperatures. Secondly, incorporation of phosphorus from the phosphate salt introduces more defects in the carbon core. And lastly, carbonization temperature is optimized to 90 °C for blue emissive dots, as higher temperatures were not favourable for the biodot synthesis.38
2.2. Biopolymers
Biological macromolecules such as DNA and proteins have been used to synthesize biodots. Generally, biodots derived from biopolymeric precursors do not require additional passivating agents or dopants, possibly due to the presence of a variety of molecular moieties in the polymeric structure, which enables self-passivation or self-doping. Table 2 summarizes the properties of biodots synthesized from various biopolymers including enzymes, polysaccharides, etc., mainly through hydrothermal treatment and/or microwave heating.| Precursor | Passivating agent | Sizea (nm) | QYb (%) | Treatment method | Application | λ ex/λem (nm) | Ref. | |
|---|---|---|---|---|---|---|---|---|
| a In cases where the average size of the biodot was not reported, and the range of sizes are reported. b QY is reported in reference to quinine sulfate, unless otherwise stated. | ||||||||
| DNA | E. coli DNA | — | 6 | N/A | Hydrothermal | Drug delivery | 366/445 | 44 |
| Salmon sperm DNA powder | — | 30 | N/A | Hydrothermal | Ag+ detection | 275/∼460 | 45 | |
| Poly-cytosine DNA | — | 12–20 | N/A | Hydrothermal | Biothiols detection, glutathione reductase activity detection | 300/390, 290/420 | 46 | |
| Proteins | BSA | TTDDA | 2–6 | 11 | Hydrothermal | Bioimaging | 360/448 | 47 |
| BSA, formic acid | — | 8.32 | 17.10 | Hydrothermal | Bioimaging (nucleus imaging) | 320/407 | 48 | |
| BSA | — | 5.4 ± 2.3 | 14 | Microwave irradiation | Ag+ detection | 360/400 | 49 | |
| Lipase | ||||||||
| Pepsin | ||||||||
| Haemoglobin | — | 4.0 | 3.9 | Pyrolysis and sonication | Bioimaging | 230/380 | 53 | |
| Polysaccharides | Starch | — | 4–5 | N/A | Heating in 0.1 M KOH | F− detection | 350/450 | 54 |
| Citrus pectin hydrogel | — | 2.7 | 1.1 | Hydrothermal | Bioimaging | 360/460 | 52 | |
| Xylan | NH4OH | 7.89 | 4.0 | Microwave | Tetracycline detection | 360/442 | 50 | |
| Xylan | BPEI | 8.2 | 8.0 | Microwave | Tannic acid detection | 340/463 | 51 | |
DNA extracted from different natural resources may give rise to biodots of varying sizes and photoluminescence properties. For example, Song et al. obtained relatively large size biodots (around 30 nm) from the double-stranded DNA of salmon sperm via hydrothermal treatment (160 °C for 4 h).45 The salmon sperm DNA-dots still retained the cytosine and thymine residues, which are highly specific to Ag+ and Hg2+.45 Similarly, Qiu's group reported biodots with an average size of 16 nm and with a fluorescence QY of 1.1% prepared by low temperature heating (80 °C for 12 h) of poly-cytosine (poly-C) DNA.46 The chemical composition of poly-C dots closely resembled that of its DNA precursor. Same as the previous two cases, cytosine rings of the DNA remained intact in the poly-C dots, which enable selective detection of Ag+ and Hg2+ ions in solution. This study shows the precursor sequence dependent-photoluminescence properties of DNA-dots. It is postulated that DNA precursors with higher cytosine content or successive cytosine bases may increase the photoluminescence centers of the resultant DNA-dots.46 Thus, the DNA sequence with more cytosine bases would lead to the formation of DNA dots with higher PL intensity in general.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond formation in the biodots.48
N bond formation in the biodots.48
Microwave treatment is another popular method for protein-dot synthesis. Zhang's group demonstrated the microwave synthesis of biodots using various protein precursors including BSA, lipase and pepsin.49Fig. 4d shows the proposed mechanism for the protein-dot formation. Prior to microwave treatment, proteins were fragmented into peptide chains using denaturing agents such as urea and guanidine hydrochloride. Upon microwave treatment, the peptide chains were cross-linked through intermolecular dehydration. Further heating resulted in intramolecular dehydration, producing C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds, thus creating aromatic centers. Supersaturation of the aromatic centers resulted in nucleation, giving rise to fluorescent protein-dots with a carbon core. No fluorescent biodots were observed if the protein precursors were not denatured first. This is possibly due to the formation of hydrophobic aggregates which hindered the aromatization and nucleation processes that are essential to the formation of fluorescent carbon dots.49
N bonds, thus creating aromatic centers. Supersaturation of the aromatic centers resulted in nucleation, giving rise to fluorescent protein-dots with a carbon core. No fluorescent biodots were observed if the protein precursors were not denatured first. This is possibly due to the formation of hydrophobic aggregates which hindered the aromatization and nucleation processes that are essential to the formation of fluorescent carbon dots.49
2.3. Biomass
Recently, biomass conversion to useful materials has been a hot research topic. This emerging “green chemistry” concept has led to the development of biodots from biomass wastes such as food wastes (e.g., fruit peels55–60 and used coffee grounds61), floral wastes (rose petals62), and even human wastes (e.g., urine63 and hair64–66) as shown in Fig. 3. In addition to the efforts to recycle or upcycle biomass wastes, passivating agents and dopants are less frequently used in the synthesis of biodots, thus saving the use of natural resources. This is due to the fact that biomass contains plenty of heteroatoms such as nitrogen, phosphorus and sulphur that can passivate the surface of biodots. However, as different sources of biomass (e.g., straws, fruits, etc.) have different chemical compositions and concentrations of molecular building blocks, biodots produced from different sources of biomass exhibit vastly different surface functionalities and optical properties.67Table 3 summarizes various treatment methods, such as pyrolysis, sonication, calcination, etc., used to prepare biomass-derived fluorescent dots using plant- or animal-based materials as precursors.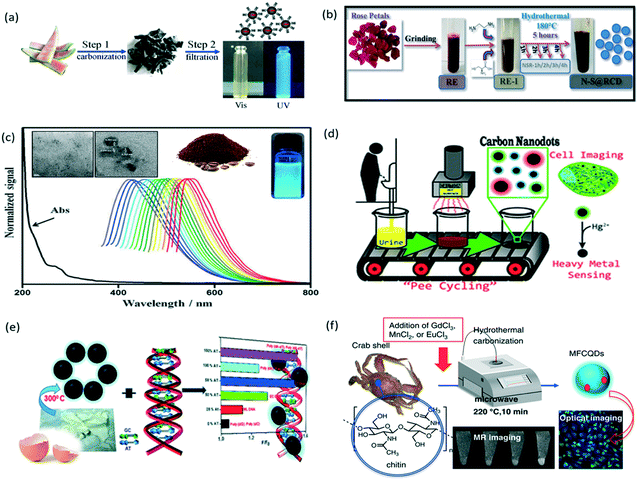 | ||
| Fig. 3 Green synthesis of biomass-derived photoluminescent nanodots from recycling or upcycling bio-wastes: (a) watermelon peel, reproduced, adapted and reprinted with permission from ref. 56; (b) rose petal, reproduced, adapted and reprinted with permission from ref. 62; (c) used coffee ground, reproduced, adapted and reprinted with permission from ref. 61; (d) urine, reproduced, adapted and reprinted with permission from ref. 63; (e) egg shell membrane, reproduced, adapted and reprinted with permission from ref. 68; and (f) crab shell, reproduced, adapted and reprinted with permission from ref. 69. | ||
| Precursor | Passivating agent/surface modifier | Sizea (nm) | QYb (%) | Treatment method | Application | λ ex/λem (nm) | Ref. | |
|---|---|---|---|---|---|---|---|---|
| a In cases where the average size of the biodot was not reported, and the range of sizes are reported. b QY is reported in reference to quinine sulfate, unless otherwise stated. | ||||||||
| Tea, coffee & spices | Used green tea | — | 3.4 ± 0.8 | 4.3 | Calcination | Therapeutic | 345/420 | 82 |
| Used coffee grounds | — | 5 ± 2 | 3.8 | Calcination | Bioimaging | 365/440 | 61 | |
| Saffron | — | 6 | 23.6 | Hydrothermal | Bioimaging | 400/485 | 91 | |
| Prilocaine detection | ||||||||
| Fruit peel | Pomelo peel | — | 2 to 4 | 6.9 | Hydrothermal | Hg2+ detection | 365/444 | 55 |
| Watermelon peel | — | 2.0 ± 0.5 | 7.1 | Low-temperature carbonization | Bioimaging | — | 56 | |
| Lemon peel | — | 9.5 | 11 | Hydrothermal | Carmine quantification | 425/505 | 57 | |
| Orange peel | — | 2–7 | 36 | Oxidation & hydrothermal | Photocatalyst | 340/431 | 58 | |
| Pineapple peel | — | 2–3 | 42 | Hydrothermal | Hg2+ detection | 380/∼460 | 59 | |
| Keypad lock device | ||||||||
| Memory device | ||||||||
