Open Access Article
Open Access ArticleA novel receptor-free polydiacetylene nanofiber biosensor for detecting E. coli via colorimetric changes†
Abhishek
Bhattacharjee
a,
Richard
Clark
b,
Claudia
Gentry-Weeks
b and
Yan Vivian
Li
 *acd
*acd
aDepartment of Design and Merchandising, Colorado State University, Fort Collins, USA. E-mail: yan.li@colostate.edu
bDepartment of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins, USA
cSchool of Biomedical Engineering, Colorado State University, Fort Collins, USA
dSchool of Advanced Materials Discovery, Colorado State University, Fort Collins, USA
First published on 27th October 2020
Abstract
A nanofiber-based polydiacetylene (PDA) biosensor was prepared with polyurethane (PU) to detect Escherichia coli. The main advantage of this PU-PDA nanofiber biosensor was the simplicity of the preparation method via electrospinning where no complex receptors were required to detect E. coli via colorimetric properties. Diacetylene monomers self-assembled during the electrospinning process that significantly enhanced the interaction between the PDA macromolecules and E. coli, resulting in high sensitivity of bacterial detection. Experiments showed that the PU-PDA nanofiber system was highly sensitive and could quickly detect E. coli by a colorimetric transition that is visible to the naked eye. The colorimetric property of the PU-PDA nanofiber was highly sensitive to organic solvents, suggesting potential for application in fingerprinting devices capable of distinguishing organic solvents. Additional experiments indicated that the color-changing substances (CCS) in the complex E. coli culture were not cell-associated, rather E. coli secretion associated. Further experiments were conducted to understand the nature of the CCS in E. coli culture and the results showed that the CCS are free extracellular polymeric substances (EPS) in E. coli culture.
Introduction
π-Conjugated polymers (CP) with unique optical and electrochemical properties have been used in several applications including solar cells,1 light-emitting diodes,2 photovoltaic cells,3 sensors,4 artificial muscles,5 and laser technology.6 Due to the delocalized π electrons along the macromolecular backbone, CPs can transfer nonspecific interactions in the molecular structures into transducible responses,7 resulting in characteristic optical and electrical properties. Polydiacetylene (PDA) is an attractive CP due to its quick chromatic responses upon exposure to various external stimuli such as solvents, temperature, electricity, light, mechanical stress, chemical stress, viruses, and bacteria.8 PDAs have also been used in microorganism sensing,9 drug delivery,10 supercapacitors,11 and oil–water separations.12 From our lab, Alam et al. and Yapor et al. reported that PDA nanofiber composites show rapid blue-to-red color transition responding to bacteria, heat, and pH, suggesting their potential for use in biosensor applications, especially in developing point-of-care devices such as smart wound dressings that can be used to detect bacterial infection in wound care.13,14 Point-of-care devices are recently of great interest in hospitals, battlefields, and nursing homes owing to many advantages over traditional testing technologies, such as simplicity, affordability, and effective and efficient patient care.1510,12-Pentacosadiynoic acid (PCDA) is a common monomer to prepare PDAs via the 1,4-addition reaction of PCDAs followed by photo-polymerization via UV or γ irradiation. Mainly, substrate-based and solution-based PDA sensors have been reported for sensor applications due to easy fabrication, cell-mimicry, stability, and enhanced sensitivity.8 Although solution-based PDA sensors provide advantages in biomedical applications especially due to a high degree of cell mimicry, they are often unstable due to the aggregation and precipitation of PDA molecules.16 The substrate-based PDA sensors overcome this drawback because they are relatively stable and sensitive in many sensing applications.16 Nevertheless, to detect pathogens such as bacteria via PDA based sensors, both substrate and solution-based PDAs were reported that can sense either the bacterial cell or the materials bacteria secrete into the environment during the growth phase. Complex receptors such as specific bacterial antibodies were incorporated into the PDA sensing system that reacted with the bacterial cell and thus detected the presence of bacteria. Park et al. reported a glass-supported PDA based sensing system where different bacterial antibodies were used to detect bacteria via colorimetric transition.17 An indirect approach is to detect the bacterial secretion where amine functionalization of the PDA monomer and incorporation of complex receptors were performed to facilitate the reaction between the amine-functionalized PDA and bacterial secretion materials. For instance, Villalobos et al. prepared a PDA based sensing system via amine functionalization of the diacetylene monomer that could detect bacterially secreted materials (supernatant fluid from a bacterial culture) by colorimetric transition after 4 h of incubation.18 In addition to the receptors, lipids such as DMPC (dimyristoyl-sn-glycero-3-phosphocholine) are often used in the PDA based sensors that mimic the host cell surface and enhance the sensitivity of the system. Kim et al. reported the effect of lipid insertion in the PDA system where an increased amount of lipids exhibited higher chromatic response when bacteria were introduced into the system.19 dos Santos Pires et al. reported a similar trend, where no significant color change was found when pure diacetylene monomer PCDA (10,12-pentacosadiynoic acid) was used, but a significant color change was observed when TDER (N-[(2-tetradecanamide)-ethyl]-ribonamide) was incorporated into the PCDA.20 However, the preparation of these PDA sensing systems using complex functionalization with receptors and lipids is complicated and often demands sophisticated machinery and personnel, which is a shortcoming of the existing PDA based biosensors.
