Open Access Article
Open Access ArticleCovalent attachment of chitosan to graphene via click chemistry for superior antibacterial activity†
Jing
Huang
 *ab,
Zhenyao
Yin
a and
Jinggao
Wu
c
*ab,
Zhenyao
Yin
a and
Jinggao
Wu
c
aState Key Laboratory of Silkworm Genome Biology, Key Laboratory of Sericultural Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs, College of Biotechnology, Southwest University, Chongqing 400715, P. R. China. E-mail: hj41012@163.com
bInstitute for Clean Energy & Advanced Materials, Faculty of Materials and Energy, Southwest University, Chongqing 400715, P. R. China
cKey Laboratory of Rare Earth Optoelectronic Materials & Devices, College of Chemistry and Materials Engineering, Huaihua University, Huaihua 418000, P. R. China
First published on 2nd June 2020
Abstract
Chemically converted graphene sheets are functionalized by treatment with aryl diazonium salts via click chemistry. Characteristic results indicate that chitosan is anchored onto the graphene surface by way of benzyl 1,2,3-triazole. In view of the biological appraisal, the prepared sample demonstrates higher antimicrobial activity than that of pure chitosan.
Introduction
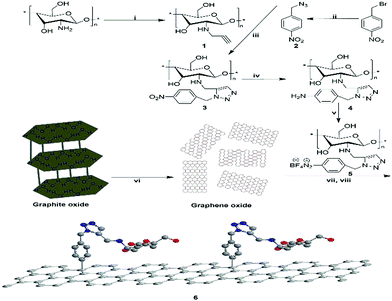
Drug resistance in pathogenic bacteria is increasing throughout the world, leading to higher mortality and increased healthcare costs.1 This has created an urgent need to devote continuous efforts to the discovery and development of new antimicrobials with broader spectrum of activity and lower toxicity. Many structural frameworks have been described as privileged structures2 and in particular the five membered heterocyclic imidazole nucleus is endowed with various biological activities due to the presence of ring nitrogens.3 Hence, with the emergence of resistance mechanisms, there is a need for newer antimicrobial agents to combat resistance developed against widely used antimicrobial drugs.4,5 Click chemistry provides a powerful tool in chemical biology and drug discovery, as it allows efficient and clean creation of compounds under extremely mild conditions.6 The Huisgen thermal cycloaddition between an organic azide and an alkyne was rendered clickable through copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC),7 which results in the exclusive formation of 1,4-disubstituted 1,2,3-triazoles. CuAAC click chemistry has gained particular significance and popularity from its wide range of applications in drug discovery,8 bioconjugation,9 and materials science.10 A close examination on their inhibition of hTK-2 revealed that a bulkier substituent, preferably a large aromatic ring, may provide key interactions for target binding.11,12 The topic of graphene chemistry has been reviewed extensively. The covalent functionalization of pristine graphene involves mostly reactive species that can form covalent adducts with the sp2 carbon structures in graphene. The diazonium chemistry was introduced to graphene by the Haddon group,13 which could be employed to increase the conductivity14 and to control the potential15 as well as the solubility of the graphene.16,17 One of the most versatile and facile methods of covalently grafting organic molecules onto graphene surfaces is the reduction of aryl diazonium salts through either electrochemical or spontaneous reduction with the substrate.18–21 In particular, the reduction of 4-nitrophenyl diazonium tetrafluoroborate could lead to a fully ordered close-packed monolayer of vertically oriented nitrophenyl groups on graphene.22,23Herein we report, for the first time, the design and development of a sample (Scheme 1) of chitosan anchored on graphene via the group of 1,2,3-triazole for an antibacterial agent. The biocompatibility of chitosan and the cooperation with 1,2,3-triazole toward antibacterial activity were investigated in detail.