| Mango peel | — | 3 | 8.5 ± 0.2 | Calcination | Bioimaging | 310/425 | 60 | |
| Fe2+ detection | ||||||||
| Lemon peel | — | 1–3 | ∼14 | Hydrothermal | Cr6+ detection | 360/∼450 | 81 | |
| Photocatalytic activity enhancer | ||||||||
| Traditional Chinese medicine herbs | Pollen Typhae Carbonisata | — | 4.2 ± 1.4 | N/A | Pyrolysis | Therapeutic | 352/442 | 70 |
| Phellodendri cortex | — | 1.2–4.8 | 9.62 | Pyrolysis | Therapeutic | 363/438 | 71 | |
| Schizonepetae Herba Carbonisata | — | 0.8–4.0 | 2.26 | Pyrolysis | Therapeutic | 355/454 | 72 | |
| Fruit, vegetable, tuber, and root crops | Fresh banana juice | — | 3 | 8.95 | Heating with ethanol | — | 360/460 | 73 |
| Fresh Lantana camara juice | EDA | 5 ± 3 | 33.15 | Hydrothermal | Pb2+ detection | 360/450 | 74 | |
| Fresh lemon juice | — | 5.8 | 10.20 | Hydrothermal | Bioimaging | 365/450 | 92 | |
| Fresh sweet lemon juice | — | 2–4 | N/A | Solvothermal | Antibiofilm agent | 380/∼460 (water) | 76 | |
| 460/∼520 (ethanol) | ||||||||
| Coconut water | — | 2 ± 0.5 (140 °C) | 2.8 | Microwave assisted hydrothermal | Thiamine detection | 290/346 | 75 | |
| 4 ± 1 (160 °C) | 363/435 | |||||||
| 4 ± 2 (180 °C) | 467/519 | |||||||
| Spinach | — | 1.0–5.0 | 53 | Hydrothermal | Bioimaging | 330/405 | 93 | |
| 2-,4-Nitrophenol detection | ||||||||
| Fluorescent ink | ||||||||
| Fresh carrot juice | — | 5.5 | 5.16 | Hydrothermal | Bioimaging | 360/442 | 94 | |
| Sweet potato | — | 3.39 | 8.64 | Hydrothermal | Fe3+ detection | 360 / 442 | 95 | |
| Radish | 4.4 | 15 | Hydrothermal | Cu2+ detection | 360/450 | 96 | ||
| Acetic acid vapour detection | ||||||||
| Tender ginger juice | — | 4.3 ± 0.8 | 13.4 | Hydrothermal | Therapeutic | 325/400 | 97 | |
| Tamarind | — | 1 to 3 | 4 | Hydrothermal | — | 360/442 | 98 | |
| Seeds, flower parts & leaves | Chia seeds | — | 2 to 6 | — | Pyrolysis | Hydrazine detection | — | 77 |
| Acacia concinna seeds | — | 2.5 ± 0.7 | 10.20 | Microwave | Cu2+ detection | 390/468 | 78 | |
| Almond husk | — | 5 to 20 | ∼1.6 | Carbonization & oxidative treatment | Removal of p-nitrophenol dye | 400/550 | 99 | |
| Eggplant sepal | — | 6.11 ± 1.20 | 11 | Hydrothermal | Bioimaging | 390/∼470 | 100 | |
| Bamboo leaves | BPEI | 3.6 | 7.1 | Hydrothermal | Cu2+ detection | 365/440 | 101 | |
| Rosa indica | Ethylenediamine | 4.51 ± 1.46 | 9.6 | Hydrothermal | — | 320/397 | 62 | |
| L-Cysteine | ||||||||
| Rapeseed pollen | — | 5.2 | 7.7 | Sonication & hydrothermal | Fertilizer | 360/432 | 79 | |
| Bee pollen | — | 2.01 | 7.7 | Sonication & hydrothermal | Fertilizer | 340/∼440 | 80 | |
| Other plant biomass | Wheat straw | — | 1.7 ± 0.23 | 9.2 | Hydrothermal | Bioimaging | 304/418 | 102 |
| Fe3+ detection | ||||||||
| Plant soot | TTDDAPoly lysine Cysteine Chitosan |
3.1 | 0.72 (bare)1.10 4.28 1.44 2.05 |
Chemical nitric acid oxidation | Bioimaging | 320/430 | 103 | |
| Larch | — | 20.35 | 13.4 | Hydrothermal | Photosensitizer (photocatalyst) | 350/440 | 83 | |
| 16.4 | 14.9 | 330/420 | ||||||
| 11.9 | 16.6 | 320/380 | ||||||
| 6.48 | 18.1 | 280/365 | ||||||
| Processed material wastes | Paper | — | 4.8 | 10.8 | Hydrothermal | Bioimaging | 360 / 449 | 84 |
| Paper | 3-Aminopropyl | 4.5 | ∼5.1 | Hydrothermal | Trinitrotoluene detection | 350/450 | 85 | |
| Triethoxylane | ||||||||
| Egg | Egg white | — | 2.1 | 43 | Hydrothermal | Bioimaging | 315/420 | 86 |
| Fe3+ detection | ||||||||
| Egg yolk oil | — | <10 | 5.01 | Pyrolysis | Bioimaging | — | 87 | |
| Egg shell membrane | — | 3.35 ± 0.5 | 8 | Pyrolysis & ultrasonication | Bioimaging | 350/420 | 68 | |
| Cow Milk | Milk | — | 3 | NA | Microwave cooking | H2O2 sensing | 365/∼450 | 89 |
| Pro-drug | ||||||||
| Milk | — | <10 | 12 | Hydrothermal | Therapeutic | 360/444 | 16 | |
| Milk | — | 5 ± 0.07 | 9.68 | Hydrotherm al | DNA recognition | 380/466 | 88 | |
| L-Cysteine | 4 ± 0.07 | 10.38 | 340/430 | |||||
| Urea | 3 ± 0.07 | 15.39 | 320/435 | |||||
| Body wastes | Urine | — | 20.6 ± 8.4 | 5.3 | Thermal | Bioimaging | 325/392 | 63 |
| Vitamin C | 11.4 ± 6.6 | 4.3 | Hg2+ detection | 350/427 | ||||
| Asparagus | 38.8 ± 20.6 | 2.7 | 425/500 | |||||
| Human hair | — | 2.3 | 17.3 | Pyrolysis & ultrasonication | Preparation of microbeads & microfibers | 360/430 | 65 | |
| Human hair | — | 49 | 24.8 | Hydrothermal | Hg2+ detection | 340/433 | 64 | |
| Human hair | — | 4.56 | 10.75 | Pyrolysis & ultrasonication | Hg2+ detection | 330/415 | 66 | |
| Seafood waste crab shell | Gd3+ | 4.0 ± 0.7 | 19.84 | Microwave assisted pyrolysis (hydrothermal) | Theranostic agents | 350/425 | 69 | |
| Mn2+ | 4.5 ± 1.0 | 12.86 | ||||||
| Eu3+ | 4.2 ± 0.6 | 4.97 | ||||||
2.3.1.1. Fruit peels and floral wastes. Fruit peels are also popular biomass waste materials that have been explored for the synthesis of photoluminescent biodots. For example, pomelo peels,55 watermelon peels (Fig. 3a),56 and two kinds of lemon peels57,81 were used as sole precursors to synthesize fruit-peel biodots with fluorescence QYs of 6.9%, 7.1%, 11%, and 14%, respectively. When dried orange peels had been oxidized with sodium hypochlorite before hydrothermal treatment, fluorescent biodots with a QY of 36% was produced.58 Using ethanol as a solvent, Vandarkuzhali and co-workers recently reported the synthesis of biodots from pineapple peels with a fluorescence QY as high as 42%,59 showing the possibility to improve the brightness of biodots by changing reaction conditions or pre-treatment methods.
For the utilization of floral waste, Sharma et al. reported the bioinspired synthesis of photoluminescent nanodots from rose petals.62 As shown in Fig. 3b, the petals of rose flower were first crushed in a domestic mixer to obtain rose extract (RE), which was then mixed with ethylenediamine and L-cysteine to form a homogeneous RE-1 solution, followed by 5 h hydrothermal treatment at 180 °C to form the rose-dots with interesting optical properties viz. appreciable fluorescence QY, good photostability and wavelength tuned emission.
2.3.1.2. Coffee and tea. Chang's group synthesized amorphous biodots from used coffee grounds61 and used green tea leaves82via autoclave calcination. The biodots derived from coffee grounds have a mean size of (5.0 ± 2.0) nm and a fluorescence QY of 3.8% (Fig. 3c). Interestingly, C–N stretching and C–H stretching peaks, which are the characteristic peaks for the caffeine structure, were identified by FTIR analysis of the coffee-ground dots. It is suggested that the removal of hydrophobic molecules from coffee grounds prior to the synthesis would enhance the fluorescence QY of biodots.61 This observation hints the importance of extraction and purification of the basic building blocks from plant wastes. On the other hand, fluorescent biodots derived from used green tea exhibited an average size of (3.4 ± 0.8) nm and a QY of 4.3%. Most importantly, these green tea-dots show good efficacy in inhibiting the growth of breast cancer cells. FTIR analysis indicated that the catechins (form more than 30 wt% of green tea) were adsorbed on the surface of green tea-derived biodots, rendering them with excellent anti-cancer properties.82 These two examples demonstrated the successful integration of the biological functions and molecular structure of natural precursors to the as-synthesized biodots.
2.3.1.3. Herbs. Traditional Chinese medicine (TCM) ingredients are another popular choice of precursors for biodot synthesis with good biocompatibility and therapeutic effects. Charcoal drugs have long been used as a medicine for haemorrhagic conditions.72 Having noticed that biodots are usually formed as a by-product during the processing of herbs to produce charcoal drugs, several groups have employed a modified pyrolysis method to obtain fluorescent biodots from herbs such as Schizonepetae Herba Carbonisata,72Typhae Carbonisata70 and Phellodendri Cortex.71 In these studies, biodots were obtained from the aqueous extract of the herbs calcined at 350 °C. More importantly, these TCM-derived biodots exhibited good haemostatic effect, indicating the inheritance of intrinsic medicinal functions from the herbal precursors.