This paper reports a simple, receptor-free fabrication strategy of preparing PDA biosensors capable of detecting E. coli via a blue-to-red colorimetric transition. PDA nanofiber composites were prepared using an electrospinning method and the stretching on the nanofibers due to electrostatic forces enhanced the self-assembly of the PCDA monomer clusters, resulting in a flexible, powerless, receptor-free PDA nanofiber biosensor potentially for many medical applications. The color change of the PDA nanofibers in response to the E. coli grown in LB media was visible to the naked eye with a response time of 0.5–3 min that was significantly short, suggesting a sensitive detection. The critical bacterial concentration (CBC) that was defined as a concentration initiating a colorimetric transition in the PDA nanofibers was found to be approximately 9 × 108 CFU mL−1, suggesting an effective and sufficient detection limit for practical uses in sensors. When the nanofibers were tested with two distinct components of E. coli culture, it was found that the colorimetric response (CR%) in the nanofibers tested with supernatant fluid was significant (CR = 10%). The colorimetric response of the nanofibers was further increased to 28% by the autoclaved supernatant fluid. In comparison, no color change was found in testing with E. coli cell pellets. The results suggested that the color-changing substances (CCS) in E. coli culture were not cell-associated rather secretion associated, which was in good agreement with the receptor-free fabrication strategy of preparing PDAs. When a minimum nutrition medium (M9 minimal medium) was used to grow E. coli, no color change was found in the PDA nanofiber. In addition, the PDA nanofibers demonstrated colorimetric transition behaviors in response to twelve commonly used organic solvents. The percentage of colorimetric response was dependent on the nature of the solvent. Further analysis confirmed that the CCS were the extracellular polymeric substances (EPS) that E. coli produced in the process of biofilm formation. The results may expand the application of current PDA biosensors in detecting EPS that is generally the primary component of bacterial biofilms.
Experimental section
Materials
10,12-Pentacosadiynoic acid (PCDA, 98%, GFS Organics, Columbus, OH) was used as a monomer for preparing PDA. Thermoplastic polyurethane (PU) was purchased from The Lubrizol Corporation (Wickliffe, OH) to be used as a matrix polymer. Tetrahydrofuran (THF, 99%) and N,N-dimethylformamide (DMF, 99.8%, extra dry, AcroSeal) were purchased from Fisher Scientific (Waltham, MA) for preparing the solvent system to synthesize PDA nanofibers.E. coli ATCC25922 (1.0 × 104 CFU per pellet) was purchased from ATCC (Manassas, VA). A lysogeny Broth (LB) and M9 minimal media were used for the growth of E. coli. For the size-fractionation test, Amicon® Ultra-4 Centrifugal Filter Units having molecular weight cutoffs of 3k, 30k, and 100k Da were purchased from Millipore Sigma (Burlington, MA). The prepared lipopolysaccharide (LPS) from E. coli was purchased from Millipore Sigma (Burlington, MA). Organic solvents that were used to study solvatochromism included hexanes, xylenes, isopropyl alcohol, 1-butanol, tetrahydrofuran (THF), methanol, ethanol (200 proof), glacial acetic acid, N,N-dimethylformamide (DMF), chloroform (CHCl3), acetone, and dimethyl sulfoxide (DMSO). Distilled water was used as a control. All the chemicals and solvents were used without further purification.
Methods
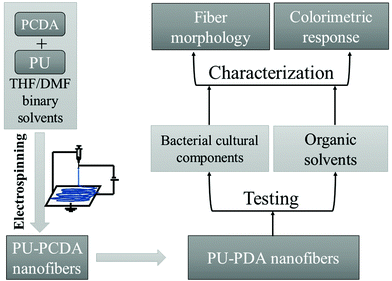
PU-PDA nanofiber biosensors were prepared via electrospinning and then tested with E. coli culture components as well as with organic solvents. Fig. 1 shows a schematic of the experimental design.PDA preparation
PDA powders were prepared from 10,12-pentacosadiynoic acid (PCDA) monomers. 6.4 g of PCDA was dissolved in 35 mL chloroform and stirred to make a homogenous solution. The solution was then filtered to remove any contaminants. The solvent was removed from the filtered solution by continuous stirring under the hood until dry powders were isolated. Millipore water was added to make a 1.29% w/v suspension and the suspension was sonicated for 30 min. After the sonication, the suspension was allowed to cool down to room temperature and stored at 4 °C for 24 h. Then, the suspension was transferred to a glass beaker with a magnetic stir bar and irradiated with UV light at 254 nm for 10 min. The color of the suspension changed to dark blue, resulting in PDA. 7 mL of the dark blue suspension was taken into a centrifuge tube and centrifuged for 10 min at 6000 rpm. The dark blue precipitation at the bottom of the tube was taken out and kept in an open glass vial to allow the solid to completely dry, resulting in blue PDA powders.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 984 × g for 10 min at 27 °C. The supernatant fluid was collected, filtered (0.2 μm syringe polyethersulfone filters), and divided evenly into two flasks. One half of the supernatant fluid was autoclaved at 121.11 °C at 23 psi for 20 min and the other half was kept in the incubator at room temperature. On the other hand, the obtained bacterial cell pellet after centrifugation was washed twice using a 0.2% sterile saline solution and centrifuged again at 11
984 × g for 10 min at 27 °C. The supernatant fluid was collected, filtered (0.2 μm syringe polyethersulfone filters), and divided evenly into two flasks. One half of the supernatant fluid was autoclaved at 121.11 °C at 23 psi for 20 min and the other half was kept in the incubator at room temperature. On the other hand, the obtained bacterial cell pellet after centrifugation was washed twice using a 0.2% sterile saline solution and centrifuged again at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 984 × g for 10 min at 27 °C. The cell pellets were then dispersed in a 0.9% saline solution and separated into two flasks. One half of the cell pellet solution was autoclaved at 121.11 °C at 23 psi for 20 min and the other half was kept at room temperature. Besides, two supplementary batches of supernatant fluid were prepared following the same procedures described above. One batch of supernatant fluid (including both autoclaved and non-autoclaved) was used to treat PU-PDA nanofibers at 24 h (ts = 24 h) storage time at room temperature. The other batch was kept for 36 h (ts = 36 h) storage time. E. coli culture in M9 minimal media was prepared and divided into four component solutions following the procedure described for LB. To summarize, the E. coli culture components included (1) autoclaved supernatant fluid, (2) non-autoclaved supernatant fluid, (3) autoclaved bacterial cell pellets, and (4) non-autoclaved bacterial cell pellets from both LB and M9 minimal media. LB and M9 minimal medium solutions were used as controls.
