Open Access Article
Open Access ArticleMicrofluidic chip with pillar arrays for controlled production and observation of lipid membrane nanotubes†
Juan Manuel
Martinez Galvez‡
 a,
Maite
Garcia-Hernando‡
a,
Maite
Garcia-Hernando‡
 bc,
Fernando
Benito-Lopez
bc,
Fernando
Benito-Lopez
 c,
Lourdes
Basabe-Desmonts
c,
Lourdes
Basabe-Desmonts
 *bd and
Anna V.
Shnyrova
*bd and
Anna V.
Shnyrova
 *a
*a
aBiofisika Institute (CSIC, UPV/EHU) and Department of Biochemistry and Molecular Biology, University of the Basque Country, Leioa, Spain. E-mail: anna.shnyrova@ehu.eus
bBIOMICs-microfluidics, Microfluidics Cluster UPV/EHU, University of the Basque Country UPV/EHU, Spain. E-mail: lourdes.basabe@ehu.eus
cAnalytical Microsystems & Materials for Lab-on-a-Chip (AMMa-LOAC) Group, Microfluidics Cluster UPV/EHU, Analytical Chemistry Department, University of the Basque Country UPV/EHU, Spain
dIKERBASQUE, Basque Foundation for Science, Spain
First published on 29th June 2020
Abstract
Lipid membrane nanotubes (NTs) are a widespread template for in vitro studies of cellular processes happening at high membrane curvature. Traditionally NTs are manufactured one by one, using sophisticated membrane micromanipulations, while simplified methods for controlled batch production of NTs are in growing demand. Here we propose a lab-on-a-chip (LOC) approach to the simultaneous formation of multiple NTs with length and radius controlled by the chip design. The NTs form upon rolling silica microbeads covered by lipid lamellas over the pillars of a polymer micropillar array. The array's design and surface chemistry set the geometry of the resulting free-standing NTs. The integration of the array inside a microfluidic chamber further enables fast and turbulence-free addition of components, such as proteins, to multiple preformed NTs. This LOC approach to NT production is compatible with the use of high power objectives of a fluorescence microscope, making real-time quantification of the different modes of the protein activity in a single experiment possible.
Introduction
Various transformations of the cellular endomembrane system, such as the formation of transport carriers, tubular networks, and transmembrane contact sites, require the creation of high membrane curvature. Lipid nanotubes (NTs), 1D hollow membrane cylinders made of biologically relevant lipids, have proven to be a versatile and convenient template for quantitative reconstitution and analyses of the high membrane curvature creation and its effects on the membrane properties and protein behavior. Particularly, lipid NTs have been extensively used to study protein and lipid sorting,1,2 mechanisms of membrane fission,3,4 and membrane remodeling by specialized proteins.5Traditionally, these NTs are formed by applying a local pulling force to a parent reservoir membrane.6 Such a membrane can be a planar bilayer lipid membrane or a giant unilamellar vesicle (GUV), a membrane sphere of several microns in diameter. The pulling is done using a micron-sized object, e.g., a microsphere or a tip of a glass pipette positioned by a precise manipulator system or optical tweezers.6,7 The NT radius, ranging from a few to hundreds of nanometers, is set by the ratio between the bending rigidity of the parent membrane, function of its lipid composition, and its lateral tension.6 The NT length, usually in the micrometer range, is set by the experimentalist. Long NTs can be micro-manipulated into sophisticated web-like structures.8,9 Hurtig and co-authors showed for the first time that the NTs pulled from a GUV membrane can be weaved around SU-8 photoresist micropatterns.10,11 They found that the SU-8 polymer was a suitable supporting material for the lipid NT structures and their reservoirs. However, while such micromanipulation enables precise control over the NT extension and geometry, it also requires expensive equipment as well as lengthy procedures and training. Besides, only one NT is analyzed at a time, making the acquisition of enough datasets for statistical analysis a tedious task.
Critical simplification of NT production was proposed by Pucadyil and coauthors,12,13 who achieved the formation of multiple NTs by rolling 20 μm silica beads covered with lipids over a passivated coverglass. This procedure allowed for the fast batch production of the NTs of several microns in length. Further development of this system was proposed in 2015 by Dar and co-authors, who produced NTs by applying liquid flux to prehydrated lipid multilamellas.4,14 In this case, the sticking of the membrane to several defects on a passivated coverglass stabilized the NT position and geometry. While this approach has been instrumental in researching the membrane remodeling mechanism by several proteins,4,15 the NT lengths and radii attain somewhat random values as the means of controlling these critical parameters are lacking. Besides, the NT positioning close to the glass substrate might impede the protein interactions with the NT membrane.