2.3.1.4. Larch. Liu's group conducted a systematic study to examine the effect of the hydrothermal reaction temperature on the photoluminescent properties of biodots using larch as a precursor.83 The as-produced biodots exhibited different sizes, lattice structures and fluorescence QYs. It was observed that the biodots synthesized at 200 °C exhibited a mean diameter of 20.4 nm with an obvious stripe-like hexagonal structure on the surface, while smaller biodots (6.5 nm) were produced at 260 °C, showing an ordered cubic structure. An increase in temperature not only resulted in a decrease in average size and carbon content, but also an increase in fluorescence QY and oxygen content of the biodots. These may be due to the promotion of carbonization at higher reaction temperatures, leading to formation of a more compact nanostructure, which may also affect the surface properties of the larch-derived dots.83
2.3.2.1. Chicken eggs. Various parts of eggs have been utilized for the synthesis of biodots. Zhang et al. used egg white in hydrothermal treatment (220 °C, 48 hours) to obtain biodots with an average diameter of 2.1 nm and a high fluorescence QY of 43%.86 Zhao and co-workers obtained biodots from egg yolk by mimicking the conditions for preparing egg yolk oil in Chinese medicine.87 The egg yolk powder was heated up to 260 °C in a furnace over an hour, and the temperature was maintained for another hour. Upon cooling, egg yolk biodots were extracted with water and purified through dialysis, exhibiting a fluorescence QY of 5.01%.87 On the other hand, Devi's group made use of the egg shell membrane to synthesize biodots through a combination of pyrolysis and sonochemical method (Fig. 3e).68 The extracted egg shell membrane was heated in a muffle furnace for 4 hours at 300 °C, ground and dispersed in water, and then ultrasonicated for 2 hours, before removing the large particles by centrifugation. The as-synthesized egg shell biodots have an average size of 3.4 ± 0.5 nm and a fluorescence QY of 8%.68
2.3.2.2. Cow milk. Milk is an important source of food which contains nutrients like proteins and lactose. The use of milk has attracted much attention as a precursor to tune colour fluorescence of biodots. Wang, L. et al. reported the synthesis of blue emissive biodots from milk with a size of 3 nm and with a good fluorescence QY of 12% by hydrothermal treatment (180 °C, 2 h).16 In the same year, Qin's group reported the use of L-cysteine (sulfur-dopant) or urea (i.e. nitrogen-dopant) with milk for the hydrothermal synthesis of blue-green emissive biodots (180 °C, 8 h).88 Results show that doping helps to enhance the PL intensity, where the N-doped biodots exhibited the highest fluorescence QY (15%) among those synthesized from milk, L-Cys + milk and urea + milk, respectively. The fluorescence QY is found to be inversely correlated to the size of milk-dots (i.e., higher QY, smaller size) with a blue-shift in excitation and emission maxima as shown in Table 3.88 Besides, a rapid microwave heating method (with time intervals ranging from 30–150 s) has also been developed to form milk-dots with sizes between 1 and 7 nm.89
2.3.2.3. Human body wastes. In addition to animal sources, human body wastes such as urine and hair have been reported as useful precursors for biodot synthesis. As shown in Fig. 3d, Essner et al. reported the synthesis of “pee-dots” via thermal upcycling of urine (containing urea and uric acid), which involved heating dehydrated urine at 200 °C for 12 h followed by reconstitution in water.63 Dietary modifications, such as inclusion of ascorbic acid and asparagus, have also been found to affect the size, sensory and fluorescence QY of the pee-dots. In 2014, Hou et al. reported the use of human hair as a precursor to form photoluminescent biodots with a QY of 24.8% from hydrothermal synthesis.64 In the same year, Liu and co-workers reported the synthesis of biodots from thermal treatment of hair at 300 °C for 2 h in a nitrogen atmosphere, followed by resuspension in different solvents using ultrasonication.65 It was revealed that the PL intensity of the hair-dots increased with increasing solvent polarity.65 In another case, Guo et al. obtained hair-dots via thermal treatment (QY of 10.75%), and noted that the solid-phase synthesis produced biodots with a lower fluorescence QY than the hydrothermal synthesis, possibly due to inferior interactions during the carbonization process.66
2.3.2.4. Seafood wastes. Seafood wastes such as fish scales90 and crab shells69 have been utilized for biodot synthesis. Notably, crab shells in combination with transition metal ions such as Gd3+, Mn2+ and Eu3+ produced magnetofluorescent biodots (Fig. 3f). The crab shell derived biodots synthesized together with the transition metals (i.e., Gd3+, Mn2+ and Eu3+) showed good magnetic properties and relatively high fluorescence QYs (19.84%, 12.86%, and 14.97%, respectively), which enable dual-modality bioimaging (fluorescence magnetic resonance imaging).69
2.4. Formation mechanisms of biodots
The formation mechanisms of biodots derived from different natural sources of precursors are summarized in Fig. 4. The well-accepted formation mechanism of biodots derived from small biomolecules typically consists of a series of reactions (Fig. 4a and b).21,67,104,105 Firstly, intermolecular condensation reactions occur to link up small molecules. Secondly, the linked molecules are further polymerized through formation of covalent bonds or via non-covalent interactions, which are subsequently carbonized to produce a carbon core. Sometimes, passivating agents are used to passivate the carbon surface in enhancing the photoluminescence of the as-synthesized biodots.21,67,104,105 The synthesis of biodots from basic biomolecules, i.e., nucleotides and amino acids may offer better control over the reactions to produce more uniform biodots.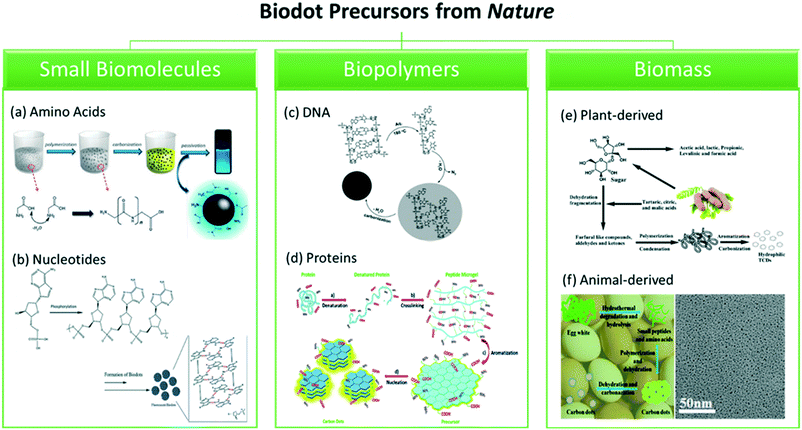 | ||
| Fig. 4 Proposed formation mechanism of biodots derived from different natural resources. (Left) Small biomolecules: (a) amino acid, glycine, reproduced, adapted and reprinted with permission from ref. 105, and (b) nucleotides, reproduced, adapted and reprinted with permission from ref. 21. (Middle) Biopolymers: (c) DNA, reproduced, adapted and reprinted with permission from ref. 44, and (d) proteins, reproduced, adapted and reprinted with permission from ref. 49. (Right) Biomass: (e) tamarind, reproduced, adapted and reprinted with permission from ref. 98, and (f) egg white, reproduced, adapted and reprinted with permission from ref. 86. | ||
As compared to small molecules, polymeric precursors such as DNA (Fig. 4c) and proteins (Fig. 4d) usually require a dehydration or denaturation process to unfold their original structures, while still keeping majority of the covalent bonds. The scaffolds then cross-link and polymerize before aromatization occurs. From the previously discussed biopolymer studies, there are distinctive differences (in terms of molecular structures) between the DNA and protein precursors used in the synthesis of biodots. In the cases of DNA-dots, heterocyclic rings of the DNA precursors remain intact upon forming the biodots under tested reaction conditions, whereas protein precursors are usually highly degraded into reaction products, and subsequently aromatized to form fluorescent biodots. Since denaturation is required for the protein dot synthesis, it might be more cost-effective to design the biodots by selecting appropriate combination of amino acids and reaction conditions.
In general, formation of biodots from biomass mainly involves polymerisation of molecules present in the biomass followed by carbonization.67 However, the top-down synthesis of biodots involving the use of plant or animal sources is more sophisticated and complex because the formation process requires a greater extent of decomposition to release the required intermediates to form the carbon core (Fig. 4e and f). Due to the rich functionalities and heteroatoms present in the biomass, passivating agents and dopants are seldom used in the synthesis of biodots. Furthermore, plenty of biomass resources could provide greater opportunities to form multifunctional biodots with biomimetic functions or intrinsic therapeutics effects inherited from their original sources of natural precursors such as herbs, and could turn the biomass wastes into useful nanomaterials for various applications which will be discussed in the following sections.
3. Biomedical applications of biodots
Biodots exhibit bright and stable photoluminescence with tunable emissions, which render them extremely suitable for bioimaging applications. Owing to their ultrasmall size, excellent biocompatibility and good aqueous solubility, they are also considered a good option as drug or gene carriers. Most notably, many of these biodots inherit the therapeutic properties from their natural precursors (e.g., hemostasis) which makes them great candidates for theranostic applications. In the following sections, we will illustrate the successful utilization of biodots in various biomedical applications.3.1. Bioimaging
Biodots have emerged as new fluorescent probes for biological imaging due to their inherent biocompatibility, excellent water solubility and photostability, as well as ultra-small size which enable fast cellular uptake. Different types of cells, including bacteria cells (e.g., E. coli,102A. niger75 and C. elegans102), animal cells (e.g., rat bone marrow,91 rat olfactory mucosa,91 porcine LLC-PK1,61 CHO53,103 and mice embryonic fibroblast63) and human cells (e.g., HeLa,53,56,86,89,100 mammary Hs-578,37 mammary gland breast/duct carcinoma BT-474,63 breast cancer Bcap-37,37 brain glioma tumor U87 (ref. 16)) as well as the transport system of Rome lettuce (Latuca sativa L.)79 have been successfully imaged using various biodots derived from different natural resources. Ex vivo imaging of guppy fish has also been reported.103 These biodots are believed to enter the cells through endocytosis.37,61,100 More specifically, the positively-charged biodots utilize the clathrin-mediated and/or caveolae-mediated endocytosis pathways to enter the cells.42One of the important considerations in synthesizing biodots for bioimaging applications is the choice of passivating agents, especially when small molecules are used as precursors. It has been reported that the use of passivating agents such as branched polyethyleneimine (BPEI) and poly(acrylic acid) (PAA) could cause cytotoxicity to the biodots,25 which may pose limitations on some applications such as in vivo imaging. Nevertheless, some passivating agents are non-toxic. For example, biodots passivated with polyethylene glycol (PEG) and poly(propionylethyleneimine-co-ethyleneimine) (PPEI-EI) were safe to use at the levels required for in vivo animal studies.15 Furthermore, the surface charge of biodots could be tuned by using passivating agents. For instance, Sachdev et al. showed that hydrothermal treatment of chitosan with either PEI or PEG produced biodots with positive (CD-PEI) and negative charges (CD-PEG), respectively.106 Since the cell membrane is negatively charged, it was found that the CD-PEI was able to bind to the cell membrane more efficiently than the CD-PEG, demonstrating that positively-charged passivating agents could be used to improve the uptake of biodots by cells for bioimaging.106
In addition, some biodots have demonstrated the potential for nuclear-targeting bioimaging. Jung et al. reported zwitterionic biodots which could first enter the cytoplasm of HeLa cells by endocytosis via their positively-charged surface moieties, then translocate into the nucleus by utilizing the negatively-charged moieties.42 On the other hand, Wang et al. reported biodots with zwitterionic surface properties that could enter the HeLa cells through passive penetration without utilizing the endocytotic pathway.100 Chemical modifications have also been made to endow the biodots with specific nuclear-targeting abilities. For example, Song and co-workers reported one- and two-photon nucleus imaging of mouse melanoma B16-F10 cells by conjugating biodots derived from tryptophan and formic acid to trans-activator of transcription (TAT) peptides.41 TAT peptide-conjugated biodots (TAT-CDs) effectively entered the nucleus, enabling nucleus imaging (Fig. 5).