984 × g for 10 min at 27 °C. The cell pellets were then dispersed in a 0.9% saline solution and separated into two flasks. One half of the cell pellet solution was autoclaved at 121.11 °C at 23 psi for 20 min and the other half was kept at room temperature. Besides, two supplementary batches of supernatant fluid were prepared following the same procedures described above. One batch of supernatant fluid (including both autoclaved and non-autoclaved) was used to treat PU-PDA nanofibers at 24 h (ts = 24 h) storage time at room temperature. The other batch was kept for 36 h (ts = 36 h) storage time. E. coli culture in M9 minimal media was prepared and divided into four component solutions following the procedure described for LB. To summarize, the E. coli culture components included (1) autoclaved supernatant fluid, (2) non-autoclaved supernatant fluid, (3) autoclaved bacterial cell pellets, and (4) non-autoclaved bacterial cell pellets from both LB and M9 minimal media. LB and M9 minimal medium solutions were used as controls.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 984 × g and 4 °C, decanted, and ethanol was mixed with the decanted solution. After the container was inverted 24 times and stored at −20 °C for 18 h, the precipitate was pelleted by centrifugation at 11,984 × g for 15 min at 4 °C, resulting in free EPS. The free EPS pellet was suspended in 70% ethanol and the suspension was washed with distilled water. Residual ethanol was evaporated from free EPS suspension by keeping it at room temperature for 48 h. 5 mL distilled water was added to the suspension and vortexed. The resultant solution was used to test on PU-PDA nanofibers.
984 × g and 4 °C, decanted, and ethanol was mixed with the decanted solution. After the container was inverted 24 times and stored at −20 °C for 18 h, the precipitate was pelleted by centrifugation at 11,984 × g for 15 min at 4 °C, resulting in free EPS. The free EPS pellet was suspended in 70% ethanol and the suspension was washed with distilled water. Residual ethanol was evaporated from free EPS suspension by keeping it at room temperature for 48 h. 5 mL distilled water was added to the suspension and vortexed. The resultant solution was used to test on PU-PDA nanofibers.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio for each solution. Then, the solutions were used to test the PU-PDA nanofibers in a tissue culture well. The nanofiber mats were immersed in the LPS solutions for 72 h.
10 ratio for each solution. Then, the solutions were used to test the PU-PDA nanofibers in a tissue culture well. The nanofiber mats were immersed in the LPS solutions for 72 h.
Results and discussion
Electrospinning and PU-PDA nanofiber morphology
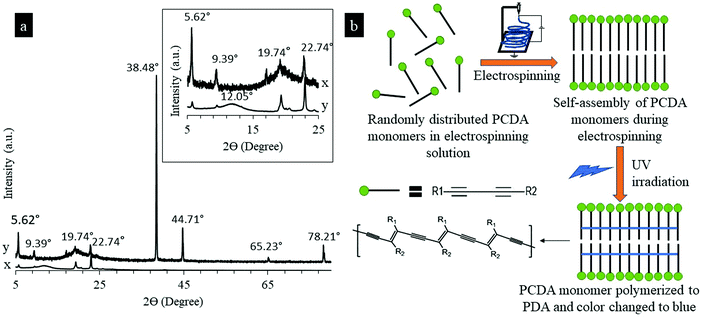
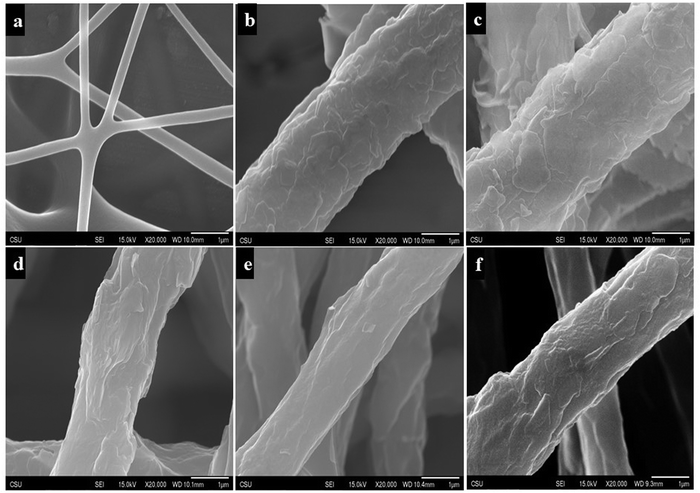
In the electrospinning of nanofibers, large electrostatic forces due to high voltage enable molecular orientation in a nanofiber when the nanofiber is drawn and produced. The large electrostatic forces have been reported to be highly efficient in enhancing fiber orientation and hence improving the mechanical performance of the fibers.22–24 In the electrospinning of PCDA and PU mixture solution, PCDA monomers were randomly distributed in the PU solution. Fig. 2a shows X-ray diffraction (XRD) spectra of blue electrospun PU-PDA nanofibers and non-electrospun PDA powders. Significant differences were found between the XRD crystallography of electrospun PDA nanofibers and non-electrospun PDA powders. A broad peak at a 2θ value of 12.05° was observed in the XRD spectra of the PDA powder, indicating a large amorphous region,25 which was absent in the spectra of PU-PDA nanofiber, confirming improved crystallinity in the fiber compared to the powder (Fig. 2a). Sharp and strong peaks at 2θ values of 5.62°, 9.39°, 38.48°, and 44.71° found in the XRD spectra of the PDA nanofiber suggesting significant enhancement of crystallinity