Results and discussion
The successful attachment of the chitosan groups on graphene is confirmed by FT-IR (Fig. S1, ESI†). New peaks are observed in the spectrum of 6 at 851–899, 1021, and 1367 cm−1, which corresponds respectively to the stretching vibrations of C–O and the heterocycle as well as –CH2–, in good agreement with the spectrum of chitosan. The shape, position, and intensity of other bands do not change significantly in comparison to the spectrum of the starting material and are characteristic of thermally reduced GO (Graphene Oxide). In addition, the peak at 1602 cm−1 which is ascribed to phenyl group stretching vibrations and the band at 1279 cm−1 which is put down to the vibration of a triazole ring, further verify the successful immobilization of chitosan.Diffuse reflectance UV-vis spectra are also acquired in order to probe any change in the structure of the graphene sheets upon functionalization. UV-visible spectra of 6 show a high absorption in the entire spectral region, which is characteristic of graphene (Fig. S2, ESI†). The mass attenuation coefficients for 5 and 6 (taking into account the contribution from the grafted chitosan chains) are found to be at 560 nm. We also observe a red shift from 220 nm to 240 nm upon anchoring on graphene, which might be attributed to the p–p* transition of the phenyl groups.
The solubilities of 5 and 6 are evaluated by water contact angle measurements. The water contact angles of the samples are 25.8° and 82.3°, respectively, which indicates that the water contact angles increase after immobilizing on graphene (5vs.6, 25.8° vs. 82.3°) and that the solubilities vary from hydrophilicities to hydrophobicities. The result clearly demonstrate that the alteration of the solubility of the samples due to the graphene and diazonium functionalization improves the solubility of pristine graphene without using an external stabilizer.
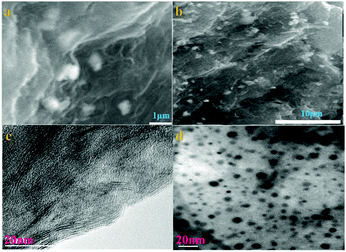
Shown in Fig. 1a and c, the inerratic neat plate layers are ascribed to the aggregation of the parts of graphene in sample 6, while the anomalous suborbicular segments are attributed to the congregation of proportions of aryl groups in sample 6. Large amounts of nanoparticles with a hierarchical diameter are distributed on the surface of the layered graphene, indicating that the functional groups of aryl diazonium salt anchored on the graphene successfully. The chemical reactivity of graphene is influenced by its physical structure, including the number of layers, the type and structure of edges, the degree of strain and grain boundaries. Mechanical strain in the graphene lattice has been theoretically shown to increase chemical reactivity, and graphene is corrugated with nanometer-scale ripples. The curvature helps to accommodate the movement of carbon atoms out of the plane when changing from sp2 to sp3 hybridization upon covalent functionalization. Due to the combined effect of the disordered atomic structures and enhanced DOS (density of states), an increased chemical reactivity is expected.24
After the aryl diazonium salt 5 anchored on the graphene surface, the intensities of all peaks of 6 obviously increase (Fig. S3, ESI†). Meanwhile, XRD is an efficient tool to characterize the d-space of graphene materials. Sample 6 displays seven characteristic peaks centered at 22.62°, 43.38°, 43.4°, 50.5°, 64.96°, 74.2° and 78.06°, providing evidence that both of them comprise a similar layered structure with an interlayer space of around 3.93 Å. The intensities of the peaks of 6 are clearly stronger than those of 5, owing to the introduction of graphene. Meanwhile, the XRD pattern of graphene exhibits a broad peak centered at 2θ = 23.4°, indicating a d-space of 3.74 Å. On account of this, the immobilization on graphene contributes to the augmentation of the interlayer space of 6.
The morphology and structure of the prepared materials are characterized by transmission electron microscope (TEM). The TEM image of 6 shows that the nanoparticles are distributed on the surface of the graphene with the hierarchical particle size from a few nm to ∼10 nm. Fig. 1b and d show that sample 6 has a unique network nanostructure and clearly illustrate that the aryl diazonium salts in the nanostructure are composed of nanoparticles that are uniformly distributed on graphene.