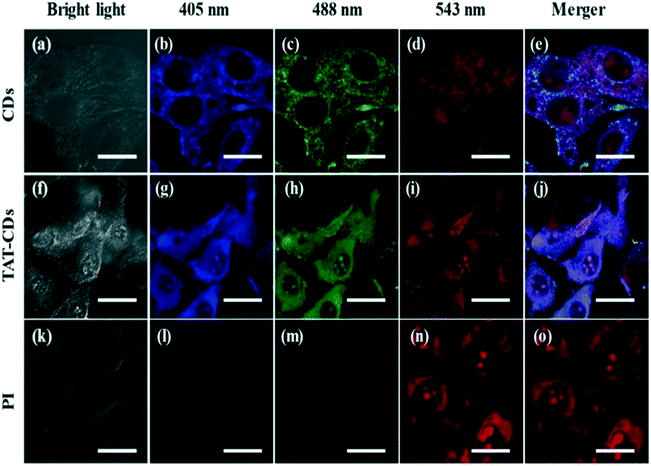 | ||
| Fig. 5 One-photon photoluminescence imaging of mouse melanoma B16-F10 cells using (a–e) biodots (CDs) derived from tryptophan and formic acid, (f–j) TAT-peptide-conjugated biodots (TAT-CDs), and (k–o) propidium iodide (PI) at various excitation wavelengths. PI served as a positive control for nucleus imaging. Reproduced, adapted and reprinted with permission from ref. 41. | ||
Other than targeting specific intracellular organelles, biodots can also be molecularly engineered to target specific cell types. Recently, our team has successfully synthesized two types of blood cell selective biodots by mixing a hydroxyl containing amino acid and a charged polymeric precursor at an optimized ratio in a hydrothermal reaction (Fig. 6a).107 The type I biodots are passivated by polyethylenimine (PEI), which have been shown to label white blood cells (WBCs) with much higher fluorescence intensity than the red blood cells (RBCs). This excellent contrast between WBCs and RBCs in both confocal microscopy and flow cytometry enables lysis-free counting of WBCs and three-part leukocyte differential analysis in whole blood. On the other hand, the chitosan-passivated type II biodots selectively lyse RBCs with >98% efficiency to allow simultaneous fluorescence labelling of WBCs for efficient total leucocyte counting without a tedious lysis step. The multifunctional amino acid-polymer biodots developed in this work achieved high efficiency in WBC labelling, imaging and differential counting in whole blood towards faster and cheaper disease diagnosis in one test.107
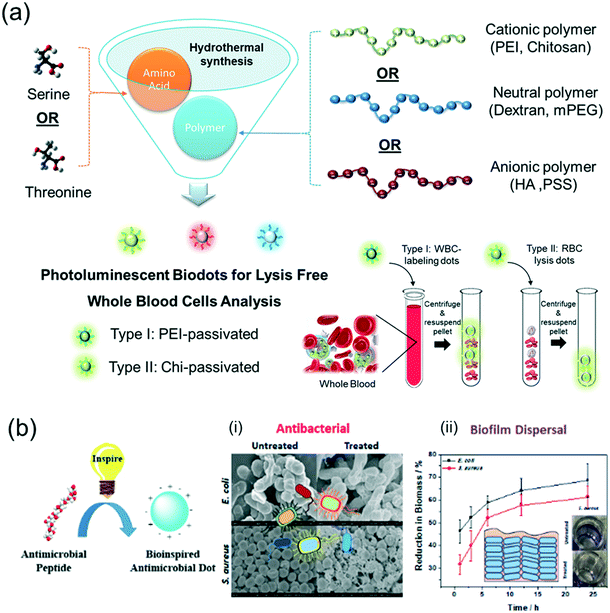 | ||
| Fig. 6 Molecular engineering of multifunctional amino acid-polymer biodots with intrinsic biological and/or biomimetic properties via facile hydrothermal reactions. (a) Blood-cell selective biodots for lysis-free whole blood analysis. Reproduced, adapted and reprinted with permission from ref. 107. (b) Bioinspired antimicrobial nanodots (BAM-dots). (i) SEM images of untreated E. coli and S. aureus, and BAM-dot treated E. coli and S. aureus for 1 hour at MIC. (ii) Biofilm dispersal study of the BAM-dots against E. coli and S. aureus at 2× MIC. Inset: Photos of untreated and BAM-dot treated bacteria stained with crystal violet. Reproduced, adapted and reprinted with permission from ref. 17. | ||
3.2. Theranostics
Theranostic agents are those that could be used for diagnosis of a medical condition while exhibiting therapeutic efficacy.108 On top of the excellent photoluminescence properties of biodots for medical imaging, they can also be molecularly engineered to exhibit therapeutic functions such as singlet oxygen generation for photodynamic therapy21 or drug/gene nanocarriers42,109–111 that enable monitoring of therapeutics distribution within the human body. Some herb-derived biodots have also been reported to possess effective pharmacological properties. Fig. 7 shows some examples of photoluminescent biodots with intrinsic or engineered antimicrobial, anticancer and pharmacological functions.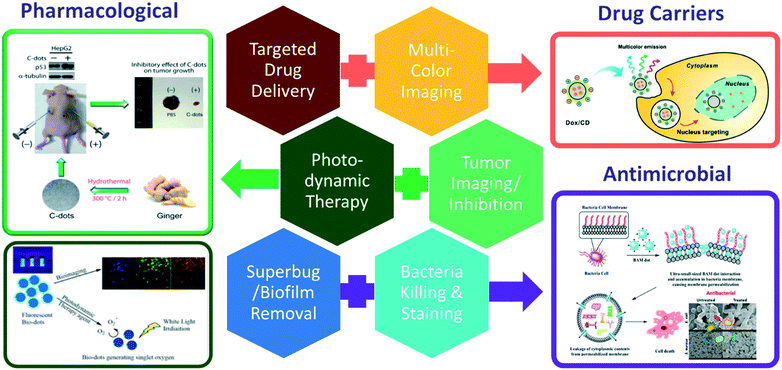 | ||
| Fig. 7 Theranostic applications of biodots as drug carriers (targeted drug delivery and nucleus tracking reproduced, adapted and reprinted with permission from ref. 42), antimicrobial agents (bacterial staining, killing and biofilm removal, reproduced, adapted and reprinted with permission from ref. 17) and pharmacological agents (top: tumor inhibition and imaging, reproduced, adapted and reprinted with permission from ref. 97; bottom: Photodynamic therapy and multicolor imaging, reproduced, adapted and reprinted with permission from ref. 22). | ||
3.2.1.1. Doxorubicin drug carriers. Doxorubicin (Dox) is an anti-cancer drug used for cancer treatment by inhibiting the synthesis of nucleic acids.109 There are several reports of using biodots as Dox carriers to improve cellular and nucleus uptake of Dox.42,69,109–111 In these reports, Dox molecules were either conjugated covalently onto the biodots,69,109 or loaded through non-covalent interactions with the biodots.42,110,111 All these reports clearly show that biodots are effective carriers for Dox to enhance its delivery into the nucleus of cancer cells as these biodots could penetrate into the nucleus effectively (Fig. 7, right top).109–111
3.2.1.2. Platinum-based drug carriers. Platinum-based drugs have also been loaded onto biodots. For example, Xie's group conjugated a platinum-based anticancer prodrug, Oxa(IV)-COOH, onto biodots derived from citric acid and polyenepolyamine (passivating agent) in order to produce theranostic nanoparticles (CD-Oxa) to enable drug monitoring.19 Since the theranostic biodots did not have specific selectivity towards cancer cells, CD-Oxa was injected directly into the hepatocarcinoma tumor xenografted on the Chinese Kunming mice, and monitored through fluorescence imaging. It was observed that the PL intensity of theranostic dots in the tumour site faded over time (no fluorescence observed after 72 hours). The tumor volume was also monitored and found to be reduced by 82.4% after 72 hours.19
3.2.1.3. siRNA gene carriers. Multifunctional theranostic biodots have been developed by Wu and coworkers as effective gene carriers to release small interference RNAs (siRNAs) preferentially in lung cancer cells.20 First, positively charged biodots with a mean size of 9.0 nm were produced from glycerol and PEI via microwave pyrolysis. The PEI molecules were then conjugated with disulfide-containing N,N′-bis(acryloyl)cystamine (BAC) to produce reducible PEI-Cdots (rPEI-Cdots). Once taken into the cancer cells, disulfide bonds could be reduced to release siRNA. In order to impart cancer cell selectivity to nanocarriers, rPEI-Cdots were further functionalized with folic acid to produce fc-rPEI. Folic acid acted as a targeting agent because one of the characteristics of cancer cells is the increase in the expression of the folate receptor. The negatively charged siRNA was then loaded onto the positively charged rPEI-Cdots via electrostatic attraction. From PEI-Cdots to fr-rPEI-Cdots, there was a significant increase in both the particle size from 9.0 nm to 143.1 nm and surface charge from 4.4 mV to 25.7 mV. The increased positive charge further enhanced the loading of siRNA onto the fr-rPEI-Cdots. As anticipated, significantly greater cellular uptake of fr-rPEI-Cdots@siRNA was observed in lung cancer cells via receptor mediated endocytosis in contrast to that in healthy fibroblasts. Upon absorption, the endosomes fuse with lysosomes, causing fr-rPEI-Cdots@siRNA to break down. The siRNAs then undergo endosomal escape. In vivo study in nude mouse models showed that the mean size of the H460 tumour planted on the nude mice treated with the biodot-based gene carrier was significantly smaller than that of the control, with a large amount of nanocarriers accumulated in the lungs.20
3.2.1.4. Mitochondria-targeting drug carriers. Wu's group reported the hydrothermal synthesis of biodots with a mean size of 2.1 nm from chitosan, ethylenediamine and mercaptosuccinic acid which could target mitochondria without the need for conjugation with other mitochondria-targeting molecules.112 The as-synthesized biodots possess a positively charged surface (+28.1 mV) and lipophilic characteristic with an adequate degree of delocalization within the conjugated structure, which enhanced their absorption due to the negative potential inside the transmembrane of mitochondria. Fluorescence imaging of MCF-7 breast cancer cells showed that the biodots were concentrated in mitochondria and only a small amount was found in other organelles such as lysosomes, Golgi body and endoplasmic reticulum. It was reported that the biodots were absorbed via energy-dependent cave-in-involved endocytosis. The mitochondria-targeting biodots were then loaded with Rose Bengal (RB), a common photosensitizer, for photodynamic therapy (PDT). For MCF-7 cells treated with RB-loaded biodots, there was a sharp decrease in cell viability (from 100 to 11%) when irradiated with a 30 mW laser. When MCF-7 cells were treated with a free form of RB, the cells remained 100% viable upon laser irradiation. This result shows that the RB-loaded biodots could significantly increase the photodynamic efficacy of the drug, which might be due to the enhanced cellular uptake to the targeted action site (i.e., mitochondria).112
3.2.2.1. Anticancer and tumor inhibition. Some biodots have shown excellent anticancer properties to inhibit the growth of cancer cells or tumors by encouraging apoptosis.82,97 For example, biodots synthesized from used green tea can inhibit up to 80% (relative to catechin) of cancer cell growth tested on MCF-7 and MDA-MB-231 breast cancer cell lines, respectively, while maintaining low toxicity in normal MCF-10A cells. The inhibitory effect was achieved by increasing the production of reactive oxygen species (ROS), which in turn encouraged apoptosis.82 As shown in Fig. 7 (left top), biodots synthesized from fresh tender ginger juice demonstrated selective tumor growth inhibition (IC50 = 0.35 mg mL−1) in hepatocellular carcinoma (HepG2) cells by increasing the p53 expression in HepG2 cells, leading to apoptosis.97 Most importantly, the inhibitory effect was not observed in healthy hepatocytes (FL83B) and mammary epithelial cells (MCF-10A), as well as in other tested human cancer cell lines including lung cancer cells (A549), breast cancer cells (MDA-MB-231) and cervical cancer cells (HeLa), demonstrating good selectivity for hepatocellular carcinoma cells. Moreover, the ginger-dots have successfully inhibited the HepG2 tumor growth in nude mice.97
The possibility of using biodots as anticancer prodrugs has also been reported. Chakraborty et al. synthesized blood-dots to cause apoptosis of cancer cells three-fold higher than that in normal cells.53 This pharmacological effect is due to an increased level of hydrogen peroxide (H2O2), which is one of the biomarkers of cancer due to overexpression of superoxide dismutase in cancer cells. Since the biodots derived from blood contained Fe2+, the Fenton reaction converted H2O2 into reactive oxygen species (ROS), which killed cancer cells by causing oxidative damage to the cancer cell's DNA.53
3.2.2.2. Haemostatic effects. As mentioned in the previous section, traditional Chinese medicine (TCM) ingredient-derived biodots exhibit intrinsic pharmacological functions inherited from their precursors.70–72,87 For instance, egg yolk oil,87Pollen Typhae Carbonisata (Pu Huang),70 and Phellodendri Cortex (Huang Bo)71 synthesized biodots can all be used as haemostatic agents. Tail amputation and liver laceration studies in mouse models found that the aforementioned TCM-derived biodots were as effective as haemocoagulase in inducing hemostasis.70–72,87 Examinations of coagulation parameters further revealed that these biodots were able to exert haemostatic effects by triggering intrinsic coagulation and fibrinogen systems.70,71,87 Zhang et al. observed similar results in biodots derived from Schizonepetae Herba Carbonisata (Jingjietan), except that their haemostatic effects were exerted through extrinsic coagulation and fibrinogen systems.72 These findings indicate the enormous potential of developing multifunctional biodots from medicinal plants such as charcoal drugs for theranostic applications.
3.2.2.3. Photodynamic therapeutics. In photodynamic therapy, cancer cells are locally killed by ROS produced by a photosensitizer under white light illumination. Very recently, our group has developed a new type of fluorescent biodots derived from nucleotides (N-dots) with intrinsic singlet oxygen (1O2) generation capability for PDT treatment and multicolour bioimaging.21 Among the N-dots, it was found that the deoxyadenosine monophosphate (dAMP)-dots display high photoluminescence properties (QY = 12.4% and photostability = 91.4%) with a remarkable 1O2 quantum yield of 1.20, surpassing the 1O2 quantum yield of many conventional photosensitizers, including Rose Bengal (0.75), riboflavin (0.54), porphyrins (0.44–0.85) and Ce6 (∼0.63). Incubating dAMP-dots with HeLa cancer cells reveal that 10 min of white light illumination is sufficient to kill more than 60% of the cancer cells. This work provides a facile way to develop multifunctional biodots with intrinsic PDT functions for non-invasive cancer treatment.
Recently, biodots derived from Citrus limetta (pathogenic yeast) have demonstrated antibiofilm ability in Candida albicans.76 Infections by Candida begin when the yeast adheres to a hydrophobic surface, followed by formation of biofilms. As such, patients who have prosthetic devices or catheters are more vulnerable to the infection. Shaikh et al. showed that when polystyrene surfaces were coated with biodots, Candida albicans were not able to adhere to the surface of the coated surface, thereby preventing biofilm formation.76
4. Agricultural and environmental applications of biodots
Apart from biomedical applications, biodots derived from natural precursors with negligible toxicity, good photostability and water solubility are also good for agricultural and environmental applications, ranging from ultrasensitive fluorescent sensors for detecting water pollutants or industrial wastes (e.g., heavy metals, pesticides, pyridine, etc.) to effective biodot based fertilizers for plant growth enhancement (Fig. 8).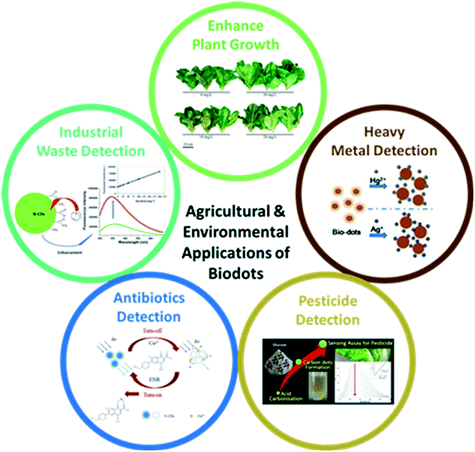 | ||
| Fig. 8 Agricultural and environmental applications of biodots for (clockwise) enhanced plant growth (reproduced, adapted and reprinted with permission from ref. 79), heavy metal detection (reproduced, adapted and reprinted with permission from ref. 45), pesticide detection (reproduced, adapted and reprinted with permission from ref. 113), antibiotics detection (reproduced, adapted and reprinted with permission from ref. 114) and industrial waste detection (reproduced, adapted and reprinted with permission from ref. 115). | ||
4.1. Biodots as fertilizers to promote plant growth
A good fertilizer should not only enhance plant growth, but also possess qualities such as negligible toxicity, good bioavailability, low cost and large-scale production. Biodots which contain significant amounts of carbon and nitrogen are small enough to be absorbed by plant cells. In addition, biodots are water-soluble, non-toxic and biocompatible. Most importantly, they are cheap to produce on a large scale from a variety of carbon sources available in Nature. As such, biodots may become promising alternatives to commercial fertilizers in use today.For the biodots derived from bee pollen, a lower concentration of the biodot-based fertilizer (i.e., 3.5 mg L−1) is sufficient to significantly promote the cabbage growth, which is measured through parameters such as fresh and dry weight, amount of chlorophyll a and b, and soluble sugar content. The rate of photosynthesis was also increased due to the ribulose bisphosphate regeneration, which was correlated with the nitrogen content in the leaves. The bee pollen-dots also improve the permeability of the cell membrane in these plants, as evidenced by an increase in the potassium content. Since nitrate uptaken by plants is influenced by potassium ions, greater absorption of potassium may have caused a decrease in the nitrate content (5.62% lower as compared to that in the control group).80
4.2. Biodots for environmental sensing
In addition to bioimaging applications, biodots with tailored target specificity have been used to detect chemical and biological species in the environment. Typically, these target analytes (metal ions or antibiotic molecules) are quantified by measuring photoluminescence (PL) signal changes, through fluorescence “turn-off”, “turn-on”, and/or enhancement mechanisms. In a fluorescence “turn-off” assay, the analyte itself is a quencher of the biodots, leading to quenching of the PL intensity in a concentration dependent manner. While for the case of “turn-on” assays, the fluorescence of biodots is first quenched by a suitable quencher (e.g., metal nanoparticle) to form the sensing probes. The analyte then interacts with either the quencher or biodots to competitively displace the quencher in a concentration-dependent manner. In some cases, the analyte of interest causes the enhancement of fluorescence signal in a linear correlation. In the following sections, we will discuss different examples utilizing biodots as the sensing elements for detecting environmental hazards.In addition, there are methods which employed more intricate designs to detect target-specific metal ions based on unique recognition properties of biomolecules. For example, Zhu's group45 used biodots synthesized from DNA to detect environmental pollutants, including Ag+ and Hg2+ in water samples based on their affinity interactions with specific DNA bases. In this case, fluorescence quenching occurred when Ag+ and Hg2+ interacted with DNA bases thymine (T) and cytosine (C) in the DNA-dots to form T–Hg2+–T and C–Ag+–C complexes, leading to highly selective detection of metal ions due to the complexation-caused quenching mechanism.45
Other analytical methods, such as UV-VIS spectrometry and turbidimetric analysis, have also been employed for metal ion detection using biodots as the sensing elements. For example, Kolekar's group reported that addition of Fe3+ to the sucrose-derived biodots not only causes the fluorescence quenching, but also increases the absorption signal that enables quantification via simple UV-VIS spectroscopy.31 On the other hand, Renuka's group used biodots synthesized from table sugar in turbidimetric analysis for quantification of Pb2+ ions in water samples.116 The presence of lead ions causes the table sugar-dots to aggregate, leading to the quantification of Pb2+via turbidity measurements. This turbidity method is more sensitive than the previously discussed fluorescence quenching method74 for detecting Pb2+ ions in a low ppb range (i.e., LOD of 14 ppb).116
4.2.2.1. Detection of pesticides. Chang et al. reported the detection of paraoxon-ethyl pesticide extracted from spiked plant leaves using biodots derived from sucrose via acid carbonization.113 In order to record the conditions of pesticide-sprayed plants in farming, the authors placed the spiked plants in a fume hood overnight, allowing evaporation of the pesticides to take place. It was found that only 0.02% of pesticides remained in the extract as compared to the original spiked concentration in the leaves. Upon addition of the extracted pesticide, the PL intensity of the sucrose-dots quenched in a concentration-dependent manner up to 5.80 mM, with a LOD of 0.22 μM. The real-sample recovery calculated with the aid of the HPLC method was 94.8%.113
4.2.2.2. Detection of herbicides. Wang et al. successfully designed a fluorescence “turn-on” assay for the detection of glyphosate herbicides in cereals using wool-derived biodots.117 The biodots with an average size of 2.8 nm and a fluorescence QY of 16.3% were synthesized from the microwave pyrolysis of wool. Silver nanoparticles (AgNPs) were used to quench the wool-dots via the inner filter effect. In the presence of glyphosate, the PL intensity of AgNP quenched wool-dots was recovered due to the strong electrostatic interactions of glyphosate (negatively charged) with the AgNPs (positively charged) to form aggregates, releasing the biodots from AgNPs. The linear range of detection occurs at 0.025–2.5 μM mL−1 of glyphosate with a detection limit of 12 ng mL−1. When tested with the cereal samples that had been spiked with glyphosate for 5–30 days, the calculated fluorescence recovery ranged from 89.5 to 110.0%.117