in the PDA nanofiber in comparison with PDA powders. It has been previously reported that large external forces could overcome the molecular attraction between PCDA monomers and PU molecules, thus promoting molecular alignment in PCDA monomers, resulting in PCDA self-assembly structures.26 During electrospinning, large electrostatic forces were able to significantly align the PCDA monomers and therefore enhance the self-assembly structures of PCDA. An illustration scheme shown in Fig. 2b demonstrates the molecular alignment and self-assembly of PCDA, as well as the PDA molecular vesicles after the UV polymerization.Fig. 3 shows the SEM images of the PU nanofiber, PU-PDA nanofiber, and the nanofibers interacting with different components from E. coli culture. The PU nanofibers had a diameter ranging from 200–300 nm. The addition of PDA in the PU nanofibers increased the resultant fiber diameter ranging from 200–2000 nm. A flake-like structure on the PU-PDA fiber surface (Fig. 3b–f) was found compared to the PU nanofibers.27 No significant change in fiber morphology (Fig. 3c–f) was found in the nanofibers after the colorimetric detection of E. coli occurred in later testing.
Sensitivity of detecting E. coli-critical bacterial concentration
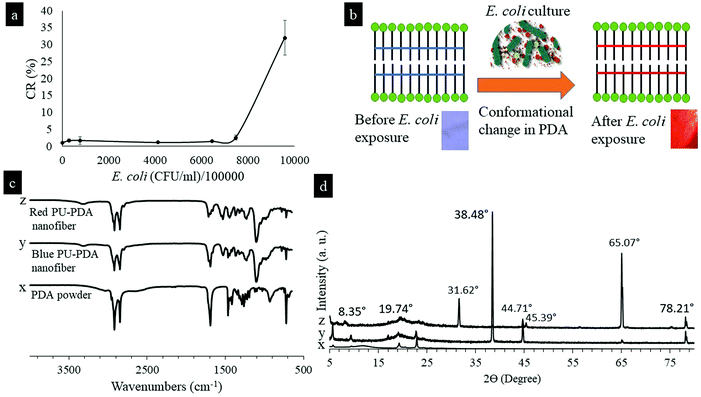
Critical bacterial concentration (CBC) is defined as the concentration of bacteria at which colorimetric transition in PDA occurs. The CBC determines the sensitivity of using the PU-PDA nanofiber for the identification of bacterial presence in point-of-care devices such as smart wound dressings. The colony-forming unit per volume (CFU mL−1) of E. coli is a common indicator of bacterial concentration. Fig. 4a shows a plot of colorimetric response (CR%) of the PU-PDA nanofiber as a function of E. coli concentration in CFU mL−1. The color change in PU-PDA nanofibers began to occur immediately when exposed to the E. coli concentration that was approximately 9 × 108 CFU mL−1. The nanofiber mat started changing color in some spots immediately at the CBC and then completely changed in red after a couple of minutes. The response time was nearly consistent with an increase in bacterial concentration. The bacterial concentration in wounds causing infection generally lies in the range of 105 and 109 CFU mL−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) .28,29 If the detection limit were too low, the detection response in practical applications would occur even at a bacterial level normally found in healthy skins, resulting in a false signal of sensing. Therefore, the PU-PDA nanofiber composites demonstrated sufficient sensitivity for practical infection detection in wound care. The high sensitivity of the PDA nanofibers most likely contributed to the highly ordered and self-assembled PDA vesicle nanostructures that were sensitive to external stimuli, resulting in a conformational change in the PDA vesicles and eventually a colorimetric transition from blue-to-red in the PDA nanofibers. Fig. 4b shows an illustration scheme of conformational change in a PDA vesicle and the blue-to-red color transition in the PDA nanofibers.
.28,29 If the detection limit were too low, the detection response in practical applications would occur even at a bacterial level normally found in healthy skins, resulting in a false signal of sensing. Therefore, the PU-PDA nanofiber composites demonstrated sufficient sensitivity for practical infection detection in wound care. The high sensitivity of the PDA nanofibers most likely contributed to the highly ordered and self-assembled PDA vesicle nanostructures that were sensitive to external stimuli, resulting in a conformational change in the PDA vesicles and eventually a colorimetric transition from blue-to-red in the PDA nanofibers. Fig. 4b shows an illustration scheme of conformational change in a PDA vesicle and the blue-to-red color transition in the PDA nanofibers.
Previous reports on PDA suggested a conformational change in the π conjugated electron backbone structure of PDA when the color changes from blue to red.13,14 The conformational change in PDA nanofibers was confirmed by FTIR spectra shown in Fig. 4c. A shift of characteristic peaks on the FTIR spectra between the blue nanofibers and the red nanofibers was found in carbonyl stretch from 1691 to 1715 cm−1. The carbonyl bond strength increased and the bond length shortened in the red nanofibers, suggesting a decrease in hydrogen bond strength and resulting in a disruption in the hydrogen bond along the side chain of PDAs. The disruption in the hydrogen bond initiated a conformational change in the π electron cloud of PDA and resulted in a color change in the nanofiber composite. In addition, Fig. 4c shows the FTIR spectra of non-electrospun PDA powders. All of the characteristic peaks were present in both PDA nanofibers and powders, suggesting no differences in the chemical structures of PDA in nanofibers and powders. Fig. 4d shows XRD spectra of the PU-PDA nanofibers after color change. The intense peak at 38.48° found in the blue nanofibers was reduced drastically in the red nanofibers. New peaks at 8.35°, 31.62°, 45.39°, and 65.07° were found in the red nanofibers in a comparison with the blue nanofibers. The XRD results suggested changes in crystallography during color shift from blue-to-red, which is most likely the result of a conformational change in the electron cloud of PDA molecules in the nanofibers.
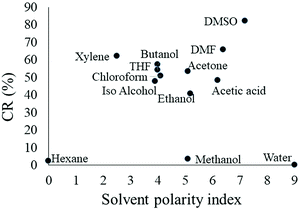
Colorimetric response of nanofibers in organic solvents
Solvatochromism is a property that indicates a color change in molecules or materials in organic solvents. PDAs exhibit solvatochromism where the molecules turn blue-to-red when different organic solvents are introduced. Several PDA based systems have been reported in the literature to identify organic solvents30 and volatile organic compounds.31 To assess the sensitivity of the PU-PDA nanofiber biosensor, the nanofibers were immersed in 12 common organic solvents and the colorimetric responses (CR%) were calculated. Fig. 5 shows the CR% generated by each organic solvent as a function of the solvent polarity index. 10 out of 12 solvents were able to cause a color change in the nanofibers instantly. Hexane and methanol could not generate a significant colorimetric response. With an increase of the polarity index from 0 (hexane) to 7.2 (DMSO), the CR% of the nanofibers increased exponentially by almost 80%, resulting in the highest CR% (82%) in DMSO. As different solvents generated a different CR%, the PU-PDA nanofiber biosensor offers the potential to differentiate and detect specific organic solvents. However, it was also found from the tests that the fiber mats began to disrupt with an increase in the solvent polarity index. The solvents with a polarity index higher than 4 disassembled the fibers, resulting in serious damages at the edges of the fiber mats. The fiber mats were nearly completely dissolved in THF, acetic acid, and DMF after 30 min of exposure. The exception of methanol to change the color while having a polarity index the same as acetone indicates that the solvent polarity may not be the sole responsible factor for a color change in PU-PDA nanofibers. The different levels of colorimetric responses to different solvents could be very helpful in developing sensors for solvent identification.Colorimetric response of the nanofiber for E. coli culture components
E. coli strain of ATCC25922 was cultured in a highly nutritious LB medium and then the E. coli cells were separated as pellets by centrifugation. The supernatant fluid after the centrifugation process was collected which contained all the secretion materials E. coli produced during the growth phase. A significant colorimetric transition from blue to red was found when the nanofiber was treated with supernatant fluid. The color change began within 30 s at the edges of the nanofiber mats and progressed towards the center of the mats. The response time within 30 s was significantly short in comparison with those reported previously, suggesting a sensitive detection offered by the PU-PDA nanofibers.18,32,33 After 3 min, the red covered most of the nanofiber mat decorated with some blue patches. The blue patches were due to the inability of the supernatant fluid to penetrate the nanofiber mat. PU-PDA is a superhydrophobic material12 and hence the wettability of the nanofiber mat was poor. The blue patches eventually became red after 24 h. On the other hand, no color change was observed when the nanofibers were tested with E. coli cell pellets or LB culture medium solution (control). The results are in good agreement with the receptor-free fabrication method of PDA nanofibers. The color change by supernatant fluid occurred most likely because the color changing substances (CCS) present in the E. coli secretion reacted with the highly ordered PDA vesicles in the nanofiber and triggered a molecular configuration change in the PDA vesicles, resulting in a visible colorimetric response in the PDA nanofibers. Interestingly, the observation of color change in supernatant fluid but not in cell pellets was previously reported in the PDA that was purposefully functionalized and attached with a lipid receptor to detect E. coli, Shigella sonnei, and Salmonella typhimurium.18 In comparison, the current method of electrospinning to fabricate PDA biosensors was in favor because it was a simple and cost-effective strategy in developing receptor-free PDA biosensors. Table 1 shows images of PU-PDA nanofiber mats before and after testing with E. coli cell pellets, supernatant fluid, and LB media (control).Autoclaving is a sterilization process where high-temperature steam is used with high pressure to denature essential structural and enzymatic proteins, DNA, and RNA, resulting in the death of the bacteria. Proteins and nucleic acids in the bacterial supernatant fluid are denatured by the extreme heat and pressure during the autoclaving process.34 It was found that the PDA nanofiber tested with autoclaved supernatant fluid changed its color significantly faster than that tested with non-autoclaved supernatant fluid, suggesting that structural changes of CCS in the E. coli supernatant fluid occurred during autoclaving and accelerated the colorimetric response in the PDA molecules. The autoclaving experiment also indicated that the CCS may not be a protein, DNA, or RNA since they typically denature during high heat and pressure. Additionally, the colorimetric response of the PDA nanofibers was tested with lipopolysaccharide (LPS). LPS is a surface molecule that is commonly found in Gram-negative bacteria such as E. coli, especially in supernatant fluid, and maintains the structural integrity of the bacterial outer membrane.35 LPS was prepared separately with distilled water and LB broth to test with PU-PDA nanofibers and no color change was observed.
To quantify the colorimetric response of the PU-PDA nanofibers, reflectance data were taken for each of the nanofiber mats before and after exposure to each E. coli culture component at certain time intervals (t). Two representative reflectance spectra of the PU-PDA nanofibers are shown in Fig. 6a and b. The spectra of the nanofibers tested in LB media showed no significant changes from t = 0 to t = 180 min during the test as shown in Fig. 6a, which agrees with the lack of color change in the photographs shown in Table 1. In the case of autoclaved supernatant fluid, a peak at 540 nm (red) developed progressively and significantly on the spectra from t = 0 to t = 180 min during the test as shown in Fig. 6b, suggesting a colorimetric transition from blue to red in the nanofiber mat as shown in the photographs. The reflectance intensity at blue (640 nm) and red (540 nm) on the spectra was used in calculating colorimetric response percentages (CR%) for the nanofibers tested with different E. coli culture components. Fig. 6c shows the CR% as a function of testing time for the nanofibers tested. To see a significant color transition in the nanofiber, it took 3 min which confirms the sensitivity of the system discussed previously. The colorimetric transition was visible at t = 3 min, continued increasing, and plateaued, resulting in a peak colorimetric transition of 28% for the autoclaved supernatant fluid and 10% for the non-autoclaved supernatant fluid, respectively. The CR% was negligible for the PU-PDA nanofiber mats tested with E. coli cell pellets from t = 0 to t = 24 h.
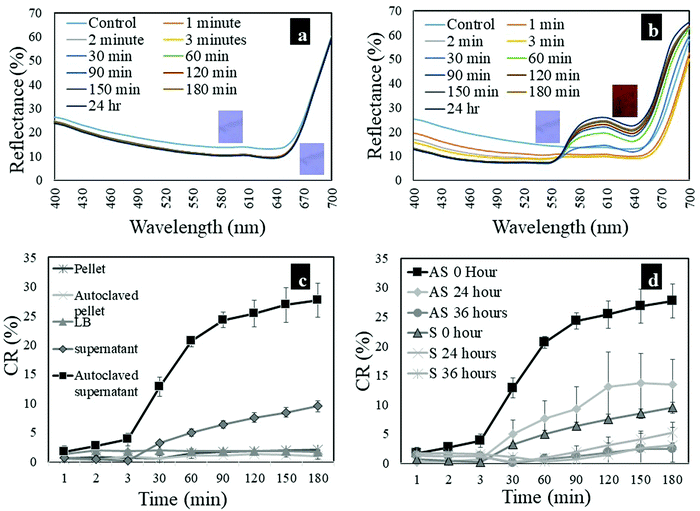 | ||
| Fig. 6 Representative reflectance spectra of PU-PDA nanofibers treated with (a) LB medium (control) and (b) autoclaved supernatant fluid; (c) CR% of PU-PDA nanofibers treated with bacterial components and (d) CR% of PU-PDA nanofibers treated with supernatant fluid following storage for 0 h, 24 h and 36 h (AS = autoclaved supernatant fluid; S = non-autoclaved supernatant fluid). A whole set of reflectance spectra are provided in Fig. S1 in the ESI.† | ||
Colorimetric response of nanofibers to supernatant fluid with increased storage time
Supernatant fluids with different storage times (ts = 0, 24, and 36 h) were evaluated for the ability to activate a colorimetric transition in the PU-PDA nanofiber. Components in the supernatant fluid such as carbohydrates and proteins can undergo structural changes such as degradation over time as no new metabolites are produced in the absence of bacterial cells. Table 2 shows the photographs of PU-PDA nanofiber mats before and after testing with supernatant fluid with different storage times (ts). Supernatant fluid with increased storage time changed the color of nanofibers significantly slower than the supernatant fluid with no storage time, suggesting degradation in CCS with increased storage. In addition, the degradation of CCS in autoclaved supernatant fluid with ts = 36 h was significant because the vibrant red color was absent in the nanofibers, which was not observed in non-autoclaved supernatant fluid with ts = 36 h (Table 2).Fig. 6d shows CR% of the nanofibers tested with supernatant fluid with different storage times (ts). Generally, color change started at t = 3 min and then increased in CR% followed by a plateau at t = 180 min. A reduction in CR% was found with an increase in the storage time of supernatant fluid, which agrees with the photographs presented in Table 2. The CR% at the plateau in the case of autoclaved supernatant fluid was 28% at ts = 0 h, 14% at ts = 24 h, and 2.5% at ts = 36 h, respectively. The CR% at the plateau decreased by almost 25.5% from ts = 0 h to ts = 36 h. On the other hand, the non-autoclaved supernatant fluid demonstrated CR% at the plateau of 10% at ts = 0 h, 5% at ts = 24 h, and 3% at ts = 36 h, respectively. CR% at the plateau decreased by almost 7%.