In order to further investigate the structure change of graphene after functionalization, XPS analysis of 6 is performed, which demonstrates the presence of C, N and O (Fig. S4A, ESI†). Two strong peaks at 285.0 eV (C1s) and 532.6 eV (O1s) (Fig. S4D, ESI†) are observed for sample 6. Meanwhile, a new but comparatively weak signal is also shown at 399.65 eV (N1s) (Fig. S4C, ESI†). The C1s spectrum of 6 shows a significant decrease of the signals at 282–300 eV, which indicates loss of C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functionalities (Fig. S4B, ESI†). After the chemical attachment of aryl groups, the single peak at 284.63 eV, which is due to the sp2 hybridized C atoms, is transformed into a broad envelope and deconvolution reveals that it consists of several components. In addition, the XPS survey spectrum reveals N1s peaks and the high resolution N1s spectrum shows two peaks centered at 399.7 and 406 eV (Fig. S3C, ESI†). The peak at higher BE is assigned to the 1,2,3-triazole which is formed by a click coupling reaction. The lower BE N1s peak at 399.7 eV is associated with imine groups and confirms the presence of aryl groups on the graphene surface. The relative atomic concentration of nitrogen in sample 6 is evaluated to be 7.79%, authenticating the presence of nitrogenous groups in the sample.
O functionalities (Fig. S4B, ESI†). After the chemical attachment of aryl groups, the single peak at 284.63 eV, which is due to the sp2 hybridized C atoms, is transformed into a broad envelope and deconvolution reveals that it consists of several components. In addition, the XPS survey spectrum reveals N1s peaks and the high resolution N1s spectrum shows two peaks centered at 399.7 and 406 eV (Fig. S3C, ESI†). The peak at higher BE is assigned to the 1,2,3-triazole which is formed by a click coupling reaction. The lower BE N1s peak at 399.7 eV is associated with imine groups and confirms the presence of aryl groups on the graphene surface. The relative atomic concentration of nitrogen in sample 6 is evaluated to be 7.79%, authenticating the presence of nitrogenous groups in the sample.
Hence, successful functionalization supports the assumption that partial rearomatization of the nanosheets under hydrazine reduction conditions has occurred and thereby provided a surface for aryl grafting using the diazonium species.25
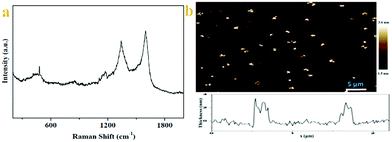
Raman spectra of 6 are acquired at an excitation wavelength of 532 nm (Fig. 2a). Two first-order bands are observed in the 1000–1800 cm−1 region, which correspond to vibrations of carbon atoms at defect sites (D band, ≈1340 cm−1) and vibrations of carbon atoms in the ideal graphitic lattice (G band, ≈1585 cm−1).26 The difference spectrum, calculated by subtracting both spectra, reveals that the G band became sharper and the D band intensity decreased slightly upon grafting. This observation implies a small increase in the size of the sp2 domains. We consider two factors that decrease the 2D peak intensity after diazonium functionalization, as illustrated in Fig. 2. First, graphene loses an electron to reduce the diazonium salt to an aryl radical. Second, physisorbed diazonium molecules and oligomers can dope the graphene via surface charge transfer.24
The Raman spectrum of 6 shows a similar profile to that of GO with a diamondoid (D) to graphitic (G) ratio close to 1, confirming incomplete recovery of the graphene structure, similar to what is observed for thermally reduced graphene. There are two features in the Raman spectra of pristine graphene, the G mode at ∼1585 cm−1 and the 2D mode at∼2700 cm−1. The G mode's position and shape do not change after the chemical modification, implying the reserve of the backbone of the graphene lattice. Otherwise, the G mode will split into two distinct peaks because the 2-fold degeneracy of the TO and LO phonons is lifted by symmetry breaking, as found in carbon nanotubes27 and the aromatic molecule decorated the graphene monolayer.28 But one must have noticed that in the Raman spectra of modified graphene, the most significant change is the appearance of the D mode at ∼1350 cm−1. The D mode is arising from the defect-involved double resonant Raman process at the K point in the Brillouin zone.29 Since the backbone of graphene is preserved, we attribute the presence of the D mode to the transition from sp2 to sp3 hybridization of the graphene carbon atoms. In fact, the presence of the D mode is generally used to monitor the covalent bonds formed in graphene and widely accepted.30 These spectroscopic features shown herein fully demonstrate that we successfully modified graphene via a click reaction, and the result of the corresponding reaction should be the aryl groups connected to the graphene sheets via σ-bonds.
Morphological insights into the chitosan–graphene hybrid materials come from AFM. Individual functionalized graphene sheets are imaged using tapping mode AFM. Fig. 2b shows images of graphene sheets spin coated onto a mica surface using a 0.1 mg mL−1 dispersion of 6 in deionized water. The theoretical height for a graphene sheet functionalized on both sides is ∼2.2 nm, assuming that the height of the bare graphene sheets is 1 nm31 with the substituted aromatic groups contributing ∼0.6 nm in height. On average, the height of functional graphene sheets ranges from 1.8 to 3.2 nm. Meanwhile, analysis of numerous AFM images obtained in tapping mode, reveals the presence of graphene-oxide sheets with heights ranging between 1.6 and 2.5 nm. Considering the height of a single graphene sheet as 0.8–1.0 nm and the added contribution from the linked positively charged imidazolium moieties as well as the coulombic repulsions between the modified layers, it is logical to expect that the obtained images are representative of single- and/or bi-layers of exfoliated modified graphene sheets (vide supra Raman).
The presence of functional groups on the graphene sheets is further analyzed using TGA (Fig. S5, ESI†). TG analysis of 6 shows a weight loss of 8.5% for 6 below 200 °C. Meanwhile, 6 begins a sharp weight loss at 210 °C. The thermal stabilities of 6 are higher than that of pure chitosan. Obviously, sample 6 is proceeding in two major steps: the first desorption is attributed to physisorbed reaction byproducts that do not bind covalently to the graphite/graphene substrate and are thus removed at low temperatures; the second, covalently bound addends are cleaved from the carbon surface, as is apparent from the significant reduction in mass. In addition, the onset temperature and the fact that the removal of organic groups occurs over 200 °C confirms that the groups are chemically anchored and not simply adsorbed. This underscores the efficacy of the functionalization method described here.
The results of antimicrobial activities indicate that the samples show moderate activities against certain tested stains in vitro. As seen in Table 1, samples 5 and 6 exhibit comparable antibacterial activities with MIC values varying from 4 to 32 mg mL−1, which shows higher properties than that of chitosan.
| Minimum inhibitory concentration (MIC, mg mL−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fungal strains | Bacterial strains | ||||||||
| Compds | C. albicans | C. mycoderma | S. aureus | B. subtilis | P. aeruginosa | E. coli | S. dysenteriae | MRSA | E. typhosa |
| a Minimum inhibitory concentrations were determined by a two-fold serial dilution method for microdilution plates. b A = chitosan; B = chloramphenicol; C = fluconazole. c C. albicans, Candida albicans (ATCC76615); C. mycoderma, Candida mycoderma; S. aureus, Staphylococcus aureus (ATCC25923); B. subtilis, Bacillus subtilis (ATCC6633); P. aeruginosa, Pseudomonas aeruginosa; E. coli, Escherichia coli (JM109); S. dysenteriae, Shigella dysenteriae; MRSA, Methicillin-resistant Staphylococcus aureus (N315); E. typhosa, Eberthella typhosa. | |||||||||
| 5 | >512 | >512 | 16 | 32 | 16 | 32 | 8 | 32 | 32 |
| 6 | >512 | >512 | 8 | 16 | 8 | 16 | 4 | 16 | 16 |
| A | >512 | >512 | 128 | 256 | 128 | 256 | 128 | 256 | 256 |
| B | — | — | 4 | 2 | 8 | 4 | 4 | 2 | 2 |
| C | 0.5 | 4 | — | — | — | — | — | — | — |
Among all the tested strains, sample 6 displays superior antibacterial activities against S. dysenteriae with an MIC value of 4 mg mL−1, which is comparable to that of the reference drug Chloramphenicol (4 mg mL−1) and is higher than that of chitosan (128 mg mL−1). Meanwhile, sample 5 indicates lower properties against S. dysenteriae than that of sample 6 (5vs.6: 8 vs. 4 mg mL−1), which is ascribed to the effect of the graphene. A similar phenomenon occurs for other tested strains such as against S. aureus (5vs.6: 16 vs. 8 mg mL−1); MRSA (5vs.6: 32 vs. 16 mg mL−1); and E. typhosa (5vs.6: 32 vs. 16 mg mL−1). In view of these facts, it could be deduced that anchoring aryl diazonium salt on the graphene contributes to enhancing the antibacterial activity.