4.2.2.3. Detection of antibiotics. Unabsorbed antibiotics released into the environment as wastes are of great concern. One such example is enrofloxacin, an antibiotic used to treat animals which has comparatively low bioavailability (i.e., 72.12% when a dosage of 7.5 mg kg−1 was administered to the Thai swamp buffalos).114 Guo et al. developed a fluorescence “turn-on” assay to detect enrofloxacin using biodots (QY = 20.5%) derived from solid pyrolysis of DL-malic acid and glycine.114 The fluorescence of biodots was first turned off using Cu2+ ions as the quencher via electron transfer. Upon addition of enrofloxacin, the fluorescence was turned on due to the displacement of Cu2+ from the biodots, as coordination of Cu2+ with enrofloxacin was more favourable than the binding of Cu2+ with biodots. The optimised sensing probe exhibited a linear detection range of enrofloxacin from 1.0–15.0 μg mL−1, with a fluorescence recovery ranging from 96.5% to 109% when tested on antibiotic spiked real water samples.114
4.2.2.4. Detection of pyridine. Pyridine is an industrial waste, which is used in the production of drugs and agrochemicals. It is harmful to human beings as it can cause respiratory problems, infertility, and nausea.115 A biodot based pyridine detection method has been developed by Campos and co-workers. They found that pyridine caused fluorescence enhancement of nitrogen-doped biodots derived from lactose (N-CDs).115 The N-CDs were synthesized using lactose as the carbon source and aqueous ammonia as the nitrogen source. The resultant biodots have an average size of 50 nm and a fluorescence QY of 11%. Upon addition of pyridine, fluorescence enhancement of the N-CDs was observed in a concentration-dependent manner, exhibiting a linear detection range of 0.13–2.53 mM, with a LOD of 0.03 mM. It was suggested that the fluorescence enhancement was attributed to the change in the surface state of N-CDs due to the complex formation between negatively charged N-CDs and positively charged pyridine, which induced the aggregation of nanodots involving an energy transfer process.115
4.2.2.5. Detection of hydrazine. Hydrazine is an organic compound used in the production of several pharmaceuticals and pesticides, and is highly toxic and dangerously unstable. Apart from the popular fluorometric methods, biodots have also been used in amperometric assays. Sha and co-workers used biodot coated glassy carbon electrodes (GCEs) to quantify hydrazine in drinking water.77 The biodots were synthesized from chia seeds to enable oxidation of hydrazine on the surface of GCEs. The oxygen-containing functional groups of the chia seed-dot coated GCEs oxidize the hydrazine to allow sensitive quantification by amperometry.77
4.2.2.6. Detection of hydrogen peroxide. H2O2 is a chemical heavily used in the food industry as an oxidiser or bleaching agent. It is also found in the human body as a short-lived product in biochemical processes and is toxic to cells. Most recently, our group has developed a versatile method to form biodot@silver nanoparticle (AgNP) hybrids as dual optical sensing probes (i.e., colorimetric and fluorescent) for detecting H2O2 and H2O2-producing species such as glucose and cholesterol in complex samples.118 As shown in Fig. 9, the fluorescent biodots derived from serine and histamine precursors (Ser-His dots) were first synthesized and used as templates to anchor Ag+ ions, which were then irradiated under UV light to form AgNPs on the biodots, causing the quenching of fluorescence via inner filter effects. The resulting Bio@AgNPs possess unique redox capability which enable highly sensitive H2O2 detection through oxidative AgNPs etching, triggering both colorimetric changes of AgNPs (from yellowish to colourless) and “turn-on” blue-emissive fluorescence signal of biodots. This work demonstrated an effective and facile nanodot-templating method to fabricate highly functional nanohybrids for sensing applications.
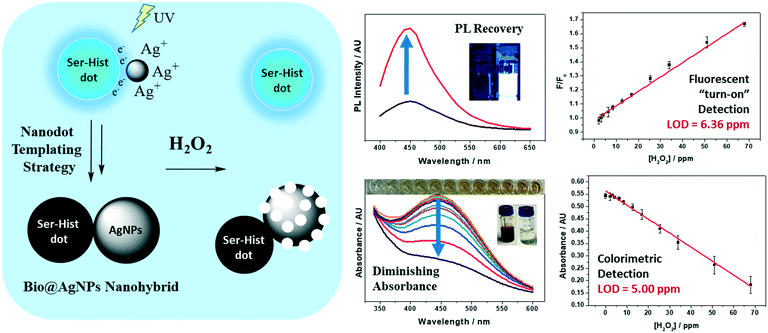 | ||
| Fig. 9 Biodot-templating synthesis of plasmonic-fluorescent (Bio@AgNPs) nanohybrids for dual optical (fluorescence “turn on” and colorimetric) detection of hydrogen peroxide and H2O2-producing species or reactions. Reproduced, adapted and reprinted with permission from ref. 118. | ||
5. Conclusions and future outlook
The emergence of bioinspired synthesis in recent years has led to the widespread investigation of various natural materials for molecular engineering of biodots. The natural precursors including (1) small biomolecules such as amino acids and sugars, (2) biopolymers (e.g., proteins, nucleic acids and polysaccharides) and also (3) complex biomass like fruits and milk have gained paramount interest as attractive sustainable carbon sources for the synthesis of biodots, due to their easy accessibility, affordability, green nature and renewability, as well as abundant functional groups for heteroatom doping to enable fine-tuning of the properties of biodots at the molecular level.In this report, the latest synthesis strategies and formation mechanisms of biodots derived from three distinct classes of natural precursors have been systematically reviewed and discussed. It is revealed that the small biomolecule precursors often act as the basic units to link up, polymerize and carbonize into the resultant biodots, while biopolymeric precursors such as proteins usually dehydrate or denature first before reorganizing into defined aromatic structures. These bottom-up approaches not only offer high programmability of biomolecular building blocks for the bioinspired synthesis, but also allow greater control over the molecular structure and fine-tuned properties of the biodots. In contrast, the derivation of biodots from the top-down approaches involves more sophisticated processes depending on the sources of biomass (e.g., food, floral or human wastes), which include extractions and interactions of multiple active ingredients from plants or animals. Thus, it is usually more difficult to tune the final properties of resultant biodots through rational molecular design. However, biomass, particularly unwanted materials such as fruit peels or used coffee grounds, can serve as a renewable carbon source to turn wastes into useful biodots for many applications ranging from bioimaging, biosensing, promoting plant growth to environmental pollutant detection.
Thousands of years of history in traditional medicine gives mankind vast choices of natural herbs with various therapeutic functions. Recently, this has been investigated for biodot synthesis, which will marry the benefits of traditional medicine and the unique properties of biodots for targeted anticancer therapy. Most crucial in these applications is the fine control of the molecular structures of the as-synthesized biodots so that the original functions especially therapeutic effects of the precursors (e.g., traditional herbs) can be well-retained and imparted to the biodots. On the other hand, some biodots inherit the specific recognition abilities from the molecular structures of precursors (e.g., DNA sequences), which offer high selectivity and ultrasensitivity for nanosensor development. Notably, all of these biodots are inherently biocompatible and environmentally-friendly, some of which even exhibit extraordinary biomimetic functions (e.g., bioinspired antimicrobial nanodots)17 not seen in their original precursors, leading to multiple functionalities similar to the molecular machine in cells.
We believe that learning from nature and using modern synthetic technologies to retain or mimic natural functions in designing new biodots is the next prominent frontier of biodot research. With the widespread use of artificial intelligence (AI) in recent years, it is believed that machine learning algorithms may be very useful in speeding up biodot design and synthesis processes. One potential direction is the AI-assisted searching and analysis of current databases for natural resources to guide the selection of suitable combinations of natural precursors to customize biodots with desirable biomimetic functions. Another emerging application would be the development of near-infrared (NIR)-emissive biodots for biomedical imaging due to the significantly lower auto-fluorescence and deeper tissue penetration. However, to date, very few NIR-emitting biodots have been reported in the literature.119 Analysing the current database of biodots derived from various natural resources using AI-enabled technology could aid in the understanding of the molecular design strategies for emission control and consequently accelerate the design and fine-tuning of the emission of biodots to the NIR region. We expect that coupling modern AI-technology with more experimental work inspired by nature will allow us to gain more insights into the formation mechanisms and crucial control parameters for synthesis to rationally tailor the design of multifunctional biodots for different application needs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This review is supported by the Newcastle University (RSA/CCEAMD5010) and the Institute of Materials Research and Engineering under the Biomimetic and Biomedical Program (IMRE/16-1P1401).Notes and references
- X. T. Zheng, A. Ananthanarayanan, K. Q. Luo and P. Chen, Small, 2015, 11, 1620–1636 CrossRef CAS PubMed.
- T. Yuan, T. Meng, P. He, Y. Shi, Y. Li, X. Li, L. Fan and S. Yang, J. Mater. Chem. C, 2019, 7, 6820–6835 RSC.
- X. Sun and Y. Lei, TrAC, Trends Anal. Chem., 2017, 89, 163–180 CrossRef CAS.