Here, we propose a simple and user-friendly microfluidics approach for the fast formation of many free-standing lipid NTs with controlled geometry. Our system consists of polymer micropillars standing at set distances over a coverglass bottom of a microfluidic chip. The NTs and NT networks are produced by rolling silica microspheres covered with membrane lamellas over the pillar array, thus avoiding complex micropipette manipulations of the membrane. The resulting NTs are attached to the top of the micropillars, with their extension being set by the interpillar distance. The relatively small height of the micropillars allows for real-time observation of the NT behavior with high-resolution objectives of an inverted bright-field fluorescence microscope. Moreover, the radii and lamellarity of the NTs can be controlled by changing the extension of the lipid reservoir on the pillars, making the system tunable by design.
Experimental
Reagents
Ethanol, isopropanol, methanol and acetone were from Scharlabs. SU-82005 photoresist and developer were from MicroChem. Polydimethylsiloxane (PDMS) (SYLGARD™ 184 Silicone elastomer) was purchased from Ellsworth Adhesives. N-Isopropylacrylamide (NIPAAm), the crosslinker N,N′-methylene-bis(acrylamide) (mBAAm), the photoinitiator 2,2-dimethoxy-2-phenylacetophenone (DMPA), the silanizing agent 3-(trimethoxysilyl)propyl methacrylate, and the ionic liquid trihexyltetradecyl-phosphonium dicyanamide (DCA) were from Sigma-Aldrich.1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), cholesterol, L-α-phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (RhPE) were from Avanti Lipids.
Bovine serum albumin (BSA) was from Fisher Scientific.
Fabrication of the SU-8 micropillar pattern over the coverglass
Arrays of micropillars were produced directly on a 25 mm round coverglass (0.13–0.16 mm thickness, Hecht Assistent) by photolithography. A photomask was designed to create three arrays of cylindrical pillars of 15 μm in diameter, each array composed of 4 discrete zones with 10, 20, 30, and 40 μm separation between the pillars (Fig. S1A and B†). For the production of SU-8 pillars, we used previously described procedures (see the ESI,† Methods). Briefly, a 5 μm thick layer of SU-82005 (MicroChem Inc.) photoresist was spin-coated directly onto the coverglass, soft-baked, aligned with a high-resolution film photomask and exposed to UV in 4 pulses of 20 s of 5 mJ s−1 UV light (365 nm) in contact mode. After a post-bake, the non-polymerized photoresist was removed by developer immersion, followed by an isopropanol wash and a hard bake of the coverglass. The height and the structures of the micropillars of the imprinted SU8 pattern were confirmed by optical profilometry (4.2 ± 0.3 μm (n = 10)) and SEM, respectively (Fig. S1B†).Fabrication of the ionogel (IO) micropillar pattern over the coverglass
The production of DCA IO pillar microarrays was performed by spreading a 10 μm layer of prepolymer over a functionalized coverglass followed by exposure to different UV doses of 365 nm wavelength by contact photolithography (ESI† Methods). The prepolymer IO solution has a monomer (NIPAAm), a photoinitiator (DMPA), a crosslinker (mBAAm), and an ionic liquid (DCA)16–18 mixed at a fixed molar ratio (ESI† Methods). Importantly, the ionic liquid used during the prepolymer preparation may act as a lipid membrane solvent. In order to avoid this effect, the micropatterns obtained after IO polymerization were thoroughly washed with isopropanol and water. To further assure the absence of the ionic liquid in the final product, the pillars were heated to 45 °C to induce material shrinkage and provoke unbound DCA expelling from the polymer network. Although the IO patterns were obtained correctly, they had lower resolution as compared to the SU-8 pillars, rendering a pillar height of 2 ± 1 μm, as measured by optical profilometry (n = 10). The resolution decrease could be due to the viscosity of the prepolymer solution and its interaction with the glass. Using light pulses instead of constant light exposure increased the resolution of the IO micropattern (for further details, see the ESI†).Production of the microfluidic devices
To complete the microfluidic device, we designed three parallel channels with a 2 mm width per 8 mm length, which were transferred to a PSA (127 μm thickness, ARcare 8939, Adhesive Research Inc.) and attached to the bottom of a 55 mm Pyrex™ borosilicate glass Petri dish. After that, PDMS, mixed at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, was poured on top of the PSA mold at the bottom of the Petri dish, degassed and cured at 60 °C for 40 min. The polymerized PDMS was then peeled out from the dish and further baked at 60 °C for 2 h. After punching 0.5 mm inlet and outlet orifices for fluidic connections, the PDMS section was attached to the previously micropatterned coverglass, so that the three channels were aligned with the three arrays of micropillars on the coverglass (Fig. S1C†).