Notably, chitosan personally displays antibacterial activities to a certain extent as well in Table 1, accompanied by MIC values varying from 128 to 256 mg mL−1. When the 1,2,3-triazole is introduced into chitosan, the antibacterial activity of 5 is better than that of chitosan such as against MRSA (5vs. chitosan: 32 vs. 256 mg mL−1) and further increases up to 16 mg mL−1 after the immobilization of 5 onto the surface of graphene. Moreover, the antibacterial properties of 1,2,3-triazole have been reported.32 These results strongly suggest that the bulkiness provided by the aromatic substituent is crucial to target binding and that its separation from the triazole ring through a linkage tremendously compromises the antiviral potency.
Above all, this result displays that the antimicrobial effect of nanocomposite 6 is very effective owing to its special configuration and confirms that the symbiotic effect of chitosan and 1,2,3-triazole as well as functionalized graphene is responsible for the observed strong bacterial efficiency. Moreover, the absence of oxide functional groups on the edges of the graphene sheets presents a stronger interaction between the more sharpened edges of the graphene (oxide) nanowalls and the cell membrane of the bacteria.
Unfortunately, the antifungal evaluation reveals that almost all of the synthesized 5 and 6 exhibit poor antifungal activities against C. albicans and C. mycoderm, which are relatively weak in comparison with their antibacterial activities. These results also validate the fact that chitosan derivatives 6 have no obvious antifungal activity in vitro, although a 1,2,3-triazole ring is incorporated into the scaffold.
Conclusions
In summary, we employed a convenient procedure, originally developed for functionalization of graphene, to functionalize graphene with chitosan via a click reaction, allowing these nanosheets to be provided with effective antimicrobials. The sample based on graphene with highly porous structures might have potential applications in pharmaceutical and materials science. The 1,2,3-triazole group in the configuration demonstrated in this study may be important for designing and preparing the sample based on graphene.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the financial support from Faculty of Materials and Energy and Institute for Clean Energy & Advanced Materials, Southwest University and Chongqing Key Laboratory for Advanced Materials and Technologies of Clean Electrical Power Sources and the Recruitment Program of Southwest University (SWU117023).References
- K. M. Cao, X. Ding, Y. P. Sheng, Y. Wang and Y. Z. Liu, Chem. Commun., 2020, 56, 4599 RSC.
- X. D. Wen and J. Q. Guan, Nanoscale, 2020, 12, 8065 RSC.
- K. Yamamoto and K. Takatsuka, Phys. Chem. Chem. Phys., 2020, 22, 7912 RSC.
- C. H. N. Barros, H. Devlin, D. W. Hiebner, S. Vitale, L. Quinn and E. Casey, Nanoscale Adv., 2020, 2, 1694 RSC.
- R. M. Tikhov and N. Y. Kuznetsov, Org. Biomol. Chem., 2020, 18, 2793 RSC.
- M. Jaiswal, S. Y. Zhu, W. J. Jiang and Z. W. Guo, Org. Biomol. Chem., 2020, 18, 2938 RSC.
- O. V. Nesterova, O. E. Bondarenko, A. J. L. Pombeiro and D. S. Nesterov, Dalton Trans., 2020, 49, 4710 RSC.
- C. E. Colwell, T. W. Price, T. Stauch and R. Jasti, Chem. Sci., 2020, 11, 3923 RSC.
- T. Ding and J. J. Baumbergl, Nanoscale Adv., 2020, 2, 1410 RSC.
- P. S. O. Ozgen, S. Atasoy, B. Z. Kurt, Z. Durmus, G. Yigit and A. Dag, J. Mater. Chem. B, 2020, 8, 3123 RSC.
- G. H. Lu, L. P. Zuo, J. F. Zhang, H. S. Zhu, W. R. Zhuang, W. Wei and H. Y. Xie, Biomater. Sci., 2020, 8, 2283 RSC.
- T. X. Wang, H. P. Liang, D. A. Anito, X. S. Ding and B. H. Han, J. Mater. Chem. A, 2020, 8, 7003 RSC.
- T. Torres, Chem. Soc. Rev., 2017, 46, 4385 RSC.
- G. Bottari, M. Herranz, L. Wibmer, M. Volland, L. R. Pérez, D. M. Guldi, A. Hirsch, N. Martín, F. D'Souza and T. Torres, Chem. Soc. Rev., 2017, 46, 4464 RSC.
- P. S. Fernández, M. Bissett and H. Ago, Chem. Soc. Rev., 2017, 46, 4572 RSC.
- R. Samal, S. Mondal, A. S. Gangan, B. Chakraborty and C. S. Rout, Phys. Chem. Chem. Phys., 2020, 22, 7903 RSC.
- T. T. T. Hanh, N. M. Phi and N. V. Hoa, Phys. Chem. Chem. Phys., 2020, 22, 7210 RSC.
- T. P. Zhou, N. Zhang, C. Z. Wu and Y. Xie, Energy Environ. Sci., 2020, 13, 1132 RSC.
- S. A. Hashemi, S. Ramakrishna and A. G. Aberle, Energy Environ. Sci., 2020, 13, 685 RSC.
- C. Y. Zhang, S. Wang, H. Zhang, Y. L. Feng, W. M. Tian, Y. Yan, J. M. Bian, Y. C. Wang, S. Y. Jin, S. M. Zakeeruddin, M. Grätzel and Y. Shi, Energy Environ. Sci., 2019, 12, 3585 RSC.
- M. M. Liu, L. L. Wang, K. N. Zhao, S. S. Shi, Q. S. Shao, L. Zhang, X. L. Sun, Y. F. Zhao and J. J. Zhang, Energy Environ. Sci., 2019, 12, 2890 RSC.
- K. V. Kumar, S. Gadipelli, B. Wood, K. A. Ramisetty, A. A. Stewart, C. A. Howard, D. J. L. Brett and F. R. Reinoso, J. Mater. Chem. A, 2019, 7, 10104 RSC.
- J. D. Qin, Y. B. Zhang, S. E. Lowe, L. X. Jiang, H. Y. Ling, G. Shi, P. Liu, S. Q. Zhang, Y. L. Zhong and H. J. Zhao, J. Mater. Chem. A, 2019, 7, 9646 RSC.
- G. L. Wang, L. l. Zou, Q. H. Huang, Z. Q. Zou and H. Yang, J. Mater. Chem. A, 2019, 7, 9447 RSC.
- X. Wang, L. Chen, Z. T. Yuan, J. Rong, J. Feng, I. Muzammil, X. H. Yu, Y. N. Zhang and Z. L. Zhan, J. Mater. Chem. A, 2019, 7, 8967 RSC.
- S. Chen, Y. f. Xiang, C. Peng, W. J. Xu, M. K. Banks and R. Wu, Inorg. Chem. Front., 2019, 6, 903 RSC.
- K. L. Zhang, L. Wang, W. L. Cai, L. F. Chen, D. Wang, Y. H. Chen, H. L. Pan, L. B. Wang and Y. T. Qian, Inorg. Chem. Front., 2019, 6, 955 RSC.
- N. Deka, R. Patidar, S. Kasthuri, N. Venkatramaiah and G. K. Dutta, Mater. Chem. Front., 2019, 3, 680 RSC.
- M. Chen, Y. Luo, C. Zhang, L. Guo, Q. T. Wang and Y. Wu, Org. Chem. Front., 2019, 6, 116 RSC.
- S. Godlewski, M. Engelund, D. Peña, R. Zuzak, H. Kawai, M. Kolmer, J. Caeiro, E. Guitián, K. P. C. Vollhardt, D. S. Portal, M. Szymonski and D. Pérez, Phys. Chem. Chem. Phys., 2018, 20, 11037 RSC.
- E. Rozhina, S. Batasheva, A. Danilushkina, M. Kryuchkova, M. Gomzikova, Y. Cherednichenko, L. Nigamatzyanova, F. Akhatova and R. Fakhrullin, Med. Chem. Commun., 2019, 10, 1457 RSC.
- J. Shin, S. Lee and M. Cha, Med. Chem. Commun., 2019, 8, 625 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, FT-IR, and TG. See DOI: 10.1039/d0ma00082e |
| This journal is © The Royal Society of Chemistry 2020 |