- Y. Wang, Y. Zhu, S. Yu and C. Jiang, RSC Adv., 2017, 7, 40973–40989 RSC.
- J. Wang, G. Liu, K. C.-F. Leung, R. Loffroy, P.-X. Lu and Y. X. J. Wang, Curr. Pharm. Des., 2015, 21, 5401–5416 CrossRef CAS PubMed.
- B. Yao, H. Huang, Y. Liu and Z. Kang, Trends Chem., 2019, 1, 235–246 CrossRef.
- X. T. Zheng, H. V. Xu and Y. N. Tan, in Advances in Bioinspired and Biomedical Materials Volume 2, American Chemical Society, 2017, ch. 7, vol. 1253, pp. 123–152 Search PubMed.
- H. V. Xu, X. T. Zheng, B. Y. L. Mok, S. A. Ibrahim, Y. Yu and Y. N. Tan, J. Mol. Eng. Mater., 2016, 04, 1640003 CrossRef CAS.
- Y. Yu, B. Y. L. Mok, X. J. Loh and Y. N. Tan, Adv. Healthcare Mater., 2016, 5, 1844–1859 CrossRef CAS PubMed.
- J. Xie, Y. N. Tan and J. Y. Lee, in Nanotechnologies for the Life Sciences, 2012, DOI:10.1002/9783527610419.ntls0209.
- X. Liu, J. Pang, F. Xu and X. Zhang, Sci. Rep., 2016, 6, 31100 CrossRef CAS PubMed.
- M. K. Barman and A. Patra, J. Photochem. Photobiol., C, 2018, 37, 1–22 CrossRef CAS.
- R. Jelinek, in Carbon Quantum Dots, Springer International Publishing, Cham, 2017, ch. 2, pp. 5–27, DOI:10.1007/978-3-319-43911-2_2.
- Y.-W. Zeng, D.-K. Ma, W. Wang, J.-J. Chen, L. Zhou, Y.-Z. Zheng, K. Yu and S.-M. Huang, Appl. Surf. Sci., 2015, 342, 136–143 CrossRef CAS.
- Y. Wang and A. Hu, J. Mater. Chem. C, 2014, 2, 6921–6939 RSC.
- L. Wang and H. S. Zhou, Anal. Chem., 2014, 86, 8902–8905 CrossRef CAS PubMed.
- H. V. Xu, X. T. Zheng, C. Wang, Y. Zhao and Y. N. Tan, ACS Appl. Nano Mater., 2018, 1, 2062–2068 CrossRef CAS.
- P. Karfa, E. Roy, S. Patra, S. Kumar, A. Tarafdar, R. Madhuri and P. K. Sharma, RSC Adv., 2015, 5, 58141–58153 RSC.
- M. Zheng, S. Liu, J. Li, D. Qu, H. Zhao, X. Guan, X. Hu, Z. Xie, X. Jing and Z. Sun, Adv. Mater., 2014, 26, 3554–3560 CrossRef CAS PubMed.
- Y.-F. Wu, H.-C. Wu, C.-H. Kuan, C.-J. Lin, L.-W. Wang, C.-W. Chang and T.-W. Wang, Sci. Rep., 2016, 6, 21170 CrossRef CAS PubMed.
- X. T. Zheng, Y. C. Lai and Y. N. Tan, Nanoscale Adv., 2019, 1, 2250–2257 RSC.
- H. V. Xu, X. T. Zheng, Y. Zhao and Y. N. Tan, ACS Appl. Mater. Interfaces, 2018, 10, 19881–19888 CrossRef CAS PubMed.
- F. Nawaz, L. Wang, L.-f. Zhu, X.-j. Meng and F.-S. Xiao, Chem. Res. Chin. Univ., 2013, 29, 401–403 CrossRef CAS.
- J. Jiang, Y. He, S. Li and H. Cui, Chem. Commun., 2012, 48, 9634–9636 RSC.
- Y. Choi, N. Thongsai, A. Chae, S. Jo, E. B. Kang, P. Paoprasert, S. Y. Park and I. In, J. Ind. Eng. Chem., 2016, 47, 329–335 CrossRef.
- M. Zhang, L. Hu, H. Wang, Y. Song, Y. Liu, H. Li, M. Shao, H. Huang and Z. Kang, Nanoscale, 2018, 10, 12734–12742 RSC.
- S. Hill and M. C. Galan, Beilstein J. Org. Chem., 2017, 13, 675–693 CrossRef CAS PubMed.
- Z. Ma, H. Ming, H. Huang, Y. Liu and Z. Kang, New J. Chem., 2012, 36, 861–864 RSC.
- Z. Chen, J. Wang, H. Miao, L. Wang, S. Wu and X. Yang, Sci. China: Chem., 2015, 59, 487–492 CrossRef.
- X. Liu, J. Liu, B. Zheng, L. Yan, J. Dai, Z. Zhuang, J. Du, Y. Guo and D. Xiao, New J. Chem., 2017, 41, 10607–10612 RSC.
- V. M. Naik, D. B. Gunjal, A. H. Gore, S. P. Pawar, S. T. Mahanwar, P. V. Anbhule and G. B. Kolekar, Diamond Relat. Mater., 2018, 88, 262–268 CrossRef CAS.
- N. Li, S. G. Liu, Y. Z. Fan, Y. J. Ju, N. Xiao, H. Q. Luo and N. B. Li, Anal. Chim. Acta, 2018, 1013, 63–70 CrossRef CAS PubMed.
- X. Zeng, L. Zhang, J. Yang, Y. Guo, Y. Huang, H. Yuan and Y. Xie, New J. Chem., 2017, 41, 15216–15228 RSC.
- R. Zhang and W. Chen, Biosens. Bioelectron., 2014, 55, 83–90 CrossRef CAS PubMed.
- J. Qin, L. Zhang and R. Yang, Spectrochim. Acta, Part A, 2019, 207, 54–60 CrossRef CAS PubMed.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- J. Gong, X. An and X. Yan, New J. Chem., 2014, 38, 1376–1379 RSC.
- S. K. Bhunia, N. Pradhan and N. R. Jana, ACS Appl. Mater. Interfaces, 2014, 6, 7672–7679 CrossRef CAS PubMed.
- S. Lu, X. Zhao, S. Zhu, Y. Song and B. Yang, Nanoscale, 2014, 6, 13939–13944 RSC.
- B. Sinduja and S. Abraham John, Spectrochim. Acta, Part A, 2018, 193, 486–491 CrossRef CAS PubMed.
- Y. Song, X. Li, S. Cong, H. Zhao and M. Tan, Colloids Surf., B, 2019, 180, 449–456 CrossRef CAS PubMed.
- Y. K. Jung, E. Shin and B. S. Kim, Sci. Rep., 2015, 5, 18807 CrossRef CAS PubMed.
- C. X. Guo, J. Xie, B. Wang, X. Zheng, H. B. Yang and C. M. Li, Sci. Rep., 2013, 3, 2957 CrossRef PubMed.
- H. Ding, F. Du, P. Liu, Z. Chen and J. Shen, ACS Appl. Mater. Interfaces, 2015, 7, 6889–6897 CrossRef CAS PubMed.
- T. Song, X. Zhu, S. Zhou, G. Yang, W. Gan and Q. Yuan, Appl. Surf. Sci., 2015, 347, 505–513 CrossRef CAS.
- Q. H. Li, L. Zhang, J. M. Bai, Z. C. Liu, R. P. Liang and J. D. Qiu, Biosens. Bioelectron., 2015, 74, 886–894 CrossRef CAS PubMed.
- Z. Zhang, J. Hao, J. Zhang, B. Zhang and J. Tang, RSC Adv., 2012, 2, 8599–8601 RSC.
- M. Tan, X. Li, H. Wu, B. Wang and J. Wu, Colloids Surf., B, 2015, 136, 141–149 CrossRef CAS PubMed.
- X. Liu, T. Li, Y. Hou, Q. Wu, J. Yi and G. Zhang, RSC Adv., 2016, 6, 11711–11718 RSC.
- P. Yang, Z. Zhu, M. Chen, W. Chen and X. Zhou, Opt. Mater., 2018, 85, 329–336 CrossRef CAS.
- P. Yang, Z. Zhu, M. Chen, X. Zhou and W. Chen, Spectrochim. Acta, Part A, 2019, 213, 301–308 CrossRef CAS PubMed.
- X. J. Zhao, W. L. Zhang and Z. Q. Zhou, Colloids Surf., B, 2014, 123, 493–497 CrossRef CAS PubMed.
- D. Chakraborty, S. Sarkar and P. K. Das, ACS Sustainable Chem. Eng., 2018, 6, 4661–4670 CrossRef CAS.
- A. Basu, A. Suryawanshi, B. Kumawat, A. Dandia, D. Guin and S. B. Ogale, Analyst, 2015, 140, 1837–1841 RSC.
- W. Lu, X. Qin, S. Liu, G. Chang, Y. Zhang, Y. Luo, A. M. Asiri, A. O. Al-Youbi and X. Sun, Anal. Chem., 2012, 84, 5351–5357 CrossRef CAS PubMed.
- J. Zhou, Z. Sheng, H. Han, M. Zou and C. Li, Mater. Lett., 2012, 66, 222–224 CrossRef CAS.
- A. Su, D. Wang, X. Shu, Q. Zhong, Y. Chen, J. Liu and Y. Wang, Chem. Res. Chin. Univ., 2018, 34, 164–168 CrossRef CAS.
- A. Prasannan and T. Imae, Ind. Eng. Chem. Res., 2013, 52, 15673–15678 CrossRef CAS.
- S. A. A. Vandarkuzhali, S. Natarajan, S. Jeyabalan, G. Sivaraman, S. Singaravadivel, S. Muthusubramanian and B. Viswanathan, ACS Omega, 2018, 3, 12584–12592 CrossRef CAS PubMed.
- X.-Y. Jiao, L.-s. Li, S. Qin, Y. Zhang, K. Huang and L. Xu, Colloids Surf., A, 2019, 577, 306–314 CrossRef CAS.
- P.-C. Hsu, Z.-Y. Shih, C.-H. Lee and H.-T. Chang, Green Chem., 2012, 14, 917–920 RSC.
- V. Sharma, S. K. Singh and S. M. Mobin, Nanoscale Adv., 2019, 1, 1290–1296 RSC.
- J. B. Essner, C. H. Laber, S. Ravula, L. Polo-parada and G. A. Baker, Green Chem., 2016, 18, 243–250 RSC.