Colorimetric response of nanofibers to size-fractionated supernatant fluid
The supernatant fluid is a complex mixture of monomers and biopolymers having different molecular weights. To determine a specific molecular weight range, the E. coli supernatant fluid was size-fractionated and tested with PU-PDA nanofibers. Table 3 shows the photographs of PU-PDA nanofibers tested with size-fractionated supernatant fluid. The color change was found for all fluids that passed through a filter with a molecular weight cutoff of 3k, 30k, and 100k Da. The color change was also observed for the fluid that could not pass through the filter having a molecular weight cutoff of 100k Da. This result suggested that the CCS was composed of multimetric substances that were not a single component in the fluid, but multiple components with different molecular weights.Colorimetric response of nanofibers to extracellular polymeric substances (EPS)
The bacterial supernatant fluid is essentially made of materials produced by bacteria that are collectively known as extracellular polymeric substances (EPS). The EPS produced by E. coli can broadly be classified as bound and free EPS. The bound EPS remains attached to the E. coli cell and the free EPS independently floats in the biofilm.21 As discussed previously, E. coli cell pellets did not induce any color change in PU-PDA nanofibers, which indicates that the CCS was not the bound EPS. Therefore, free EPS were extracted from both autoclaved and non-autoclaved supernatant fluid to test with nanofibers. Table 4 shows the color change behaviors of the nanofibers immersed in free EPS solutions. The free EPS extracted from both autoclaved and non-autoclaved supernatant fluid changed the color of nanofibers. Due to the superhydrophobic nature of the PU-PDA nanofiber mat, the free EPS solution could not penetrate the nanofiber fiber mat completely. In addition, the concentration of CCS in free EPS might not be as high as in the supernatant fluid. For these reasons, some portion of the nanofiber remained blue after free EPS exposure as shown in the photographs. The difference in the red color between the nanofibers treated with free EPS collected from autoclaved and non-autoclaved supernatant fluids can be a result of structural differences in CCS due to the autoclaving process. The response time of color changes was 30 min in the free EPS extracted from autoclaved supernatant fluid and 3h for the non-autoclaved fluid, respectively. To the best of our knowledge, it is the first report of testing PDAs with free EPS extracted from bacterial culture, which enhanced the understanding of the response mechanism in bacteria-sensing PDA biosensors.Colorimetric response of nanofibers to E. coli culture components from M9 minimal media
There are different secretion pathways for a Gram-negative bacterium such as E. coli and these secretion pathways can change with variations in the surrounding environment such as culture media.36 M9 minimal medium contains only salts and a nitrogen source and is often used to study the nutritional aspects of E. coli growth when supplemented with different carbon sources and amino acids. M9 minimal medium is a defined growth medium that lacks the peptides, amino acids, vitamins, and trace elements found in LB media. In this experiment, M9 minimal medium was supplemented with arabinose and thiamine (due to the auxotrophy of E. coli) and different E. coli culture components were tested with the PU-PDA nanofibers. The photographs from the result are shown in Table 5. No color changes were promoted by any of the culture components from M9 minimal media. The result is significantly different from the experiment with LB media and most likely, the minimal nutritional components that E. coli received from M9 minimal media were not enough to produce the CCS responsible for a color change in the PU-PDA nanofibers.Conclusions
A simple method of electrospinning was reported to fabricate molecular receptor-free PDA nanofiber biosensors that demonstrated a rapid blue-to-red change in detecting E. coli with a response time of 0.5 min to 3 min and a critical bacterial concentration (CBC) of 9 × 108 CFU mL−1. The receptor-free PDA nanofiber biosensors attributed high sensitivity of bacterial detection via colorimetric transition to a highly ordered and self-assembled PDA molecular vesicle structure that was promoted and enhanced in the electrospinning process due to large electrostatic forces. The PDA nanofiber composites also showed solvatochromism in response to different organic solvents with different colorimetric response percentage, suggesting a potential sensor application in fingerprinting organic solvents. The nature of the color-changing substances (CCS) in the complex E. coli culture was investigated. The colorimetric response of the nanofibers was found with the supernatant fluid and free EPS. The CCS that was produced in rich culture media such as LB media was unlikely a protein, DNA, RNA, or LPS and was likely the composition of multimetric components with different molecular weights. Interestingly, when E. coli received minimum nutrition, it changed the secretion system and thus altered the colorimetric transition behavior of the PDA nanofiber system. The CCS was not present in E. coli grown in a minimum nutrition medium (M9 minimal medium) because no color change was found in the PDA. On the other hand, the color-changing rate of the nanofibers decreased significantly with increasing storage time, suggesting that the CCS degraded or lost the ability to interact with the PU-PDA nanofibers with an increase of storage time.Conflicts of interest
The authors declare no potential conflict of interest concerning the research, authorship, and/or publication of this article.Acknowledgements
This work was supported by the funding from the Colorado State University College Research Council and the AATCC Graduate Student Research Support Grant provided by the American Association of Textile Chemists and Colorists (AATCC).References
- S. Günes, H. Neugebaur and N. Sariciftci, Chem. Rev., 2007, 107, 1324–1338 CrossRef.
- Q. Pei, G. Yu, C. Zhang, Y. Yang and A. Heeger, Science, 1995, 269, 1086–1088 CrossRef CAS.
- K. M. Coakley and M. D. McGehee, Chem. Mater., 2004, 16, 4533–4542 CrossRef CAS.
- X. Feng, L. Liu, S. Wang and D. Zhu, Chem. Soc. Rev., 2010, 39, 2411–2419 RSC.
- E. Smela, Adv. Mater., 2003, 15, 481–494 CrossRef CAS.
- C. Kallinger, M. Hilmer, A. Haugeneder, M. Perner, W. Spirkl, U. Lemmer, J. Feldmann, U. Scherf, K. Müllen, A. Gombert and V. Wittwer, Adv. Mater., 1999, 10, 920–923 CrossRef.
- D. T. McQuade, A. E. Pullen and T. M. Swager, Chem. Rev., 2000, 100, 2537–2574 CrossRef CAS.
- X. Qian and B. Städler, Chem. Mater., 2019, 31, 1196–1222 CrossRef CAS.
- D. H. Charych, J. O. Nagy, W. Spevak and M. D. Bednarski, Science, 1993, 261, 585–588 CrossRef CAS.
- L. Li, X. An and X. Yan, Colloids Surf., B, 2015, 134, 235–239 CrossRef CAS.
- M. Ulaganathan, R. V. Hansen, N. Drayton, H. Hingorani, R. G. Kutty, H. Joshi, S. Sreejith, Z. Liu, J. Yang and Y. Zhao, ACS Appl. Mater. Interfaces, 2016, 8, 32643–32648 CrossRef CAS.
- L. Chen, F. Wu, Y. Li, Y. Wang, L. Si, K. I. Lee and B. Fei, J. Membr. Sci., 2018, 547, 93–98 CrossRef CAS.
- A. K. M. M. Alam, J. P. Yapor, M. M. Reynolds and Y. V. Li, Materials, 2016, 9, 202 CrossRef.
- J. P. Yapor, A. Alharby, C. Gentry-Weeks, M. M. Reynolds, A. K. M. M. Alam and Y. V. Li, ACS Omega, 2017, 2, 7334–7342 CrossRef CAS.
- A. St John and C. P. Price, Clin Biochem Rev, 2014, 35, 155–167 Search PubMed.
- S. U. Son, S. B. Seo, S. Jang, J. Choi, J. Lim, D. K. Lee, H. Kim, S. Seo, T. Kang, J. Jung and E.-K. Lim, Sens. Actuators, B, 2019, 291, 257–265 CrossRef CAS.
- C. H. Park, J. P. Kim, S. W. Lee, N. L. Jeon, P. J. Yoo and S. J. Sim, Adv. Funct. Mater., 2009, 19, 3703–3710 CrossRef CAS.
- P. Villalobos, M. I. Chavez, Y. Olguín, E. Sánchez, E. Valdés, R. Galindo and M. E. Young, Electron. J. Biotechnol., 2012, 15, 4–15 Search PubMed.
- K.-W. Kim, H. Choi, G. S. Lee, D. J. Ahn and M.-K. Oh, Colloids Surf., B, 2008, 66, 213–217 CrossRef CAS.
- A. C. dos Santos Pires, N. de Fatima Ferreira Soares, L. H. M. da Silva, M. do C. H. da Silva, M. V. De Almeida, M. Le Hyaric, N. J. de Andrade, R. F. Soares, A. B. Mageste and S. G. Reis, Sens. Actuators, B, 2011, 153, 17–23 CrossRef.
- K. E. Eboigbodin and C. A. Biggs, Biomacromolecules, 2008, 9, 686–695 CrossRef CAS.
- X. Zhang, Fundamentals of Fiber Science, DEStech Publications Inc., Lancaster, PA, 2014 Search PubMed.
- N. Bhardwaj and S. C. Kundu, Biotechnol. Adv., 2010, 28, 325–347 CrossRef CAS.
- F. Ko, Y. Gogotsi, A. Ali, N. Naguib, H. Ye, G. L. Yang, C. Li and P. Willis, Adv. Mater., 2003, 15, 1161–1165 CrossRef CAS.
- C. F. Holder and R. E. Schaak, ACS Nano, 2019, 13, 7359–7365 CrossRef CAS.
- S. K. Chae, H. Park, J. Yoon, C. H. Lee, D. J. Ahn and J.-M. Kim, Adv. Mater., 2007, 19, 521–524 CrossRef CAS.
- F. Hassan, C. Gentry-Weeks, M. M. Reynolds and Y. V. Li, J. Appl. Polym. Sci., 2019, 136, 47877–47890 CrossRef.
- M. F. Freshwater and C. T. Su, Ann. Plast. Surg., 1980, 4, 216–218 CrossRef CAS.
- M. C. Robson, R. J. Mannari, P. D. Smith and W. G. Payne, Am. J. Surg., 1999, 178, 399–402 CrossRef CAS.
- S. Pumtang, W. Siripornnoppakhun, M. Sukwattanasinitt and A. Ajavakom, J. Colloid Interface Sci., 2011, 364, 366–372 CrossRef CAS.
- H. Jiang, Y. Wang, Q. Ye, G. Zou, W. Su and Q. Zhang, Sens. Actuators, B, 2010, 143, 789–794 CrossRef CAS.
- W. Wu, J. Zhang, M. Zheng, Y. Zhong, J. Yang, Y. Zhao, W. Wu, W. Ye, J. Wen, Q. Wang and J. Lu, PLoS One, 2012, 7, e48999, DOI:10.1371/journal.pone.0048999.
- T. V. de Oliveira, N. de F. F. Soares, D. J. Silva, N. J. de Andrade, E. A. A. Medeiros and A. T. Badaró, Sens. Actuators, B, 2013, 188, 385–392 CrossRef CAS.
- H. Neurath, J. P. Greenstein, F. W. Putnam and J. A. Erickson, Chem. Rev., 1944, 34, 157–265 CrossRef CAS.
- M. A. Valvano, O Antigen Biosynthesis, in Comprehensive Natural Products II, ed. H.-W. B. Liu and L. Mander, Elsevier, USA, 2010, pp. 297–314 DOI:10.1016/B978-008045382-8.00683-3.
- A. W. Maresso, Bacterial Secretion Systems, in Bacterial Virulence- A Conceptual Primer, Springer Nature Switzerland AG, Cham, Switzerland, 2019, pp. 103–114 DOI:10.1007/978-3-030-20464-8_9.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ma00619j |
| This journal is © The Royal Society of Chemistry 2020 |