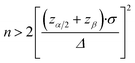Open Access Article
Open Access ArticleOrgan-on-chip model shows that ATP release through connexin hemichannels drives spontaneous Ca2+ signaling in non-sensory cells of the greater epithelial ridge in the developing cochlea†‡
Flavia
Mazzarda
 ab,
Annunziata
D'Elia
ab,
Annunziata
D'Elia
 ab,
Roberto
Massari
ab,
Roberto
Massari
 a,
Adele
De Ninno
a,
Adele
De Ninno
 c,
Francesca Romana
Bertani
c,
Francesca Romana
Bertani
 c,
Luca
Businaro
c,
Luca
Businaro
 c,
Gaia
Ziraldo
c,
Gaia
Ziraldo
 ad,
Veronica
Zorzi
ad,
Veronica
Zorzi
 ad,
Chiara
Nardin
ad,
Chiara
Nardin
 a,
Chiara
Peres
a,
Chiara
Peres
 a,
Francesco
Chiani
a,
Francesco
Chiani
 a,
Abraham
Tettey-Matey
a,
Abraham
Tettey-Matey
 a,
Marcello
Raspa
a,
Marcello
Raspa
 a,
Ferdinando
Scavizzi
a,
Ferdinando
Scavizzi
 a,
Alessandro
Soluri
a,
Alessandro
Soluri
 a,
Anna Maria
Salvatore
a,
Anna Maria
Salvatore
 a,
Jun
Yang
a,
Jun
Yang
 ef and
Fabio
Mammano
ef and
Fabio
Mammano
 *ag
*ag
aCNR Institute of Biochemistry and Cell Biology, Monterotondo, Rome, Italy. E-mail: fmazzarda@gmail.com; nunzia_delia@yahoo.it; roberto.massari@cnr.it; zira13@gmail.com; veronicazorzi.vimm@gmail.com; chiara.nardin@ibbc.cnr.it; chiara.peres@ibbc.cnr.it; francesco.chiani@cnr.it; abraham.matey@ibbc.cnr.it; marcello.raspa@cnr.it; ferdinando.scavizzi@cnr.it; alessandro.soluri@cnr.it; annamaria.salvatore@cnr.it
bDepartment of Science, Università degli Studi di Roma3, Rome, Italy
cCNR Institute for Photonics and Nanotechnology, Rome, Italy. E-mail: adele.deninno@ifn.cnr.it; francesca.bertani@cnr.it; luca.businaro@ifn.cnr.it
dInstitute of Otorhinolaryngology, Università Cattolica del Sacro Cuore, Rome, Italy
eDepartment of Otorhinolaryngology Head and Neck Surgery, Xinhua Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China. E-mail: dryangjunxinhua@163.com
fShanghai Key Laboratory of Translational Medicine on Ear and Nose diseases, Shanghai, China
gDepartment of Physics and Astronomy “G. Galilei”, University of Padova, Padua, Italy. E-mail: fabio.mammano@unipd.it
First published on 23rd July 2020
Abstract
Prior work supports the hypothesis that ATP release through connexin hemichannels drives spontaneous Ca2+ signaling in non-sensory cells of the greater epithelial ridge (GER) in the developing cochlea; however, direct proof is lacking. To address this issue, we plated cochlear organotypic cultures (COCs) and whole cell-based biosensors with nM ATP sensitivity (ATP-WCBs) at the bottom and top of an ad hoc designed transparent microfluidic chamber, respectively. By performing dual multiphoton Ca2+ imaging, we monitored the propagation of intercellular Ca2+ waves in the GER of COCs and ATP-dependent Ca2+ responses in overlying ATP-WCBs. Ca2+ signals in both COCs and ATP-WCBs were inhibited by supplementing the extracellular medium with ATP diphosphohydrolase (apyrase). Spontaneous Ca2+ signals were strongly depressed in the presence of Gjb6−/− COCs, in which connexin 30 (Cx30) is absent and connexin 26 (Cx26) is strongly downregulated. In contrast, spontaneous Ca2+ signals were not affected by replacement of Panx1−/− with Panx1+/+ COCs in the microfluidic chamber. Similar results were obtained by estimating ATP release from COCs using a classical luciferin–luciferase bioluminescence assay. Therefore, connexin hemichannels and not pannexin 1 channels mediate the release of ATP that is responsible for Ca2+ wave propagation in the developing mouse cochlea. The technological advances presented here have the potential to shed light on a plethora of unrelated open issues that involve paracrine signaling in physiology and pathology and cannot be addressed with standard methods.
Introduction
Extracellular ATP modulates the function of both sensory and non-sensory cells in the inner ear (reviewed in ref. 1–3). In the developing cochlea, ATP triggers cytosolic Ca2+ concentration oscillations and propagation of intercellular Ca2+ waves that carry crucial biochemical information to the cochlear sensory epithelium.4–6 The latter is subdivided into two embryologically distinct regions, the lesser epithelial ridge (LER) and the greater epithelial ridge (GER), which are separated by a pair of immature inner and outer pillar cells.7–9 The GER encompasses the nascent inner sulcus, the tall columnar cells of the Kölliker organ,10,11 the inner hair cells (IHCs) and their abutting supporting cells.12,13 Ca2+ waves arise spontaneously in the GER14–19 and appear to play a crucial role in normal development of the cochlear sensory epithelium, hearing acquisition, functional maturation of hair cells,20–23 redox homeostasis and age-related hearing loss.24 A wealth of experimental data and modeling studies support the notion that Ca2+ wave propagation in the developing cochlea is sustained by the release of ATP from the cytosol of non-sensory cells to the endolymph,18,25–27 in which baseline ATP concentration ([ATP]) is in the low nM range.28,29 Genetic interference experiments support a crucial role for Cx26 and Cx30, the two major cochlear connexins,30–32 in intercellular Ca2+ wave propagation in the developing cochlea17,18,25,33–36 (reviewed in ref. 2 and 37). However, direct proof that ATP release through connexin hemichannels drives spontaneous Ca2+ wave propagation in the GER is lacking.To address this critical issue, we used two global knock out (KO) mouse strains, namely connexin 30 KO (Gjb6−/−; MGI: 2447863) and pannexin 1 KO (Panx1−/−; MGI: 5310802). Gjb6−/− mice fail to acquire hearing and do not develop endocochlear potential.17,38 Furthermore, they show defects of the endothelial barrier in capillaries of the stria vascularis39 and impaired gap junction-mediated transfer of the fluorescent D-glucose derivative 2-NBDG.40 Accordingly, not only is Cx30 not expressed, but also Cx26 levels are severely reduced in the developing cochlea of this KO strain.33 In contrast, Panx1−/− mice25,41 have an increased susceptibility for atrial fibrillation and show a QT-prolongation cardiac phenotype.42 However, their hearing sensitivity, outer hair cell-based “cochlear amplifier” and cochlear nerve function, analyzed by auditory brainstem response (ABR) and distortion product otoacoustic emission (DPOAE) recordings, are normal, together with connexin expression and gap-junction coupling in the developing organ of Corti.34
As detailed below, we developed an innovative organ-on-chip model which, by the coupling of microfluidic technology, whole cell-based biosensors (WCBs)43 with nM sensitivity to extracellular ATP25 and multiphoton imaging, allowed systematic addressing of purinergic signaling in COCs obtained from the above mentioned KO mice. Our results provide direct evidence that ATP release through connexin hemichannels drives spontaneous Ca2+ wave propagation in the GER.