- J. Hou, J. Li, J. Sun, S. Ai and M. Wang, RSC Adv., 2014, 4, 37342–37348 RSC.
- S.-S. Liu, C.-F. Wang, C.-X. Li, J. Wang, L.-H. Mao and S. Chen, J. Mater. Chem. C, 2014, 2, 6477–6483 RSC.
- Y. Guo, L. Zhang, F. Cao and Y. Leng, Sci. Rep., 2016, 6, 35795 CrossRef CAS PubMed.
- M. L. Liu, B. B. Chen, C. M. Li and C. Z. Huang, Green Chem., 2019, 21, 449–471 RSC.
- S. Pramanik, S. Chatterjee, G. S. Kumar and P. S. Devi, Phys. Chem. Chem. Phys., 2018, 20, 20476–20488 RSC.
- Y. Y. Yao, G. Gedda, W. M. Girma, C. L. Yen, Y. C. Ling and J. Y. Chang, ACS Appl. Mater. Interfaces, 2017, 9, 13887–13899 CrossRef CAS PubMed.
- X. Yan, Y. Zhao, J. Luo, W. Xiong, X. Liu, J. Cheng, Y. Wang, M. Zhang and H. Qu, J. Nanobiotechnol., 2017, 15, 60 CrossRef PubMed.
- X. Liu, Y. Wang, X. Yan, M. Zhang, Y. Zhang, J. Cheng, F. Lu, H. Qu, Q. Wang and Y. Zhao, Nanomedicine, 2018, 13, 391–405 CrossRef CAS PubMed.
- M. Zhang, Y. Zhao, J. Cheng, X. Liu, Y. Wang, X. Yan, Y. Zhang, F. Lu, Q. Wang and H. Qu, Artif. Cells, Nanomed., Biotechnol., 2017, 1–10, DOI:10.1080/21691401.2017.1379015.
- B. De and N. Karak, RSC Adv., 2013, 3, 8286–8290 RSC.
- R. Bandi, R. Dadigala, B. R. Gangapuram and V. Guttena, J. Photochem. Photobiol., B, 2018, 178, 330–338 CrossRef CAS PubMed.
- R. Purbia and S. Paria, Biosens. Bioelectron., 2016, 79, 467–475 CrossRef CAS PubMed.
- A. F. Shaikh, M. S. Tamboli, R. H. Patil, A. Bhan, J. D. Ambekar and B. B. Kale, J. Nanosci. Nanotechnol., 2019, 19, 2339–2345 CrossRef CAS PubMed.
- R. Sha, S. S. Jones, N. Vishnu, B. Soundiraraju and S. Badhulika, Electroanalysis, 2018, 30, 2228–2232 CrossRef CAS.
- J. R. Bhamore, S. Jha, T. J. Park and S. K. Kailasa, Sens. Actuators, B, 2018, 277, 47–54 CrossRef CAS.
- Y. Zheng, G. Xie, X. Zhang, Z. Chen, Y. Cai, W. Yu, H. Liu, J. Shan, R. Li, Y. Liu and B. Lei, ACS Omega, 2017, 2, 3958–3965 CrossRef CAS PubMed.
- Y. Zheng, H. Zhang, W. Li, Y. Liu, X. Zhang, H. Liu and B. Lei, RSC Adv., 2017, 7, 33459–33465 RSC.
- A. Tyagi, K. M. Tripathi, N. Singh, S. Choudhary and R. K. Gupta, RSC Adv., 2016, 6, 72423–72432 RSC.
- P.-C. Hsu, P.-C. Chen, C.-M. Ou, H.-Y. Chang and H.-T. Chang, J. Mater. Chem. B, 2013, 1, 1774–1781 RSC.
- Q. Wu, W. Li, P. Wu, J. Li, S. Liu, C. Jin and X. Zhan, RSC Adv., 2015, 5, 75711–75721 RSC.
- J. Wei, L. Qiang, J. Ren, X. Ren, F. Tang and X. Meng, Anal. Methods, 2014, 6, 1922–1927 RSC.
- S. Devi, R. K. Gupta, A. K. Paul and S. Tyagi, Mater. Res. Express, 2018, 6, 25605 CrossRef.
- Z. Zhang, W. Sun and P. Wu, ACS Sustainable Chem. Eng., 2015, 3, 1412–1418 CrossRef CAS.
- Y. Zhao, Y. Zhang, X. Liu, H. Kong, Y. Wang, G. Qin, P. Cao, X. Song, X. Yan, Q. Wang and H. Qu, Sci. Rep., 2017, 7, 4452 CrossRef PubMed.
- D. Wang, X. Wang, Y. Guo, W. Liu and W. Qin, RSC Adv., 2014, 4, 51658–51665 RSC.
- D. Wang, L. Zhu, C. Mccleese, C. Burda and J.-f. Chen, RSC Adv., 2016, 6, 41516–41521 RSC.
- Y. Zhang, Z. Gao, X. Yang, J. Chang, Z. Liu and K. Jiang, RSC Adv., 2019, 9, 940–949 RSC.
- A. A. Ensafi, S. Hghighat Sefat, N. Kazemifard, B. Rezaei and F. Moradi, Sens. Actuators, B, 2017, 253, 451–460 CrossRef CAS.
- A. Tadesse, D. Rama Devi, M. Hagos, G. Battu and K. Basavaiah, Asian J. Nanosci. Mater., 2018, 1, 36–46 Search PubMed.
- G. Ren, M. Tang, F. Chai and H. Wu, Eur. J. Inorg. Chem., 2018, 2018, 153–158 CrossRef CAS.
- Y. Liu, Y. Liu, M. Park, S.-J. Park, Y. Zhang, M. R. Akanda, B.-Y. Park and H. Y. Kim, Carbon Lett., 2017, 21, 61–67 CrossRef.
- J. Shen, S. Shang, X. Chen, D. Wang and Y. Cai, Mater. Sci. Eng., C, 2017, 76, 856–864 CrossRef CAS PubMed.
- J. Praneerad, N. Thongsai, P. Supchocksoonthorn, S. Kladsomboon and P. Paoprasert, Spectrochim. Acta, Part A, 2019, 211, 59–70 CrossRef CAS PubMed.
- C.-L. Li, C.-M. Ou, C.-C. Huang, W.-C. Wu, Y.-P. Chen, T.-E. Lin, L.-C. Ho, C.-W. Wang, C.-C. Shih, H.-C. Zhou, Y.-C. Lee, W.-F. Tzeng, T.-J. Chiou, S.-T. Chu, J. Cang and H.-T. Chang, J. Mater. Chem. B, 2014, 2, 4564–4571 RSC.
- M. Asha Jhonsi and S. Thulasi, Chem. Phys. Lett., 2016, 661, 179–184 CrossRef CAS.
- K. M. Tripathi, A. Tyagi, M. Ashfaq and R. K. Gupta, RSC Adv., 2016, 6, 29545–29553 RSC.
- Y. Wang and X. Jiang, Sci. China: Chem., 2016, 59, 836–842 CrossRef CAS.
- Y. Liu, Y. Zhao and Y. Zhang, Sens. Actuators, B, 2014, 196, 647–652 CrossRef CAS.
- M. Yuan, R. Zhong, H. Gao, W. Li, X. Yun, J. Liu, X. Zhao, G. Zhao and F. Zhang, Appl. Surf. Sci., 2015, 355, 1136–1144 CrossRef CAS.
- M. Tan, L. Zhang, R. Tang, X. Song, Y. Li, H. Wu, Y. Wang, G. Lv, W. Liu and X. Ma, Talanta, 2013, 115, 950–956 CrossRef CAS PubMed.
- P. Roy, P.-C. Chen, A. P. Periasamy, Y.-N. Chen and H.-T. Chang, Mater. Today, 2015, 18, 447–458 CrossRef CAS.
- P. C. Hsu and H. T. Chang, Chem. Commun., 2012, 48, 3984–3986 RSC.
- A. Sachdev, I. Matai and P. Gopinath, RSC Adv., 2014, 4, 20915–20921 RSC.
- X. T. Zheng and Y. N. Tan, Small, 2019, e1903328, DOI:10.1002/smll.201903328.
- J. Xie, S. Lee and X. Chen, Adv. Drug Delivery Rev., 2010, 62, 1064–1079 CrossRef CAS PubMed.
- L. Yang, Z. Wang, J. Wang, W. Jiang, X. Jiang, Z. Bai, Y. He, J. Jiang, D. Wang and L. Yang, Nanoscale, 2016, 8, 6801–6809 RSC.
- Y. Yuan, B. Guo, L. Hao, N. Liu, Y. Lin, W. Guo, X. Li and B. Gu, Colloids Surf., B, 2017, 159, 349–359 CrossRef CAS PubMed.
- T. Kong, L. Hao, Y. Wei, X. Cai and B. Zhu, Cell Proliferation, 2018, 51, e12488 CrossRef PubMed.
- X.-W. Hua, Y.-W. Bao, Z. Chen and F.-G. Wu, Nanoscale, 2017, 9, 10948–10960 RSC.
- M. M. F. Chang, I. R. Ginjom and S. M. Ng, Sens. Actuators, B, 2017, 242, 1050–1056 CrossRef CAS.
- X. Guo, L. Zhang, Z. Wang, Y. Sun, Q. Liu, W. Dong and A. Hao, Spectrochim. Acta, Part A, 2019, 219, 15–22 CrossRef CAS PubMed.
- B. B. Campos, C. Abellan, M. Zougagh, J. Jimenez-Jimenez, E. Rodriguez-Castellon, J. C. Esteves da Silva, A. Rios and M. Algarra, J. Colloid Interface Sci., 2015, 458, 209–216 CrossRef CAS PubMed.
- V. A. Ansi and N. K. Renuka, Sens. Actuators, B, 2018, 264, 67–75 CrossRef CAS.
- L. Wang, Y. Bi, J. Hou, H. Li, Y. Xu, B. Wang, H. Ding and L. Ding, Talanta, 2016, 160, 268–275 CrossRef CAS PubMed.
- H. V. Xu, Y. Zhao and Y. N. Tan, ACS Appl. Mater. Interfaces, 2019, 11, 27233–27242 CrossRef CAS PubMed.
- H. Ding, X. Zhou, B. Qin, Z. Zhou and Y. Zhao, J. Lumin., 2019, 211, 298–304 CrossRef CAS.
Footnote |
| † These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |