Open Access Article
Open Access ArticleFast antimicrobial susceptibility testing on Escherichia coli by metabolic heat nanocalorimetry†
Yang
Liu
 ,
Thomas
Lehnert
and
Martin A. M.
Gijs
,
Thomas
Lehnert
and
Martin A. M.
Gijs
 *
*
Laboratory of Microsystems, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland. E-mail: martin.gijs@epfl.ch
First published on 13th July 2020
Abstract
Fast spreading of antimicrobial resistance is now considered a major global health threat. New technologies are required, enabling rapid diagnostics of bacterial infection combined with fast antimicrobial susceptibility testing (AST) for evaluating the efficiency and dosage of antimicrobial compounds in vitro. This work presents an integrated chip-based isothermal nanocalorimetry platform for direct microbial metabolic heat measurements and evaluates its potential for fast AST. Direct detection of the bacteria-generated heat allows monitoring of metabolic activity and antimicrobial action at subinhibitory concentrations in real-time. The high heat sensitivity of the platform enables bacterial growth detection within only a few hours of incubation, whereas growth inhibition upon administration of antibiotics is revealed by a decrease or the absence of the heat signal. Antimicrobial stress results in lag phase extension and metabolic energy spilling. Oxygen consumption and optical density measurements provide a more holistic insight of the metabolic state and the evolution of bacterial biomass. As a proof-of-concept, a metabolic heat-based AST study on Escherichia coli as model organism with 3 clinically relevant antibiotics is performed and the minimum inhibitory concentrations are determined.
Introduction
Steadily increasing antimicrobial resistance (AMR) of medically relevant microorganisms is an important threat to global health.1–3 Multidrug resistance of common pathogens, such as Staphylococcus aureus, Enterobacteriaceae and Pseudomonas aeruginosa, brings about severe therapeutic challenges, associated with elevated morbidity and mortality rates.4 An often cited AMR review report, based on detailed scenario analyses of the impact of AMR spreading on health care and macroeconomics, estimated to 10 million the number of deaths per year attributable to AMR in 2050, i.e. more than for most of the other major causes of disease-related death.5 An European cross-national database study established a correlation between AMR and outpatient antibiotic use.6 Moreover, clinical studies revealed a high percentage of unnecessary prescriptions of antibiotics for therapeutic and prophylactic purposes. This fact has been recognized as one of the main causes of emerging AMR. In many cases prescribed drugs are broad-spectrum antimicrobial agents, which further promotes AMR development and spreading.Rapid diagnosis of bacterial infection combined with fast and accurate antimicrobial susceptibility testing (AST) is one of the keys to counteract AMR progression, in particular by optimizing and personalizing the therapeutic management of infected patients.7 AST in vitro techniques aim to predict the efficiency of an antimicrobial compound for treating an infection. Ideally, an AST protocol should provide the minimum inhibitory concentration (MIC) for microbial growth, i.e. the lowest drug concentration that inhibits bacterial growth after overnight incubation.8 Standard AST methods in clinical microbiological diagnostics, in particular the disk diffusion method and broth microdilution (BMD), are based on established, robust and often manual protocols. A commercial diffusion-based gradient method (Etest®, BioMérieux) may be used for MIC determination.9 Current clinical protocols, however, suffer from important limitations, in particular extended time to result or the requirement of relatively large amounts of viable microorganisms. Automated AST systems based on turbidity measurements aim to reduce time to result, typically to below 24 h.10 Moreover, a range of new AST technologies is under development to tackle the AMR problem,10,11 either in a 96-well plate or microfluidic format, including molecular diagnostic tools to detect the presence of resistance genes12 and fast AST assays based on single-cell imaging, for instance.13 Nevertheless, most of these technologies still face major barriers with respect to general acceptance by the health care system and market penetration.7
In this context, we explore microbial heat production as a direct indicator of metabolic activity and viability of bacterial populations. Metabolic heat/heat flow curves provide real-time information on microbial growth dynamics and can be used for detecting bacterial growth inhibition in the presence of antimicrobials in vitro. The potential of isothermal microcalorimetry (IMC) for microbial activity monitoring, metabolic studies and drug assays has already been demonstrated by means of commercial devices. The latter use sealed mL-size sample ampoules and have very low heat power detection limits, typically in the range of a few μW down to 0.2 μW.14,15 For instance, different metabolic phases of Escherichia coli (E. coli) bacterial growth have been identified by analyzing heat flow profiles.16 Highly sensitive and fast detection of bacterial contamination (≥1–10 CFU mL−1) was also demonstrated by recording heat flow of artificially contaminated blood platelets samples.17 The potential of IMC for rapid differentiation between methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) was evaluated by von Ah et al. by using the antibiotics cefoxitin and oxacillin.18 Subsequently, Baldoni et al. reported a study using a 48-channel batch calorimeter for the detection of methicillin-resistance in S. aureus including genetically distinct clinical isolates.19 MRSA isolates were correctly identified after 5 h by testing against cefoxitin, however, very high bacterial inocula (McFarland turbidity of 5) were used to reduce time to result. Yang et al. analyzed the growth rate of E. coli and the time to maximum heat power as a function of concentration for two different cephalosporins.20 The two antibiotics tested affected the heat power curves in a clearly different manner. A more extensive AST study was performed by von Ah et al., analyzing heat flow curves for reference strains of E. coli and S. aureus for 12 different antibiotics. The results have been grouped by mode of antimicrobial action, revealing different thermal signatures for each compound family.21
Different microfabricated and microfluidic integration formats have been proposed for high-sensitivity nanocalorimeters. A miniaturized chip-based calorimeter, similar to the one presented in the present work, has been developed earlier by Higuera-Guisset et al. The authors studied the effect of two different growth media and of the culture temperature on the metabolic heat curves of E. coli.22 Another group used a flow-through chamber chip module, enclosed by two high-precision thermostats, for the study of biofilm inactivation by predatory bacteria and for monitoring biofilm eradication with antibiotics.23–25 Johannessen et al. implemented a sub-nL open-reservoir assay for heat measurements on a small number of isolated living cells.26 Torres et al. reported the microfabrication of 96-detector enthalpy arrays on polyimide membranes and implemented rapid electrostatic merging/mixing of droplets.27 The technology enabled studying kinetic parameters of enzyme-catalyzed reactions.28 Lee et al. proposed a more complex closed-chamber microfluidic approach with precise nL sample manipulation capabilities for measuring the heat of reaction of urea hydrolysis, for instance.29 Inomata et al. developed a picocalorimeter for detection of heat produced by a brown fat cell attached to a Si microstage connected to a cantilevered Si sensor. Heat is sensed by the resulting shift in the resonant frequency.30 An ultrasensitive micro-DSC (differential scanning calorimeter) for liquid protein sample characterization was demonstrated by Wang et al. The device is based on vanadium oxide thermistors and a microfluidic dual-chamber calorimeter design.31 Kim et al. recently reported a thin-film parylene microfluidic calorimeter with on-chip vacuum insulation. Measurement of cellular metabolic power changes upon controlled stimuli was demonstrated.32 A more recent commercial microcalorimetry instrument (Symcel AB) uses a well plate format for tracking the activity of living cells and biological processes.33
In this work, we present an isothermal nanocalorimetry platform designed for monitoring microbial growth dynamics with high sensitivity, thus enabling fast AST based on metabolic heat measurements. The INCfAST system (acronym for “isothermal nanocalorimetry platform for fast AST”), which is operated in a microincubator format, was optimized with respect to high thermal stability, a prerequisite for fast detection of bacterial infection or establishing accurate antibiograms. We evaluated microbial heat generation under different culture conditions. More importantly, we performed a proof-of-concept study comprising metabolic heat AST assays of E. coli exposed to 3 commonly used antimicrobial drugs (ciprofloxacin, ampicillin and gentamicin). Specific features of the metabolic heat/heat flow curves showed clear variations as a function of the antimicrobial concentration and MIC values could be determined for the 3 compounds. Furthermore, in a modified version of the isothermal platform, an oxygen sensor was implemented, enabling heat and oxygen consumption measurements for identical culture conditions in parallel. This offered a more detailed phenotypic fingerprint of the microbial metabolic activity upon drug exposure, while optical density measurements over time of the bacterial cultures provided additional insight in the development of bacterial biomass.
Results and discussion
Nanocalorimetry platform and oxygen consumption measurements
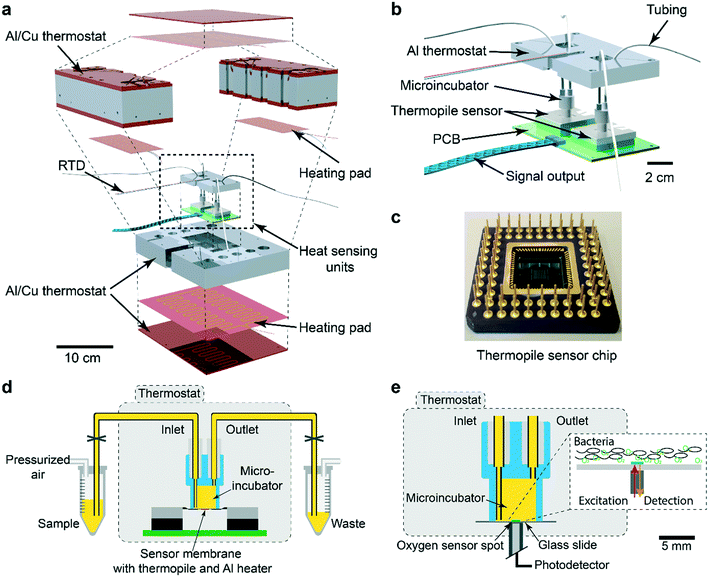
The nanocalorimetry platform is constructed around a sensing unit comprising a microincubator for bacterial culture and a thermopile sensor chip that detects heat flow corresponding to the metabolic heat power generated by the microbial sample during incubation. A dual chip arrangement allows increasing assay throughput or differential/control measurements. Several important modifications have been implemented with respect to a previous version of the system,34 mainly aiming at improving specific requirements for allowing high-sensitivity detection and accurate recording of metabolic heat profiles. These features include high thermal stability and fluidic integration suitable for fast thermalization of the sample and drug solutions. Fig. 1a shows an exploded 3D view of the INCfAST platform. Sensor chip and microincubator are inserted in the central part of the Al/Cu thermostat. A proportional–integral–derivative (PID) temperature control system with several heating pads provides uniform temperature distribution and stabilization at the sample location within ±250 μK at 37 °C (Fig. S1,† mK-range for the previous version of the platform34). Consequently, a limit of detection (LOD) for heat power of microbial suspensions as low as 750 nW was achieved (Fig. S2†). The core of the platform comprises the dual chip system with microincubators, fluidic connections and electrical read-out for independent parallel measurements (Fig. 1b). Voltage signals generated by the thermal sensors are recorded by a two-channel nanovoltmeter (34420A, Keysight, USA). Commercially available sensor chips were used in this study (XEN NCM9924, Xensor Integration, Delft, The Netherlands) (Fig. 1c). The sensing mechanism is based on a thin-film thermopile distributed around the center region of a Si membrane. The relatively thick membrane (45 μm according to the data sheet) provides sufficient robustness for liquid nanocalorimetry applications.More details of the integrated system are shown in Fig. 1d. The microincubator that receives the bacterial suspension is bonded onto the sensor surface. The liquid sample is in direct contact with the Si sensor membrane (no polymer membrane on the bottom of the microincubator), thus maximizing heat transfer to the sensing elements. The microincubator has one sample inlet and one waste outlet. The corresponding reservoirs are located outside the thermostat at room temperature for convenient operation. Nevertheless, thermalization of compound solutions and bacterial suspensions during injection is fast and not limiting the performance of the platform for the present application involving relatively slow heat signal variations. Optionally, the reservoirs could be placed inside the thermostat for reducing thermal stabilization time. Placing the whole setup inside a foam/polystyrene housing (not shown) further reduced perturbations due to environmental temperature variations.
The INCfAST nanocalorimetry measurements were combined with real-time oxygen consumption measurements of a microbial suspension under culture conditions that are equivalent to the heat assays. For that purpose, the thermopile sensor was replaced by a glass substrate holding a sputter-deposited thin-film optical oxygen sensor spot (Fig. 1e). The sensor features detection of oxygen-dependent luminescence in the near-infrared (REDFLASH Technology, PyroScience GmbH). The microincubator was fixed directly over the sensor and optical access was provided from the bottom of the thermostat. The same thermal control and fluidic systems were used for both platforms, ensuring identical assay conditions. Parallel heat and oxygen measurements were performed by injecting simultaneously identical bacterial samples into each platform.
Analysis of microbial metabolic heat profiles
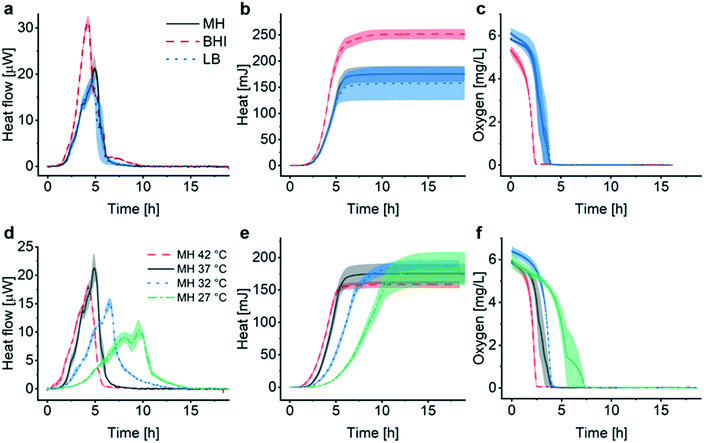
Fig. 2a (black curve) shows a representative metabolic heat flow curve (i.e. heat power P(t) [W]) for E. coli in Mueller–Hinton broth (MH) recorded on the INCfAST platform (V = 150 μL, inoculum size 2.8 ± 0.5 × 105 CFU mL−1, 37 °C). The P(t) curve displays the time-dependent metabolic heat flow generated by the bacterial population in the center region of the Si chip membrane towards the Si bulk edge, thus depends on the actual number of viable cells and their metabolic activity. In the initial segment the signal is below LOD, corresponding to an apparent lag phase or delay time (tdelay ≈ 1.5 h) associated with low biomass and/or low metabolic activity. Subsequently, exponential growth generates a rapidly rising signal up to a transient maximum rate of heat production Pmax (at ∼5 h, 21.3 ± 2.5 μW) followed by a sharp decline. Assay curves have been recorded up to 20 h, however only the most relevant time interval [0, 7 h] is shown in Fig. 2a. At longer incubation times (≥10–11 h) metabolic heat production ceased due to nutrient and/or oxygen depletion in the microincubator. At this stage, most of the bacteria are still alive as revealed by optical microscopy inspection or are possibly in a dormant state. The aggregate metabolic heat curve (energy Q(t) [J]) was obtained by integration of the heat flow curve over time (Fig. 2a, red curve). Accordingly, the Q(t) curve features three phases, namely the initial lag phase, the exponential growth region (for t ∼ [1.5 h; 5.5 h]) and a stationary phase (for t ≳ 7 h), in which the total heat does not increase any more (Qmax ≈ 175 ± 15 mJ). For the metabolic assays discussed below, the P(t) and Q(t) curves shown in Fig. 2a were used as reference for normal culture conditions on the INCfAST platform.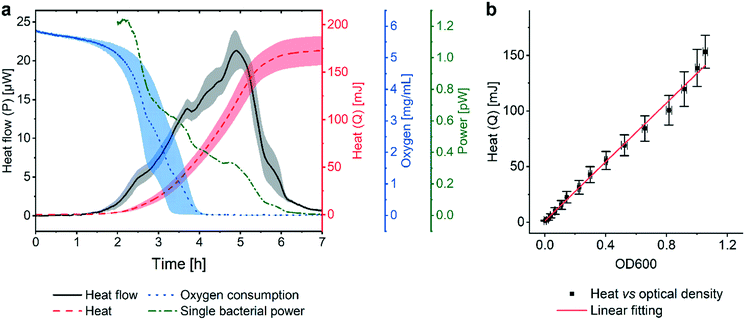 | ||
| Fig. 2 Metabolic heat and oxygen consumption measurements performed on the nanocalorimetry platform. (a) Metabolic heat flow curve P(t) (black) for E. coli ATCC 25922 with typical incubation conditions on the INCfAST platform (Mueller–Hinton broth, 37 °C). The heat power signal was obtained by read-out of the thermopile sensor voltage output after calibration. Integration of the heat flow curve over time provides the corresponding metabolic heat curve Q(t) (red). The single bacteria heat power (green curve) was estimated by normalization of Q(t) by the biomass at a given time point, as determined from optical density (OD600) measurements. Synchronized parallel on-chip oxygen consumption measurements revealed complete oxygen depletion in the microincubator after 4 h (blue curve), indicating the transition from an aerobic to an anaerobic metabolic state. The assay interval for t = [0, 7 h] is emphasized in the plot. For t > 7 h heat values remain constant, all other values tend to zero. (b) Plot of the metabolic heat signal vs. OD600 of a bacterial suspension up to the exponential growth phase (using data shown in (a) and Fig. S3a† for t = [0, 5 h]). The proportional relationship (coefficient 134 mJ/1.0 OD600) indicates that the heat signal is directly related to the amount of bacteria in this regime. All curves represent mean ± SE (n = 3). | ||
We performed optical density measurements (OD600, i.e. at 600 nm) under culture conditions comparable to the BMD method on a plate reader. As shown in Fig. S3a† for culture in MH, the exponential phases of the Q(t) and the OD600 curves overlap well. A scaling or heat/OD600 conversion coefficient of 134 mJ/1.0 OD600 for MH was determined by linear regression of the Q(t) vs. OD600 plot shown in Fig. 2b. This observation indicates that heat curves recorded with our system can be safely correlated with bacterial growth or the actual biomass density in the microincubator, respectively. Mean slopes Q/t in the exponential region may be considered as indicative for bacterial growth rates, whereas heat flow profiles P(t) are more sensitive to transient phenomena of the growth behavior. Furthermore, P(t) can be normalized with respect to biomass simply by dividing through the converted Q(t) values. The resulting curve (Fig. 2a, green curve), showing a continuous decline (∼0 pW for t > 7 h), provides a rough estimate of the time evolution of the metabolic heat production per bacterium for the specific culture conditions. For the calculation we applied a conversion equivalence of 1.0 OD600 corresponding to 5.0 × 108 CFU mL−1.35
In order to further analyze the metabolic phenotype, we measured the oxygen concentration during bacterial growth in the thermalized microincubator by modifying the nanocalorimetry platform into a luminescence detection-based oxygen sensor platform, taking care to keep culture conditions identical to those of the heat flow assay. For aerobic bacteria such as E. coli, oxygen consumption is an important indicator of metabolic activity and viability. The microincubator itself can be considered as gas non-permeable, thus the oxygen concentration directly reflects the actual oxygen consumption of the bacterial population. In the case study discussed in Fig. 2, the oxygen signal started declining sharply at the onset of exponential growth (i.e. at ∼1.5 h) and reached a non-detectable level after 4 h (Fig. 2a, blue curve). Interestingly, the oxygen consumption curve indicates that depletion of oxygen occurs earlier than the time point were heat flow is maximum (Pmax at ∼5 h, black curve in Fig. 2a), suggesting that the bacteria population continues growing normally for at least ∼1 h in an anaerobic metabolic state. As a facultative anaerobic bacterium, E. coli thus appears not to be immediately sensitive to oxygen depletion. Indicative heat power/bacterium mid-range values are 1.1 pW (at t = 2.5 h) for the aerobic metabolic state and 0.32 pW (at t = 5 h) for the anaerobic state, respectively (Fig. 2a, green curve). Transitions between different metabolic states may in principle result in more complex heat flow curves (see Discussion). In the present case, a small crinkle-like feature can be identified at ∼3.7 h (Fig. 2a, black curve), possibly corresponding to the transition from aerobic to anaerobic conditions. Nevertheless, no significant impact on the overall shape of the heat flow curve is observed, suggesting a fast shift between the two metabolic states.
Metabolic activity of E. coli for different culture conditions
Assessment of the metabolic activity of E. coli in different culture media (Fig. 3a–c), as well as for several culture temperatures (Fig. 3d–f), was performed as a first evaluation of the INCfAST platform performance. Heat flow curves have been recorded in brain heart infusion medium (BHI), MH and lysogeny broth (LB) (Fig. 3a). These media have different compositions and amounts of nutrients (Table S1†), affecting growth rate and metabolic activity of the bacteria. We found that rich media, in particular BHI, promotes transient metabolic heat production reflected by the Pmax values of the heat flow curves, yielding 31.3 ± 1.2 μW in BHI (highest amount of carbon compounds), 21.3 ± 2.5 μW in MH (intermediate nutrient level) and 18.7 ± 1.4 μW in LB (low amount of nutrients) (Fig. 3a). Consequently, culture in BHI also enhanced the total heat generated during incubation (Qmax ≈ 252 ± 10 mJ in Fig. 3b), whereas Q(t) reached comparable levels for MH and LB in the stationary phase (175 ± 15 mJ and 157 ± 32 mJ, respectively). Oxygen depletion occurs earliest for BHI, indicating fastest growth and/or highest metabolic activity, whereas oxygen curves are comparable for MH and LB (Fig. 3c).For all media, OD600 curves overlap well with the corresponding heat curves in the exponential region (after y-axis scaling and time synchronization, Fig. S3a–c†). OD600 measurements reveal that the increase of the bacterial biomass is fastest in BHI compared to MH or LB. As already shown in Fig. 2b for MH, heat/OD600 coefficients were determined by linear regression in the exponential region for all media tested (Fig. S3d†). These coefficients vary for the different culture media (BHI 84 mJ/1.0 OD600, MH 134 mJ/1.0 OD600 and LB 162 mJ/1.0 OD600), indicating that heat curves cannot be converted to growth curves based on biomass in a universal manner for all conditions. Considering the Pmax values stated above, maximum growth rates of 1.9 × 105 CFU s−1 for BHI, 8.1 × 104 CFU s−1 for MH and 6.0 × 104 CFU s−1 for LB, respectively, can be derived for culture in the microincubator of the INCfAST platform (1.0 OD600 = 5.0 × 108 CFU mL−1). Incubation of E. coli in BHI yields the fastest growth rate but the lowest heat production rate. This finding is in agreement with a previous statement in literature, claiming that the heat production rate per unit weight is inversely proportional to the growth rate in the exponential phase.36 Interestingly, for all 3 media conditions in the microincubator Qmax values are reached on the same time scale (at t ≈ 6 h, Fig. 3b).
We also compared the effect of culture temperature in MH on the heat flow curves (Fig. 3d) and the heat curves (Fig. 3e). As expected, bacterial populations show the highest transient growth rate (heat production rate) under most favorable conditions (37 °C, Pmax ≈ 21.3 ± 2.5 μW) and lower values for higher (42 °C) or lower temperatures (in particular for 27 °C with Pmax ≈ 10.3 ± 1.8 μW). The apparent lag phase tdelay (∼1.5 h at 37 °C) is shortest for 42 °C (<1 h) and becomes more important for decreasing temperatures (∼3 h at 27 °C). Further alterations of the heat flow curves, comprising a general broadening or flattening of the profiles and emerging secondary transient peaks, is observed for culture at low temperatures (Fig. 3d, 32 °C and 27 °C). P(t) broadening also results in lower mean slopes Q/t in the exponential phase (Fig. 3e, in particular at 27 °C). Heat signals reach comparable Qmax levels for all culture temperatures (160–190 mJ), but on longer time scales for decreasing temperature. Corresponding OD curves evolve similarly (Fig. S4†). The temperature effect on oxygen consumption is in line with the heat curves as well, showing a clear delay of oxygen depletion at most unfavorable growth conditions (27 °C) (Fig. 3f).
Antimicrobial susceptibility testing based on metabolic heat assays
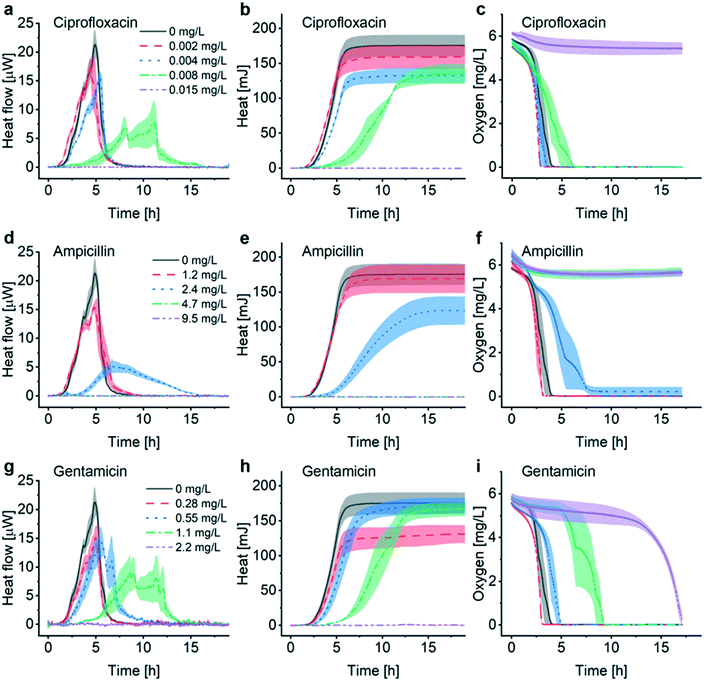
The INCfAST platform was primarily designed for probing the efficacy of antimicrobial drugs and possibly for monitoring specific modes of action by means of metabolic heat assays. The presence or absence of a heat signal during a preset incubation period provides information on antimicrobial susceptibility or resistance of a bacterial strain. We performed AST on E. coli ATCC 25922 by analyzing the metabolic heat profiles for a selection of clinically relevant antibiotics and estimated MIC values. Heat flow curves P(t) for ciprofloxacin, ampicillin and gentamicin are shown in Fig. 4a, d and g and the corresponding heat curves Q(t) in Fig. 4b, e and h, respectively. P(t) and Q(t) reference curves without antibiotic (0 mg L−1, MH, 37 °C) are the same as for the previous assays (Fig. 2a and 3). Fig. 4c, f and i are the oxygen consumption curves recorded for these assays. In the low drug concentration range and for all 3 antimicrobial agents tested, heat flow curves approximately maintain a shape similar to culture in pure medium (Fig. 4a, d and g, black and red curves, respectively), indicating normal exponential bacterial growth in the microincubator. A decrease of the maximum heat power Pmax values and the appearance of bump-like features may be noted. Stronger alterations occur with increasing drug concentrations, especially for values approaching MICs. Heat signals disappear at concentrations above MIC (Fig. 4a, d and g, purple curves).For ciprofloxacin, for instance, heat flow curves are relatively confined for drug concentrations ≤0.004 mg L−1, whereas at concentrations ≥0.015 mg L−1 heat production is inhibited (Fig. 4a). In the intermediate concentration range, i.e. for 0.008 mg L−1 in this case, the heat flow curve flattens significantly and secondary transient structures become more apparent. On the corresponding heat curve the mean slope Q/t in the exponential growth region decreases (Fig. 4b, green curve). The total heat signal disappears at ≥0.015 mg L−1 for the assay duration of 20 h. Moreover, a clear prolongation of the apparent lag phase tdelay from ∼1.5 h at 0 mg L−1 to ∼4 h at 0.008 mg L−1 occurs. Based on this series of experiments, we assume that the MIC value for ciprofloxacin falls into the concentration interval 0.008 mg L−1 < MIC ≤ 0.015 mg L−1. Clinical reference values established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) are summarized in Table 1 for the antibiotics studied in this work.37 EUCAST provides a MIC range of 0.004–0.016 mg L−1 for ciprofloxacin, thus the INCfAST result overlaps well with the reference range.
| Antibiotic | INCfAST MIC interval (mg L−1) | EUCAST MIC range (mg L−1) |
|---|---|---|
| a The European Committee on Antimicrobial Susceptibility Testing. | ||
| Ciprofloxacin | 0.008 < MIC ≤ 0.015 | 0.004–0.016 |
| Ampicillin | 2.4 < MIC ≤ 4.7 | 2–8 |
| Gentamicin | 1.1 < MIC ≤ 2.2 | 0.25–1 |
We performed similar AST assays for ampicillin (Fig. 4d and e) and gentamicin (Fig. 4g and h). Ampicillin heat flow curves showed prolonged lag phase and a strongly modified profile at 2.4 mg L−1 (i.e. close to MIC). Secondary transient structures are not visible in this case. The mean slope Q/t in the exponential growth region decreases strongly for this concentration (Fig. 4e, blue curve). Heat production was inhibited for higher concentrations (≥4.7 mg L−1) providing an INCfAST interval of 2.4 mg L−1 < MIC ≤ 4.7 mg L−1, which falls within the EUCAST reference value range 2–8 mg L−1 (Table 1). Heat profiles for bacterial culture with increasing concentrations of gentamicin evolved in a similar way as for ciprofloxacin. Interestingly, close to MIC the secondary P(t) transient features are strongly pronounced but the mean slope Q/t in the exponential growth region seems to be less affected (Fig. 4g and h, green curve). In the case of gentamicin, however, the INCfAST interval 1.1 mg L−1 < MIC ≤ 2.2 mg L−1 was just above the EUCAST range (Table 1). As a control, we performed AST using the BMD method on a plate reader (Fig. S5†). OD600 results for ciprofloxacin and ampicillin are consistent with heat measurements in terms of MIC values. The OD600 interval 0.55 mg L−1 < MIC ≤ 1.1 mg L−1 for gentamicin is lower compared to the heat assay and overlaps well with the EUCAST range of 0.25–1 mg L−1.
Oxygen consumption as a viability indicator can be applied as another method to determine metabolism and MIC for non-anaerobic bacteria. For concentrations below MIC, oxygen is gradually consumed and the time to full oxygen depletion in the microincubator increases clearly for concentrations approaching MIC (Fig. 4c, f and i). For ciprofloxacin and ampicillin oxygen levels remain constant above MIC because of metabolic inactivity or death of the bacteria (purple curves in Fig. 4c and f). In the specific case of gentamicin, oxygen consumption at subinhibitory concentrations evolves in principle as for ciprofloxacin and ampicillin, even if delays are more pronounced close to the expected MIC (e.g. green curve in Fig. 4f, 1.1 mg L−1). Surprisingly, oxygen depletion is also observed at a concentration of 2.2 mg L−1, i.e. above the MIC value range derived from the heat assay. The oxygen concentration in the microincubator decreased sharply only after a prolonged incubation period of ∼13 h (purple curve in Fig. 4i), indicating that a small population of bacteria was able to survive in the microincubator. The antimicrobial action of gentamicin is based on an oxygen-dependent process.38 It is possible that diffusion of oxygen to bacteria clusters attached on the surface of the oxygen-sensing spot is limited thus inhibiting temporarily the action of gentamicin.
Energy spilling of E. coli due to antimicrobial stress
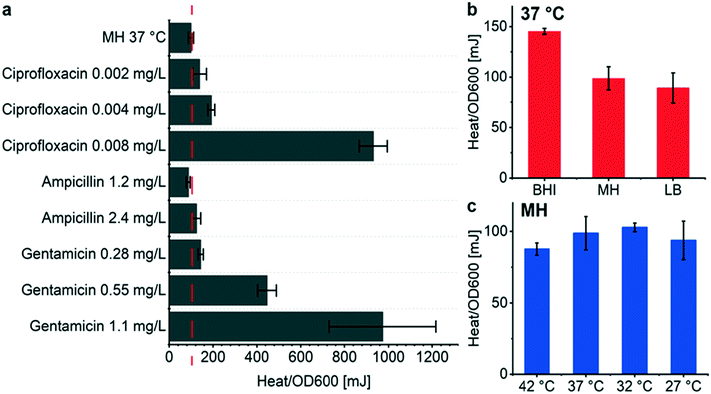
OD600 growth curves recorded with the BMD method on a plate reader show that the overall biomass density decreases for increasing antimicrobial concentration in the considered assay duration for the 3 compounds tested (Fig. S5†), whereas observing a systematic behavior of the heat signals Qmax is less obvious (Fig. 4b, e and h). This observation implies that the metabolic heat production per biomass unit may be enhanced under antimicrobial stress. In order to analyze the correlation of heat production and biomass in the presence of antimicrobials more accurately, we measured OD600 values of samples directly collected from the microincubator at the end of each metabolic heat assay (at t = 20 h). By this means, we may evaluate precisely the Qmax to OD600 ratio of the final bacterial suspension, thus providing an average value resulting from the incubation process on the INCfAST platform. Fig. 5a summarizes the heat/OD600 coefficients for different antimicrobial exposures below MIC and compares these values to bacterial growth without antibiotics (98 ± 12 mJ/1.0 OD600 for MH, dashed line). The plot indicates that for ciprofloxacin and gentamicin the bacterial heat production per biomass unit is strongly promoted by antimicrobial stress, reaching a more than 9-fold enhancement close to MICs (931 ± 64 mJ/1.0 OD600 at 0.008 mg L−1 for ciprofloxacin and 970 ± 243 mJ/1.0 OD600 at 1.1 mg L−1 for gentamicin, respectively). The effect is much less pronounced for ampicillin. These values, taken at the end of the assay, are indicative for comparing the impact of the different antibiotics, but absolute heat/OD600 coefficients may be different if taken during the growth phase at an earlier time point than in the stationary phase, as the biomass may still evolve.A similar analysis based on assay endpoint detection of OD600 and Qmax has been done for different culture media (Fig. 5b). In this case, the heat/OD600 coefficients decreased from BHI, MH to LB, corresponding to the order of the respective nutrient levels (98 ± 12 mJ/1.0 OD600 for MH). This was not the case with the coefficients derived previously by linear regression during the exponential growth phase (Fig. 2b and S3d,† 134 ± 3 mJ/1.0 OD600 for MH). OD600 values have been determined either with a sample retrieved from the closed microincubator (Fig. 5b) or cultured by BMD on a shaking plate reader (Fig. 2b and S3d†). Both methods evaluate the heat/OD600 coefficients in a methodically different way, with focus on averaging over the assay duration or on the exponential growth region, respectively. As outlined above, the endpoint method is better suited for evaluating energy spilling over the whole assay duration. For the average heat per biomass at different culture temperatures in MH, no significant differences were observed (Fig. 5c).
Discussion
Current AST protocols in clinical microbiological laboratories generally require several incubation cycles for preparing standardized inoculums of isolated microorganisms and/or for pathogen identification. Reliable detection of growth inhibition typically again takes up to ∼20 h, or even longer depending on the bacterial species. As a consequence, AST results may be available to clinicians only after several days.9,10 Hence, in order to improve antimicrobial prescription and treatment practice, there is a clear need for implementing new technologies enabling fast and fully automated AST protocols.7,10,11IMC has high potential for biomedical applications in general and for microbial studies in particular.14–16 In this view IMC may also be considered as one of the near-future alternatives for AST in clinical settings.11 For instance, among other specific advantages, calorimetric assays can in principle be performed directly with opaque clinical samples, thus reducing significantly time to result. The non-specificity of the heat signal, however, requires carefully designed protocols in order to extract the information of interest and interpretation of calorimetric data in terms of meaningful microbiological parameters.15 Despite their performance, commercial IMC systems have drawbacks related to the use of sealed mL-size ampoules, requiring relatively large inoculum size and impeding fluidic manipulations during the assay. The chip-based nanocalorimetry platform developed in this work aims to overcome such bottlenecks, mainly through miniaturization and improved fluidic integration. The INCfAST platform was designed for direct injection of bacterial suspensions or drug solutions into the isothermal microincubator. This feature reduces delay times (only the small sample/drug volume needs to be thermalized), enables automated operation and eventually enhances assay versatility. Minimizing thermal time constants by reducing sample volumes and thermostat size, combined with high sensitivity, is a prerequisite for implementing fast AST protocols.
In the present proof-of-concept study, AST assays were performed on purified samples with volumes of only 150 μL. In order to estimate the lower limit of time to detection, we consider the LOD of 750 nW of the platform, corresponding to ∼106 CFU (assuming 1.1 pW per E. coli bacterium). For an inoculum size of 4.2 × 104 CFU (2.8 × 105 CFU mL−1) and taking a typical doubling time of 15–20 min for E. coli in laboratory conditions, the limit of quantification (i.e. 3 × LOD ≈ 2.2 μW or ∼2.2 × 106 CFU per microincubator) should be reached after ∼2 h. In our study, heat flow was indeed reliably detectable after a time lapse of ∼2 h (corresponding to ∼2 μW) under normal culture conditions (MH, no antibiotics, 37 °C) (Fig. 2a). An additional conservative time lapse of 30 min for thermalization was allowed after the injection before starting data recording of each experimental curves. This estimation is based on a fast-growing laboratory strain of E. coli as bacterial model.
Based on the heat flow curves in Fig. 4, AST on E. coli ATCC 25922 with ciprofloxacin, ampicillin and gentamicin could be safely performed in approximately 5–6 hours using the INCfAST platform. After this incubation duration, a significant difference between the heat flows of the lag phase with antibiotic and the “no growth” of cultures at the MIC occurs in all 3 cases. Nevertheless we have to consider that most clinical isolates, for instance Klebsiella pneumonia,39Acinetobacter baumannii,40 and Pseudomonas aeruginosa41 have doubling times in the range of 40 to 140 min, i.e. longer than the E. coli strain in the present study. Accordingly, the amount of microbials required for reliable detection might only be reached on a longer time scale. On the other hand, the heat power per bacterium also varies for different species. Higher heat power relieves partly the adverse effect of slow growth in terms of time to result. Further investigations will be needed to evaluate the performance of the present approach in real clinical settings and to determine actual realistic time scales of the metabolic heat AST assays.
Compared to other AST techniques, the thermal profiles of metabolic heat assays reveal significantly more information on the mode and kinetics of action of antimicrobial compounds. In the frame of this study, we observed clear variations of growth-related patterns in the presence of antibiotics. One important parameter is the time lapse to reach a detectable heat signal tdelay (apparent lag phase). Extension of tdelay, in particular for concentrations close to MICs, was observed for the 3 compounds tested (Fig. 4). This feature may be interpreted as a transient bacteriostatic effect, especially in the case of ciprofloxacin and gentamicin, where the mean growth rate Q/t in the delayed exponential phase is less affected than for ampicillin. On the other hand, ciprofloxacin, ampicillin and gentamicin are bactericidal antibiotics and in principle two reasons may be considered for longer apparent lag phases: (i) the drug compound kills most of the bacteria population, but a small amount of surviving cells (persisters) can be detected only with delay,42 or (ii) elongation of the lag phase of the bacterial growth cycle attributed to enzymatic adaptations or genetic regulations in response to changing micro-environmental biochemical conditions.43,44 Single-cell analysis would be necessary for discriminating between these two possibilities. It has been shown previously that lag phase extension in the presence of antibiotics is an important criterion that has to be taken into account, in addition to MIC or IC50 (concentration at 50% growth inhibition) values, in order to evaluate the efficacy of a drug.45 For instance, bacterial strains were found to increase the lag time as a strategy for developing compound tolerance in response to antibiotic stress.46 Such findings are important for the clinical evaluation of antimicrobial treatments. As outlined above, the high sensitivity of the INCfAST platform allows bacterial growth detection using a standard inoculum size in pure medium after only ∼2.5 h for E. coli. Detection of infection in clinical samples should in principle be possible on the same time scale. Lag phase extension in the presence of antibiotics, however, counteracts this advantage. Nevertheless, even if antimicrobial action delays the onset of metabolic heat production, heat signals could still be detected as early as ∼4 h just below MICs for the specific conditions of our assays (Fig. 4). Actual time windows in clinical settings for differentiation between growth/growth inhibition based on standard methods are set to 16–20 h. For implementing a new AST approach this time frame has to be reconsidered in order to take full advantage of the new technology, but also for defining an acceptable minimum assay time with respect to reliability of the results from the clinician's point of view.
Antimicrobial action not only delays the onset of heat production, but also has a significant impact on the transient features of the heat flow curves. Progressively emerging secondary peaks and a general broadening of the curves with increasing antimicrobial concentration is most evident in Fig. 4a and g for ciprofloxacin and gentamicin, in particular close to MICs (green curves). Similar features were also observed in heat profiles without drug exposure and are generally attributed to metabolic transitions. Metabolizing sequentially two different carbon sources results in successive growth cycles with multiple exponential phases, possibly separated by a phase with reduced growth rate or transient growth inhibition, giving rise to diauxic shifts.47,48 As observed in Fig. 3d, such effects may be enhanced at low culture temperatures, possibly due to reduced cellular enzymatic activity. Transitions between respiration and a fermentation states (Fig. 2a, black curve) may also generate more or less pronounced features or peaks, depending on the respective time scales.49 It was noted previously that deconvolution of heat profiles yields individual Gaussians that can be used to estimate the metabolic heat produced by different mechanisms.16
Comprehensive understanding of the dynamic growth behavior at subinhibitory concentrations revealed by specific metabolic heat flow profiles might enable anticipation of antimicrobial efficacy and possibly extrapolation to MIC values, provided that quantification of growth parameters through adequate growth models can be applied.21,50 Interestingly, it was demonstrated by von Ah et al., based on an IMC study with 12 antibiotics on E. coli and S. aureus that it could be possible to distinguish between different modes of antimicrobial action. In addition to alterations of transient features, variations in tdelay and mean growth rates Q/t were observed. The 3 antibiotics tested in our nanocalorimetric study have different modes of action. Ciprofloxacin targets topoisomerase II (DNA gyrase), which inhibits bacterial DNA synthesis, ampicillin inhibits cell wall synthesis and gentamicin inhibits bacterial protein synthesis through binding to the A site of 16S ribosomal RNA.51 As discussed before, we observed a strong impact on the heat profiles for all compounds. Nevertheless, no conclusions can be drawn from our results with respect to different modes of action. Heat flow curves for ciprofloxacin (Fig. 4a) and gentamicin (Fig. 4g) close to MIC are very similar, for ampicillin (Fig. 4d) the overall shape is different. A more systematic study is necessary for further going interpretations. Investigation of bactericidal and bacteriostatic modes of action would also be of interest.
Total heat curves Q(t) as integrals of heat flow curves allow a better evaluation of mean growth rates Q/t in the exponential region. The aggregated heat value Qmax in the stationary phase is determined by growth-limiting factors, such as oxygen/nutrient depletion and waste production in the microincubator. Interestingly, comparing heat curves (Fig. 4) and OD600 values of samples after incubation in the microincubator revealed that the metabolic heat production may be enhanced under antimicrobial stress (Fig. 5), an observation that can be understood as energy spilling. Inefficient use or spilling of adenosine triphosphate (ATP) as energy source is known as a possible survival strategy for many bacterial species. In E. coli, energy spilling could be mediated by a futile cycle of potassium or ammonium ions.52 As far as can be deduced from our nanocalorimetric assays (Fig. 5a), ciprofloxacin and gentamicin induced strong energy spillage close to MICs, whereas the effect was only weakly pronounced for ampicillin. Previously, an initial increase of the heat production rate upon antibiotic exposure was observed in Pseudomonas putida biofilms and related to energy-dependent resistance mechanisms.53 In addition, there is evidence that antibiotics can perturb bacterial respiration and central metabolism. A recent publication demonstrated that antibiotic-induced adenine limitation increases purine biosynthesis and ATP demand, which might relate antimicrobial stress to enhanced bacterial heat production.54
Combining oxygen consumption measurements with metabolic heat recordings of bacteria under antimicrobial stress provides a more holistic phenotypic characterization of metabolic activity. Time to full oxygen depletion in the microincubator increases clearly close to MIC (Fig. 4c, f and i). In our assays, oxygen depletion appeared on a shorter time scale than significant features of the heat flow profiles (e.g. earlier than the maximum of heat production Pmax). Oxygen consumption may therefore be explored as another potential parameter for fast AST on aerobic bacteria. Nevertheless, oxygen depletion in the microincubator has to be considered carefully when evaluating the results. For instance, the MIC interval for gentamicin determined with the INCfAST platform was above EUCAST reference values (Table 1). The oxygen-dependent antimicrobial action of gentamicin may be at the origin of this observation.38 Progressive oxygen depletion in the microincubator during the assay possibly reduced the antimicrobial efficacy of gentamicin, resulting in somewhat enhanced MIC values with respect to conventional protocols. Accordingly, for OD measurements (Fig. S5c†), where shaking of the plate reader ensures oxygen replenishment of the culture solution, no deviation from the standard MIC range was observed. On the other hand, the enclosed microincubator system might be suitable for performing fast AST on anaerobic bacteria, which can be challenging with the conventional agar-based culture methods.
Experimental
Microincubator fabrication
Polycarbonate microincubators (V = 150 μL, Ø = 5.7 mm, height = 6 mm) fabricated by micromilling were employed for the metabolic heat assays on the INCfAST platform. Polycarbonate offers good mechanical stability and high chemical resistance with respect to the sterilization protocols applied for the experiments.Sensor calibration
Sensor calibration was performed by means of the Al thin-film resistive heater on the sensing area of the chip membrane. A deionized (DI) water-filled microincubator was positioned on the sensor during calibration in order to simulate actual assay conditions. Calibration of the thermopile voltage output was performed by generating defined heat power signals via an external electrical power source. We determined a power/voltage coefficient of 0.88 W V−1 for the present system configuration (Fig. S6†). This value is about 3 times higher than in the previous version of the platform (0.29 W V−1).34 This improvement was mainly achieved by optimizing the heat transfer from the sample in the microincubator to the sensor membrane.Microbial sample preparation
Assays in this work were carried out using the E. coli ATCC 25922 strain (E. coli WDCM 00013 Vitroids™, Sigma-Aldrich). This organism is a typical AST control strain. We used a standard BMD protocol for bacterial culture and sample preparation. E. coli inoculum was first introduced into liquid MH culture medium and incubated overnight at 37 °C. Subsequently, 10 μL of this suspension was cultured on MH agar plates (Thermo Scientific, CM0337) and diluted by streaking. Single colonies were picked to prepare a bacterial suspension with an OD value of 0.2 measured at 600 nm (0.5 McFarland standard), i.e. a concentration of ∼1 × 108 CFU mL−1. By dilution in pure culture medium we obtained the primary samples for the heat flow assays with an initial bacterial concentration of 2.8 ± 0.5 × 105 CFU mL−1 for all experiments (confirmed by 3 serial quality control experiments). A purity, viability and concentration check prior to each measurement series was carried out by inoculation on a non-selective agar plate (after further 100-fold dilution) and colony counting after overnight culture. With our quality control experiments, we determined a concentration of 3.7–5.5 × 108 CFU mL−1 for 1.0 OD600, which is consistent with the value given by Stevenson et al.35 The bacterial suspension was stored in a small flacon and used within 15 min as inoculum on the INCfAST platform. For some of the metabolic heat assays in this work different culture media were compared, including BHI (Thermo Scientific, CM1135), MH (Thermo Scientific, CM0405), and LB (Sigma Aldrich, L3522). All culture media and agar plates were prepared according to the recommended protocols and concentrations. Microbial suspensions and antimicrobial solutions for OD600 measurements were prepared in the same way as for the heat assays. We performed OD600 measurements on a plate reader (24-well plate, inoculum 2.8 ± 0.5 × 105 CFU mL−1, 37 °C). The microbial suspension in each well was sealed with 500 μL mineral oil (Sigma-Aldrich, M8410) to prevent evaporation.Antimicrobial solutions
Ciprofloxacin, ampicillin sodium salt, and gentamicin were purchased from Sigma-Aldrich. Ampicillin sodium salt and gentamicin are soluble in DI water. 1 mol L−1 hydrogen chloride solution was used for ciprofloxacin. Ciprofloxacin, ampicillin, and gentamicin stock solutions were prepared with a concentration of 25.5 mg mL−1, 51.9 mg mL−1 and 36.0 mg mL−1, respectively. Stock solutions were stored at 4 °C and used within maximum 1 week to avoid a risk of declining activity. Prior to the measurements stock solutions were thermalized at room temperature and gradually diluted in DI water to prepare a series of concentrations according to the potency of each antibiotic (10 times higher than the final target concentrations). Test solution aliquots were prepared by further 10-fold dilution in MH. Target concentrations were chosen according to the range suggested by EUCAST. The bacterial inoculum was added to the test solutions immediately before sample injection into the microincubator.Platform sterilization
In order to avoid damage of the fragile sensor membrane during operation, different key components, in particular the microincubator and the sensor, were generally reused in successive measurements. After each experiment, a cleaning and sterilization protocol was applied to the whole fluidic circuit of the nanocalorimetry platform, including tubing, the microincubator and the sensor surface. First, the circuit was thoroughly rinsed with ethanol in order to remove and elute most of the bacteria in the system, followed by soaking all exposed surfaces in RAPICIDE™ OPA/28 High-Level Disinfectant (Cantel Medical Corp.) for 20 min for sterilization and repeated washing with sterilized DI water. Control experiments without bacterial inoculum confirmed the efficiency of this protocol (no microbial heat signal was detected for ≤15 h, Fig. S2†). The platform and all auxiliary devices were kept under a biosafety cabinet to avoid contamination from the environment during the experiment.Heat flow measurement protocol
Metabolic heat generation of E. coli ATCC 25922 was studied under different culture conditions and upon exposure to antimicrobial compounds. First, the microincubator was installed in the platform and connected to the reservoirs for the bacterial sample/drug suspension and waste, respectively (Fig. 1d). The platform was then stabilized to the temperature set point (normally 37 °C) and kept at that temperature for consecutive measurements. The sample solution was injected into the microincubator by pressure control of the sample reservoir (Fluigent MFCS pressure pump). The microincubator was then isolated by closing the valves in the inlet/outlet tubing and recording of the heat flow curves was started. Different assay conditions were successively tested by changing the external sample reservoir and refilling the microincubator after applying the cleaning/sterilization protocol described above. The platform thermalization was not interrupted. From the initial part of the experimental recordings, we found that thermal stabilization of the injected solution and the sensor can be safely achieved within 30 min. On the heat flow curves P(t) and heat curves Q(t) shown in this paper, t = 0 h defines the time point when signal recording was actually started, i.e. 30 min after solution injection. Heat flow curves were generally recorded through the whole culture cycle (sampling frequency 0.5 Hz) until no heat signal could be detected anymore (usually up to 20 h).Statistical analysis and baseline correction
All nanocalorimetric experiments shown in the paper were performed 3 times. Heat flow P(t) raw data was first filtered by an average filter to minimize the noise level. Mean values are indicated by lines on the figures and the shaded regions correspond to the standard error (SE), calculated using OriginLab® software. For baseline correction of the P(t) curves, a spline fit was generated by the baseline correction function of the OriginLab® software (Fig. S7†). Oxygen consumption and OD measurements were performed 3 times as well. Calibration of the oxygen sensor was performed according to a protocol provided by PyroScience GmbH.Conclusions
There is a general consensus among health care actors involved in infectious diseases management and microbiologists that the implementation of new technologies aiming to reduce the time to result of current clinical AST protocols is a key issue in combatting the increasingly important global health problem related to AMR of clinically important bacterial strains. In this regard, we developed a miniaturized isothermal nanocalorimetry platform (INCfAST) providing a direct method for measuring bacterial metabolic heat of E. coli. Considering bacterial heat production as a new phenotype opens the way to a more comprehensive understanding of fundamental metabolic processes, especially upon drug exposure. High heat detection sensitivity and signal stability enabled detection of bacterial growth in the microincubator under normal conditions after only ∼2–3 hours. Eventually infection in clinical samples could be monitored on a similar time scale. The integrated fluidic design facilitates sample handling and the implementation of automated assay protocols. The versatility of the method was first demonstrated by analyzing metabolic heat profiles for various culture conditions over extended durations. More importantly, we could demonstrate that our nanocalorimetry platform has high potential for evaluating the efficacy and activity of antimicrobial compounds, in particular with respect to the implementation of fast AST protocols. Based on our experimental heat flow curves, AST could be performed in 5–6 hours using a fast-growing laboratory strain of E. coli as bacterial model. Heat flow profiles provided a detailed real-time insight of the dynamics of antimicrobial effects for all 3 antibiotics tested (ciprofloxacin, ampicillin and gentamicin). We observed notable lag phase extensions close to MICs and alterations of transient features in the heat flow curves. MIC values obtained with the INCfAST platform are consistent with accepted clinical values for E. coli in the case of ciprofloxacin and ampicillin. Gentamicin showed a slightly higher value range, most likely due to its oxygen-dependent activity. In contrast to the standard BMD method, the calorimetric approach does not rely on optical parameters, in principal raw patient samples could therefore be used, omitting culture steps for sample purification. Discrimination between growth/growth inhibition in the presence of antibiotics was possible on a time scale of only a few hours. In this view, we anticipate that with the present nanocalorimetry approach, AST assays can be performed on a faster time scale and with enhanced information content than current conventional clinical methods.Author contributions
Y. L. designed and characterized the new nanocalorimetry platform and performed the microbiological assays. Y. L., T. L. and M. G. wrote or revised the paper. T. L. and M. G. supervised this work.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors gratefully acknowledge the support of EPFL-ATMX and EPFL-AFA workshops for the fabrication of the nanocalorimetry thermostats and the 3D-printed microincubators, respectively, as well as Peter Brühlmeier (EPFL-ACI workshop) for the fabrication of the PCB for the sensor chip. We thank Prof. John McKinney and Dr. Neeraj Dhar for useful discussions. This work was carried out within the framework of the EU project “New Diagnostics for Infectious Diseases” (http://www.nd4id.eu/). This project received funding from the European Union's Horizon 2020 program under the Marie Skłodowska-Curie grant agreement No. 675412.Notes and references
- L. Cantas, S. Q. A. Shah, L. M. Cavaco, C. M. Manaia, F. Walsh, M. Popowska, H. Garelick, H. Bürgmann and H. Sørum, Front. Microbiol., 2013, 4, 96 CrossRef CAS PubMed.
- R. Sugden, R. Kelly and S. Davies, Nat. Microbiol., 2016, 1, 16187 CrossRef CAS PubMed.
- S. B. Zaman, M. A. Hussain, R. Nye, V. Mehta, K. T. Mamun and N. Hossain, Cureus, 2017, 9, e1403 Search PubMed.
- F. D. Lowy, J. Clin. Invest., 2003, 111, 10 CrossRef PubMed.
- J. O'Neill, Tackling Drug-Resistant Infections Globally: Final Report and Recommendations, in The Review on Antimicrobial Resistance, Chaired by Jim O’Neill, 2016 Search PubMed.
- H. Goossens, M. Ferech, R. Vander Stichele, M. Elseviers and ESAC Project Group, Lancet, 2005, 365, 579–587 CrossRef.
- A. van Belkum, G. Lüdke, J. G. Lisby, G. Kahlmeter, A. Mohess, K. Becker, J. P. Hays, N. Woodford, K. Mitsakakis, J. Moran-Gilad, J. Vila, H. Peter, J. H. Rex and W. M. Dunne, Nat. Rev. Microbiol., 2019, 17, 51–62 CrossRef CAS PubMed.
- J. M. Andrews, J. Antimicrob. Chemother., 2001, 48, 5–16 CrossRef CAS PubMed.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6, 71–79 CrossRef PubMed.
- S. Puttaswamy, S. K. Gupta, H. Regunath, L. P. Smith and S. Sengupta, Arch. Clin. Microbiol., 2018, 9, 83 Search PubMed.
- A. van Belkum and W. M. Dunne, J. Clin. Microbiol., 2013, 51, 2018–2024 CrossRef PubMed.
- P. Athamanolap, K. Hsieh, L. Chen, S. Yang and T.-H. Wang, Anal. Chem., 2017, 89, 11529–11536 CrossRef CAS PubMed.
- Ö. Baltekin, A. Boucharin, E. Tano, D. I. Andersson and J. Elf, Proc. Natl. Acad. Sci. U. S. A., 2017, 201708558 Search PubMed.
- O. Braissant, D. Wirz, B. Göpfert and A. U. Daniels, Sensors, 2010, 10, 9369–9383 CrossRef CAS PubMed.
- O. Braissant, A. Bachmann and G. Bonkat, Methods, 2015, 76, 27–34 CrossRef CAS PubMed.
- O. Braissant, D. Wirz, B. Göpfert and A. U. Daniels, FEMS Microbiol. Lett., 2010, 303, 1–8 CrossRef CAS PubMed.
- A. Trampuz, S. Salzmann, J. Antheaume and A. U. Daniels, Transfusion, 2007, 47, 1643–1650 CrossRef PubMed.
- U. von Ah, D. Wirz and A. U. Daniels, J. Clin. Microbiol., 2008, 46, 2083–2087 CrossRef PubMed.
- D. Baldoni, H. Hermann, R. Frei, A. Trampuz and A. Steinhuber, J. Clin. Microbiol., 2009, 47, 774–776 CrossRef PubMed.
- L. N. Yang, F. Xu, L. X. Sun, Z. B. Zhao and C. G. Song, J. Therm. Anal. Calorim., 2008, 93, 417–421 CrossRef CAS.
- U. von Ah, D. Wirz and A. Daniels, BMC Microbiol., 2009, 9, 106 CrossRef PubMed.
- J. Higuera-Guisset, J. Rodríguez-Viejo, M. Chacón, F. J. Muñoz, N. Vigués and J. Mas, Thermochim. Acta, 2005, 427, 187–191 CrossRef CAS.
- F. Buchholz, J. Lerchner, F. Mariana, U. Kuhlicke, T. R. Neu, H. Harms and T. Maskow, Biofouling, 2012, 28, 351–362 CrossRef CAS PubMed.
- J. Lerchner, A. Wolf, F. Buchholz, F. Mertens, T. R. Neu, H. Harms and T. Maskow, J. Microbiol. Methods, 2008, 74, 74–81 CrossRef CAS PubMed.
- F. Mariana, F. Buchholz, J. Lerchner, T. R. Neu, H. Harms and T. Maskow, Int. J. Med. Microbiol., 2013, 303, 158–165 CrossRef CAS PubMed.
- E. A. Johannessen, J. M. R. Weaver, L. Bourova, P. Svoboda, P. H. Cobbold and J. M. Cooper, Anal. Chem., 2002, 74, 2190–2197 CrossRef CAS PubMed.
- F. E. Torres, P. Kuhn, D. D. Bruyker, A. G. Bell, M. V. Wolkin, E. Peeters, J. R. Williamson, G. B. Anderson, G. P. Schmitz, M. I. Recht, S. Schweizer, L. G. Scott, J. H. Ho, S. A. Elrod, P. G. Schultz, R. A. Lerner and R. H. Bruce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 9517–9522 CrossRef CAS PubMed.
- M. I. Recht, F. E. Torres, D. D. Bruyker, A. G. Bell, M. Klumpp and R. H. Bruce, Anal. Biochem., 2009, 388, 204–212 CrossRef CAS PubMed.
- W. Lee, W. Fon, B. W. Axelrod and M. L. Roukes, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15225–15230 CrossRef CAS PubMed.
- N. Inomata, M. Toda, M. Sato, A. Ishijima and T. Ono, Appl. Phys. Lett., 2012, 100, 154104 CrossRef.
- S. Wang, S. Yu, M. S. Siedler, P. M. Ihnat, D. I. Filoti, M. Lu and L. Zuo, Rev. Sci. Instrum., 2016, 87, 105005 CrossRef PubMed.
- J. Kim, S. Seo, J. Kim, S. Nam and W. Lee, IEEE 32nd International Conference on Micro Electro Mechanical Systems (MEMS), 2019, pp. 544–546 Search PubMed.
- I. Wadsö, D. Hallén, M. Jansson, J. Suurkuusk, T. Wenzler and O. Braissant, Thermochim. Acta, 2017, 652, 141–149 CrossRef.
- R. Krenger, T. Lehnert and M. A. M. Gijs, Lab Chip, 2018, 18, 1641–1651 RSC.
- K. Stevenson, A. F. McVey, I. B. N. Clark, P. S. Swain and T. Pilizota, Sci. Rep., 2016, 6, 38828 CrossRef CAS PubMed.
- E. Rosenberg and I. Zilber-Rosenberg, Gut Microbes, 2016, 7, 283–285 CrossRef CAS PubMed.
- EUCAST, The European Committee on Antimicrobial Susceptibility Testing, in Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST. Version 9.0, 2019, 2019, http://www.eucast.org Search PubMed.
- P. D. Walker and S. V. Shah, Am. J. Physiol., 1987, 253, C495–C499 CrossRef CAS PubMed.
- M. Regué, B. Hita, N. Piqué, L. Izquierdo, S. Merino, S. Fresno, V. J. Benedí and J. M. Tomás, Infect. Immun., 2004, 72, 54–61 CrossRef PubMed.
- J. H. Moffatt, M. Harper, P. Harrison, J. D. F. Hale, E. Vinogradov, T. Seemann, R. Henry, B. Crane, F. St. Michael, A. D. Cox, B. Adler, R. L. Nation, J. Li and J. D. Boyce, Antimicrob. Agents Chemother., 2010, 54, 4971–4977 CrossRef CAS PubMed.
- L. Yang, J. A. J. Haagensen, L. Jelsbak, H. K. Johansen, C. Sternberg, N. Høiby and S. Molin, J. Bacteriol., 2008, 190, 2767–2776 CrossRef CAS PubMed.
- A. Harms, E. Maisonneuve and K. Gerdes, Science, 2016, 354, aaf4268 CrossRef PubMed.
- I. Swinnen, Int. J. Food Microbiol., 2004, 94, 137–159 CrossRef CAS PubMed.
- M. D. Rolfe, C. J. Rice, S. Lucchini, C. Pin, A. Thompson, A. D. S. Cameron, M. Alston, M. F. Stringer, R. P. Betts, J. Baranyi, M. W. Peck and J. C. D. Hinton, J. Bacteriol., 2012, 194, 686–701 CrossRef CAS PubMed.
- B. Li, Y. Qiu, H. Shi and H. Yin, Analyst, 2016, 141, 3059–3067 RSC.
- O. Fridman, A. Goldberg, I. Ronin, N. Shoresh and N. Q. Balaban, Nature, 2014, 513, 418–421 CrossRef CAS PubMed.
- W. F. Loomis and B. Magasanik, J. Bacteriol., 1967, 93, 1397–1401 CrossRef CAS PubMed.
- A. Solopova, J. van Gestel, F. J. Weissing, H. Bachmann, B. Teusink, J. Kok and O. P. Kuipers, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7427–7432 CrossRef CAS PubMed.
- P. Johansson and I. Wadsö, Thermochim. Acta, 1999, 342, 19–29 CrossRef CAS.
- O. Braissant, G. Bonkat, D. Wirz and A. Bachmann, Thermochim. Acta, 2013, 555, 64–71 CrossRef CAS.
- M. A. Kohanski, D. J. Dwyer and J. J. Collins, Nat. Rev. Microbiol., 2010, 8, 423–435 CrossRef CAS PubMed.
- J. B. Russell, J. Mol. Microbiol. Biotechnol., 2007, 13, 1–11 CrossRef CAS PubMed.
- F. Buchholz, A. Wolf, J. Lerchner, F. Mertens, H. Harms and T. Maskow, Antimicrob. Agents Chemother., 2010, 54, 312–319 CrossRef CAS PubMed.
- J. H. Yang, S. N. Wright, M. Hamblin, D. McCloskey, M. A. Alcantar, L. Schrübbers, A. J. Lopatkin, S. Satish, A. Nili, B. O. Palsson, G. C. Walker and J. J. Collins, Cell, 2019, 177, 1649–1661.e9 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc00579g |
| This journal is © The Royal Society of Chemistry 2020 |