Methods
Animals
All animals (Mus musculus) used in this study (see Table 1) were bred at the National Research Council-Institute of Biochemistry and Cell Biology (CNR-IBBC), Infrafrontier/ESFRI-European Mouse Mutant Archive (EMMA), Specific Pathogen-Free (SPF) barrier unit (Monterotondo Scalo, Rome, Italy). Mice were housed in individually ventilated caging systems (Tecniplast, Gazzada, Italy) at a temperature (T) of 21 ± 2 °C, relative humidity (RH) of 55 ± 15% with 50–70 air changes per hour (ACH) and under controlled (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 hour) light–dark cycles (7 am–7 pm). Mice had ad libitum access to water and a standard rodent diet (Emma 23, Mucedola, Settimo Milanese, Italy). Both male and female homozygous Gjb6−/− [EMMA ID (EM): 00323] or Panx1−/− (EM: 11476) pups at post-natal day 5 (P5) and their wild type P5 siblings (Gjb6+/+, Panx1+/+) were used. The background strains of these mice were C57BL/6J for Gjb6−/− and C57BL/6N for Panx1−/− mice. Note that there is no discernible difference in the auditory phenotype of the closely related C57BL/6J and C57BL/6N mouse strains.44
12 hour) light–dark cycles (7 am–7 pm). Mice had ad libitum access to water and a standard rodent diet (Emma 23, Mucedola, Settimo Milanese, Italy). Both male and female homozygous Gjb6−/− [EMMA ID (EM): 00323] or Panx1−/− (EM: 11476) pups at post-natal day 5 (P5) and their wild type P5 siblings (Gjb6+/+, Panx1+/+) were used. The background strains of these mice were C57BL/6J for Gjb6−/− and C57BL/6N for Panx1−/− mice. Note that there is no discernible difference in the auditory phenotype of the closely related C57BL/6J and C57BL/6N mouse strains.44
| Genotype | Gjb6 +/+ | Gjb6 −/− | Panx1 +/+ | Panx −/− | C57BL6/N |
|---|---|---|---|---|---|
| Male | 5 | 5 | 7 | 7 | 2 |
| Female | 5 | 5 | 6 | 6 | 1 |
| Total | 10 | 10 | 13 | 13 | 3 |
Experimental animals were culled by trained personnel using gaseous anaesthesia followed by a rising concentration of CO2. All the experimental procedures were agreed upon, reviewed and approved by local animal welfare oversight bodies and were performed with the approval and direct supervision of the CNR-IBBC/Infrafrontier—Animal Welfare and Ethical Review Body (AWERB), in accordance with general guidelines regarding animal experimentation, approved by the Italian Ministry of Health, in compliance with the Legislative Decree 26/2014 (ref. Project licence no. 68/2016-PR), transposing the 2010/63/EU Directive on protection of animals used in research. This work was also conducted based on recommendations from both ARRIVE and PREPARE guidelines.45,46
Genotyping
Panx1 mice were genotyped according to published protocols by standard PCR on extracted mouse tail tips using the following primers:Panx1 f: 5′-GGAAAGTCAACAGAGGTACCC-3′.
Panx1 r: 5′-CTTGGCCACGGAGTATGTGTT-3′.
LacZ: 5′-GTCCCTCTCACCACTTTTCTTACC-3′.
Mice with normal Panx1 alleles (Panx1+/+) were targeted by the above f and r primers and identified by a 330 bp band, whereas Panx1−/− were targeted by primers Panx1 f and LacZ, and were identified by a 630 bp band. Panx1± mice were identified by the simultaneous presence of a 330 bp and a 630 bp band. Primer pairs for Gjb6−/− mice were specific for the wild type alleles:
Gjb6 f: 5′-GGTACCTTCTACTAATTAGCTTGG-3′,
Gjb6 r: 5′-AGGTGGTACCCATTGTAGAGGAAG-3′.
To visualize the deletion, primers specific for the lacZ region (that flanks the deleted allele) were used in combination with the corresponding wild type forward primer:
Gjb6lac 5′-AGCGAGTAACAACCCGTCGGATTC-3′.
Mice with normal Gjb6 alleles (Gjb6+/+) were identified by a 544 bp band, whereas Gjb6−/− mice were identified by a 460 bp band. See also Table 1.
Study design
In order to construct the optimal experimental design and estimate the minimum number of animals necessary for the experiments (sample size of the groups), for each type of experiment and for each genetically modified and control strain (Gjb6−/−, Gjb6+/+, Panx1−/−, Panx1+/+), we set probability α = 5% = 0.05 for the type I error in the t test. Then, fixing β = 4α = 20% = 0.2 to obtain a test power of 1 − β = 80% = 0.8, we computed the number n of each of the two samples to be compared using the formula:47with zα/2 = 1.96 and zβ = 0.842. Based on experiments of the same type carried out in prior work, we quantified the variability of the data (variance, σ2) and established the minimum difference Δ = μ1 − μ2 between averages that had a biological significance. By expressing both parameters as percentages and setting σ = 12.5% = 0.125 and Δ = 16% = 0.16, we obtained n > 9.58. The actual number of mice is provided in Table 1. To minimize subjective bias, sample identity (e.g. genotypes) was randomized by associating an identification number to each sample before processing. No sample was excluded from the analysis.
Preparation of COCs
P5 mouse pups were humanely euthanized, both cochleae were carefully removed, quickly dissected in ice-cold HEPES buffered (pH 7.2) HBSS (Cat. No. 14025050, Thermo Fisher Scientific, Waltham, MA, USA) and placed onto 5 mm round glass coverslips coated with Cell-Tak (Cat. No. 354240, Corning, Corning, NY, USA). COCs were incubated overnight in DMEM/F12 (Cat. No. 11320-074, Thermo Fisher Scientific) supplemented with 5% FBS (Cat. No. 10270-106, Thermo Fisher Scientific) and 100 μg ml−1 ampicillin (Cat. No. A0166, Merck, St. Louis, MO, USA) at 37 °C in a humid air atmosphere enriched with 5% CO2. The tectorial membrane was carefully removed prior to each experiment.Generation and culture of ATP-WCBs
To generate ATP-WCBs, we transduced HEK293-T cells (CRL-3216™, ATCC, Manassas, VI, USA) with a lentiviral vector encoding P2Y purinoceptor 2 (P2Y2R), a high affinity G-protein coupled receptor that mobilizes Ca2+ from intracellular stores upon binding extracellular ATP.48,49 By using the Gibson assembly technique,50 we inserted the P2Y2R complementary DNA (a gift of Marta Fumagalli, Laboratory of Molecular and Cellular Pharmacology of Purinergic Transmission, University of Milan) into a 3rd generation backbone vector (pUltra-hot, Addgene plasmid #24130, Addgene, Watertown, MA, USA; a gift from Malcolm Moore) that uses 2A self-cleaving peptide technology51 to express the protein of interest (P2Y2R) and the mCherry fluorescent reporter52 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio under the UbC promoter. Viral particles based on this transfer vector were produced using a standard protocol;53 thereafter the virus-containing supernatant was harvested and stored into cryovials in 1 ml aliquots at −80 °C.
1 ratio under the UbC promoter. Viral particles based on this transfer vector were produced using a standard protocol;53 thereafter the virus-containing supernatant was harvested and stored into cryovials in 1 ml aliquots at −80 °C.
Prior to viral transduction, parental HEK293-T cells were maintained in DMEM/F12 (Cat. No. 11320-074, Thermo Fisher Scientific) containing 10% heat inactivated FBS (Cat. No. 10270-106, Thermo Fisher Scientific), 1% penicillin/streptomycin (Cat. No. 15070063, Merck), 1% L-glutamine (Cat. No. 25030024, Merck) and 1% sodium pyruvate (Cat. No. 11360070, Merck). Twenty-four hours before viral transduction (day 1), HEK293-T cells were trypsinized and plated onto 35 mm Petri dishes such that they would be 30–50% confluent at the time of transduction. On the day of transduction (day 2), the culture medium was removed and replaced with the thawed virus-containing supernatant. Sequa-brene (8 μg ml−1, Cat. No. S2667, Merck; synonym: hexadimethrine bromide, polybrene) was added to each Petri dish, followed by gentle swirl to enhance transduction efficiency. HEK293-T cells were incubated overnight, at 37 °C under a controlled atmosphere of CO2 (5%), in the virus-containing supernatant. The following day (day 3), the transduction medium was removed and replaced with 2 ml of complete culture medium. The percentage of HEK293-T transduced cells (>95%), henceforth referred to as WBCs, was estimated 48 hours post-transduction by fluorescence microscopy of the mCherry reporter.
Construction of the ATP dose–response curve for ATP-WCBs
ATP-WCBs were re-plated in black 96-well plates (Cat No. 655090, Greiner Bio-One Italia S.r.l, Cassina de Pecchi, Italy) and cultured for 24 hours as detailed above. Thereafter, ATP-WCBs were incubated for 2 hours at 37 °C and 5% CO2 with the loading solution of the FLIPR Calcium 6 Assay Kit (Cat. No. R8190, Molecular Devices, San Jose, CA, USA). Ca2+-Dependent fluorescence changes promoted by ATP binding to the P2Y2R were measured using a Varioskan LUX multimode microplate reader (Cat. No. VL0L00D0, Thermo Fisher Scientific) after injection of known amounts of ATP (from 0.1 nM to 1 μM) in the wells. To achieve the desired [ATP], a 1 mM stock solution was prepared by dissolving ATP (Cat. No. A1852, Merck) in PBS and diluting the stock in the loading solution of the FLIPR Calcium 6 Assay Kit.Ca2+ imaging in individual ATP-WCBs
To visualize Ca2+ responses evoked by ATP in individual ATP-WCBs, the latter were plated on 12 mm ∅ coverslips and incubated for 30 min at 37 °C in serum-free DMEM (Cat. No. 10-017-CV, Corning, Corning, NY, USA) supplemented with the Ca2+-selective fluorescent probe Fluo 8H™ AM (5 μM, dissociation constant Kd = 232 nM, Cat. No. 21091, AAT Bioquest, Sunnyvale, CA, USA). The incubation medium also contained Pluronic F-127 (0.1% w/v, Cat. No. P2443, Merck) and sulfinpyrazone (250 μM, Cat. No. S9509, Merck) to prevent dye sequestration and secretion.54 To allow for de-esterification, coverslips with loaded ATP-WCBs were transferred to the stage of a custom-made spinning disk microscope55 and perfused for 10 min at a flow rate of 0.1 mL min−1 with extracellular medium (EXM) containing (in mM): NaCl 135, KCl 5.8, CaCl2 1.3, NaH2PO4 0.7, MgCl2 0.9, HEPES–NaOH 10, D-glucose 6, pyruvate 2, amino acids, and vitamins (pH 7.48, 307 mOsm). ATP dissolved in EXM was applied by pressure (5 pounds per square inch, PSI) using a PV820 pneumatic pico-pump (World Precision Instruments Inc., Sarasota, FL, USA) coupled to a glass microcapillary with an opening diameter of 4 μm placed under a 60× water immersion microscope objective (NIKON FLUOR, NA = 1.0, Nikon Corporation, Tokyo, Japan) near the center of the field of view and in close proximity to ATP-WCBs. For Ca2+ imaging, Fluo 8H™ fluorescence was excited using a 488 nm diode laser (Cat. No. COMPACT-150G-488-SM, World Star Tech, Markham, Ontario, Canada) by 45° reflection off a triband dichroic mirror (Cat. No. FF395/527/610-Di01, Semrock, Rochester, NY, USA). Fluo 8H™ fluorescence emission was filtered through a triband filter (Cat. No. FF425/527/685-25, Semrock) followed by a green band pass filter (Cat. No. ET535/30M, Chroma Technology Corp., Bellows Falls, VT, USA) located in front of a cooled s-CMOS camera (pco.edge, PCO AG, Kelheim, Germany) coupled to the microscope.55 Suramin (150 μM; Cat. No. S2671, Merck) was used to reversibly inhibit the ATP-evoked Ca2+ responses (see Results).Dual focal plane multiphoton Ca2+ imaging in COCs and ATP-WCBs
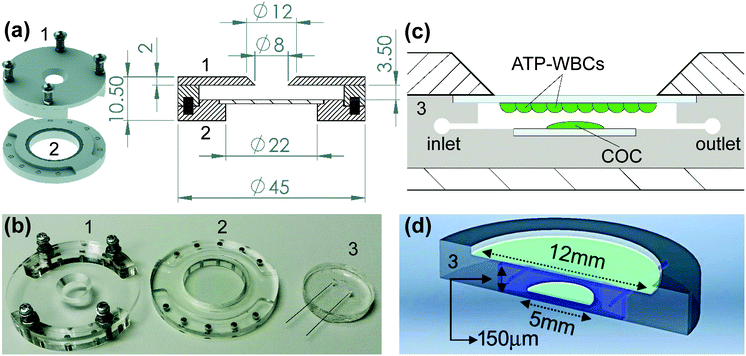
The day before the experiment, ATP-WCBs were plated on 12 mm ∅ coverslips at a density of 2 × 104 cells per cm2. At experiment time, they were loaded with Fluo 8H™ AM as described above. COCs were plated on 5 mm ∅ coverslips the day before the experiment, and incubated for 45 min at 37 °C in serum-free DMEM supplemented with 20 μM Fluo 8H™ AM, Pluronic F-127 (0.1% w/v) and sulfinpyrazone (250 μM) at experiment time. To allow for de-esterification, both cultures were transferred into an ad hoc designed microfluidic chamber (Fig. 1), perfused for 10 min with EXM and placed on the stage of a custom-made two-photon microscope (Fig. 2) based on a Bergamo II architecture (Thorlabs Imaging System, Sterling, VI, USA). The polydimethylsiloxane (PDMS) insert of the chamber (Fig. 1b–d) included microfluidic channels connected by 25 mm stainless-steel tubing (AISI 316L, 0.25/0.12 mm, Unimed SA, CH-1007 Lausanne, Switzerland) and flexible external tubing (Tygon ST R-3603/R3607, Harvard Apparatus, Holliston, MA 01746 United States) to a proximally located peristaltic pump (P-70, Harvard Apparatus). This arrangement reduced the effective chamber volume to 10 μl and the total volume in the fluid path to 200 μl. The flow rate was kept ≤100 μl min−1 to minimize the effects of fluid shearing stress,56 and perfusion was stopped during the image-recording session (which generally lasted 1 min). EXM (containing 2 mM Ca2+) was replaced by LCS, a medium with an endolymph-like Ca2+ concentration (20 μM)57 and an otherwise identical composition. LCS was supplemented either with the ATP analogue 6-N,N-diethyl-D-beta-gamma-dibromomethylene adenosine triphosphate (ARL67156, 100 μM, Cat. No. A265, Merck), or apyrase (40 U ml−1, Cat. No. A6535, Merck), depending on the type of experiment (see Results).The multiphoton system was equipped with two scanning heads, one with resonant-galvo (RG) mirrors and the other with galvo–galvo (GG) mirrors, and was coupled to a mode-locked titanium–sapphire (Ti:Sa) fs pulsed laser (Chameleon Vision II Laser, Coherent, Inc., Santa Clara, CA, USA). The RG scanner was used for imaging, whereas the GG scanner was used to focally photodamage a pre-defined spot in the GER of the cochlea by focusing the collimated laser beam onto the sample through a 25× water-immersion objective (XLPLN25XWMP2, NA 1.05, Olympus Corporation, Tokyo, Japan; the same objective was also used to image both preparations). Multiphoton excitation of Fluo 8H™ was performed at 920 nm, whereas its emission signal was filtered in the range 505–545 nm by a single band-pass filter (Cat. No. FF02-525/40-25, Semrock/IDEX, Rochester, NY, USA) placed in front of a non-descanned GaAsP detector (Cat. No. H7422-50, Hamamatsu Photonics K.K., Shizuoka, Japan, Fig. 2). Electro-optical modulators (EOM) and mechanical ultra-fast shutters were used to control both photodamage light dose (920 nm, power 150 mW, 100 ms) and imaging light exposure using the ThorImage LS 3.1 software (Thorlabs). Sequences of 512 × 512 pixel-frames were acquired at a final rate of 1 frame per second (after frame averaging) by oscillating the objective between the focal planes of the COC and ATP-WCBs (separated by a distance comprised between 20 μm and 100 μm, average 50 μm) using a piezo-electric actuator and paired controller (PFM450, Thorlabs). Laser excitation intensity and frame averaging were adjusted to minimize photobleaching and phototoxicity, while achieving enough signal to noise ratio and temporal resolution. All experiments were performed at room temperature (22–25 °C).
Luciferin–luciferase ATP bioluminescence assay
After overnight incubation under standard culture conditions (37 °C, 5% CO2), COCs plated onto 5 mm glass coverslips were transferred into a black 96-well plate containing 200 μl serum-free DMEM/F12 in each well. Before starting the ATP release-stimulation protocol, COCs were washed once with a solution (NCS) containing a normal (1.8 mM) extracellular Ca2+ concentration and (in mM): 137 NaCl, 5.36 KCl, 0.81 MgSO4, 0.44 KH2PO4, 0.18 Na2HPO4, 25 HEPES and 5.55 D-glucose (pH 7.3). COCs were then washed a second time either with NCS or with a zero Ca2+ solution (ZCS) containing (in mM): 137 NaCl, 5.36 KCl, 0.44 KH2PO4, 0.18 Na2HPO4, 0.1 EGTA, 25 HEPES and 5.55 D-glucose (pH 7.3).To quantify ATP release, COCs were incubated in either NCS or ZCS for 20 minutes at 37 °C, 5% CO2. To limit degradation of the released ATP, both solutions were supplemented with ARL67156 at a final concentration of 100 μM. The amount of ATP released under these conditions was measured with a bioluminescent ATP assay kit (Cat. No. A22066, ThermoFisher Scientific) using a luminometer (Victor Light 1420, Perkin Elmer, Waltham, MA, USA). All bioluminescence measurements reported in this article fell within the linearity range of the ATP standard curve generated according to the manufacturer's instructions. All experiments were performed at room temperature (22–25 °C).
Image processing and data analysis
Image processing and data analysis were carried out using the open source ImageJ software and MATLAB (R2019, The MathWorks, Inc., Natick, MA, USA). Ca2+ signals were quantified either as pixel-by-pixel relative changes of fluorescence emission intensity ΔF(t), where t is time, F(t) is fluorescence at time t, ΔF(t) = F(t) − F0 and F0 is the pre-stimulus fluorescence,58 or as 100 × ΔF(t)/(Fmax − F0) where Fmax = max[F(t)] during the time interval of the recording.Statistics
For statistical data analysis, the normality of distribution was assayed using the Kolmogorov–Smirnov test and statistical comparisons of means were made using the Student t test. The Mann–Whitney U test was used for data which were not normally distributed and/or had dissimilar variance. All statistical analyses were performed using MATLAB (R2019). Mean values are quoted ± standard error of the mean (s.e.m.) where p = p-value < 0.05 was assumed as statistically significant.Results
Sensitivity of ATP-WCBs to ATP
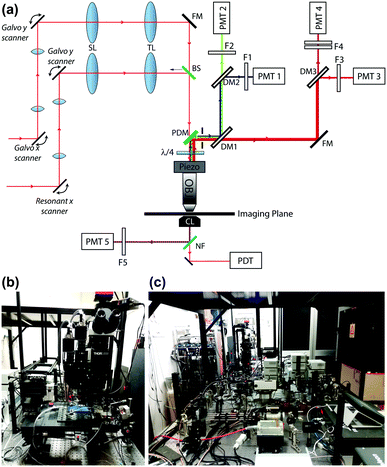
To characterize the sensitivity of ATP-WCBs to ATP, we loaded cells with Ca2+ selective dyes and measured fluorescence intensity changes (ΔF) evoked by different [ATP] in the range from 0.1 nM to 1 μM (see Methods). Fig. 3a shows dose–response data fitted with a Hill function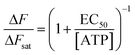 | (1) |
Solving eqn (1) for [ATP] with EC50 = 6.2 nM yields
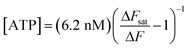 | (2) |
Together, these experiments suggested that ATP-WCBs, if placed sufficiently close to the ATP source in a suitable environment, had the required sensitivity to detect physiologically relevant levels of ATP.
Design, construction and test of a microfluidic chamber suitable to detect ATP release from COCs
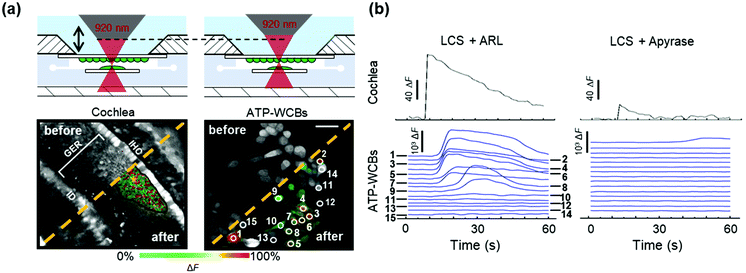
To determine whether ATP-WCBs responded to the ATP released in the extracellular medium by COCs, we exploited the intrinsic optical sectioning capabilities of multiphoton microscopy63 and performed dual focal plane Ca2+ imaging in an ad hoc designed, closed and transparent micro-fluidic chamber hosting ATP-WCBs at the top and a proximally located COC at the bottom (see Methods and Fig. 1 and 2). To increase detection sensitivity, hydrolysis of ATP was reduced by supplementing the extracellular medium with ARL67156 (100 μM), a widely used ectonucleotidase inhibitor.64,65Initially, we used the second scanning head and fast beam shuttering of the multiphoton microscope (Fig. 2) to rapidly photodamage a single GER cell (see Methods) in COCs from wild type C57BL6/N pups, resulting in the time-controlled spreading of a Ca2+ wave around the damage spot (Fig. 4a and Video S1 in the ESI‡). The overlying ATP-WCBs in the microfluidic chamber responded with clearly detectable and time-delayed Ca2+ signals (n = 3 out of 3 independent experiments). No signal was detected in ATP-WCBs if the same procedure was performed without including a COC in the chamber (n = 3 independent experiments), indicating that the observed responses were not due to photodamage of ATP-WCBs or other spurious effects.
Replacing ARL67156 in the extracellular medium with apyrase (40 U ml−1), an ATP-hydrolyzing enzyme,66 caused a dramatic reduction of Ca2+ wave amplitude and extent in COCs. Ca2+ responses in the overlying ATP-WCBs were correspondingly reduced, confirming that they were due to the diffusion of the released ATP in the narrow fluid gap (20–100 μm) intercalated between the two cellular systems. For the representative experiment in Fig. 4b, the responses, integrated over time and summed over all ATP-WCBs within the field of view, were 3.36 × 106 A.U. in the presence of ARL67156 vs. 7.00 × 104 A.U. in the presence of apyrase (a 48-fold reduction). By using eqn (2) to estimate the average [ATP] that reached ATP-WCBs in the presence of ARL67156, we obtained values in the range from 5 nM to 48 nM. Similar results were obtained in two other independent experiments.
Together, these results suggested that ATP-WCBs in the microfluidic chamber could detect ATP release during stimulated Ca2+ wave propagation in the GER.
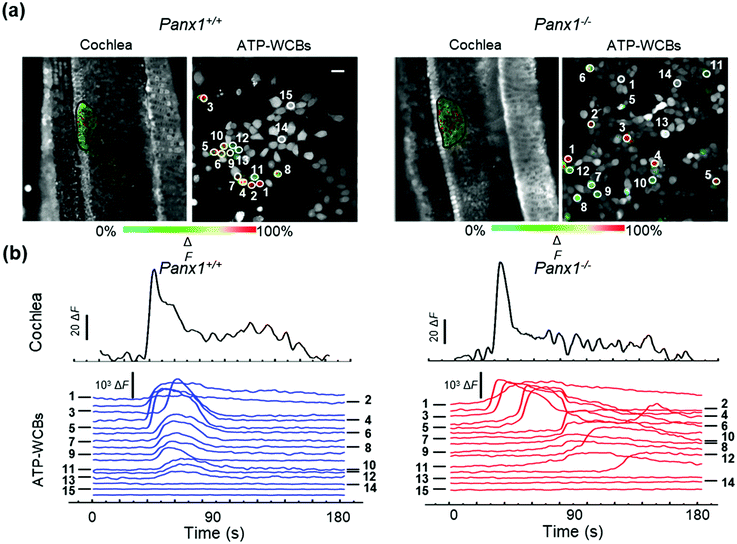
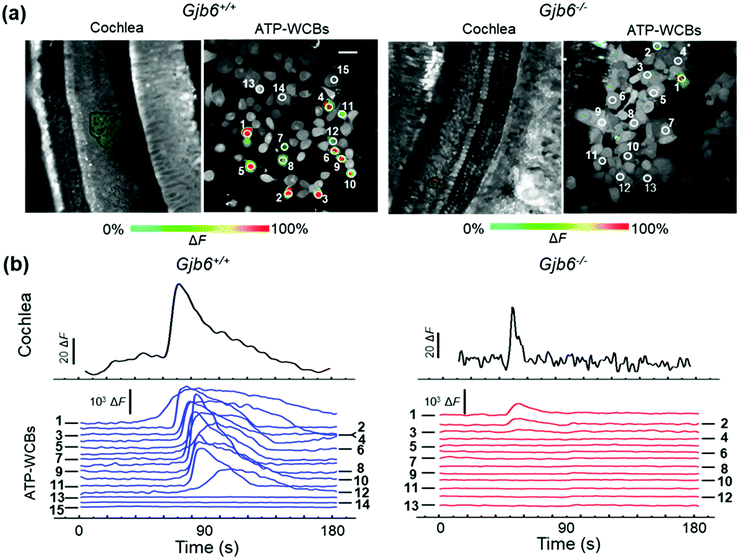
Analysis of spontaneous Ca2+ wave propagation and ATP release from the GER of genetically modified mice by dual multiphoton imaging in the microfluidic chamber
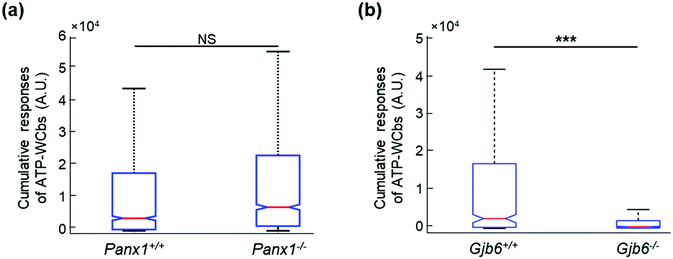
Based on the positive outcomes of the aforementioned experiments, we decided to investigate spontaneous Ca2+ wave propagation in the GER of pups with global deletion of Panx1 (Fig. 5 and Video S2 in the ESI‡) or Gjb6 (Fig. 6 and Video S3 in the ESI‡) using the microfluidic chamber. The distributions of ATP-WCB Ca2+ responses, integrated over time and driven by spontaneous Ca2+ wave propagation in the underlying GER from Panx1−/− and Panx1+/+ COCs, were statistically indistinguishable (Fig. 7a: Panx1−/−: mean = 2.38 × 104, median = 7.33 × 103, I.Q.R. = 2.19 × 104 A.U.; Panx1+/+: mean = 2.07 × 104 A.U., median = 3.88 × 103, I.Q.R. = 1.74 × 104 A.U.; m = 1695 ATP-WCBs in each case; p = 0.105, Mann–Whitney U test; pooled results from n = 7 pups of each genotype). By using eqn (2) to estimate the average [ATP] near ATP-WCBs, we obtained values in the range from 7 nM to 30 nM for Panx1−/− as well as Panx1+/+ COCs.In prior work, the frequency of GER spontaneous Ca2+ waves was dramatically reduced in Gjb6−/− animals compared to their siblings with normal Gjb6 alleles (Gjb6+/+).18,20 Here, in addition, we report significantly different distributions of Ca2+ responses obtained from ATP-WCBs overlying Gjb6−/− COCs in the microfluidic chamber (Fig. 7b; mean = 2.90 × 103, median = 460, I.Q.R. = 1.97 × 103 A.U.) vs. Gjb61+/+ COCs (mean = 2.30 × 104, median = 2.59 × 103, I.Q.R. = 1.67 × 104 A.U.; m = 606 ATP-WCBs in each case; p = 3.04 × 10−27, Mann–Whitney U test; pooled results from n = 4 pups of each genotype, aged P5).
Independent estimate of ATP release from Panx1−/− and Cx30−/− cochlear organotypic cultures
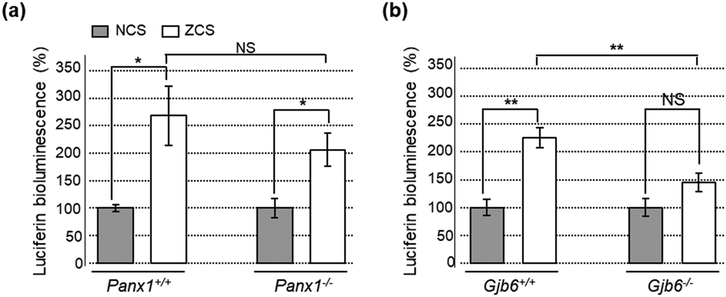
As an additional control, we used also a standard luciferin–luciferase ATP bioluminescence assay67 to measure ATP release from COCs in 96-well plates (see Methods). In COCs obtained from Panx1+/+ and Panx1−/− P5 pups, ATP release was equally enhanced by lowering the concentration of extracellular Ca2+ from 1.8 mM (NCS) to 0 mM (ZCS), a procedure that increases the open probability of connexin hemichannels68 without affecting pannexin channels.69–71 Specifically, the amount of ATP released in the extracellular medium in ZCS was 205% ± 30% of the NCS control in Panx1−/− COCs (p = 0.015, t test) and 268% ± 54% in Panx1+/+ COCs (p = 0.011, t test). The ATP released in the extracellular medium was not significantly different between the two genotypes, either in ZCS (p = 0.265, t test) or in NCS (p = 0.93, t test, n = 6 cultures of either genotype; Fig. 8a). In contrast, the ATP released in the extracellular medium in ZCS was 225% ± 18% of the NCS control in Gjb6−/− COCs (p = 0.0001, t test) and 145% ± 16% in Gjb6+/+ COCs (p = 0.074). In this case, the amount of released ATP in ZCS was significantly different between the two genotypes (p = 0.001, t test; Fig. 8b).Together, the results of Fig. 4–8 indicate that: (i) ATP-WCBs in the microfluidic environment had the required sensitivity to detect the ATP released by stimulated as well as spontaneous Ca2+ wave propagation in the GER; (ii) ATP release was unaffected by Panx1 genetic ablation (iii) but strongly reduced by Gjb6 ablation.
Discussion and conclusions
Prior work has unveiled a critical role for connexin hemichannels in Ca2+ wave propagation in both the LER25–27 and the GER.17,18,22 The goal of this work was to determine whether ATP release through connexin hemichannels underlies Ca2+ wave propagation also in the GER of the developing cochlea (rather than only in the LER, as abundantly documented). To address this key issue, we used microfluidics, which is capable of manipulating cells and organ models in a highly controllable microenvironment (Fig. 1), coupled with multiphoton imaging (Fig. 2). This disruptive combination of key enabling technologies is creating breakthroughs in current understanding of cell and developmental biology and is providing insights into disease diagnosis in numerous fields.72WCBs were the third essential element of our design (Fig. 3). For more than a quarter century, WCBs have provided critical insight into the physiological effect of numerous analytes (reviewed in ref. 43, 73 and 74). To create ATP-WCBs, we infected HEK-293T cells with a lentiviral vector encoding the highly sensitive P2Y2R, one of the two major P2Y receptors expressed in the developing cochlea.3,6 The binding of ATP to P2Y2R promotes the generation of diacylglycerol and InsP3 from PLC-dependent hydrolysis of PIP2.18 Intracellular diffusion of InsP3 and subsequent binding to InsP3 receptors triggers Ca2+ release from the endoplasmic reticulum (ER), increasing the cytosolic Ca2+ concentration.75 By optically monitoring these Ca2+ fluctuations with Ca2+-selective dyes,58 we determined that our ATP-WCBs had the required sensitivity to detect [ATP] fluctuations in the sub-micromolar range, with EC50 close to the baseline [ATP] measured in the rodent endolymph in vivo.28,29
By oscillating the objective of the multiphoton microscope between the focal planes of COC and overlying ATP-WCBs in the microfluidic chamber, we correlated [ATP]-dependent Ca2+ fluctuations in ATP-WCBs to stimulated (Fig. 4) as well as spontaneous Ca2+ wave propagation in the GER of genetically modified mice (Fig. 5 and 6). Ca2+ signals where inhibited by apyrase, as previously reported for ATP-dependent Ca2+ wave propagation in the LER of wild type mice.4 Importantly, spontaneous Ca2+ wave propagation in the GER resulted in time-delayed ATP-WCB responses that were strongly reduced by global deletion of Gjb6, but unaffected by global deletion of Panx1 (Fig. 7). Quantification of ATP release from COCs by means of a standard luciferin–luciferase bioluminescence assay corroborated this conclusion (Fig. 8). However, the luciferin-luciferase approach, the main strategy to measure ATP nowadays, which focuses on supernatants, falls short of providing the kind of information that is made available by our organ-on-chip/ATP-WCBs system and optical technology.
Although pannexin 1 channels71 are thought to mediate ATP release in other cellular systems,76–81 the function of cochlear pannexins remains obscure.34,82–84 Prior work with the Panx1−/− strain used here confirmed (i) successful ablation of Panx1 both in the cochlea and in the brain; (ii) normal hearing sensitivity, normal function of the outer hair cell-based “cochlear amplifier” and absence of cochlear nerve defects; (iii) normal expression of inner ear connexins and gap junction communication in the organ of Corti.34 These published results and the data presented here together rule out that pannexin 1 channels contribute to the purinergic signaling that underlies Ca2+ wave propagation in the developing cochlea.2
Gjb6 −/− mice, in which Cx30 is deleted globally38 and Cx26 is dramatically down-regulated in the sensory epithelium of the developing cochlea,33 are a model of non-syndromic hearing loss and deafness (DFNB1, reviewed in ref. 37 and 85). Prior work revealed impaired Ca2+ wave propagation in the developing cochlea of these mice,18,33 which could be restored by viral transduction with a bovine adeno-associated vector (BAAV) encoding Cx30.18,86 The expression of connexin hemichannels at the endolymphatic surface of the cochlear sensory epithelium has been confirmed by immunolabeling with antibodies that bind the extracellular loops of Cx26 and Cx30.25,26,87 In addition, spontaneous Ca2+ wave propagation at the surface of the GER was inhibited by (i) flufenamic acid (FFA),17,18,22 a nonspecific blocker of connexin hemichannels88,89 which, unlike carbenoxolone,90 is ineffective on pannexin 1 channels at concentrations <300 μM (ref. 80 and 91) and (ii) the abEC1.1 antibody, which does not interfere with pannexin 1 channels, is selective for Cx26, Cx30 and Cx32 hemichannels, reduces connexin hemichannel currents and abrogates ATP release.35,36,92 This vast body of experimental results, together with the data presented here, demonstrate that ATP-dependent ATP release through connexin hemichannels sustains Ca2+ wave propagation not only in the LER,25–27 but also in the GER.
The ATP released during Ca2+ wave propagation in the GER has been proposed to trigger Ca2+ action potential (AP) activity in immature IHCs, driving bursts of APs in the auditory nerve fibers.14 However, subsequent work has shown AP firing in the complete absence of Ca2+ activity in the GER.20 Therefore, it remains to be established if and how these two types of spontaneous activities correlate with each other. This stands out a key open question for cochlear physiopathology, as connexin expression and spontaneous Ca2+ signaling in the GER are essential for normal development of the sensory epithelium, hair cell functional maturation, hearing acquisition,20–23 redox homeostasis and age-related hearing loss.24 We are confident that the technological advances presented here have the potential to shed light on this as well as a plethora of other unrelated open issues that concern the role of paracrine signaling in physiology and pathology93 and cannot be addressed with standard methods.
Author contributions
Conceptualization, F. M. (Fabio Mammano); methodology, F. M., Fl. M. (Flavia Mazzarda), A. D'E., R. M., A. D. N., F. R. B., A. T. M., L. B., F. C.; resources F. M., F. R. B., L. B., A. S., M. R., F. S.; investigation, Fl. M., C. P., G. Z., V. Z., C. N.; formal analysis, data curation, visualization and writing—original draft preparation, Fl. M.; supervision, writing—review and editing, project administration and funding acquisition A. M. S., M. R., J. Y., F. M. All the authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.Acknowledgements
This project has received funding from Fondazione Telethon (grant GGP13114), the National Research Council of Italy (CNR) Grant No. DSB.AD008.370.003\TERABIO-IBCN and the University of Padova Grant SID\BIRD187130 to F. M., and from the Italian Ministry of Research and University, grant PRIN 2017FTJ5ZE Sensory decay and aging to M. R. The authors thank Mr. Fabrizio Bonaventura for technical assistance, as well as Prof. Mario Bortolozzi and two anonymous reviewers for constructive criticism and helpful suggestions.References
- G. D. Housley, A. Bringmann and A. Reichenbach, Trends Neurosci., 2009, 32, 128–141 CrossRef CAS PubMed.
- F. Mammano, Semin. Cell Dev. Biol., 2013, 24, 31–39 CrossRef CAS PubMed.
- E. Berekmeri, J. Szepesy, L. Koles and T. Zelles, Brain Res. Bull., 2019, 151, 109–118 CrossRef CAS PubMed.
- J. E. Gale, V. Piazza, C. D. Ciubotaru and F. Mammano, Curr. Biol., 2004, 14, 526–529 CrossRef CAS PubMed.
- M. Beltramello, V. Piazza, F. F. Bukauskas, T. Pozzan and F. Mammano, Nat. Cell Biol., 2005, 7, 63–69 CrossRef CAS PubMed.
- V. Piazza, C. D. Ciubotaru, J. E. Gale and F. Mammano, Cell Calcium, 2007, 41, 77–86 CrossRef CAS PubMed.
- D. J. Lim and M. Anniko, Acta Oto-Laryngol., Suppl., 1985, 422, 1–69 CAS.
- D. J. Lim and J. Rueda, in Development of auditory and vestibular systems - 2, ed. R. Romand, Elsevier Science Publishing Co., New York, 1st edn, 1992, pp. 33–58 Search PubMed.
- A. A. Eggston and D. Wolff, in Histopathology of the ear, nose, and throat, Williams and Wilkins Co., Baltimore, 1947, pp. 37–64 Search PubMed.
- R. Hinojosa, Acta Oto-Laryngol., 1977, 84, 238–251 CrossRef CAS PubMed.
- S. Hou, J. Chen and J. Yang, Eur. J. Histochem., 2019, 63, 102–109 Search PubMed.
- M. W. Kelley, Int. J. Dev. Biol., 2007, 51, 571–583 CrossRef CAS PubMed.
- J. Bryant, R. J. Goodyear and G. P. Richardson, Br. Med. Bull., 2002, 63, 39–57 CrossRef CAS PubMed.
- N. X. Tritsch, E. Yi, J. E. Gale, E. Glowatzki and D. E. Bergles, Nature, 2007, 450, 50–55 CrossRef CAS PubMed.
- N. X. Tritsch, Y. X. Zhang, G. Ellis-Davies and D. E. Bergles, Purinergic Signalling, 2010, 6, 155–166 CrossRef CAS PubMed.
- H. C. Wang, C. C. Lin, R. Cheung, Y. Zhang-Hooks, A. Agarwal, G. Ellis-Davies, J. Rock and D. E. Bergles, Cell, 2015, 163, 1348–1359 CrossRef CAS PubMed.
- M. Schutz, P. Scimemi, P. Majumder, R. D. De Siati, G. Crispino, L. Rodriguez, M. Bortolozzi, R. Santarelli, A. Seydel, S. Sonntag, N. Ingham, K. P. Steel, K. Willecke and F. Mammano, Hum. Mol. Genet., 2010, 19, 4759–4773 CrossRef PubMed.
- L. Rodriguez, E. Simeonato, P. Scimemi, F. Anselmi, B. Cali, G. Crispino, C. D. Ciubotaru, M. Bortolozzi, F. G. Ramirez, P. Majumder, E. Arslan, P. De Camilli, T. Pozzan and F. Mammano, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14013–14018 CrossRef CAS PubMed.
- T. Eckrich, K. Blum, I. Milenkovic and J. Engel, Front. Mol. Neurosci., 2018, 11, 264–264 CrossRef PubMed.
- S. L. Johnson, F. Ceriani, O. Houston, R. Polishchuk, E. Polishchuk, G. Crispino, V. Zorzi, F. Mammano and W. Marcotti, Journal of Neuroscience, 2017, 37, 258–268 CrossRef CAS PubMed.
- F. Ceriani, A. Hendry, J. Y. Jeng, S. L. Johnson, F. Stephani, J. Olt, M. C. Holley, F. Mammano, J. Engel, C. J. Kros, D. D. Simmons and W. Marcotti, EMBO J., 2019, 38(9), e99839 CrossRef PubMed.
- F. Mammano and M. Bortolozzi, Cell Calcium, 2018, 70, 117–126 CrossRef CAS PubMed.
- G. Crispino, G. Di Pasquale, P. Scimemi, L. Rodriguez, F. Galindo Ramirez, R. D. De Siati, R. M. Santarelli, E. Arslan, M. Bortolozzi, J. A. Chiorini and F. Mammano, PLoS One, 2011, 6, e23279 CrossRef CAS PubMed.
- A. R. Fetoni, V. Zorzi, F. Paciello, G. Ziraldo, C. Peres, M. Raspa, F. Scavizzi, A. M. Salvatore, G. Crispino, G. Tognola, G. Gentile, A. G. Spampinato, D. Cuccaro, M. Guarnaccia, G. Morello, G. Van Camp, E. Fransen, M. Brumat, G. Girotto, G. Paludetti, P. Gasparini, S. Cavallaro and F. Mammano, Redox Biol., 2018, 19, 301–317 CrossRef CAS PubMed.
- F. Anselmi, V. H. Hernandez, G. Crispino, A. Seydel, S. Ortolano, S. D. Roper, N. Kessaris, W. Richardson, G. Rickheit, M. A. Filippov, H. Monyer and F. Mammano, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18770–18775 CrossRef CAS PubMed.
- P. Majumder, G. Crispino, L. Rodriguez, C. D. Ciubotaru, F. Anselmi, V. Piazza, M. Bortolozzi and F. Mammano, Purinergic Signalling, 2010, 6, 167–187 CrossRef CAS PubMed.
- F. Ceriani, T. Pozzan and F. Mammano, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E7194–E7201 CrossRef CAS PubMed.
- D. J. Munoz, P. R. Thorne, G. D. Housley and T. E. Billett, Hear. Res., 1995, 90, 119–125 CrossRef CAS PubMed.
- S. M. Vlajkovic, P. R. Thorne, G. D. Housley, D. J. Munoz and I. S. Kendrick, NeuroReport, 1998, 9, 1559–1565 CAS.
- J. Lautermann, H. G. Frank, K. Jahnke, O. Traub and E. Winterhager, Dev. Genet., 1999, 25, 306–311 CrossRef CAS PubMed.
- J. Lautermann, W. J. ten Cate, P. Altenhoff, R. Grummer, O. Traub, H. Frank, K. Jahnke and E. Winterhager, Cell Tissue Res., 1998, 294, 415–420 CrossRef CAS PubMed.
- A. Forge, D. Becker, S. Casalotti, J. Edwards, N. Marziano and G. Nevill, J. Comp. Neurol., 2003, 467, 207–231 CrossRef PubMed.
- S. Ortolano, G. Di Pasquale, G. Crispino, F. Anselmi, F. Mammano and J. A. Chiorini, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18776–18781 CrossRef CAS PubMed.
- V. Zorzi, F. Paciello, G. Ziraldo, C. Peres, F. Mazzarda, C. Nardin, M. Pasquini, F. Chiani, M. Raspa, F. Scavizzi, A. Carrer, G. Crispino, C. D. Ciubotaru, H. Monyer, A. R. Fetoni, A. M. Salvatore and F. Mammano, Front. Mol. Neurosci., 2017, 10, 379 CrossRef PubMed.
- L. Xu, A. Carrer, F. Zonta, Z. Qu, P. Ma, S. Li, F. Ceriani, D. Buratto, G. Crispino, V. Zorzi, G. Ziraldo, F. Bruno, C. Nardin, C. Peres, F. Mazzarda, A. M. Salvatore, M. Raspa, F. Scavizzi, Y. Chu, S. Xie, X. Yang, J. Liao, X. Liu, W. Wang, S. Wang, G. Yang, R. A. Lerner and F. Mammano, Front. Mol. Neurosci., 2017, 10, 298 CrossRef PubMed.
- G. Ziraldo, D. Buratto, Y. Kuang, L. Xu, A. Carrer, C. Nardin, F. Chiani, A. M. Salvatore, G. Paludetti, R. A. Lerner, G. Yang, F. Zonta and F. Mammano, Front. Physiol., 2019, 10, 392 CrossRef PubMed.
- F. Mammano, Cold Spring Harbor Perspect. Med., 2019, 9(7), a033233 CrossRef CAS PubMed.
- B. Teubner, V. Michel, J. Pesch, J. Lautermann, M. Cohen-Salmon, G. Sohl, K. Jahnke, E. Winterhager, C. Herberhold, J. P. Hardelin, C. Petit and K. Willecke, Hum. Mol. Genet., 2003, 12, 13–21 CrossRef CAS PubMed.
- M. Cohen-Salmon, B. Regnault, N. Cayet, D. Caille, K. Demuth, J. P. Hardelin, N. Janel, P. Meda and C. Petit, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6229–6234 CrossRef CAS PubMed.
- Q. Chang, W. Tang, S. Ahmad, B. Zhou and X. Lin, PLoS One, 2008, 3, e4088 CrossRef PubMed.
- P. Bargiotas, A. Krenz, S. G. Hormuzdi, D. A. Ridder, A. Herb, W. Barakat, S. Penuela, J. von Engelhardt, H. Monyer and M. Schwaninger, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 20772–20777 CrossRef CAS PubMed.
- S. Petric, S. Klein, L. Dannenberg, T. Lahres, L. Clasen, K. G. Schmidt, Z. Ding and B. C. Donner, Cell. Physiol. Biochem., 2016, 38, 487–501 CrossRef CAS PubMed.
- Q. Gui, T. Lawson, S. Shan, L. Yan and Y. Liu, Sensors, 2017, 17(7), pii: s17071623 CrossRef PubMed.
- K. L. Kane, C. M. Longo-Guess, L. H. Gagnon, D. Ding, R. J. Salvi and K. R. Johnson, Hear. Res., 2012, 283, 80–88 CrossRef CAS PubMed.
- C. Kilkenny, W. J. Browne, I. C. Cuthill, M. Emerson and D. G. Altman, PLoS Biol., 2010, 8, e1000412 CrossRef PubMed.
- A. J. Smith, R. E. Clutton, E. Lilley, K. E. A. Hansen and T. Brattelid, Lab. Anim., 2018, 52, 135–141 CrossRef CAS PubMed.
- S. Lemeshow, D. W. Hosmer, J. Klar and S. K. Lwanga, Adequacy of sample size in health studies, Wiley, New York, NY, USA, 1990 Search PubMed.
- M. P. Abbracchio, G. Burnstock, J. M. Boeynaems, E. A. Barnard, J. L. Boyer, C. Kennedy, G. E. Knight, M. Fumagalli, C. Gachet, K. A. Jacobson and G. A. Weisman, Pharmacol. Rev., 2006, 58, 281–341 CrossRef CAS PubMed.
- C. E. Parr, D. M. Sullivan, A. M. Paradiso, E. R. Lazarowski, L. H. Burch, J. C. Olsen, L. Erb, G. A. Weisman, R. C. Boucher and J. T. Turner, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 3275–3279 CrossRef CAS PubMed.
- D. G. Gibson, L. Young, R. Y. Chuang, J. C. Venter, C. A. Hutchison, 3rd and H. O. Smith, Nat. Methods, 2009, 6, 343–345 CrossRef CAS PubMed.
- J. H. Kim, S. R. Lee, L. H. Li, H. J. Park, J. H. Park, K. Y. Lee, M. K. Kim, B. A. Shin and S. Y. Choi, PLoS One, 2011, 6, e18556 CrossRef CAS PubMed.
- N. C. Shaner, M. Z. Lin, M. R. McKeown, P. A. Steinbach, K. L. Hazelwood, M. W. Davidson and R. Y. Tsien, Nat. Methods, 2008, 5, 545–551 CrossRef CAS PubMed.
- M. Sena-Esteves and G. Gao, Cold Spring Harb. Protoc., 2018, 2018(4) DOI:10.1101/pdb.prot095687.
- F. Di Virgilio, T. H. Steinberg and S. C. Silverstein, Methods Cell Biol., 1989, 31, 453–462 CrossRef CAS PubMed.
- F. Ceriani, C. D. Ciubotaru, M. Bortolozzi and F. Mammano, Methods Mol. Biol., 2016, 1427, 223–241 CrossRef CAS PubMed.
- M. J. Levesque and R. M. Nerem, J. Biomech. Eng., 1985, 107, 341–347 CrossRef CAS PubMed.
- S. K. Bosher and R. L. Warren, Nature, 1978, 273, 377–378 CrossRef CAS PubMed.
- F. Mammano and M. Bortolozzi, in Calcium Measurement Methods, ed. A. Verkhratsky and O. Petersen, Humana Press, New York, 2010, vol. 43, pp. 57–80 Search PubMed.
- I. von Kugelgen, Pharmacol. Ther., 2006, 110, 415–432 CrossRef PubMed.
- R. Janssens, P. Paindavoine, M. Parmentier and J. M. Boeynaems, Br. J. Pharmacol., 1999, 127, 709–716 CrossRef CAS PubMed.
- J. B. Schachter and T. K. Harden, Br. J. Pharmacol., 1997, 121, 338–344 CrossRef CAS PubMed.
- S. Langlois, K. N. Cowan, Q. Shao, B. J. Cowan and D. W. Laird, Mol. Biol. Cell, 2008, 19, 912–928 CrossRef CAS PubMed.
- W. Denk, J. H. Strickler and W. W. Webb, Science, 1990, 248, 73–76 CrossRef CAS PubMed.
- S. A. Levesque, E. G. Lavoie, J. Lecka, F. Bigonnesse and J. Sevigny, Br. J. Pharmacol., 2007, 152, 141–150 CrossRef CAS PubMed.
- T. D. Westfall, C. Kennedy and P. Sneddon, Br. J. Pharmacol., 1996, 117, 867–872 CrossRef CAS PubMed.
- A. M. Kettlun, L. Uribe, V. Calvo, S. Silva, J. Rivera, M. Mancilla, M. A. Valenzuela and A. Traverso-Cori, Phytochemistry, 1982, 21(3), 551–558 CrossRef CAS.
- H.-W. Yeh and H.-W. Ai, Annu. Rev. Anal. Chem., 2019, 12, 129–150 CrossRef CAS PubMed.
- I. Fasciani, A. Temperan, L. F. Perez-Atencio, A. Escudero, P. Martinez-Montero, J. Molano, J. M. Gomez-Hernandez, C. L. Paino, D. Gonzalez-Nieto and L. C. Barrio, Neuropharmacology, 2013, 75, 479–490 CrossRef CAS PubMed.
- P. Whyte-Fagundes and G. Zoidl, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 65–71 CrossRef CAS PubMed.
- D. Patel, X. Zhang and R. D. Veenstra, FEBS Lett., 2014, 588, 1372–1378 CrossRef CAS PubMed.
- Z. Ruan, I. J. Orozco, J. Du and W. Lü, Nature, 2020 DOI:10.1038/s41586-020-2357-y.
- J. V. Rocheleau and D. W. Piston, Methods Cell Biol., 2008, 89, 71–92 CAS.
- L. Bousse, Sens. Actuators, B, 1996, 34, 270–275 CrossRef CAS.
- J. J. Pancrazio, J. P. Whelan, D. A. Borkholder, W. Ma and D. A. Stenger, Ann. Biomed. Eng., 1999, 27, 697–711 CrossRef CAS PubMed.
- C. Giorgi, A. Danese, S. Missiroli, S. Patergnani and P. Pinton, Trends Cell Biol., 2018, 28, 258–273 CrossRef CAS PubMed.
- G. A. Ransford, N. Fregien, F. Qiu, G. Dahl, G. E. Conner and M. Salathe, Am. J. Respir. Cell Mol. Biol., 2009, 41, 525–534 CrossRef CAS PubMed.
- M. Sridharan, S. P. Adderley, E. A. Bowles, T. M. Egan, A. H. Stephenson, M. L. Ellsworth and R. S. Sprague, Am. J. Physiol., 2010, 299, H1146–H1152 CAS.
- E. Dolmatova, G. Spagnol, D. Boassa, J. R. Baum, K. Keith, C. Ambrosi, M. I. Kontaridis, P. L. Sorgen, G. E. Sosinsky and H. S. Duffy, Am. J. Physiol., 2012, 303, H1208–H1218 CAS.
- J. K. Sandilos, Y. H. Chiu, F. B. Chekeni, A. J. Armstrong, S. F. Walk, K. S. Ravichandran and D. A. Bayliss, J. Biol. Chem., 2012, 287, 11303–11311 CrossRef CAS PubMed.
- G. Dahl, F. Qiu and J. Wang, Neuropharmacology, 2013, 75, 583–593 CrossRef CAS PubMed.
- J. M. Beckel, S. L. Daugherty, P. Tyagi, A. S. Wolf-Johnston, L. A. Birder, C. H. Mitchell and W. C. de Groat, J. Physiol., 2015, 593, 1857–1871 CrossRef CAS PubMed.
- J. M. Abitbol, J. J. Kelly, K. Barr, A. L. Schormans, D. W. Laird and B. L. Allman, Biochem. J., 2016, 473, 4665–4680 CrossRef CAS PubMed.
- J. M. Abitbol, B. L. O'Donnell, C. B. Wakefield, E. Jewlal, J. J. Kelly, K. Barr, K. E. Willmore, B. L. Allman and S. Penuela, J. Mol. Med., 2019, 97, 723–736 CrossRef CAS PubMed.
- P. Whyte-Fagundes, R. Siu, C. Brown and G. Zoidl, Neurosci. Lett., 2019, 695, 32–39 CrossRef CAS PubMed.
- F. J. Del Castillo and I. Del Castillo, Front. Mol. Neurosci., 2017, 10, 428 CrossRef PubMed.
- G. Crispino, F. Galindo Ramirez, M. Campioni, V. Zorzi, M. Praetorius, G. Di Pasquale, J. A. Chiorini and F. Mammano, Sci. Rep., 2017, 7, 6567 CrossRef PubMed.
- C. Clair, L. Combettes, F. Pierre, P. Sansonetti and G. Tran Van Nhieu, Exp. Cell Res., 2008, 314, 1250–1265 CrossRef CAS PubMed.
- E. G. Harks, A. D. de Roos, P. H. Peters, L. H. de Haan, A. Brouwer, D. L. Ypey, E. J. van Zoelen and A. P. Theuvenet, J. Pharmacol. Exp. Ther., 2001, 298, 1033–1041 CAS.
- R. Guinamard, C. Simard and C. Del Negro, Pharmacol. Ther., 2013, 138, 272–284 CrossRef CAS PubMed.
- K. Michalski and T. Kawate, J. Gen. Physiol., 2016, 147, 165–174 CrossRef CAS PubMed.
- A. W. Lohman and B. E. Isakson, FEBS Lett., 2014, 588, 1379–1388 CrossRef CAS PubMed.
- Y. Kuang, V. Zorzi, D. Buratto, G. Ziraldo, F. Mazzarda, C. Peres, C. Nardin, A. M. Salvatore, F. Chiani, F. Scavizzi, M. Raspa, M. Qiang, Y. Chu, X. Shi, Y. Li, L. Liu, Y. Shi, F. Zonta, G. Yang, R. A. Lerner and F. Mammano, EBioMedicine, 2020, 102825, DOI:10.1016/j.ebiom.2020.102825.
- Z. Wu and Y. Li, Neurosci. Res., 2020, 152, 35–43 CrossRef PubMed.
Footnotes |
| † Data will be made available via University of Padova's Data Repository. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc00427h |
| This journal is © The Royal Society of Chemistry 2020 |