1 ratio, was poured on top of the PSA mold at the bottom of the Petri dish, degassed and cured at 60 °C for 40 min. The polymerized PDMS was then peeled out from the dish and further baked at 60 °C for 2 h. After punching 0.5 mm inlet and outlet orifices for fluidic connections, the PDMS section was attached to the previously micropatterned coverglass, so that the three channels were aligned with the three arrays of micropillars on the coverglass (Fig. S1C†).
Preparation of the microsphere-supported lipid films
Lipid multilamellas supported on silica beads were formed as reported previously for a giant supported bilayer template.19 Briefly, the desired lipid mixtures (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RhPE 99
RhPE 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol%, DOPC
1 mol%, DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPS
DOPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RhPE 49
RhPE 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol% or DOPC
1 mol% or DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE
DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPS
DOPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PI(4,5)P2
PI(4,5)P2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RhPE 46
RhPE 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol%) were prepared from lipid stocks in chloroform to 0.1 mg of total lipid. Chloroform is a hazardous solvent that causes respiratory tract, eye, and skin irritation; thus, personal protective gear should be used as indicated in the MSDS when handling lipid stock solutions. The lipid mixtures were vacuum dried for 30 min, rendering uniform lipid films at the bottom of the vials. The films were then rehydrated in 100 μL of 1 mM HEPES, pH 7.0, and subjected to light vortexing, resulting in the formation of multilamellar vesicles (MLVs). The MLV solution can be stored at 4 °C for up to three days, depending on the lipid composition. A small aliquot of 40 μm plain silica beads (Microspheres–Nanospheres Co, USA) was three times washed with ultrapure water. A 0.2 μL aliquot of the bead solution in water was transferred into a 10 μL aliquot of MLVs. The bead–MLV mixture was then divided into five even drops on a clean Parafilm surface. The drops were dried in vacuum for 15–30 min or until the water evaporated completely. The resulting multi-layered lipid films on the surface of the microspheres were used to produce the nanotubes. In order to achieve reproducibility in the next steps of the procedure, it is recommended to characterize the lipid coverage of the bead surface each time by measuring the lipid to bead ratio as described previously.12,19
1 mol%) were prepared from lipid stocks in chloroform to 0.1 mg of total lipid. Chloroform is a hazardous solvent that causes respiratory tract, eye, and skin irritation; thus, personal protective gear should be used as indicated in the MSDS when handling lipid stock solutions. The lipid mixtures were vacuum dried for 30 min, rendering uniform lipid films at the bottom of the vials. The films were then rehydrated in 100 μL of 1 mM HEPES, pH 7.0, and subjected to light vortexing, resulting in the formation of multilamellar vesicles (MLVs). The MLV solution can be stored at 4 °C for up to three days, depending on the lipid composition. A small aliquot of 40 μm plain silica beads (Microspheres–Nanospheres Co, USA) was three times washed with ultrapure water. A 0.2 μL aliquot of the bead solution in water was transferred into a 10 μL aliquot of MLVs. The bead–MLV mixture was then divided into five even drops on a clean Parafilm surface. The drops were dried in vacuum for 15–30 min or until the water evaporated completely. The resulting multi-layered lipid films on the surface of the microspheres were used to produce the nanotubes. In order to achieve reproducibility in the next steps of the procedure, it is recommended to characterize the lipid coverage of the bead surface each time by measuring the lipid to bead ratio as described previously.12,19
NT formation in the microfluidic chamber
The microfluidic chamber was first flushed for a few seconds with bovine serum albumin (BSA) solution at 0.1 g L−1 in ultrapure water to prevent lipid adhesion to the coverslip and PDMS walls. The chamber was then washed and filled with the working buffer (150 mM KCL, 20 mM HEPES, 1 mM EDTA). 40 μm silica beads containing the lipid film multilayers were picked up with a fire-closed tip of a glass micropipette and introduced into the chamber through the input port of the PDMS section. The NTs were formed by rolling the beads over the micropillar arrays. Such rolling was performed by applying negative pressure on the outlet port of the PDMS section with a syringe pump or by gently tilting the whole chamber while observing the bead line movement. Importantly, to avoid undesirable background signals, the beads were purged away from the pillar array area of the chamber at the end of the NT formation procedure. If vesicles or membrane aggregates were detected in the bulk upon the bead removal, additional perfusion of the chamber was performed to ensure low fluorescence background.Imaging and characterization of the NTs
The fluorescently labeled lipid NTs were imaged with a wide-field fluorescence inverted microscope (Nikon Eclipse Ti-E motorized inverted microscope equipped with a CoolLed pE-4000 light source and Andor Zyla sCMOS camera) using the same intensity and camera settings each time. μManager software20 was used for image acquisition. The images were further analyzed using Fiji21 and Origin 8.0 software.The number of NTs observed in a field of view depended on the objective used (TIRF oil immersion 100×/1.49 or 40×/0.75 air objectives), number of beads introduced, and the separation between the pillars. For the analysis of the NT radii, we selected NT micrographs with uniform and low background signals located near the focal plane. Note that near the focal plane, the integral fluorescence intensity of the NTs remains constant upon slight out-of-focus movements of the NTs (Fig. S2†), allowing for quantification of the NT radii as described elsewhere4,22 (Fig. S3, ESI† Methods).
Results and discussion
Generation of NTs between SU-8 micropillars
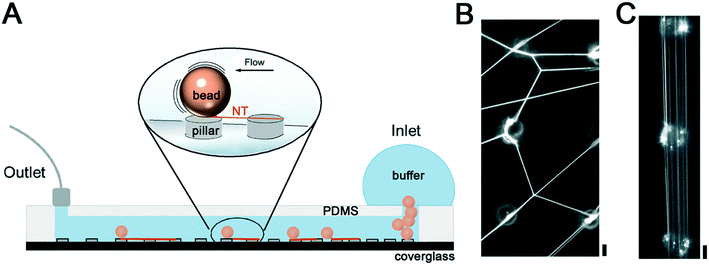
The biocompatibility of SU-8 photoresist11,23,24 makes this material a suitable choice for the production of NTs made of biomembrane mimicking lipid compositions. To induce the formation of NTs between the SU-8 pillars, we inserted lipid-coated silica beads into the inlet of the microfluidic chamber, previously filled with the working buffer (Fig. 1A). Next, we rolled the beads over the SU-8 pillar micropattern by gently tilting the chamber. The tilting caused random movements of the microbeads over the microarray resulting in the formation of an NT web between the pillar tops (Fig. 1B).We then attempted to direct the movement of the beads in the chamber and produce parallelly aligned NTs. We generated a laminar flow by applying fixed withdrawal rates at the outlet port of the chamber and monitored the flow-induced NT formation by optical microscopy. We found that individual beads traveled at 21 ± 10 and 397 ± 81 μm s−1 speed in the 40 μm separation region of the chamber when 20 and 100 μL min−1 withdrawal rates were applied, respectively (Movies S1 and S2†). In both cases, the beads moved quasi-linearly in the direction of the flow while touching the pillars. As expected, the interaction of the beads with the pillars resulted in the formation of parallel NTs stretched between the pillar tops (Fig. 1C). Importantly, the interpillar distance determined the final speed of bead displacement in the chamber, being higher for larger separation between the pillars. Withdrawal rates below 20 μL min−1 were not enough to induce bead movement, setting the lower speed threshold for the NT formation. Such a threshold should change for beads of different densities and sizes, and thus should be established experimentally if a different substrate is to be used.
We then compared the transversal plot profiles of the NT fluorescence intensity for the different NT formation methods tested (see Fig. S2†). The corresponding histogram of the areas under the fluorescence intensity peaks reveals that the NTs produced by this procedure have different lamellarity.14 For the slow (20 μL min−1 withdrawal rate) flow-induced method, the histogram revealed two NT populations with higher and lower fluorescence intensity, presumably corresponding to uni- and bilamellar NTs (Fig. 2A). We tested the unilamellar nature of the NTs from the first group by perfusing the chamber with a 5 mM solution of the fluorescence quencher sodium dithionite. A ∼50% drop of the fluorescence intensity confirmed the unilamellarity of the NT membranes (Fig. 2B).
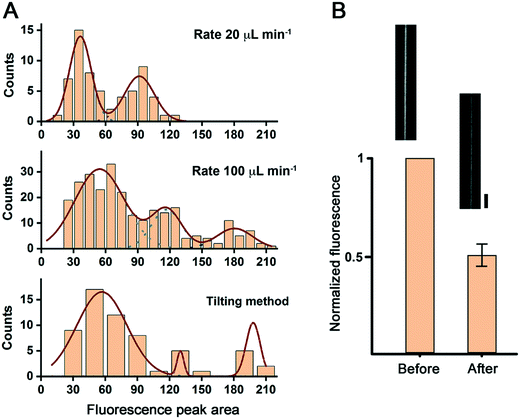 | ||
| Fig. 2 NT lamellarity in different production regimes. A. Populations of NTs observed upon analyzing the area under the fluorescence peak of the linear plot across the NTs (see the ESI,† Methods and Fig. S2). B. The fluorescence of NTs identified as unilamellar in (A) decreases to its half upon addition of sodium dithionite. The micrographs show an example of an NT before and after sodium dithionite addition. Scale bar 4 μm. | ||
For the fast (100 μL min−1 withdrawal rate) flow, as well as for the gentle tilting method, the distributions of the fluorescence intensity were broader. They had at least three populations of NTs, including NTs with three concentric lipid bilayers (Fig. 2A). Such an apparent difference in the intensity distribution may be due to the fast bead displacement in these two cases. The rapid displacement generates a significant drag force at the bead surface resulting in the detachment of several membrane lamellas at a time. The same drag force, and the flux in the case of flow-induced NT formation, may result in NT thinning and breakage, particularly for the unilamellar NTs, which translates into a right shift and the broader population distribution seen in the corresponding histograms. Indeed, the flux-induced production of NTs at the fast-flow rate was accompanied by a massive NT breakage, resulting in many membrane tubular structures floating in the chamber (data not shown). Due to the preparation procedure, we could not monitor this phenomenon for the gentle tilting method. However, the measured speed of bead displacement, in this case, was established at 667 ± 83 μm min−1 (n = 3), even higher than the rate measured for the fast-flow bead displacement method. Then, membrane breakage is also expected for this NT preparation procedure.
As the slow-flow protocol yields very distinct NT populations for the lipid composition used, it was selected for the characterization of the NT geometrical parameters. However, both the fast-flow and the gentle tilting method may be suitable for the reconstitution of membrane remodeling events. Notably, the gentle tilting method does not require any pump or syringe to generate many length-delimited NTs in a network, which may be of advantage for laboratories lacking microfluidics equipment.
Characterization and modification of the geometry of the NTs on the SU-8 pillars
Its length and radius fully describe the geometry of a unilamellar cylindrical NT. As the NTs mostly extend between adjacent pillars of the SU-8 micropattern (Fig. 1C), their length is established by the interpillar distance, a parameter tunable by the micropattern design itself.The NT radius (rNT) is set by the elastic parameters of the NT membrane as:
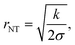 | (1) |
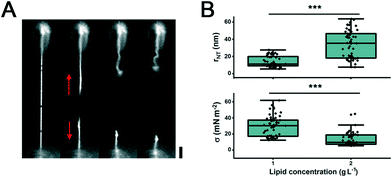
We further estimated the range of membrane tension set on the membrane while in contact with the SU-8 pillars. Considering that the bending rigidity for pure DOPC membranes at 23 °C has been reported to be 11 ± 110−20 J,26 from the eqn (1), it follows that the lateral tension acting on the NTs formed on the SU-8 pillars is 0.3 ± 0.2 mN m−2 (Fig. 3B). The tensile force F acting along the tube axis and driving the tube retraction to the reservoir can be calculated as:
| F = σ·2πrNT | (2) |
When the NTs were produced from beads with a double amount of deposited membrane material, the tension decreased, as seen by the increase in the mean value of the NT radii (Fig. 3B). These data suggest that by changing the amount of deposited lipid material and the distance between the pillars, we can control both the length and radius of the NTs.
