Open Access Article
Open Access ArticleDigital dipstick: miniaturized bacteria detection and digital quantification for the point-of-care†
Emre
Iseri
 *a,
Michael
Biggel
b,
Herman
Goossens
b,
Pieter
Moons
b and
Wouter
van der Wijngaart
*a
*a,
Michael
Biggel
b,
Herman
Goossens
b,
Pieter
Moons
b and
Wouter
van der Wijngaart
*a
aKTH Royal Institute of Technology, Stockholm, Sweden. E-mail: iseri@kth.se; wouter@kth.se
bUniversity of Antwerp, Antwerp, Belgium
First published on 10th November 2020
Abstract
Established digital bioassay formats, digital PCR and digital ELISA, show extreme limits of detection, absolute quantification and high multiplexing capabilities. However, they often require complex instrumentation, and extensive off-chip sample preparation. In this study, we present a dipstick-format digital biosensor (digital dipstick) that detects bacteria directly from the sample liquid with a minimal number of steps: dip, culture, and count. We demonstrate the quantitative detection of Escherichia coli (E. coli) in urine in the clinically relevant range of 102–105 CFU ml−1 for urinary tract infections. Our format shows 89% sensitivity to detect E. coli in clinical urine samples (n = 28) when it is compared to plate culturing (gold standard). The significance and uniqueness of this diagnostic test format is that it allows a non-trained operator to detect urinary tract infections in the clinically relevant range in the home setting.
Introduction
In digital bioassays, a reaction solution is first partitioned into a large number of isolated microreactors such that most reactors contain 0 or 1 target analyte particles, whereafter an assay in each of the microreactors reveals the presence of the analyte quantitatively and discretely. Digital bioassays are of interest for their potentially extremely low limit of detection (single molecule detection), absolute quantification, and multiplexing of the bioassays, in an uncomplicated assay format.1 Digital bioassays include digital polymerase chain reactions (dPCR),2 digital recombinase polymerase amplification (dRPA),3,4 and digital loop-mediated isothermal amplification (dLAMP)5 for nucleic acid detection; digital enzyme-linked immunosorbent assays (digital ELISA)6,7 for protein detection, and; digital colony-forming-unit (CFU) assays8,9 for bacterial detection. Platforms for digital bioassays, such as droplet microfluidic systems,10 pump-valve systems,11 planar emulsion arrays,12 SlipChip,13 lab-on-a-disc,14 and SIMPLE chip,3 either require sophisticated instrumentation4–9,11,12,14,15 or extensive sample preparation.3–5,8,10,12–14,16 The resulting cost of testing and the need for trained personnel have limited the translation of digital assays to the point-of-care (PoC) setting for common infectious diseases like urinary tract infections (UTIs).17,18Today's gold standard diagnostic test for UTI detection is culture-based phenotypic testing,19 which takes 1–3 days and requires trained personnel and laboratory facilities. Other diagnostic formats for the detection of UTIs include culture-based assays (Flexicult,17,20 dip-slides21,22), nitride sticks,23,24 nucleic acid based tests (PCR,25 LAMP5), immunoassays,26–29 and single cell imaging based tests.9 These tests are limited by their need for operation by trained personnel, lab facilities or readout instruments that add cost and complexity, or suffer from high rates of false negative results (Table 1).
| Method | Validity | Time to result | Required operator skills | Complexity/cost |
|---|---|---|---|---|
| Urine culture19 | Gold standard | 24–72 h | Trained | High |
| Flexicult (culture based)17,20 | >90% | 24 h | Trained | High |
| Nitride dipsticks23,24 | 40–70% | 2–5 min | Untrained | Low |
| Dip-slide (culture based)21,22 | 73% | 18–24 h | Trained | Low |
| Nucleic acid based5,25 | NA | 1–5 h | Trained | High |
| Immunoassays26–29 | NA | 20–120 min | Trained | High |
| Single cell imaging based9 | NA | 20–60 min | Trained | High |
| Digital dipstick (this work) | NA | 8–12 h | Untrained | Low |
Here, we provide a digital assay in dipstick format (digital dipstick) with extremely simple handling that measures Escherichia coli (E. coli) concentrations in clinical urine samples without sample preparation. Digital dipsticks have the potential to be used as a self-diagnostic test for UTIs by virtue of their three key characteristics:
1. Clinically relevant results at a potentially low cost.
2. No complicated external equipment or technical skills required during testing.
3. Easy manual readout by the naked eye or potentially automated using a cellphone camera.
Results and discussion
Digital dipsticks
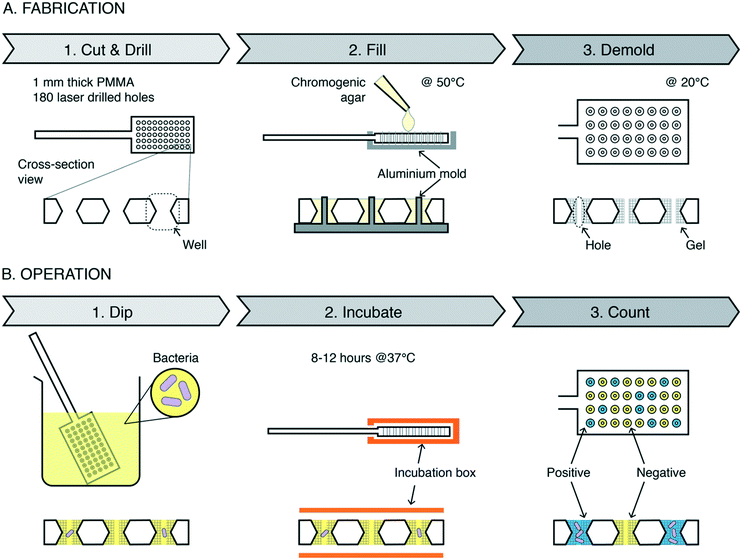
Digital dipsticks are coffee spoon-like plastic sticks made out of 1 mm thick polymethyl methacrylate (PMMA) plates (Fig. 1A and S1A†). The dipsticks contain an array of 180 small hourglass-shaped through-holes (from hereon “wells”) (Fig. S1B†). Each well lodges chromogenic agar (from hereon “gel”), which contains reagents needed for bacteria detection. Each agar gel has a central cylindrical through-hole (from hereon “hole”) with a designed 200 μm diameter and inner volume of 31.4 nL.The dipstick operation
The dipstick operation is simple (Fig. 1B and ESI† Video S1). First, the dipstick is placed in the liquid sample, e.g., raw urine, for a few seconds to capture sample in the wells. Excess sample on the dipstick surface is swiped off by inserting the dipstick into an incubation box, thereby isolating the sample in the separate wells (ESI† Video S2). The box keeps the environment humid, protecting gels from desiccation during incubation at 37 °C. In positive wells (wells containing CFU), enzymatic reactions in the growing bacteria induce a color change in the reagents in the gel.30 Bacterial growth increases the amount of enzyme and thus amplifies the color signal. Negative wells (wells without CFU) remain uncolored. The color versus no color difference of the wells forms a digital signal, and simple counting of colored wells by the naked eye or cellphone camera allows quantifying the bacterial concentration via the most probable number method.31We developed a colorimetric bioassay for miniaturized samples to detect E. coli
An investigation of the commercially available chromogenic agar (ChromAgar orientation) revealed that miniaturizing the volumes of gel during bacterial culture results in a decrease of bacterial growth and color signal (Fig. S2†). We speculate, this results from the decreased volume-per-surface ratio, resulting in a reduced availability of nutrient and reaction reagent. After testing gels with specific mixtures of signal-generating substance (ChromAgar orientation and RedGal) and nutrition broths (Rapid Coliform ChromoSelect Broth) (Fig. S3†), we selected the mixture resulting in the most vivid color change during culturing E. coli for further experiments.Sensor response
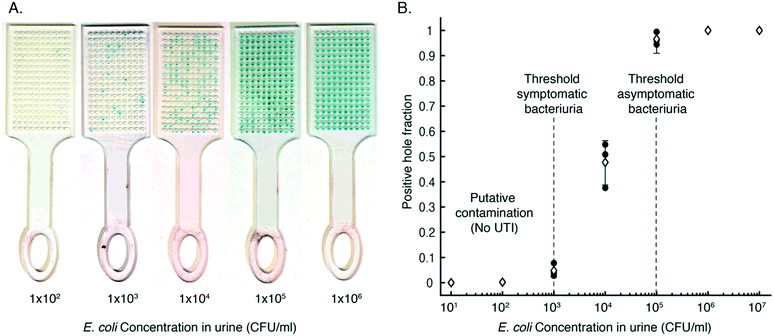
We monitored dipsticks during 24 h of incubation at 37 °C (Fig. 2A). As a sample, we used phosphate-buffered saline (PBS) spiked with E. coli in a ten-fold dilution of concentrations from 5 × 107 to 5 × 101 (n = 3). We imaged each dipstick at the start of incubation and at different time points after. Images were processed with a home-built algorithm using ImageJ and MATLAB. The time-to-color-change scales with the bacterial culture concentration. The highest tested concentration, 5 × 107 CFU ml−1, resulted in a detectable color change after 4 h, whereas concentrations below 103 CFU ml−1 needed at least 10 h of incubation. Fig. 2B shows the positive hole fraction (the number of positive holes divided by the total number of holes) of dipsticks after 12 h of incubation, which is enough to quantify samples at the clinically relevant range for UTI diagnosis. We used the most probable number method31 for curve fitting, and a best-fit analysis indicates that an average of 92 nl urine was captured in each well (between 22 nl and 155 nl with 95% CI). We defined the limit of detection (LoD) to be 253 CFU ml−1, which is the highest concentration in the sensor response curve, with 95% CI band, for which the device has the highest probability of having 1 positive well. We defined the upper limit of quantification (ULoQ) to be 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 CFU ml−1, which is the lowest concentration in the sensor response curve, with 95% CI band, for which the device has the highest probability of having 1 negative well. We also did colony counting of dilution series on agar plates to measure standard deviation in prepared samples (Fig. S4†).
500 CFU ml−1, which is the lowest concentration in the sensor response curve, with 95% CI band, for which the device has the highest probability of having 1 negative well. We also did colony counting of dilution series on agar plates to measure standard deviation in prepared samples (Fig. S4†).
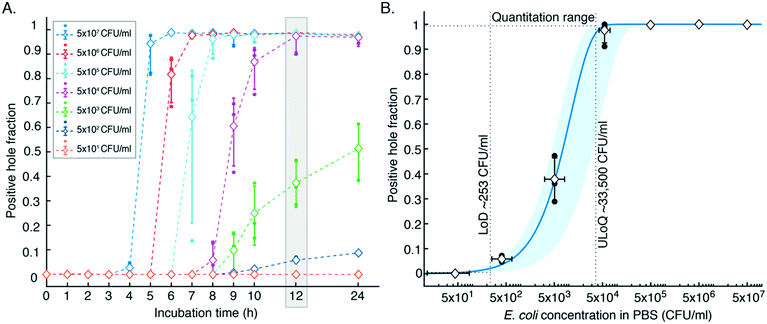 | ||
| Fig. 2 Characterization of digital dipsticks. A. Positive hole fraction vs. incubation time. B. Sensor response curve: positive hole fraction vs. E. coli concentration in PBS after 12 h incubation. Samples measured with the digital dipstick assay consisted of E. coli in phosphate-buffered saline (PBS) with a concentration between 5 × 107 and 5 × 101 CFU ml−1. Solid round dots indicate individual measurements; diamonds indicate average values (n = 3); vertical error bars indicate standard deviation; horizontal error bars indicate the standard deviation of the E. coli concentration in PBS samples; colored dashed lines connecting averages are plotted for eye guidance only. The curved solid blue line is the best-fit digital assay response curve using the most probable number method,31 indicating that an average of 92 nl urine was captured in each well. The blue band indicates the 95% confidence interval (2σ) around the sensor response curve. | ||
Validation with spiked urine samples (Fig. 3)
Validation with clinical urine samples
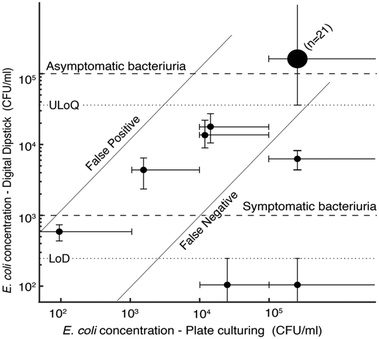
We compared digital dipstick assays to standard urine culture on agar plates (gold standard) for 28 samples of patients with confirmed E. coli infection (Fig. 4). 25 out of 28 results were correlated, whereas 3 out of 28 dipstick measurements gave false-negative results, meaning 89% sensitivity to detect E. coli in clinical samples. The digital dipsticks confirmed that one sample had a concentration below the threshold for symptomatic bacteriuria, three samples had concentrations between the thresholds for symptomatic and asymptomatic bacteriuria, and 21 samples had concentrations above the ULoQ. Two samples with concentration above 105 CFU ml−1 and one sample with concentration between 104 and 105 CFU ml−1 resulted in a false negative in which two readouts were below LoD.We also tested digital dipsticks with PBS samples spiked with species of Klebsiella pneumoniae and Proteus mirabilis. Although the assay is currently only optimized for E. coli detection, the dipsticks detect these species, showing the potential multiplexing capability of the test for different bacterial species (Fig. S5†). We foresee that our device can further be developed and optimised for bacteria identification or antibiotic susceptibility testing in future studies.
Experimental
Fabrication of digital dipstick frames
Digital dipstick frames are fabricated out of 1 mm-thick polymethyl methacrylate (PMMA) plates using a CO2 laser (VLS 2.30, Universal Laser Systems, Vienna, Austria). To get hourglass-shaped arrays, we first engraved, with 80% laser power and 50% laser speed one side of the plate with a 10 by 18 array of circles with 1.2 mm diameter and 1.5 mm center to center spacing. After that, the other side of the plate was engraved using the same laser settings after manual alignment using premade alignment markers in the laser platform. After engraving, circular holes of 0.5 mm diameter were drilled in each engraved well, and the frame was cut into a dipstick format using 90% laser power and 10% laser speed. A dipstick frame with engraved/drilled holes is shown in Fig. S1.†Development of the colorimetric bioassay for miniaturized samples
When we used the commercially available chromogenic agar (CHROMagar™ orientation, Paris, France), we observed that the color signal generated by bacterial was insufficient in miniaturized samples. We, therefore, investigated the effect of miniaturization. Chromogenic agar was prepared according to the user manual (32 g L−1 powder in DI water) and cast on glass slides in layers of various thicknesses (1 mm, 600 μm, 300 μm and 150 μm), defined with the help of spacers. Escherichia coli (E. coli) type strain ATCC 11775 was grown on the cast agar inside a humid incubator at 37 °C for 24 hours. The red colour indicating growth decreased with decreasing thickness of the agar (Fig. S2†).To improve signal generation, we modified the chromogenic agar by adding more nutrients or/and chromogenic substrates (Fig. S3†). Different combinations of chromogenic agar with added Miller's Luria broth (LB broth (Miller), Sigma-Aldrich, MO, USA) in concentrations of 20 g L−1 and 40 g L−1 and with additional Reg-Gal (6-chloro-3-indolyl-β-D-galactopyranoside, Sigma-Aldrich, MO, USA) with concentrations of 60 μg ml−1 and 120 μg ml−1, which generate red color in the presence of E. coli-specific enzyme β-galactosidase, were tested for detecting growth of E. coli. Additionally, chromogenic agar modified with E. coli specific chromogenic broth (Rapid Coliform ChromoSelect Broth, Sigma-Aldrich, MO, USA) with concentrations of 16 g L−1 and 32 g L−1, which generate blue colour in the presence of E. coli-specific enzyme β-galactosidase, were tested for E. coli growth. After 24 hours humid incubation at 37 °C, samples in small glass vials (Fig. S3A†) and 300 μm thick agar pieces (Fig. S3B†) show that the most vivid color change is provided by chromogenic agar prepared with chromogenic broth concentration of 32 g L−1. We used this modified chromogenic agar gel for the rest of our experiments in this study.
Fabrication of aluminum mold
An aluminum mold was machined by computer numerical control (CNC) milling in the shape of an array of cylindrical pins of 200 μm diameter, 1.1 mm height and 1.5 mm spacing, i.e., to fit into the laser-cut holes of dipsticks. After milling, the mold was coated with Teflon (Teflon™ AF 1600, Teflon, DE, USA) to prevent adhesion of gel during molding.Fabrication of digital dipsticks
Chromogenic agar was prepared as described above and kept at elevated temperature (>50 °C) to keep it in liquid form. The aluminum mold was placed on top of a hotplate (80 °C), and few drops of warm chromogenic agar were poured inside the mold. Then, the prefabricated digital dipstick frames were placed on the aluminium mold. Liquid agar spontaneously filled the wells of the frame. To proper filling in every well, a few drops of chromogenic agar were added on top of the dipsticks. After that, the aluminum mold was placed on a cooling block at 0 °C, resulting in the gelation of the chromogenic agar. After gelation, the dipsticks were demolded and the top and the bottom part of the dipstick were cleaned from excess agar by scalpel. Fabricated dipsticks were stored at 4 °C to prevent desiccation and used within the same day of fabrication. Devices with similar content of agarose have a reported shelf life of 4 months at 4 °C.34 Other studies show that the shelf-life of agar plates can be extended to 24 months at the room temperature by vacuum-sealed packaging under controlled atmosphere.35 Thus, we believe that long-term storage (>6 months) of digital dipsticks is possible with modified atmosphere packaging, a method frequently used in food packaging.36Fabrication of incubation boxes
Incubation boxes were fabricated in a FormLabs Form 3 printer using the standard clear transparent resin.Characterization of digital dipsticks for E. coli detection
Ten-fold dilution series of E. coli type strain ATCC 11775 in phosphate-buffered saline (PBS) (Gibco™ Dulbecco's phosphate-buffered saline, Thermo Fisher Scientific, Göteborg, Sweden) were prepared with concentrations between 5 × 107 CFU ml−1 and 5 × 101 CFU ml−1. Plate-counting experiments were performed on diluted samples to evaluate the variation in colony count at the different dilutions.| MPF = 1 − e−cv. | (1) |
We used eqn (1) to calculate the concentration, c, from a measured positive hole fraction, m, as
| c = −ln(m − 1)/v. | (2) |
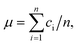 | (3) |
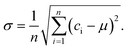 | (4) |
| M = 1 − e−v.μ, | (5) |
| M ± σ = 1 − e−v(μ±σ), | (6) |
| M ± 2σ = 1 − e−v.(μ±2σ). | (7) |
Automated positive hole fraction counting
The following automated image processing protocol was developed to determine the positive hole fraction in a dipstick automatically. Each photograph had three dipsticks measuring the same concentration next to each other. Images were first opened in ImageJ, and center coordinates of top left, top right and bottom left wells of each dipstick were manually extracted to a text file for further analysis. Photographs and text files of coordinates were imported to Matlab, and the following steps were performed to detect the blue color change and count the number of positive wells in each dipstick for every time point:1. The diameter of wells and the position of each well in pixel units for each dipstick was calculated based on the coordinate data taken from ImageJ.
2. Rectangular areas on the top and the bottom of the dipsticks were selected as a region of interest (ROI) and white balancing of the photo was processed based on these ROIs.
3. Based on the calculated position and diameters of wells, 180 circular ROI were selected for the dipstick.
4. For detecting blue color, the blue channel value of each pixel in each ROI was compared to the red channel of each pixel. Five different thresholds were set for five different brightness levels such that the difference between blue and red channel was higher for pixels to be defined as “blue”.
5. To define the well as “positive”, the number of “blue” pixels should be larger than the defined threshold.
6. Finally, the number of positive wells is counted and exported for further data analysis (Table S2†).
The Matlab script is provided in the ESI† File S1.
Validation with spiked urine samples
Ten-fold dilution series of E. coli type strain ATCC 11775 concentrations in between 107 CFU ml−1 and 101 CFU ml−1 were prepared with a urine sample of a healthy donor. Triplicate digital dipstick assays were used to measure the concentration of each sample, including a negative control, which was the untreated urine of the healthy donor. Positive hole fractions are plotted in Fig. 3B, and the data is available in Table S3.† Gel pieces in some wells were lost during the fabrication and operation process, and these wells were excluded during the hole fraction calculations.Trials to detect different species with digital dipsticks
Although our test was optimised only for E. coli detection, the chromogenic agar we used contains different substances to detect different species. We performed tests on five other common bacteria species that cause urinary tract infections (Klebsiella pneumoniae, Proteus mirabilis, Enterococcus faecium, Staphylococcus aureus, Acinetobacter baumannii) to investigate the potential multiplexing capability of digital dipsticks. We used concentrations of these bacteria >5 × 104 CFU ml−1 in PBS and ran duplicate assays. We observed color change for Proteus mirabilis (dark yellow) and Klebsiella pneumoniae (light blue/turquoise) (Fig. S5†). We did not discern colour changes for the other three species. We did not further investigate these results since our study focuses on the detection of E. coli, which cause more than 85% of uncomplicated UTIs.Validation with clinical samples
Urine samples were daily collected from the Antwerp University Hospital (UZA), and stored for more than 24 hours at 4 °C. All samples collected were verified for E. coli infection, and only E. coli positive samples were withheld. Also, samples confirmed to have multi-bacterial infection were excluded from this study. Each sample was placed on a blood agar plate and a cystine–lactose–electrolyte deficient (CLED) agar plate using the EddyJet spiral plating tool (50 μl log mode). 300 μl of samples were pipetted to EddyJet cups and were manually fed to the machine. Plates were incubated at 37 °C and colonies were counted according to the EddyJet manual instructions. Counting was performed if the growth was in the detection range of spiral plating, which is between 2 × 102 and 4 × 105 CFU ml−1. Since digital dipsticks have similar detection range, we did not plate dilution series from samples for a wider range of quantification.Simultaneously, tests with digital dipstick assays were performed on the patient samples. Due to the restricted volume of the samples, for each sample, 0.5 ml was pipetted on a disposable petri dish and the dipstick was swiped over the urine sample from both sides to ensure urine was captured in the holes of the dipstick. Dipsticks were placed in a humid chamber at 37 °C overnight (12–15 h). The number of positive holes was counted to obtain the concentration and results are plotted in Fig. 4.
Conclusions
To conclude, we presented the digital dipstick, which miniaturizes and digitizes culture-based diagnosis. The test simplicity and rapidity indicate the potential for a lower test cost and shorter turn-around time than today's gold standard, plate culturing. Sampling and compartmentalization neither require sophisticated external equipment nor technical skills. Moreover, the digital bioassay format provides straightforward quantification, for which readout can be performed via cell phone, enabling automation and easy communication of results between user and healthcare settings. We demonstrated the clinically relevant detection range for PoC diagnosis of UTI. The significance of uniquely combining microbial culture and digital assays in a simple dipstick format lays in its potential for self-diagnosing uncomplicated UTI at home or for systematically testing for asymptomatic bacteriuria in elderly patients.37Ethics statement
Ethical approval for the study was received from the ethics committee of the Antwerp University Hospital (UZA) (approval no. 19/22/281).Author contributions
EI and WvdW designed the device. EI fabricated prototypes, performed experiments for proof of concept, device characterization and did image processing. For the clinical trials, EI, MB, PM, HG and WvdW designed the experiments, EI fabricated the samples, EI and MB performed the experiments and EI analyzed the data. EI and WvdW wrote the manuscript with the input from all authors.Conflicts of interest
Authors declare no competing interests.Acknowledgements
Emre Iseri and Michael Biggel were funded through the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 675412. We thank Herland Lab at KTH for their donation of the Escherichia Coli strain ATCC 11775. We thank University Hospital Antwerp for the donation of patient urine samples.Notes and references
- Y. Zhang and H. Noji, Anal. Chem., 2017, 89, 92–101 CrossRef CAS.
- B. Vogelstein and K. W. Kinzler, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef CAS.
- E. C. Yeh, C. C. Fu, L. Hu, R. Thakur, J. Feng and L. P. Lee, Sci. Adv., 2017, 3, e1501645 CrossRef.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. Von Stetten and N. Paust, Lab Chip, 2015, 15, 2759–2766 RSC.
- N. G. Schoepp, T. S. Schlappi, M. S. Curtis, S. S. Butkovich, S. Miller, R. M. Humphries and R. F. Ismagilov, Sci. Transl. Med., 2017, 9(410) DOI:10.1126/scitranslmed.aal3693.
- D. M. Rissin, C. W. Kan, T. G. Campbell, S. C. Howes, D. R. Fournier, L. Song, T. Piech, P. P. Patel, L. Chang, A. J. Rivnak, E. P. Ferrell, J. D. Randall, G. K. Provuncher, D. R. Walt and D. C. Duffy, Nat. Biotechnol., 2010, 28, 595–599 CrossRef CAS.
- S. H. Kim, S. Iwai, S. Araki, S. Sakakihara, R. Iino and H. Noji, Lab Chip, 2012, 12, 4986–4991 RSC.
- O. Scheler, N. Pacocha, P. R. Debski, A. Ruszczak, T. S. Kaminski and P. Garstecki, Lab Chip, 2017, 17, 1980–1987 RSC.
- Ö. Baltekin, A. Boucharin, E. Tano, D. I. Andersson and J. Elf, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9170–9175 CrossRef.
- C. M. Hindson, J. R. Chevillet, H. A. Briggs, E. N. Gallichotte, I. K. Ruf, B. J. Hindson, R. L. Vessella and M. Tewari, Nat. Methods, 2013, 10, 1003–1005 CrossRef CAS.
- E. A. Ottesen, W. H. Jong, S. R. Quake and J. R. Leadbetter, Science, 2006, 314, 1464–1467 CrossRef CAS.
- K. A. Heyries, C. Tropini, M. Vaninsberghe, C. Doolin, O. I. Petriv, A. Singhal, K. Leung, C. B. Hughesman and C. L. Hansen, Nat. Methods, 2011, 8, 649–651 CrossRef CAS.
- F. Shen, W. Du, J. E. Kreutz, A. Fok and R. F. Ismagilov, Lab Chip, 2010, 10, 2666–2672 RSC.
- F. Schuler, M. Trotter, M. Geltman, F. Schwemmer, S. Wadle, E. Domínguez-Garrido, M. López, C. Cervera-Acedo, P. Santibáñez, F. Von Stetten, R. Zengerle and N. Paust, Lab Chip, 2016, 16, 208–216 RSC.
- K. Leirs, P. Tewari Kumar, D. Decrop, E. Pérez-Ruiz, P. Leblebici, B. Van Kelst, G. Compernolle, H. Meeuws, L. Van Wesenbeeck, O. Lagatie, L. Stuyver, A. Gils, J. Lammertyn and D. Spasic, Anal. Chem., 2016, 88, 8450–8458 CrossRef CAS.
- Q. Zhu, Y. Gao, B. Yu, H. Ren, L. Qiu, S. Han, W. Jin, Q. Jin and Y. Mu, Lab Chip, 2012, 12, 4755–4763 RSC.
- C. C. Butler, N. A. Francis, E. Thomas-Jones, M. Longo, M. Wootton, C. Llor, P. Little, M. Moore, J. Bates, T. Pickles, N. Kirby, D. Gillespie, K. Rumsby, C. Brugman, M. Gal, K. Hood and T. Verheij, Br. J. Gen. Pract., 2018, 68, e268–e278 CrossRef.
- L. Brookes-Howell, E. Thomas-Jones, J. Bates, M.-J. Bekkers, C. Brugman, E. Coulman, N. Francis, K. Hashmi, K. Hood, N. Kirby, C. Llor, P. Little, M. Moore, A. Moragas, K. Rumsby, T. Verheij and C. Butler, BJGP Open, 2019, 3(2) DOI:10.3399/bjgpopen18X101630.
- G. Schmiemann, E. Kniehl, K. Gebhardt, M. M. Matejczyk and E. Hummers-Pradier, Dtsch. Arztebl. Int., 2010, 107, 361–367 Search PubMed.
- M. Blom, T. L. Sørensen, F. Espersen and N. Frimodt-Møller, Scand. J. Infect. Dis., 2002, 34, 430–435 CrossRef.
- D. Guttmann and G. R. Naylor, BMJ, 1967, 3, 343–345 CrossRef CAS.
- R. Winkens, H. Nelissen-Arets and E. Stobberingh, Fam. Pract., 2003, 20(4), 410–412 CrossRef.
- M. S. Lachs, Ann. Intern. Med., 1992, 117, 135 CrossRef CAS.
- M. Juthani-Mehta, M. Tinetti, E. Perrelli, V. Towle and V. Quagliarello, Infect. Control Hosp. Epidemiol., 2007, 28, 889–891 CrossRef.
- L. E. Lehmann, S. Hauser, T. Malinka, S. Klaschik, S. U. Weber, J. C. Schewe, F. Stüber and M. Book, PLoS One, 2011, 6(2) DOI:10.1371/journal.pone.0017146.
- C. M. Shih, C. L. Chang, M. Y. Hsu, J. Y. Lin, C. M. Kuan, H. K. Wang, C. Te Huang, M. C. Chung, K. C. Huang, C. E. Hsu, C. Y. Wang, Y. C. Shen and C. M. Cheng, Talanta, 2015, 145, 2–5 CrossRef CAS.
- I. P. Alves and N. M. Reis, Biosens. Bioelectron., 2019, 145, 111624 CrossRef CAS.
- G. Kokkinis, B. Plochberger, S. Cardoso, F. Keplinger and I. Giouroudi, Lab Chip, 2016, 16, 1261–1271 RSC.
- E. Eltzov and R. S. Marks, Anal. Chem., 2016, 88, 6441–6449 CrossRef CAS.
- Z. Samra, M. Heifetz, J. Talmor, E. Bain and J. Bahar, J. Clin. Microbiol., 1998, 36, 990–994 CrossRef CAS.
- W. G. Cochran, Biometrics, 1950, 6, 105 CrossRef CAS.
- G. Bonkat, R. Pickard, R. Bartoletti, T. Cai, F. Bruyere, S. E. Geerlings, B. Köves, F. Wagenlehner, A. Pilatz, B. Pradere and R. Veeratterapillay, EAU Guidelines on Urological Infections, 2018 Search PubMed.
- M. L. Wilson and L. Gaido, Clin. Infect. Dis., 2004, 38, 1150–1158 CrossRef.
- P. Yagupsky, M. Rider and N. Peled, Eur. J. Clin. Microbiol. Infect. Dis., 2000, 19, 694–698 CrossRef CAS.
- J. V. Rogers, J. Microbiol. Exp., 2015, 2(7) DOI:10.15406/jmen.2015.02.00075.
- J. M. Farber, J. Food Prot., 1991, 54, 58–70 CrossRef CAS.
- M. Biggel, S. Heytens, K. Latour, R. Bruyndonckx, H. Goossens and P. Moons, BMC Geriatr., 2019, 19, 170 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc00793e |
| This journal is © The Royal Society of Chemistry 2020 |