Open Access Article
Open Access ArticleFully-automated and field-deployable blood leukocyte separation platform using multi-dimensional double spiral (MDDS) inertial microfluidics†
Hyungkook
Jeon
 ab,
Bakr
Jundi
c,
Kyungyong
Choi
ad,
Hyunryul
Ryu
a,
Bruce D.
Levy
c,
Geunbae
Lim
ab,
Bakr
Jundi
c,
Kyungyong
Choi
ad,
Hyunryul
Ryu
a,
Bruce D.
Levy
c,
Geunbae
Lim
 b and
Jongyoon
Han
b and
Jongyoon
Han
 *ade
*ade
aResearch Laboratory of Electronics, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA. E-mail: jyhan@mit.edu
bDepartment of Mechanical Engineering, Pohang University of Science and Technology (POSTECH), 77 Cheongam-Ro, Nam-Gu, Pohang, Gyeongbuk 37673, Republic of Korea
cDivision of Pulmonary and Critical Care Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA
dDepartment of Electrical Engineering and Computer Science, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA
eDepartment of Biological Engineering, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA
First published on 25th August 2020
Abstract
A fully-automated and portable leukocyte separation platform was developed based on a new type of inertial microfluidic device, multi-dimensional double spiral (MDDS) device, as an alternative to centrifugation. By combining key innovations in inertial microfluidic device designs and check-valve-based recirculation processes, highly purified and concentrated WBCs (up to >99.99% RBC removal, ∼80% WBC recovery, >85% WBC purity, and ∼12-fold concentrated WBCs compared to the input sample) were achieved in less than 5 minutes, with high reliability and repeatability (coefficient of variation, CV < 5%). Using this, one can harvest up to 0.4 million of intact WBCs from 50 μL of human peripheral blood (50 μL), without any cell damage or phenotypic changes in a fully-automated operation. Alternatively, hand-powered operation is demonstrated with comparable separation efficiency and speed, which eliminates the need for electricity altogether for truly field-friendly sample preparation. The proposed platform is therefore highly deployable for various point-of-care applications, including bedside assessment of the host immune response and blood sample processing in resource-limited environments.
1. Introduction
Despite their quite low proportion (about 0.1%) in human blood, leukocytes or white blood cells (WBCs) play a key role in human immune responses against pathogens. The assessment of leukocyte activation and function is crucial as the dysregulation of the host immune response underlies the pathobiology of disease.1–8 Since erythrocytes or red blood cells (RBCs) are the most abundant cell component (approximately 1000 times more than the number of WBCs) in peripheral blood, for effective and reliable leukocyte assessment, the depletion of RBCs is an essential sample preparation step before any kinds of scientific, clinical, and diagnostic tests of WBCs.1,4,6,9As the current standard for blood cell separation in the laboratory, density gradient centrifugation is the most widely used technique. Other affinity-based (labeling) methods, such as fluorescence activated cell sorting (FACS) and magnetic-activated cell sorting (MACS), have been used for precise control and separation of target cells.1,9–13 Although those methods themselves are straightforward, they entail many time-consuming steps and require well-trained technicians and well-equipped laboratories; for example, approximately 2–3 hours are needed for a typical density gradient centrifugation.9–11 In addition, for their proper operation, a large volume of blood is required (usually 30–45 mL of blood is required for density gradient centrifugation), and expensive labeling reagents should be employed for the labeling methods. More importantly, the centrifugation process and cell labeling can cause ex vivo cell activation, especially for sensitive cell types such as neutrophils and macrophages, which poses challenges to the correct assessment of the host immune response or leukocyte functions.1,4,14,15
Generally, continuous blood assessment is required for monitoring the host immune response of patients, which can cause iatrogenic anemia (anemia caused by repeated blood drawing) in critically ill patients.1,16 In the next generation of sample preparation, for more precise and effective analysis, daily or even hourly sample assessment should be processed in the bedside or in a field-deployable manner, which requires compact and portable equipment carrying out easy (without well-trained personnel), fully-automated (or even hand-operable), reliable, and fast (within 10 minutes) operation using microliter sampling without any sample damage.
In response to this critical need, several hand-held and hand-operable sample preparation devices have been demonstrated in recent years.17–21 The Prakash group reported a hand-powered and field-portable paper centrifuge (referred to as a ‘paperfuge’), inspired by a historic toy, whirligig or buzzer.17 They demonstrated that the paperfuge can achieve a high-level of centrifugal force (up to 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 r.p.m. or 30
000 r.p.m. or 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) using only hand-power, and the group successfully utilized it to separate plasma and malaria parasites. However, because the sample is processed not in a continuous but in a batch mode, it is hard to extract the separated target layer, and precise manual extraction is required for obtaining separated cells, which is susceptible to human error and unreliability (in fact, this issue is commonly shared by the standard lab-based centrifugation process.) As other interesting examples, particle focusing devices using groove-based channels and spiral channels have been introduced.18–21 The proposed devices were successfully utilized for isolation of cell components by hand-powered syringe-pushing. However, due to the relatively lower separation resolution, their applications are limited to concentration or removal of particles (or cells) from the background fluid while selective isolation of target-size particles from others is more necessary in on-site sample preparation. For example, the selective isolation of leukocytes by hand-powered (or fully hands-free) operation can enable various on-site leukocyte analyses for scientific, clinical, and diagnostic purposes, ranging from simple leukocyte counting to functional assays and nucleic acid analyses, utilizing a portable microscope22–25 and polymerase chain reaction (PCR) tools.26–29 In particular, high-quality ribonucleic acid (RNA) can be obtained and examined from the separated leukocytes for detection and identification of viruses at clinical sites, such as Ebola virus and human immunodeficiency virus (HIV), due to removal of RBCs containing hemoglobin which can compromise the amplification and detection of target RNAs from leukocytes.4,30 Indeed, from the proper extraction procedures, inactivated and non-infectious nucleic acids can be obtained so that they can be shipped across the border, to well-equipped laboratories for further sophisticated analyses such as nucleic acid sequencing.31,32
000g) using only hand-power, and the group successfully utilized it to separate plasma and malaria parasites. However, because the sample is processed not in a continuous but in a batch mode, it is hard to extract the separated target layer, and precise manual extraction is required for obtaining separated cells, which is susceptible to human error and unreliability (in fact, this issue is commonly shared by the standard lab-based centrifugation process.) As other interesting examples, particle focusing devices using groove-based channels and spiral channels have been introduced.18–21 The proposed devices were successfully utilized for isolation of cell components by hand-powered syringe-pushing. However, due to the relatively lower separation resolution, their applications are limited to concentration or removal of particles (or cells) from the background fluid while selective isolation of target-size particles from others is more necessary in on-site sample preparation. For example, the selective isolation of leukocytes by hand-powered (or fully hands-free) operation can enable various on-site leukocyte analyses for scientific, clinical, and diagnostic purposes, ranging from simple leukocyte counting to functional assays and nucleic acid analyses, utilizing a portable microscope22–25 and polymerase chain reaction (PCR) tools.26–29 In particular, high-quality ribonucleic acid (RNA) can be obtained and examined from the separated leukocytes for detection and identification of viruses at clinical sites, such as Ebola virus and human immunodeficiency virus (HIV), due to removal of RBCs containing hemoglobin which can compromise the amplification and detection of target RNAs from leukocytes.4,30 Indeed, from the proper extraction procedures, inactivated and non-infectious nucleic acids can be obtained so that they can be shipped across the border, to well-equipped laboratories for further sophisticated analyses such as nucleic acid sequencing.31,32
As a portable and electricity-free sample preparation device for on-site point-of-care testing (POCT) in resource-limited environments, in this paper, we propose a novel leukocyte separation platform based on a new type of spiral inertial microfluidic device, multi-dimensional double spiral (MDDS) device, and a check-valve-based recirculation platform. In the MDDS device composed of two spiral channels with different dimensions, we can integrate two different functions, sample focusing and separation, in a single device without sheath flow, resulting in a great increase of separation efficiency. The check-valve, which can regulate the flow direction, was used to recirculate the output into the device for more purification and concentration by repeated pushing and pulling procedures of the input syringe. The developed separation platform showed great performance in leukocyte purification (up to >99.99% RBC removal, ∼80% WBC recovery, and >85% WBC purity) with critical advantages; 1) fully-automated or even handheld operation, 2) continuous-flow separation (easy output acquisition), 3) minimal cell damage or ex vivo cell activation, 4) short operation time (4–7 minutes), 5) high reliability and repeatability (coefficient of variation, CV < 5%), 6) small sample volume requirement (50 μL of raw blood), and 7) compact and portable dimensions. Considering these performance characteristics, we expect that the developed platform could provide a valuable field-deployable sample preparation solution to point-of-care blood analyses and diagnostics.
2. Experimental section
2.1. Device fabrication
The multi-dimensional double spiral (MDDS) device was fabricated using polydimethylsiloxane (PDMS) following standard soft-lithographic techniques.33,34 The aluminum master mold with specific channel dimensions was designed using a 3D CAD software program (SolidWorks 2019) and then fabricated by a micromilling company (Whits Technologies, Singapore) for PDMS casting. The PDMS replica was made by casting degassed PDMS (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of base and curing agent of Sylgard 184, Dow Corning Inc.) onto the aluminum mold, followed by curing on a hot plate for 10 min at 150 °C. After making holes for fluidic access using disposable biopsy punches (Integra Miltex), the PDMS replica was irreversibly bonded to a glass slide using a plasma machine (Femto Science, Korea). The assembled device was placed in a 60 °C oven for at least 1 h to stabilize the bonding further.
1 mixture of base and curing agent of Sylgard 184, Dow Corning Inc.) onto the aluminum mold, followed by curing on a hot plate for 10 min at 150 °C. After making holes for fluidic access using disposable biopsy punches (Integra Miltex), the PDMS replica was irreversibly bonded to a glass slide using a plasma machine (Femto Science, Korea). The assembled device was placed in a 60 °C oven for at least 1 h to stabilize the bonding further.
2.2. Design of the recirculation system
The check-valve-based recirculation platform was designed to obtain more purified and concentrated WBCs. A connector of the platform was designed using a 3D CAD software program (SolidWorks 2019) and then fabricated using a 3D printer (Form 2, Formlabs, USA) with a specific resin (RS-F2-GPCL-04, Formlabs, USA). Three different connectors were made for three different recirculation platforms having a single-version of MDDS device, two quad-version of MDDS devices, and one quad-version of MDDS device, respectively. Two kinds of check-valves were used; one is a dual-check-valve (80183, QOSINA, USA) for regulating the flow direction on injection and extraction of the sample, and the other is a check-valve (80184, QOSINA, USA) for preventing the output in the RBC reservoir from flowing to the WBC reservoir. Using the 3D-printed connectors, we can directly connect the MDDS device, syringes (for input and output reservoirs), and check-valves through a simple and easy assembly process, resulting in recirculation platforms having high portability and minimized dead volume. To prevent the cross-contamination caused by the trapped cells on the internal membrane inside the check-valves, we used a new check-valve for each experiment; the check-valves we used are very cheap (about 1 dollar) to be used in a disposable manner.2.3. Sample preparation
For bead experiments, fluorescent polystyrene particles with diameters of 6.0 μm (18141-2, Polysciences, Inc., USA) and 10.0 μm (F8834, Invitrogen™, USA) were used after dilution in deionized water. For blood separation tests, peripheral blood samples were collected from healthy donors who reported general health and no use of medical prescription in the last 2 weeks prior to enrollment, or purchased from Research Blood Components, LLC (Boston, MA, U.S.A.); the Partners Healthcare Institutional Review Board has approved this study (IRB no.: 2002P000272) and peripheral blood (500 μL) from healthy donors was collected into heparin-containing vacutainer tubes after informed consent was obtained. For the operation of the recirculation platform, blood samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (50 μL in 25 mL with 1× phosphate-buffered saline without calcium and magnesium (PBS, Corning®)). In addition, granulocytes were isolated from peripheral blood collected from healthy donors after informed consent was obtained, using the density-based gradient over Histopaque 1077 (Sigma-Aldrich) and Histopaque 1119 (Sigma-Aldrich) as previously described.1,35
500 (50 μL in 25 mL with 1× phosphate-buffered saline without calcium and magnesium (PBS, Corning®)). In addition, granulocytes were isolated from peripheral blood collected from healthy donors after informed consent was obtained, using the density-based gradient over Histopaque 1077 (Sigma-Aldrich) and Histopaque 1119 (Sigma-Aldrich) as previously described.1,35
2.4. Device characterization
Samples were loaded to the device with the regulated flow rate by a syringe pump (Fusion 200, Chemyx Inc., USA). An inverted fluorescence microscope (IX51, Olympus Inc., USA) and a CCD camera (Sensicam QE, PCO, Germany) were used to observe the trajectories of the fluorescent particles and collect images from the device. Due to the absence of fluorescence, the trajectories of blood cells were observed by using a high-speed camera (Phantom v9.1, Vision Research Inc., USA) with a certain sample rate, 100 pictures per second (pps).2.5. Flow cytometry analysis
To determine the separation efficiency, input and output samples were collected and analyzed using a flow cytometer (Accuri C6, BD Biosciences, USA) with staining the samples with the following antibodies: fluorescein isothiocyanate (FITC)-conjugated CD45 monoclonal antibody (positive for all leukocytes) and allophycocyanin (APC)-conjugated CD66b monoclonal antibody (positive for polymorphonuclear leukocytes, PMNs); all the antibodies were purchased from eBioscience™. Considering that mononuclear leukocytes (MNLs) are composed of various cell types, and there is no efficient surface marker available to determine the total amount of MNLs, the number of MNLs was calculated as CD45-positive but CD66b-negative cells.We also used the flow cytometry method on phenotypic analysis to assess the effects of the separation process on ex vivo cell activation. Leukocytes (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 PMN) were isolated from 50 μl of peripheral blood, and molecules of leukocyte activation were assessed using flow cytometry (CD69, CD11b, and CD62L). After obtaining leukocytes, cells were incubated for 20 min at RT with the following antibodies to human proteins, with clones noted in parentheses: anti-CD45 PerCP (HI30), anti-CD66b Pacific blue (G10F5), anti-CD16 APC-Cy7 (3G8), anti-CD69 FITC (FN50), anti-CD62L Brilliant Violet 510 (DREG-56) (all from BioLegend), and anti-CD11b PE-Cy7 (ICRF44) (from Thermofisher). After staining leukocytes, cells were then lysed and fixed with 2 ml of 1
000 PMN) were isolated from 50 μl of peripheral blood, and molecules of leukocyte activation were assessed using flow cytometry (CD69, CD11b, and CD62L). After obtaining leukocytes, cells were incubated for 20 min at RT with the following antibodies to human proteins, with clones noted in parentheses: anti-CD45 PerCP (HI30), anti-CD66b Pacific blue (G10F5), anti-CD16 APC-Cy7 (3G8), anti-CD69 FITC (FN50), anti-CD62L Brilliant Violet 510 (DREG-56) (all from BioLegend), and anti-CD11b PE-Cy7 (ICRF44) (from Thermofisher). After staining leukocytes, cells were then lysed and fixed with 2 ml of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 dilution of lyse/fix buffer 5X (BD Phosflow) with dH2O for 15 min at RT. A BD LSR Fortessa flow cytometer was used to obtain data and Flowjo software version 10.1 (Tree star) was used to analyze the data. PMNs were identified by CD66b + CD45 + SSCHighFSCHigh.
4 dilution of lyse/fix buffer 5X (BD Phosflow) with dH2O for 15 min at RT. A BD LSR Fortessa flow cytometer was used to obtain data and Flowjo software version 10.1 (Tree star) was used to analyze the data. PMNs were identified by CD66b + CD45 + SSCHighFSCHigh.
2.6. PMN phagolysosome acidification assay
PMN phagolysosome formation was quantified through flow cytometry using the red E. coli pHrodo bioparticle conjugate as described in our previous work.13. Results
3.1. Design of the multi-dimensional double spiral (MDDS) device
When the fluid flows through a curved channel, fluid elements near the channel centerline with a higher flow rate, compared to the fluid near the channel wall, move outwards to the outer channel wall due to centrifugal effects, and they recirculate toward the center along the top and bottom surfaces of the channel due to conservation laws, resulting in a secondary flow, Dean flow.36–39 In the spiral inertial microfluidic device, lateral particle motion (in the cross-sectional view) is affected by inertial focusing and circulating motion which are generated by lift forces and hydrodynamic drag force of the Dean flow, respectively.36,38Depending on the particle size, the magnitudes of the applied net lift force and the Dean drag force are changed, which determine whether particles keep moving along the Dean flow or become focused on a certain equilibrium position in the channel's cross-sectional view, and the confinement ratio (CR = a/Dh, where a is the particle diameter and Dh is the hydraulic diameter of the microchannel) is a key parameter to determine the particle motion.4,19,36,40,41 Generally (for moderate flow rate conditions with a constraint of the Dean number, De = Rc(Dh/2r)1/2 < 75, where δ = Dh/2r and r represent the curvature ratio and the average radius of curvature of the channel, respectively),42 in the case of small CRs (<0.07), the net lift force applied to particles is negligible compared to the Dean drag force, resulting in the circulating motion of particles without focusing (the non-focusing mode).40,41 In the case of large CRs (≥0.07), the lift force becomes stronger and comparable with the Dean drag force, resulting in particle focusing on an equilibrium position determined by the competition between the net lift force and the Dean drag force (the focusing mode); in general, as the particle size increases, the equilibrium position gets closer to the inner wall side due to the stronger lift force. In intermediate CRs (0.01 ≤ CR < 0.07), particle motion is described as the rough focusing mode. Because only the large CR particles can be effectively focused on their equilibrium positions, the spiral device has faced a critical bottleneck in the target size range and separation performance, which limits its applicability.37,43,44
To overcome the limitation, spiral devices with multiple inlets and sheath flows were proposed.45–48 In the device, two inlets were employed for infusion of the sample and sheath flow so that all particles can be injected into the spiral channel focused on the outer wall side by the sheath flow. This allows them to start moving away from the focused flow stream to their equilibrium positions. Due to the initial focusing, particle–particle interaction can be significantly reduced during their movement to the equilibrium position with minimal particle dispersion, resulting in increased separation resolution and efficiency; particles with intermediate CR conditions can also efficiently reach their equilibrium positions while keeping their focused band, despite relatively lower lift force. However, the use of two inlets makes the flow control more complex and requires additional fluid control equipment, which significantly limits the operating flexibility and applicability of the spiral microfluidic device.
The multi-dimensional double spiral (MDDS) device proposed here was designed as a new type of spiral device to overcome the limitations of the spiral device; the initial focusing of target particles can be made in the MDDS device without sheath flow. As shown in Fig. 1a, the MDDS device is composed of sequentially connected two spiral channels having two different dimensions; the first spiral channel has a smaller dimension (rectangular cross-section with 800 μm in width and 60 μm in height) for sample focusing, and the second spiral channel has a larger dimension (trapezoidal cross-section with 800 μm in width and 80 and 120 μm in height for the inner and outer wall side, respectively) for sample separation; the trapezoidal cross-section enables effective extraction of smaller particles to the outer wall side due to strong Dean vortices at the outer half of the channel.4,49Fig. 1b shows the trajectory of particles under the optimal flow rate condition (2.3 mL min−1) in the MDDS device. Particles with diameters of 6 (green) and 10 μm (red) were used to mimic the movement of RBCs and WBCs, respectively. In the first spiral channel, both 6 and 10 μm particles are under the large CR condition and affected by strong lift forces so that both are focused into the inner wall side and then enter the outer wall side of the second spiral channel through the S-shaped transition region, like using sheath flow in the spiral device having 2 inlets (Fig. 1b); the CR values of 6 and 10 μm particles are ∼0.1 and ∼0.17, respectively. In the second spiral channel, due to the increased channel dimension, 6 μm particles no longer meet the large CR condition while 10 μm particles still remain in the large CR condition; the CR values of 6 and 10 μm particles are ∼0.06 and ∼0.1, respectively. However, benefitting from the initial focusing in the first spiral channel, not only 10 μm but also 6 μm particles can efficiently reach their equilibrium positions while keeping their focused bands, resulting in their effective separation. As shown in Fig. S1a and b,† both 6 and 10 μm particles were well-focused into the inner wall side of the first spiral channel over a certain range of flow rate conditions, 2.0–2.5 mL min−1, and the optimal flow rate condition for their separation in the second spiral channel was 2.3 mL min−1; 6 (green) and 10 μm (red) were used to mimic the movement of RBCs and WBCs, respectively. As a result, we can achieve clearer separation and obtain well-focused 10 and 6 μm particle streams from the inner and outer wall side outlets, respectively, compared to the single spiral device which has the same dimension as the second spiral channel of the MDDS device (trapezoidal cross-section with 800 μm in width and 80 and 120 μm in height for the inner and outer wall side, respectively) (Fig. 1avs. S2†).
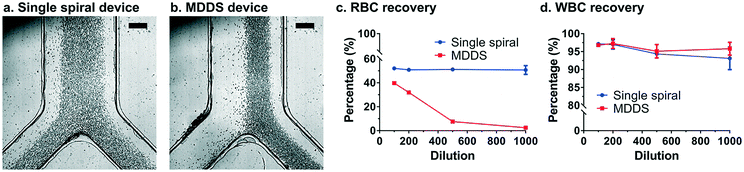
Fig. 2 shows the results of blood separation in the MDDS device compared with the single spiral device. Although the performance varied depending on the blood dilution conditions, we found that RBCs can be extracted into the outer wall side of the channel in the MDDS device more effectively compared with the single spiral device (Fig. 2avs. b, Video S1†), resulting in higher removal of RBCs in the inner wall side outlet (>92% and >97% for 500× and 1000× dilution conditions, respectively, as shown in Fig. 2c), while both devices similarly showed great recovery of WBCs (>95% in the MDDS device for all the dilution conditions, as shown in Fig. 2d); as the dilution rate decreases, the distribution of RBCs across the channel width is broadened due to the increase of the solid fraction (mainly contributed by the RBC population), which leads to a decrease in RBC removal (Fig. S3†).4 Here, the recoveries of WBCs and RBCs were calculated by comparing the cell numbers from each outlet measured by flow cytometry analysis. For example, the WBC recovery was calculated as the number of WBCs in the WBC outlet divided by sum of the WBC numbers in both RBC and WBC outlets.
3.2. Check-valve-based recirculation platform
To obtain more purified and concentrated WBCs, we also developed a new recirculation platform based on a check-valve where only one direction of flow is allowed while the opposite direction of flow is blocked by an internal membrane. Our group has previously introduced a recirculation platform using a circulatory pump.1,33,34 In the circulatory pump-based closed-loop operation, the output of the target cells was continuously fed back to the initial sample tube and re-injected into the spiral device for further purification and concentration.1,33 However, the system had an issue of flow instability caused by the mechanical fluctuation of the peristaltic pump used, which caused dispersion of particle stream and degradation of separation performance. Furthermore, human intervention would be required to stop the operation of a circulation pump at an appropriate timing to obtain well-treated output without loss; if it is stopped too early, we could not achieve enough purification and concentration, and if it is stopped too late, a significant portion of target cells could be gone to the other outlet, resulting in the loss of target cells. In contrast, in the check-valve-based recirculation platform, the target output can be recirculated to the spiral device and processed by the programmed linear (back-and-forth) motions of a syringe pump in a fully-automated and reliable manner without any flow instability issue.The dual-check-valve or rectifier we used in the platform involves two different check-valves so that once separated the WBC output can be extracted back into the input syringe at the withdrawal motion of a syringe pump and processed again through the MDDS device at the infusion motion of a syringe pump, resulting in higher purity and concentration (Fig. 3a). In our experiments, a 500× diluted blood sample (50 μL of human peripheral blood in 25 mL PBS) was used as the initial input sample considering the hematocrit-dependent separation performance (Fig. 2c). A connector was fabricated by 3D printing to directly connect the MDDS device, syringes (for input and output reservoirs), and check-valves for easier device assembly, higher portability, and minimal dead volume (Fig. S4a and S5a, Video S2†). Through the programmed back-and-forth motions of the syringe pump (three cycles of recirculation), approximately 3 mL of highly purified and concentrated WBCs can be obtained in ∼20 minutes in a fully-automated manner (>99.9% RBC removal, >80% WBC recovery, >50% WBC purity, and ∼6-fold concentrated WBCs compared to the input sample under the optimal flow rate condition, 2.3 mL min−1); for each cycle, we obtained an output having half the volume of the input sample where approximately 90% of RBCs were removed while over 90% of WBCs were recovered (Fig. S4b–e, Video S3†). To increase the throughput and reduce the operation time, we developed the quad-version of MDDS device (Fig. 3b) with a new 3D printed connector which can directly connect to two quad-version of MDDS devices (involving 8 individual MDDS devices) and syringes (for input and output reservoirs) (Fig. 3c and S5b, Video S4†); a small pressure-meter mounted connector was designed for the hand-powered operation of the platform (see section 3.3), but the simplified version of the connector without the pressure-meter was used for the general syringe-pump operation. From the three cycles of recirculation using the platform of two quad-version of MDDS devices, we can obtain approximately 3 mL of highly purified and concentrated WBCs within only 4 minutes in a fully-automated manner (>99.9% RBC removal, ∼80% WBC recovery, >40% WBC purity, and ∼6-fold concentrated WBCs compared to the input sample under the optimal flow rate condition, 2.3 × 8 = 18.4 mL min−1) (Fig. 3d–f, Video S5†).
To validate its reliability, we tested its parallel operation using three different platforms and three different blood donors (Fig. 4a, Video S6†). Our findings demonstrated that the device-dependent variation was quite small for all the blood samples and all the blood cell types as the recoveries and purity of WBCs have a coefficient of variation (CV) less than 5%; error bars of Fig. 4b represent the standard deviation of the three different platforms. In the case of the sample-dependency, the results showed that there was a significant difference in the recoveries and purity of WBCs depending on the blood donor, while the RBC removal rate was similar (∼99.9% RBC removal, 70–90% WBC recovery, and 20–50% WBC purity) (Fig. 4b). Cell type frequencies and their size distributions vary from donor to donor, which in turn leads to different solid fractions and focusing behaviors, resulting in the variation of the separation performance. In addition, we found that the PMN (polymorphonuclear leukocyte) recovery was better than the MNL (mononuclear leukocyte) recovery for all the blood samples; the recovery difference was in the range of 10–25%. This is presumably because the size of the PMN population (10–12 μm) is bigger than that of the MNL one (7–10 μm) in general, which is also in line with the results in our previous research using the single spiral device.4,50
Because the initial population of RBCs is about 1000 times more than that of WBCs, even the output with ∼99.9% RBC removal contains a similar number of RBCs to WBCs. For certain applications requiring higher WBC purity and concentration rather than fast operation, we designed another version of the recirculation platform using one quad-version of MDDS device (Fig. 5a and S5c†) to reduce the dead volume. Compared to the platform having two quad-version of MDDS devices, the platform having one quad-version of MDDS device has a lower dead volume in the channels of the 3D-printed connector (from ∼2.0 mL to ∼0.79 mL) and the MDDS device (from 2 × 0.11 mL to 1 × 0.11 mL). The reduction of dead volume makes it possible to process one more recirculation cycle, although more operation time is required; the four cycles of recirculation can be processed within 7 minutes. As shown in Fig. 5b, over 90% of RBCs were removed while over 90% of WBCs were recovered for each cycle, and about 1.6 mL volume of more purified and concentrated WBC sample was obtained from four cycles of recirculation (>99.99% RBC removal, ∼80% WBC recovery, >85% WBC purity, and ∼12-fold concentrated WBCs compared to the input sample under the optimal flow rate condition, 2.3 × 4 = 9.2 mL min−1) although there is also variation in the recoveries and purity due to donor-to-donor variation (>99.99% RBC removal, 70–80% WBC recovery, and 65–90% WBC purity) (Fig. 5c).
We made three different recirculation platforms with three different 3D-printed connectors depending on how many devices are employed (Fig. S5a–c†). Considering the trade-off among purity, concentration, recovery and operation time, the user can select a recirculation platform and the number of recirculation runs. As more devices are employed, we can reduce the operation time, but the increase of dead volume inside the device and connector can degrade the separation performance and limit the number of recirculation runs. As more recirculation steps are processed, we can achieve higher purity and concentration but lose more cells.
3.3. Hand-powered operation
The hand-powered operation (if it can be made reliable and repeatable) would be ideal for sample processing in resource-limited environments.17–21 Because the MDDS device can be operated only by a sample flow without sheath flow, the developed platform only requires a simple back-and-forth motion of an input syringe, which allows it to be operated by hand-powered syringe pushing and pulling procedures.To find out how much force is required for operating the device, we measured the applied force to the input syringe of the platform having two quad-version of MDDS devices by using a load cell, which was placed between the syringe and the pusher block of the syringe pump. The output voltage from the load cell, which varies depending on the applied force, was measured using a voltage-meter and transferred to an actual force value in real-time (Fig. 6a and b). Our findings showed that the required force for the optimal flow rate (18.4 mL min−1) was measured to be approximately 107 N, which is considered an appropriate force for hand-powered operation; the maximum pushing forces of male and female are over 300 and 200 N, respectively.51
To apply suitable force to the syringe on the hand-powered operation, a small pressure-meter was mounted on the 3D-printed connector and directly connected to the inlet channel of the 3D-printed connector, which indicates the pressure value at the inlet region in real-time. First, the pressure value was measured on the syringe pump operation under various flow-rate conditions. From the results, the load and pressure increased with a similar profile as the applied flow rate increased, and the pressure value corresponding to the optimal flow rate condition (18.4 mL min−1) was approximately 29.5 psi (Fig. 6c, Video S5†). Based on the pressure measurement from the syringe pump operation, the developed platform can be operated by simple hand-pushing and pulling motions where the input syringe should be pushed while keeping the pressure of the pressure-meter at the optimum pressure value (29.5 psi) for optimal flow rate conditions (Fig. 6d, Video S7†).
Fig. 6e and f show the separation performance on the hand-powered operation with five different trials of three cycles of recirculation using the platform of two quad-version of MDDS devices. From the results, similar to the syringe-pump-based operation, we were able to obtain approximately 3 mL of highly purified and concentrated WBCs within 4 minutes (∼99.5% RBC removal, ∼75% WBC recovery, 10–20% WBC purity, and ∼7-fold concentrated WBCs compared to the input sample under the optimal flow rate condition, 2.3 × 8 = 18.4 mL min−1) (Fig. 6e and f, Video S7†). Although the overall separation performance was slightly worse than the syringe-pump-based operation, mainly due to the inevitable flow fluctuation, the hand-operable platform could be a useful tool for blood preparation as it is a simple and fast operating process with high reliability (less than 5% of CV on the WBC recovery from the 5 different trials). In future work, the platform could be improved by replacing the pressure-meter with a flow regulator or stabilizer for more stable and easier sample infusion.18,52
3.4. Comparison with the conventional method
The developed platforms were compared to an established method for leukocyte separation by density gradient centrifugation. As shown in Fig. 7a, both platforms using two quad-version of MDDS devices (2 × MDDS with 3 cycles) and one quad version of MDDS device (1 × MDDS with 4 cycles) are able to process leukocyte separation at fixed certain times, faster than the density gradient centrifugation method by almost 25–50 fold (∼4 min at 2 × MDDS and ∼7 min at 1 × MDDS versus ∼170 min at centrifugation). Furthermore, the developed platforms showed significantly decreased RBCs (up to 2-orders lower) compared to the density gradient centrifugation method (RBC reduction down to: ∼0.1% at 2 × MDDS and ∼0.005% at 1 × MDDS compared with ∼1.4% from centrifugation) (Fig. 7b). In each recirculation cycle, because WBCs are recovered into approximately one half of the input volume, the recirculation process also has a benefit to increase WBCs' concentration. Considering the variation in the number of WBCs depending on donors (normally between 4 × 106 and 1.1 × 107 per mL),53 we calculated the concentration factor (ratio of the WBC number in the output after separation to the WBC number in the input). The results showed that WBCs were approximately 6 (at 2 × MDDS) and 12 fold (at 1 × MDDS) concentrated compared to the input sample; the actual number of WBCs in the output was in the range of 4–25 × 104 per mL (Fig. 7c).3.5. Phenotypic analysis and functional assay
PMNs are early responders of the host immune system to infection. They play a key role in preventing an overexuberant host response and limiting tissue damage and organ injury. To see if the leukocytes are activated by the isolation process using the MDDS platform, we assessed the PMN expressions of CD69 (PMN degranulation), CD11b (PMN adhesion), and CD62L (L-selectin shedding) via flow cytometry; the detailed gating strategy is described in Fig. S6.† The mean expressions of CD69 (Fig. 8a), CD11b (Fig. 8b), and CD62L (Fig. 8c) on PMNs in isolated leukocytes by the MDDS platform were similar to pre-isolation whole blood, and significantly less than cells isolated from the density gradient centrifugation method. Activation of PMNs induces the degranulation and adhesion (increase of CD69 and CD11b expressions) and increases the shedding of L-selectin (decrease of CD62L expression). To determine PMN functional responses after leukocyte isolation using the MDDS platform, we also conducted a phagocytosis assay using red E. coli pHrodo bioparticles. As shown in Fig. 8d, the result showed that a large amount of PMNs formed phagolysosomes against the E. coli pHrodo bioparticles to a similar extent both before and after isolation using the MDDS platform (over 95% of pHrodo+). Together, our results demonstrate that use of the MDDS platform for separating and enriching leukocytes from whole blood does not cause any significant change of cell properties or cell damage.4. Discussion and conclusion
Previously, many microfluidic technologies for cell sorting have been developed, providing several key advantages such as precise fluid manipulation, minimal sample volume requirement, and capability of integration with different functional devices.2–13,54,55 However, very few devices have gone through successful commercialization and transition to widespread clinical commodities,13 mainly because of the limitations of low flow rate and required meticulous device manipulation. Conventional microfluidic devices suffer from a significant volume-mismatch when connecting with macro-scale analytics instruments, which necessitates a complex experimental setup, time-consuming manual connection of fluidic lines, and technical expertise to manage these connections.12,13The inertial microfluidic device, including the spiral device, is uniquely positioned to avoid these pitfalls, due to its inherent advantages including label-free and high throughput separation (order of 1 mL min−1 per single device), simple channel design (or simple device fabrication), and straightforward and robust operation without an external force field; technically, the device can be operated by only using a fluidic pump(s) without other instruments including a microscope.4,19,33,34,36–41,43–49,56–67 Great progress has been achieved from many studies within the last decade in terms of performance improvement and expansion of its applications; for example, the technique has been applied to various clinical samples such as leukocytes,1,4,33 circulating tumor cells,46,48,59,66 mesenchymal stem cells,67 bacteria,45 and viruses.34,45 As another interesting research study, the Toner group recently reported novel separation and concentration devices based on the inertial focusing effect combined with the siphoning technique using consecutive micro-structures.68,69 Although the developed devices require complex and sophisticated channel design, benefitting from the great performance in the throughput and separation efficiency (96.6% WBC recovery and 0.0059% RBC recovery in processing whole blood at 3 mL min−1), they successfully utilized the devices for large-volume blood fractionation. While inertial microfluidic devices have been used extensively (even by non-experts) in laboratory settings, fully-automated and field-deployable (which could require high portability and electricity-free operation) implementation of inertial microfluidic systems has never been demonstrated previously. The device demonstrated here is the first implementation of a portable and fully-automated (or even handheld operable) blood fractionation system, which is critically needed especially in research environments requiring higher biosafety levels (BSL3 or higher).
The separation performance of the spiral device can be altered depending on the channel design, including the channel dimension, the number of spiral loops, and the outlet configuration. Due to the effective extraction of smaller particles by strong Dean vortices at the outer half of the channel,49 the previously proposed trapezoidal spiral channel has shown great separation performance in various applications such as isolation of circulating tumor cells,66,70 blood plasma,39 and leukocytes.4 However, because of the small difference between focused bands of separation targets, the separation resolution is limited, and the separation efficiency is very sensitive to the operation conditions (especially the flow-rate) and the outlet configuration (especially, the ratio of outlet width). Therefore, it is essential to operate the device precisely and design the outlet configuration delicately, which limits the operational flexibility of the device and requires undesired trial-and-error fabrications. The proposed MDDS device achieves sample focusing and separation in a single device. In addition to the benefits from the trapezoidal spiral channel, the initial focusing in the first spiral channel significantly reduced the particle–particle interaction during the separation process in the second spiral channel. In particular, for small particles with intermediate CR conditions, the initial focusing leads to a shorter transverse travel length to their specific equilibrium position which is relatively close to the outer wall so that they can efficiently reach their equilibrium position while keeping their focused band. As a result, the separation resolution and efficiency can be significantly improved, which was previously achieved by sheath flow in the two-inlet spiral device45–48 with more operational complexity. However, in the first spiral channel of the MDDS device, only particles in a certain size regime (large CR condition) can be well-focused by the inertial focusing effect unlike using the sheath flow. Also, the first and second spiral channels have the same flow rate because they are directly connected. Therefore, careful determination of channel dimensions (both the first and second spirals) is critical for its proper operation. Yet, the first and second spiral structures and dimensions are changed to fit the specific need of the separation process. For example, our first spiral was of rectangular cross-section with smaller dimensions (to achieve more focusing with a higher CR), while the second spiral was of trapezoidal cross-section with larger heights (to enable better separation). In the future, other parameters (such as the width of the channel) can be varied to further optimize the device.
In this paper, we focused on the isolation of WBCs from abundant RBCs which are the major inhibitors of downstream processes. However, the existence of platelets (PLT) should be considered for certain applications, and we need to determine the WBC purity when considering the PLT populations. Fig. S7† represents the PLT recoveries and WBC purities against PLTs as well as RBCs for all the three different platforms, 1) a platform having a single-version of MDDS device, 2) a platform having two quad-version of MDDS devices, and 3) a platform having one quad-version of MDDS device. Because PLTs have smaller size than RBCs, they are more difficult to be efficiently removed than RBCs in the MDDS device. As shown in Fig. S7,† we found that ∼70% of PLTs were removed in each cycle for all the platforms, and only ∼1% of PLTs were recovered in the final WBC output from 4 cycles of recirculation by the platform having one quad-version of MDDS device. Although the final recovery of PLTs is very small, (especially considering that the original count of PLTs in whole blood is generally 30–40 times higher than the WBC count), the PLT population (even at 1% or lower recovery) may significantly affect the WBC purity and downstream assay. For example, in the platform having one quad-version of MDDS device, the WBC purity was reduced from ∼85% to ∼45% by involving the PLT population.
In the recirculation platform, precise volume control is critical to avoid air infusion to the device which could degrade the separation performance and affect ex vivo cell activation. In this work, the MDDS device was designed to have the same dimensional outlets so that the ratio of output volume is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for easier sample volume handling. In the first extraction of the sample to an input syringe, air intake inevitably happens from the empty tubing parts, so we intentionally did not infuse full volume to the device but left 500 μL of spare volume in the input syringe. For example, if the sample volume is 25.0 mL, we extracted 25.0 mL from the outlet reservoir but infused only 24.5 mL to the MDDS platform to prevent air infusion to the device; we do not need to make additional spare volume in the next infusion steps because there is no further air take from the tubing parts. The non-processed spare volume could lead to degradation of separation efficiency and purity, but the 3 or 4 cycles of recirculation processes can make its effect negligible.
1 for easier sample volume handling. In the first extraction of the sample to an input syringe, air intake inevitably happens from the empty tubing parts, so we intentionally did not infuse full volume to the device but left 500 μL of spare volume in the input syringe. For example, if the sample volume is 25.0 mL, we extracted 25.0 mL from the outlet reservoir but infused only 24.5 mL to the MDDS platform to prevent air infusion to the device; we do not need to make additional spare volume in the next infusion steps because there is no further air take from the tubing parts. The non-processed spare volume could lead to degradation of separation efficiency and purity, but the 3 or 4 cycles of recirculation processes can make its effect negligible.
For reliable assessment of the host immune response or leukocyte functions, the sample preparation process must not cause any significant change of cell properties.1,4,14,15 Although various microfluidic devices have been developed and applied for leukocyte separation as clinical sample preparation, most of the research has focused on improvement of the separation performance, and only a few researchers have examined the effect of the separation process on cell properties.1,4,14,15 Because the activation status of leukocytes can be affected by the chemical environment (e.g., osmotic pressure), temperature changes, and physical manipulations,71,72 it is essential to look into the change of cell phenotypes and functions during the operation of microfluidic devices, especially in the case using external stimuli.1,4 To examine the effects of leukocyte isolation using the MDDS platform on cell phenotypes, we measured various cell surface markers and carried out a functional phagocytosis assay on sorted cells, and compared them to unprocessed cells as well as leukocytes processed by the conventional density gradient centrifugation method. The results showed that the operation of the developed platform does not cause any significant ex vivo cell activation, while the density gradient centrifugation method induces notable ex vivo cell activation.
By altering the channel dimensions of the MDDS devices, the separation cut-off size can be controlled so that the developed platform could be adaptable for various sample preparation applications. For example, the channel height can be increased or decreased to selectively isolate tumor cells from relatively smaller blood cells48,66 or bacteria cells from relatively larger blood cells,34 respectively. Also, applications of the MDDS platform are not limited to blood but can also be extended to other bio-fluids including saliva, sputum, and semen. Therefore, we anticipate that the developed separation platform could be used as an innovative tool to replace conventional sample preparation methodologies.
Author contributions
J. H. conceived and supervised the research. H. J. designed and carried out the experiments with support of K. C. K. C., H. R., and G. L. helped interpret the results. B. J. performed and analyzed the functional biological experiments under the supervision of B. D. L. The manuscript was written by H. J., B. J., K. C., H. R., B. D. L., and J. H.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grant no. U24-AI118656 (B. D. L.) and the National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIP) (No. 2017R1A6A3A11036525, 2020R1A2C2007017).References
- B. Jundi, H. Ryu, D.-H. Lee, R.-E. E. Abdulnour, B. D. Engstrom, M. G. Duvall, A. Higuera, M. Pinilla-Vera, M. E. Benson, J. Lee, N. Krishnamoorthy, R. M. Baron, J. Han, J. Voldman and B. D. Levy, Nat. Biomed. Eng., 2019, 3, 961–973 CrossRef CAS.
- J. Zhang, D. Yuan, R. Sluyter, S. Yan, Q. Zhao, H. Xia, S. H. Tan, N. T. Nguyen and W. Li, IEEE Trans. Biomed. Circuits Syst., 2017, 11, 1422–1430 Search PubMed.
- Z. Wu, Y. Chen, M. Wang and A. J. Chung, Lab Chip, 2016, 16, 532–542 RSC.
- L. Wu, G. Guan, H. W. Hou, A. A. S. Bhagat and J. Han, Anal. Chem., 2012, 84, 9324–9331 CrossRef CAS.
- A. Urbansky, F. Olm, S. Scheding, T. Laurell and A. Lenshof, Lab Chip, 2019, 19, 1406–1416 RSC.
- D. H. Kuan, C. C. Wu, W. Y. Su and N. T. Huang, Sci. Rep., 2018, 8, 1–9 CrossRef CAS.
- X. J. Hu, H. L. Liu, Y. X. Jin, L. Liang, D. M. Zhu, X. Q. Zhu, S. S. Guo, F. L. Zhou and Y. Yang, Lab Chip, 2018, 18, 3405–3412 RSC.
- Q. Guo, S. P. Duffy, K. Matthews, E. Islamzada and H. Ma, Sci. Rep., 2017, 7, 1–11 CrossRef.
- Z. T. F. Yu, K. M. Aw Yong and J. Fu, Small, 2014, 10, 1687–1703 CrossRef CAS.
- C. Petchakup, H. M. Tay, K. H. H. Li and H. W. Hou, Lab Chip, 2019, 19, 1736–1746 RSC.
- F. Cui, M. Rhee, A. Singh and A. Tripathi, Annu. Rev. Biomed. Eng., 2015, 17, 267–286 CrossRef CAS.
- X. Zhang, Z. Zhu, N. Xiang, F. Long and Z. Ni, Anal. Chem., 2018, 90, 4212–4220 CrossRef CAS.
- C. W. Shields, K. A. Ohiri, L. M. Szott and G. P. López, Cytometry, Part B, 2017, 92, 115–125 CrossRef.
- H. Ramachandraiah, H. A. Svahn and A. Russom, RSC Adv., 2017, 7, 29505–29514 RSC.
- P. Sethu, L. L. Moldawer, M. N. Mindrinos, P. O. Scumpia, C. L. Tannahill, J. Wilhelmy, P. A. Efron, B. H. Brownstein, R. G. Tompkins and M. Toner, Anal. Chem., 2006, 78, 5453–5461 CrossRef CAS.
- S. M. C. Horwitz, K. Kelleher, P. M. T. Thomas Boyce, A. Webb, A. Meier-Hellmann, G. Nollet and D. Peres-Bota, JAMA, J. Am. Med. Assoc., 2002, 288, 1499–1507 CrossRef.
- C. Chai, A. Johri, M. Prakash, M. S. Bhamla, G. Katsikis and B. Benson, Nat. Biomed. Eng., 2017, 1, 0009 CrossRef.
- N. Xiang, Y. Han, Y. Jia, Z. Shi, H. Yi and Z. Ni, Lab Chip, 2019, 19, 214–222 RSC.
- N. Xiang, X. Shi, Y. Han, Z. Shi, F. Jiang and Z. Ni, Anal. Chem., 2018, 90, 9515–9522 CrossRef CAS.
- S. Song, M. S. Kim and S. Choi, Small, 2014, 10, 4123–4129 CAS.
- S. Yan, S. H. Tan, Y. Li, S. Tang, A. J. T. Teo, J. Zhang, Q. Zhao, D. Yuan, R. Sluyter, N. T. Nguyen and W. Li, Microfluid. Nanofluid., 2018, 22, 1–10 CrossRef CAS.
- A. R. Miller, G. L. Davis, Z. M. Oden, M. R. Razavi, A. Fateh, M. Ghazanfari, F. Abdolrahimi, S. Poorazar, F. Sakhaie, R. J. Olsen, A. R. Bahrmand, M. C. Pierce, E. A. Graviss and R. Richards-Kortum, PLoS One, 2010, 5, 8–10 Search PubMed.
- A. Greenbaum, N. Akbari, A. Feizi, W. Luo and A. Ozcan, PLoS One, 2013, 8, 1–9 CrossRef.
- S. Dong, K. Guo, P. Nanda, R. Shiradkar and G. Zheng, Biomed. Opt. Express, 2014, 5, 3305 CrossRef.
- S. O. Isikman, W. Bishara, U. Sikora, O. Yaglidere, J. Yeah and A. Ozcan, Lab Chip, 2011, 11, 2222–2230 RSC.
- J. Quick, N. J. Loman, S. Duraffour, J. T. Simpson, E. Severi, L. Cowley, J. A. Bore, R. Koundouno, G. Dudas, A. Mikhail, N. Ouédraogo, B. Afrough, A. Bah, J. H. J. Baum, B. Becker-Ziaja, J. P. Boettcher, M. Cabeza-Cabrerizo, Á. Camino-Sánchez, L. L. Carter, J. Doerrbecker, T. Enkirch, I. Garciá-Dorival, N. Hetzelt, J. Hinzmann, T. Holm, L. E. Kafetzopoulou, M. Koropogui, A. Kosgey, E. Kuisma, C. H. Logue, A. Mazzarelli, S. Meisel, M. Mertens, J. Michel, D. Ngabo, K. Nitzsche, E. Pallasch, L. V. Patrono, J. Portmann, J. G. Repits, N. Y. Rickett, A. Sachse, K. Singethan, I. Vitoriano, R. L. Yemanaberhan, E. G. Zekeng, T. Racine, A. Bello, A. A. Sall, O. Faye, O. Faye, N. Magassouba, C. V. Williams, V. Amburgey, L. Winona, E. Davis, J. Gerlach, F. Washington, V. Monteil, M. Jourdain, M. Bererd, A. Camara, H. Somlare, A. Camara, M. Gerard, G. Bado, B. Baillet, D. Delaune, K. Y. Nebie, A. Diarra, Y. Savane, R. B. Pallawo, G. J. Gutierrez, N. Milhano, I. Roger, C. J. Williams, F. Yattara, K. Lewandowski, J. Taylor, P. Rachwal, D. J. Turner, G. Pollakis, J. A. Hiscox, D. A. Matthews, M. K. O'Shea, A. M. D. Johnston, D. Wilson, E. Hutley, E. Smit, A. Di Caro, R. Wolfel, K. Stoecker, E. Fleischmann, M. Gabriel, S. A. Weller, L. Koivogui, B. Diallo, S. Keita, A. Rambaut, P. Formenty, S. Gunther and M. W. Carroll, Nature, 2016, 530, 228–232 CrossRef CAS.
- G. V. Kaigala, V. N. Hoang, A. Stickel, J. Lauzon, D. Manage, L. M. Pilarski and C. J. Backhouse, Analyst, 2008, 133, 331–338 RSC.
- C. S. Liao, G. Bin Lee, H. S. Liu, T. M. Hsieh and C. H. Luo, Nucleic Acids Res., 2005, 33, 1–7 CrossRef.
- L. Liu, Z. Benyeda, S. Zohari, A. Yacoub, M. Isaksson, M. Leijon, N. Leblanc, J. Benyeda and S. Belák, Transboundary Emerging Dis., 2016, 63, e245–e250 CrossRef CAS.
- W. A. Al-Soud and P. Radstrom, J. Clin. Microbiol., 2001, 39, 485–493 CrossRef CAS.
- X. Abad, Biosafety, 2012, 1, 1–3 Search PubMed.
- J. A. Blow, D. J. Dohm, D. L. Negley and C. N. Mores, J. Virol. Methods, 2004, 119, 195–198 CrossRef CAS.
- H. Ryu, K. Choi, Y. Qu, T. Kwon, J. S. Lee and J. Han, Anal. Chem., 2017, 89, 5549–5556 CrossRef CAS.
- K. Choi, H. Ryu, K. J. Siddle, A. Piantadosi, L. Freimark, D. J. Park, P. Sabeti and J. Han, Anal. Chem., 2018, 90, 4657–4662 CrossRef CAS.
- C. Barnig, M. Cernadas, S. Dutile, X. Liu, M. A. Perrella, S. Kazani, M. E. Wechsler, E. Israel and B. D. Levy, Sci. Transl. Med., 2013, 5, 174ra26 Search PubMed.
- D. Di Carlo, Lab Chip, 2009, 9, 3038 RSC.
- D. Huang, X. Shi, Y. Qian, W. Tang, L. Liu, N. Xiang and Z. Ni, Anal. Methods, 2016, 8, 5940–5948 RSC.
- J. M. Martel and M. Toner, Annu. Rev. Biomed. Eng., 2014, 16, 371–396 CrossRef CAS.
- M. Rafeie, J. Zhang, M. Asadnia, W. Li and M. E. Warkiani, Lab Chip, 2016, 16, 2791–2802 RSC.
- N. Xiang, Z. Shi, W. Tang, D. Huang, X. Zhang and Z. Ni, RSC Adv., 2015, 5, 77264–77273 RSC.
- J. Zhou, P. V. Giridhar, S. Kasper and I. Papautsky, Lab Chip, 2013, 13, 1919 RSC.
- D. Di Carlo, J. F. Edd, D. Irimia, R. G. Tompkins and M. Toner, Anal. Chem., 2008, 80, 2204–2211 CrossRef CAS.
- A. Russom, A. K. Gupta, S. Nagrath, D. Di Carlo, J. F. Edd and M. Toner, New J. Phys., 2009, 11, 075025 CrossRef.
- S. S. Kuntaegowdanahalli, A. A. S. Bhagat, G. Kumar and I. Papautsky, Lab Chip, 2009, 9, 2973 RSC.
- H. W. Hou, R. P. Bhattacharyya, D. T. Hung and J. Han, Lab Chip, 2015, 15, 2297–2307 RSC.
- H. W. Hou, M. E. Warkiani, B. L. Khoo, Z. R. Li, R. A. Soo, D. S. W. Tan, W. T. Lim, J. Han, A. A. S. Bhagat and C. T. Lim, Sci. Rep., 2013, 3, 1–8 Search PubMed.
- A. Sarkar, H. W. Hou, A. E. Mahan, J. Han and G. Alter, Sci. Rep., 2016, 6, 1–9 CrossRef.
- M. E. Warkiani, B. L. Khoo, D. S.-W. Tan, A. A. S. Bhagat, W.-T. Lim, Y. S. Yap, S. C. Lee, R. A. Soo, J. Han and C. T. Lim, Analyst, 2014, 139, 3245–3255 RSC.
- G. Guan, L. Wu, A. A. Bhagat, Z. Li, P. C. Y. Chen, S. Chao, C. J. Ong and J. Han, Sci. Rep., 2013, 3, 1–9 Search PubMed.
- G. P. Downey, D. E. Doherty, B. Schwab, E. L. Elson, P. M. Henson and G. S. Worthen, J. Appl. Physiol., 1990, 69, 1767–1778 CrossRef CAS.
- Department of Trade and Industry, Gov. Consum. Saf. Res., 2000, p. 38 Search PubMed.
- X. Zhang, Z. Zhu, N. Xiang and Z. Ni, Biomicrofluidics, 2016, 10, 054123 CrossRef.
- J. G. Hollowell, O. W. van Assendelft, E. W. Gunter, B. G. Lewis, M. Najjar and C. Pfeiffer, Vital Heal. Stat. Ser. 11 Data from Natl. Heal. Surv. Natl. Heal. Nutr. Exam. Surv. Hisp. Heal. Nutr. Exam. Surv Search PubMed.
- H. Jeon, H. Lee, K. H. Kang and G. Lim, Sci. Rep., 2013, 3, 3483 CrossRef.
- H. Jeon, Y. Kim and G. Lim, Sci. Rep., 2016, 6, 19911 CrossRef CAS.
- A. A. S. Bhagat, S. S. Kuntaegowdanahalli and I. Papautsky, Lab Chip, 2008, 8, 1906–1914 RSC.
- J. Sun, M. Li, C. Liu, Y. Zhang, D. Liu, W. Liu, G. Hu and X. Jiang, Lab Chip, 2012, 12, 3952 RSC.
- J. Sun, C. Liu, M. Li, J. Wang, Y. Xianyu, G. Hu and X. Jiang, Biomicrofluidics, 2013, 7, 011802 CrossRef.
- J. Wang, W. Lu, C. Tang, Y. Liu, J. Sun, X. Mu, L. Zhang, B. Dai, X. Li, H. Zhuo and X. Jiang, Anal. Chem., 2015, 87, 11893–11900 CrossRef CAS.
- M. E. Warkiani, A. K. P. Tay, G. Guan and J. Han, Sci. Rep., 2015, 5, 1–10 Search PubMed.
- T. Kwon, H. Prentice, J. De Oliveira, N. Madziva, M. E. Warkiani, J. F. P. Hamel and J. Han, Sci. Rep., 2017, 7, 1–11 CrossRef CAS.
- J. Seo, M. H. Lean and A. Kole, Appl. Phys. Lett., 2007, 91, 033901 CrossRef.
- A. P. Sudarsan and V. M. Ugaz, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 7228–7233 CrossRef CAS.
- P. B. Howell, D. R. Mott, J. P. Golden and F. S. Ligler, Lab Chip, 2004, 4, 663–669 RSC.
- A. P. Sudarsan and V. M. Ugaz, Lab Chip, 2006, 6, 74–82 RSC.
- M. E. Warkiani, B. L. Khoo, L. Wu, A. K. P. Tay, A. A. S. Bhagat, J. Han and C. T. Lim, Nat. Protoc., 2016, 11, 134–148 CrossRef CAS.
- L. Yin, Y. Wu, Z. Yang, C. A. Tee, V. Denslin, Z. Lai, C. T. Lim, E. H. Lee and J. Han, Lab Chip, 2018, 18, 878–889 RSC.
- B. R. Mutlu, K. C. Smith, J. F. Edd, P. Nadar, M. Dlamini, R. Kapur and M. Toner, Sci. Rep., 2017, 7, 1–9 CrossRef CAS.
- J. M. Martel, K. C. Smith, M. Dlamini, K. Pletcher, J. Yang, M. Karabacak, D. A. Haber, R. Kapur and M. Toner, Sci. Rep., 2015, 5, 1–12 Search PubMed.
- M. E. Warkiani, G. Guan, K. B. Luan, W. C. Lee, A. A. S. Bhagat, P. Kant Chaudhuri, D. S. W. Tan, W. T. Lim, S. C. Lee, P. C. Y. Chen, C. T. Lim and J. Han, Lab Chip, 2014, 14, 128–137 RSC.
- P. Sethu, M. Anahtar, L. L. Moldawer, R. G. Tompkins and M. Toner, Anal. Chem., 2004, 76, 6247–6253 CrossRef CAS.
- C. Pelegrí, M. Rodríguez-Palmero, M. P. Morante, J. Comas, M. Castell and À. Franch, J. Immunol. Methods, 1995, 187, 265–271 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc00675k |
| This journal is © The Royal Society of Chemistry 2020 |