Controlling the NT geometry via the micropattern design
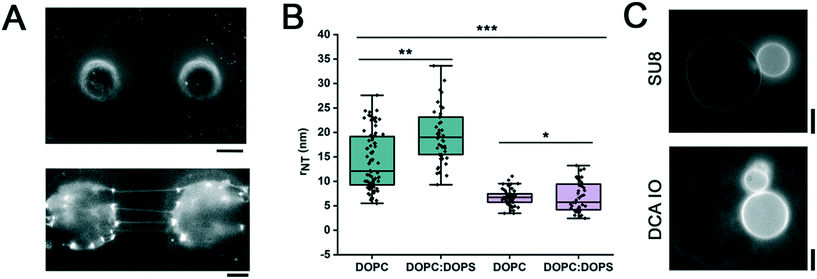
Next, we tested the conjecture that chemical modification of the pillar surface can endow the system with a defined NT geometry. We hypothesized that changing the nature of the membrane–substrate interaction at the pillars would affect the NT radius for a given lipid composition of the NT membrane.28,29 We produced pillars from DCA IO (Fig. 4A and ESI† Methods), a material with a different physicochemical behavior to the SU-8 polymer.16,30 IOs are materials composed of a polymeric network and an ionic liquid, resulting in a hybrid polymer with the physical stability of the polymer and the chemical stability of the ionic liquid.31,32 Mainly, the DCA IO is a porous material that combines its hygroscopic nature with the high hydrophobicity of the surface upon water promoted expansion.16 Strikingly, the radius of NTs made from neutral (DOPC) as well as charged (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPS) lipid compositions decreased when the NTs were formed on the DCA IO pillars as compared to the SU-8 substrate (Fig. 4B). The radius decrease indicates the higher lateral membrane tension in the DCA IO system (eqn (1)), proportional to the lipid affinity toward the pillar material.
DOPS) lipid compositions decreased when the NTs were formed on the DCA IO pillars as compared to the SU-8 substrate (Fig. 4B). The radius decrease indicates the higher lateral membrane tension in the DCA IO system (eqn (1)), proportional to the lipid affinity toward the pillar material.
The higher affinity of the lipids toward the IO pillars is evident in the pattern of the lipid coverage. While the SU8 pillars were only sparsely covered with the membrane reservoir forming only at discrete points,11 DCA IO pillars appeared to be completely covered by lipids (Fig. 4A, lower panel). Though the effect can be explained by the different BSA surface blocking efficiency for the SU-8 polymer and the DCA IO, we tend to ascribe it to the porous nature of the IO pillars having higher reservoir capacity for the lipids.
We further confirmed that the lipid–DCA IO interaction was due to the porosity/hydrophobicity of the IO surface16 and not to the residual ionic liquid, which was previously reported to act as a detergent disrupting the membrane structure.33 For that, we compared the morphologies of GUVs when added to DCA IO and SU-8 substrates. The GUVs readily adhered to the pillars made from both materials (Fig. 4C). The GUVs in contact with the DCA IO remained stable (Fig. 4C), with no apparent solvent effect. We conclude that by changing the membrane–material affinity, the geometry of the NTs can be tuned through changes in its lateral tension.
Importantly, the radius of the NTs pulled directly from the lipid reservoir on the silica beads (77 ± 2 nm)22 was significantly higher than the NT radii measured here (Fig. 4B), explaining the lipid transfer from the beads to both SU8 and IO pillars. Finally, the radii change is not significant when charged lipid species are added to the NTs formed on DCA IO (Fig. 4B), indicating that the current protocol of the NT formation can be directly applied to biomimetic lipid compositions generally containing charged lipid species.
Conclusion
In contrast to current methods for NT formation through membrane micromanipulation procedures, the LOC methodology presented here for the production of NTs and related assays does not require any sophisticated micromanipulations to qualitatively control the NT extension and radius. The combination of bead rolling with a micropillar array is a versatile and user-friendly methodology to create multiple NTs in a short time in a limited area. Such batch NT production is ideal for the analysis of protein activity in a significant population of homogeneous NTs observed under the same conditions. Micropillar array integration into the bottom plate of a microchannel enables control over the flow regime and eases the automation of the fabrication and analysis process. Besides, the whole system remains contained within a microchannel, minimizing the use of reagents, such as proteins or markers. Polymer micropatterning with different surface chemistry allows controlling the strength of the NT interaction with the polymer support and thus the NT radius set by the membrane lateral tension. Simple changes in the design, such as the interpillar distance, pillar material, or lipid loaded on the beads, enable control over the radius of the formed NTs.The NTs obtained over the pillars are free-standing, eliminating the physical contact between the NT membrane and the support observed in other high throughput systems. The low height of the pillars grown directly on a thin glass coverslip allows fluorescence microscopy monitoring of the NTs with high power objectives. We believe that this user-friendly miniaturization approach to membrane NT formation will be a valuable addition to the toolkit of in vitro reconstitution assays of processes happening at high membrane curvature.
Author contributions
Conceptualization: AVS; methodology: AVS, LBD, FBL; investigation: MGH, JMMG; formal analysis: JMMG, AVS; writing–original draft: all authors; writing–review & editing: all authors; project administration: LBD, AVS; funding acquisition: AVS, LBD, FBL; resources: AVS, LBD, FBL; supervision: AVS, LBD, FBL.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was partially supported by the Spanish Ministry of Science, Innovation, and Universities grants PGC2018-099971-B-I00, EUR2019-103830, RYC-2014-01419 and BIO2016-80417-P and the Basque Government grants IT1270-19 and IT1271-19. JMMG and MGH acknowledge the predoctoral fellowships from the University of the Basque Country (UPV/EHU). The authors are grateful for the technical support provided by SGIker (UPV/EHU and ERDF, EU) for the SEM experiments.References
- P. Singh, P. Mahata, T. Baumgart and S. L. Das, Curvature sorting of proteins on a cylindrical lipid membrane tether connected to a reservoir, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2012, 85, 051906 CrossRef PubMed.
- C. Zhu, S. L. Das and T. Baumgart, Nonlinear sorting, curvature generation, and crowding of endophilin N-BAR on tubular membranes, Biophys. J., 2012, 102, 1837–1845 CrossRef CAS PubMed.
- A. V. Shnyrova, et al. Geometric catalysis of membrane fission driven by flexible dynamin rings, Science, 2013, 339, 1433–1436 CrossRef CAS PubMed.
- S. Dar, S. C. Kamerkar and T. J. Pucadyil, A high-throughput platform for real-time analysis of membrane fission reactions reveals dynamin function, Nat. Cell Biol., 2015, 17, 1588–1596 CrossRef CAS PubMed.
- M. Simunovic, et al. How curvature-generating proteins build scaffolds on membrane nanotubes, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 11226–11231 CrossRef CAS PubMed.
- A. Roux, The physics of membrane tubes: Soft templates for studying cellular membranes, Soft Matter, 2013, 9, 6726–6736 RSC.
- V. A. Frolov, V. A. Lizunov, A. Y. Dunina-Barkovskaya, A. V. Samsonov and J. Zimmerberg, Shape bistability of a membrane neck: A toggle switch to control vesicle content release, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 8698–8703 CrossRef CAS PubMed.
- E. Evans, H. Bowman, A. Leung, D. Needham and D. Tirrell, Biomembrane Templates for Nanoscale Conduits and Networks, Science, 1996, 273, 933–935 CrossRef CAS PubMed.
- M. Karlsson, et al. Formation of geometrically complex lipid nanotube-vesicle networks of higher-order topologies, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11573–11578 CrossRef CAS PubMed.
- J. Hurtig, B. Gustafsson, M. Tokarz and O. Orwar, Electrophoretic Transport in Surfactant Nanotube Networks Wired on Microfabricated Substrates, Anal. Chem., 2006, 78, 5281–5288 CrossRef CAS PubMed.
- J. Hurtig, M. Karlsson and O. Orwar, Topographic SU-8 Substrates for Immobilization of Three-Dimensional Nanotube−Vesicle Networks, Langmuir, 2004, 20, 5637–5641 CrossRef CAS PubMed.
- S. Neumann, T. J. Pucadyil and S. L. Schmid, Analyzing membrane remodeling and fission using supported bilayers with excess membrane reservoir, Nat. Protoc., 2013, 8, 213–222 CrossRef CAS PubMed.
- T. J. Pucadyil and S. L. Schmid, Real-time visualization of dynamin-catalyzed membrane fission and vesicle release, Cell, 2008, 135, 1263–1275 CrossRef CAS PubMed.
- S. Dar, S. C. Kamerkar and T. J. Pucadyil, Use of the supported membrane tube assay system for real-time analysis of membrane fission reactions, Nat. Protoc., 2017, 12, 390–400 CrossRef CAS PubMed.
- S. C. Kamerkar, F. Kraus, A. J. Sharpe, T. J. Pucadyil and M. T. Ryan, Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission, Nat. Commun., 2018, 9, 5239 CrossRef CAS PubMed.
- N. Gil-González, T. Akyazi, E. Castaño, F. Benito-Lopez and M. C. Morant-Miñana, Elucidating the role of the ionic liquid in the actuation behavior of thermo-responsive ionogels, Sens. Actuators, B, 2018, 260, 380–387 CrossRef.
- N. Gil-González, et al. AZO Embedded Interdigitated Electrodes for Monitoring Stimuli Responsive Materials, Adv. Funct. Mater., 2018, 28, 1803127 CrossRef.
- M. Czugala, et al. Swelling and shrinking behaviour of photoresponsive phosphonium-based ionogel microstructures, Sens. Actuators, B, 2014, 194, 105–113 CrossRef CAS.
- A. Velasco-Olmo, J. Ormaetxea Gisasola, J. M. Martinez Galvez, J. Vera Lillo and A. V. Shnyrova, Combining patch-clamping and fluorescence microscopy for quantitative reconstitution of cellular membrane processes with Giant Suspended Bilayers, Sci. Rep., 2019, 9, 7255 CrossRef PubMed.
- A. Edelstein, N. Amodaj, K. Hoover, R. Vale and N. Stuurman, Computer Control of Microscopes Using μManager, Curr. Protoc. Mol. Biol., 2010, 92, 14.20.1 Search PubMed.
- J. Schindelin, et al. Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- J. Espadas, et al. Dynamic constriction and fission of endoplasmic reticulum membranes by reticulon, Nat. Commun., 2019, 10, 5327 CrossRef PubMed.
- E. Mitri, et al. SU-8 bonding protocol for the fabrication of microfluidic devices dedicated to FTIR microspectroscopy of live cells, Lab Chip, 2014, 14, 210–218 RSC.
- S. L. Tao, K. C. Popat, J. J. Norman and T. A. Desai, Surface Modification of SU-8 for Enhanced Biofunctionality and Nonfouling Properties, Langmuir, 2008, 24, 2631–2636 CrossRef CAS PubMed.
- S. L. Schmid and V. A. Frolov, Dynamin: Functional Design of a Membrane Fission Catalyst, Annu. Rev. Cell Dev. Biol., 2011, 27, 79–105 CrossRef CAS PubMed.
- R. S. Gracià, N. Bezlyepkina, R. L. Knorr, R. Lipowsky and R. Dimova, Effect of cholesterol on the rigidity of saturated and unsaturated membranes: Fluctuation and electrodeformation analysis of giant vesicles, Soft Matter, 2010, 6, 1472–1482 RSC.
- A. Upadhyaya and M. P. Sheetz, Tension in Tubulovesicular Networks of Golgi and Endoplasmic Reticulum Membranes, Biophys. J., 2004, 86, 2923–2928 CrossRef CAS PubMed.
- E. Evans and A. Yeung, Hidden dynamics in rapid changes of bilayer shape, Chem. Phys. Lipids, 1994, 73, 39–56 CrossRef CAS.
- V. Heinrich and R. E. Waugh, A piconewton force transducer and its application to measurement of the bending stiffness of phospholipid membranes, Ann. Biomed. Eng., 1996, 24, 595–605 CrossRef CAS PubMed.
- J. Le Bideau, L. Viau and A. Vioux, Ionogels, ionic liquid based hybrid materials, Chem. Soc. Rev., 2011, 40, 907–925 RSC.
- Y. Zhong, G. T. M. Nguyen, C. Plesse, F. Vidal and E. W. H. Jager, Highly Conductive, Photolithographically Patternable Ionogels for Flexible and Stretchable Electrochemical Devices, ACS Appl. Mater. Interfaces, 2018, 10, 21601–21611 CrossRef CAS PubMed.
- D. Khodagholy, et al. Organic electrochemical transistor incorporating an ionogel as a solid state electrolyte for lactate sensing, J. Mater. Chem., 2012, 22, 4440–4443 RSC.
- B. Jing, N. Lan, J. Qiu and Y. Zhu, Interaction of Ionic Liquids with a Lipid Bilayer: A Biophysical Study of Ionic Liquid Cytotoxicity, J. Phys. Chem. B, 2016, 120, 2781–2789 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc00451k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |