Open Access Article
Open Access ArticleSingle-molecule confinement with uniform electrodynamic nanofluidics†
Siddharth
Ghosh‡
 *ab,
Narain
Karedla§
ab and
Ingo
Gregor
a
*ab,
Narain
Karedla§
ab and
Ingo
Gregor
a
aIII. Institute of Physics – Biophysics and Complex Systems, University of Göttingen, Göttingen, Germany. E-mail: sg915@cam.ac.uk
bInternational Max Planck Research School for Physics of Biological and Complex Systems, Göttingen, Germany
First published on 24th July 2020
Abstract
To date, we could not engineer Nature's ability to dynamically handle diffusing single molecules in the liquid-phase as it takes place in pore-forming proteins and tunnelling nanotubes. Consistent handling of individual single molecules in a liquid is of paramount importance to fundamental molecular studies and technological benefits, like single-molecule level separation and sorting for early biomedical diagnostics, microscopic studies of molecular interactions and electron/optical microscopy of molecules and nanomaterials. We can consistently resolve the dynamics of diffusing single molecules if they are confined within a uniform dielectric environment at nanometre length-scales. A uniform dielectric environment is the key characteristic since intrinsic electronic properties of molecules were modified while interacting with any surfaces, and the effect is not the same from one dielectric surface to another. We present dynamic nanofluidic detection of optically active single molecules in a liquid. An all-silica nanofluidic environment was used to electrokinetically handle individual single-molecules where molecular shot noise was resolved. We recorded the single-molecule motion of small fragments of DNA, carbon-nanodots, and organic fluorophores in water. The electrokinetic 1D molecular mass transport under two-focus fluorescence correlation spectroscopy (2fFCS) showed confinement-induced modified molecular interactions (due to various inter-molecular repulsive and attractive forces), which have been theoretically interpreted as molecular shot noise. Our demonstration of high-throughput nanochannel fabrication, 2fFCS-based 1D confined detection of fast-moving single molecules and fundamental understanding of molecular shot noise may open an avenue for single-molecule experiments where physical manipulation of dynamics is necessary.
Introduction
Due to Brownian motion, it is difficult to keep a molecule within the detection volume for an extended period.1,2 Even after 30 years of single-molecule detection,3–5 sorting small biomolecules with a single-molecule level resolution is still a challenge.6–8 Nanofluidic9 approaches are potentially one of the means to circumvent this problem for various scientific applications.10–12 In recent years, we have seen that nanofluidic devices have a significant impact on the analytical investigation of DNA optical mapping13–16 and single virus and nanoparticle detection as well as isolation,8,17–22 ion trapping,23 and energy harvesting.24 Decomposing complex systems in nanochannels can be possible due to systematic studies of single particles from molecules to viruses over long durations by suppressing the thermal motions in two directions. Increasing detection sensitivity inside nanochannels is in high demand when electrodynamics and Debye lengths are not negligible parameters. Hence, we present a robust investigation on the dynamics of single-molecule diffusion inside a nanometric confined volume.Different methods were published describing the fabrication processes of such devices.25–29 The majority of them are methodically challenging, and single-molecule fluorescence experiments inside them may not be ‘reproducible’ due to nonuniform electrodynamic interactions with semiconductor–insulator or metal–insulator interfaces.28,30–33
Single-molecule fluorescence close to surfaces has a significant effect where the dielectric constant of a material defines the transition dipole moment.34–37 So, in nanometric confinement of a heterogeneous dielectric interface, dynamic movement of a single-molecule fluorescence signal will be inconsistent with respect to time. The fluctuation in the signal is due to dynamic orientation dependent modification of the transition dipole moment of single fluorophores. The one-sided quenching effect of a semiconductor–insulator interface shows significantly reduced emission intensity and lifetime variation of a dipole compared to a double-sided non-quenching insulator–insulator interface.32,33,38–40 For non-fluorescent single-molecule detection as well, a heterogeneous dielectric interface plays an important role since the molecular orbitals of the molecule are modified close to the surface.41 If the modification is uniform and reproducible irrespective of the molecules' orientation, this fundamental problem can be handled effectively. Uniform dielectric confinement is essential where single-molecules can be reliably identified based on their intrinsic photophysics. Therefore, we present all-silica nanochannels for multiplexed dynamic single-molecule detection of sub-3 nm-sized objects with molecular shot noise resolution.
One of the contemporary methods in our research is single-molecule recycling in a nanofluidic channel where Lesoine et al. used a 600 nm × 400 nm cross-sectional channel.42 The cross-section of the nanochannel in our case was intended to be less than the confocal diameter (given by the wavelength of the laser) to create a detection limited one-dimensional motion of single molecules inside the all-silica nanochannels of less than 100 nm cross-sectional diameter. Another notable achievement is sorting of single-molecules based on their charges by Krishnan and co-workers.43 However, their device manufacturing method since their first work on nanoparticle sorting in 2010 (ref. 44) has been a challenge for others. To date, except for them, only IBM Research (Zurich, Switzerland) reproduced a similar nanofabrication method for nanoparticle sorting in 2018.8 Such nanofabrication facilities may not be accessible to most of the academic research laboratories. Despite the technical complexity, this nanoengineering has led to single-molecule sorting to single-electron sensitivity, which is a paramount success. On that note, our work on resolving single-molecule shot noise at the nanometric level using a straightforward nanofabrication method is potentially the next avenue in this field.
The all-silica nanochannels with cross-sectional diameters ranging from 30 nm to 100 nm were used to quantify 1D flow and diffusion of single molecules, such as small DNA molecules labelled with single organic fluorophores, carbon nanodots (CNDs),45 and organic fluorophores by detecting their fluorescence, using two-focus fluorescence correlation spectroscopy (2fFCS).46–49 The unperturbed time-tagged photons due to uniform electrodynamic interaction lead to the extraction of molecular shot noise in monodispersed single-molecule concentrated liquid solutions. Dynamic 1D manipulation and 2fFCS of sub-3 nm-sized objects inside uniform dielectric nanofluidic confinement extend the single-molecule techniques towards single-molecule separation/sorting where a fundamental understanding of molecular shot-noise is required.
Fabrication of nanochannels
The process steps to create enclosed nanochannels involve fabrication of open nanochannels (trenches) using e-beam lithography (EBL) and reactive ion etching, and finally, closing them using shadow angle e-beam deposition (SAEBD). SAEBD utilises the ballistic path of the e-beam evaporation.50 When a collimated beam of an evaporated material hits an open nanotrench at a shallow incident angle, deposition does not occur in the shadowed region.51,52 Deposition on the exposed sites causes a growth of material that can enclose a large number of parallel trenches (depending on the beam diameter) leaving the shadowed regions free for a fluidic path. The process is unaffected by nanometre-sized residues of the e-beam resists and does not require an atomically clean surface, unlike any wafer bonding-based process.32 The nanochannels were prepared using pure silicon dioxide (silica, SiO2) to obtain a high photophysical signal to noise ratio of fluorophores. We demonstrate the SAEBD process by showing the SEM images of the intermediate steps. Here, we first demonstrate the process using silicon to obtain intermediate steps because silica shows low contrast in SEM due to its dielectric nature.Fig. 1a shows a schematic flowchart of creating nanochannels on Si[100] wafers (see the ESI† for sample preparation). The width of the nanotrenches can be optimised by the e-beam exposure of the EBL to the positive resist. Nanotrenches with different widths were created ranging from 30 nm to 100 nm; in Fig. 1b, 62 nm and 100 nm wide nanotrenches are shown, which were also previously used in reversible silicon-silica nanochannels.32 The lithographed e-beam resist acted as a mask for RIE to etch the final nanotrenches on silicon. We checked the depth of the trenches using AFM (ESI† Fig. S1).
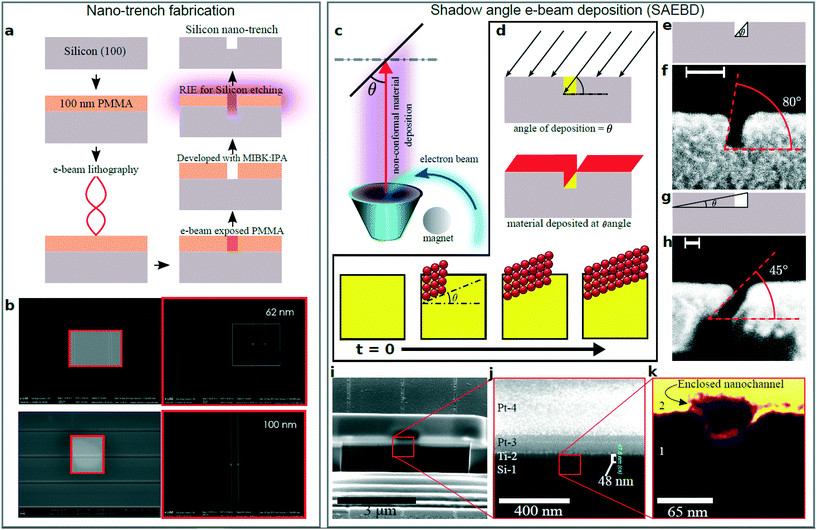 | ||
| Fig. 1 Nanochannel fabrication using SAEBD. a. Fabrication of nano-trenches on silicon with EBL and RIE. b. SEM images of the silicon nano-trenches with 62 nm and 100 nm width.32 c. SAEBD at angle θ. d. Shadows of electron beam. Arrows indicate angular e-beam evaporation. e. Schematic of high angle deposition and f. SEM image of deposition at 80°. g. Schematic of low angle deposition and h. SEM image of deposition at 45°. i. FIB cross-section of the enclosed nanochannels. Two layers of platinum were used to protect the nanochannel from high energy ions. j. Magnified view of the enclosed nanochannel. k. Further magnified view of the nanochannel where region 1 is silicon (Si-1) and region 2 is titanium (Ti-2). | ||
In the next step, SAEBD was used to enclose the nanotrenches to create closed nanochannels. Fig. 1c schematically shows the concept of the SAEBD process: a high-energy e-beam is bent by a magnet onto the reservoir of the material that sublimates and deposits on the substrate. The angular deposition (red) creates a shadowed region (yellow) that is unexposed to the depositing material depending on the deposition angle (θ) between the substrate and the vapour (Fig. 1d). To demonstrate this, an array of 5 mm long nanotrenches were cross-sectioned using a wafer sawing instrument to observe the intermediate steps while performing SAEBD. High angle deposition (Fig. 1e) has been used to show this effect; as shown in Fig. 1f, the inclined profile of the deposition is at 80°. As shown in the time evolution schematic, SAEBD closes the nanotrenches leaving a void – by decreasing θ, the unexposed area increases. At an acute angle close to 0°, the growth is nearly parallel to the surface of the substrate as shown in Fig. 1e–h. Fig. 1g schematically shows a low angle deposition and experimental demonstration of what is shown in Fig. 1h at 45°. Due to instrumental constraints, it was not possible to achieve a deposition at an acute angle close to 0°. Nevertheless, satisfactory results were obtained using θ = 45° as shown in Fig. 1i–k. Here, 60 nm titanium was deposited on the open nanotrenches at an angle of 45° with a deposition rate of 1 Å s−1 at a pressure of 2 × 10−6 mbar. We estimate that at an angle of 30°, a high-quality flat edge will be formed.
We used a focused ion beam (FIB) to investigate the cross-sections of the nanochannels. To avoid ion beam induced damage in the FIB, the top part of the cutting region was protected with thin metallic layers. We deposited two thin-films on the top surface of the enclosed nanochannels. Fig. 1i–k show the SEM images of the milled regions from low to high magnification. In Fig. 1j, the first layer (Si-1) is the silicon substrate on which nanotrenches were fabricated. The second layer is titanium (Ti-2) that was deposited at 45° by SAEBD. Layers Pt-3 and Pt-4 are platinum layers of 100 nm and 450 nm, respectively, which acted as protective layers to avoid FIB induced damage. In Fig. 1k, we observe the magnified cross-section of an enclosed nanochannel. As expected, SAEBD growth of titanium produces a well defined flat layer closing the nanochannel. The vertical thickness of the titanium film (47.8 nm) corresponds well with the experimental settings that were set to a deposition of 60 nm Ti at an angle of θ = 45°, which should result in a layer of 51 nm vertical thickness. After this proof of concept, we prepared fused silica-based nanofluidic devices (see the ESI†). The nanotrenches on the pure silica wafer were enclosed with SAEBD using pure silica at 45°. After SAEBD with silica, the substrate was annealed at 1000 °C for one hour to make sure that the evaporated silica has the same density as the substrate to obtain uniform dielectric constants at both sides.
Results and discussion
Nanofluidic device and 2fFCS detection scheme
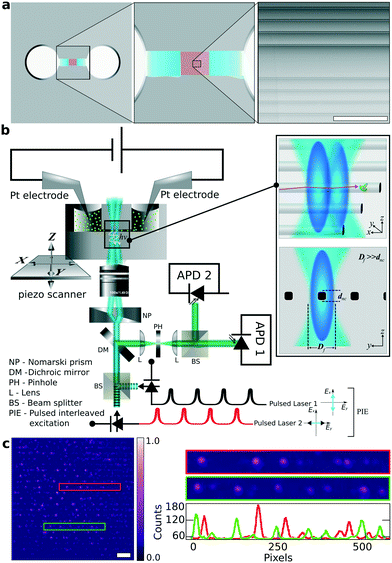
The design of the silica based nanofluidic device for performing single molecule experiments is schematically illustrated in Fig. 2a. Two reservoirs with a diameter of a few millimetres were sandblasted on the silica wafers using 70 μm silica particles before the nanofabrication process. They are separated by a distance of about 1 mm and serve as a convenient macroscopic inlet and outlet for the fluids. Each of these reservoirs is connected to an array of parallel 30 micrometre wide channels. These channels lead to either side of the final array of <100 nm wide channels. These nanochannels span over a length of l = 200 μm and connect to the microchannels. The height of the nanochannels and the microchannels is the same and they are etched in the same step using RIE.In Fig. 2a, the white and blue regions indicate the reservoir and the microchannels, respectively (ESI† Fig. S2–S4). The red stripes correspond to the array of nanochannels that are connected to the microchannels. An SEM image of these silica nanochannels is shown in the right inset of Fig. 2a. For the experiment, we filled the reservoirs with a dilute solution of fluorescent probes in the buffer. After filling one reservoir, capillary force transported the fluid through the microchannels and into the nanochannel reservoir. A relaxation time of 30 s was given to avoid the development of trapped air bubbles between two inlets before filling the second reservoir. Two 100 μm thick platinum electrodes were immersed in the reservoirs (ESI† Fig. S5), and an electric field was applied along the nanochannel. This created an electroosmotic flow53–56 that unidirectionally transported the fluid through the nanochannels as shown in Fig. 2b.
The intensity distribution in Fig. 2c shows single AlexaFluor 647 molecules (Thermo Fisher, Massachusetts, USA) lined up horizontally in all the parallel nanochannels. To immobilise the molecules, the solvent was dried at room temperature leaving behind the molecules in the channels. The photon count profile shows an average SNR of 90. The image was captured using a wide-field optical microscope by exciting the molecules with a 640 nm CW-laser (Coherent Laser Systems GmbH, Göttingen, Germany). Besides the evidence from FIB and SEM, this also proves that the nanochannels are properly enclosed and no cross-links between nanochannels are observed, unlike our previous observation.32
To restrict unwanted surface adsorption, we used fluorescent molecules carrying the same charge as the nanochannels' wall. Pure silica is negatively charged above its isoelectric point (pH(I) = 2).57,58 The buffer's pH of 8.5 leads to a considerable amount of negative charges on the walls of the silica nanochannels.
Molecular shot noise-resolved single-molecule motions
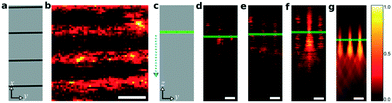
We used in-house 2fFCS47 (details in the ESI†) to study the dynamics of the molecules inside the nanochannels. The diffraction-limited focus of a laser beam in the visible range has a typical diameter of Df ≈ 300 nm to 500 nm. This is much larger than the width of the nanochannels, which is in the range of dnc ≈ 30 nm to 100 nm. Under these conditions, only a movement along the channel can be detected. Therefore, we can consider the flow inside a nanochannel as xtslquasi-1D. (Fig. 2a). In 2fFCS, two laser foci are used and are pulsed alternately with a rate of 20 MHz. By performing a temporal correlation of the signals from both foci, one can accurately determine the times that a fluorescent entity takes to move from one focus to the other. In our experiment, the displacement of the foci was ≈400 nm and they were carefully aligned with the direction of the nanochannel (Fig. 2b).47 An x–y–z piezo-scanner was used to move the device to the position of the foci and allows point measurements to be acquired as well as confocal scan images.Under the 2fFCS setup, since the nanochannels have no background fluorescence, one way to locate them is by filling them with a high concentration of fluorophores and obtain y–z confocal scan images to locate the focused plane. In this process, if we keep reducing the concentration of fluorophores to reach the single-molecule concentration, the longitudinal cross-sections of periodic point spread functions (PSFs) from the nanochannel array will turn from prominent elongated PSFs to jittery looking PSFs. The jittery nature of the PSFs is due to the movements of single molecules, which is not observable at a high concentration due to the high frequency of flowing molecules through the detection volume. The position of the maximum intensity in the cross-section PSFs refers to the focal plane. At the single-molecule concentration, this is an effective way to determine the focal plane (x–y) and the point of interest to perform 2fFCS. As the first probe of interest, we chose <2 nm-sized carbon nanodots,45 which were later renamed as graphene quantum dots (GQDs)59 and are negatively charged (see the ESI† Fig. S6). Fig. 3a shows a schematic top view of the device, where the nanochannels appear as dark lines. A confocal scan image of the x–y plane was recorded, while GQDs were flowing inside the nanochannels (Fig. 3b). The applied electric field to induce the electroosmotic flow during the measurement was 15 V mm−1. The pixel size of the image is 320 nm with a dwell time of 5 ms per pixel. This x–y confocal scan image is an important step to identify the nanochannels and point of interest for 2fFCS. We found the focused plane from the y–z scan, which also confirms the presence of dynamic molecular transport. Fig. 3c shows a scheme of a y–z section through the nanochannels. In Fig. 3d to g, we present a series of recorded y–z confocal scan images, with the volume percentage of GQDs relative to the stock solution increases from 1%, 2%, and 5% to 50%, respectively. Here, the pixel size was 100 nm with a dwell time of 2 ms per pixel. In these scans, we observe several periodic PSFs as fluorescence is emitted from sub-diffraction sized volumes. At low concentrations of GQDs, the fluorescence signals are strongly fluctuating due to the motion of the fluorophores out of the excitation focus. But as the concentration of GQDs increased, the fluctuations of the photon signal decreased and clear images of the PSFs were obtained. Here, we mark the focal plane with green lines where the intensities are maximum. We observed slightly more elongated and wider PSFs compared to standard single confocal PSFs because of two partially overlapping foci, which are orthogonally polarised from each other along with the SAEBD silica that may have a slightly different refractive index than that of the substrate. Since the GQDs were close to the size of organic fluorophores, it could not have been possible to measure the fast flow in a wide-field emCCD-based setup (ESI† Fig. S7 and the section emCCD-based detection).
Electroosmotic flow of single molecules
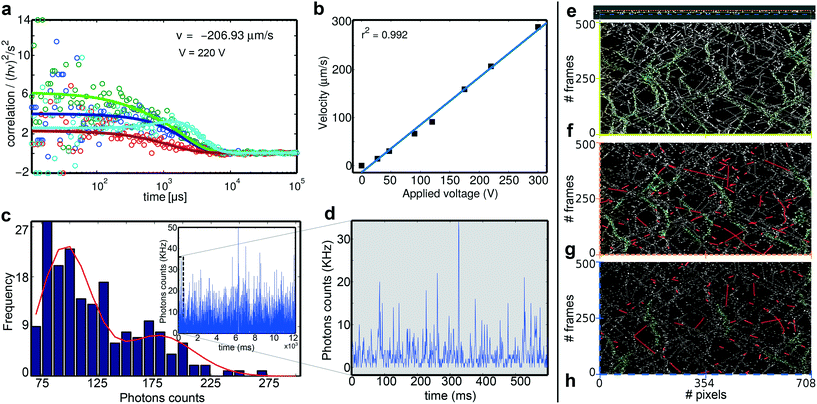
We investigated the flow velocities of single-molecules in the nanochannels using 2fFCS. In particular, we used the organic dye Atto 488 (Atto-Tec GmbH, Siegen, Germany) and the negatively charged 48 base-pair dsDNA labelled with AlexaFluor 647 (IBA GmbH, Göttingen, Germany). The molecules were diluted to a concentration of about 1 nM in aqueous buffer solution (see the ESI†). We measured the flow velocities and diffusion of the single molecules inside the nanochannels having a width of 30 nm. The effective electric field inside the nanochannel (due to the macro-, micro-, and nanochannels) configuration could not be characterised since experimentally, it was difficult to achieve at this stage. Hence, we report the calculated electric field considering an approximation of applied voltage along the reservoir, which is a mm apart. The applied electric fields range from 27 V mm−1 to 300 V mm−1 (see the ESI† Fig. S9). The temporal cross-correlation functions of the photon traces between the two excitation foci together with the well-known distance of the foci enable us to accurately determine the flow velocity of the fluorescent probes.48,49 The correlated data points from 2fFCS were fitted with the Fokker–Planck equation60 considering the 1D electroosmotic flow (see the ESI† Fig. S9 and S10). A challenge for these measurements is the tight spatial confinement of the molecules not only in the z plane but also in the y axis. To find the optimal position for the measurements, we always record a y–z confocal scan around the selected nanochannel (Fig. 3c–g), and the point that showed the highest number of photons in the scan was then chosen as the point of measurements. In wide-field microscopy (ESI† Movies S2 and S3), despite using negatively charged molecules, we have observed occasional adsorption molecules inside the nanochannels. To avoid obtaining artefacts from adsorbed molecules, before performing 2fFCS, we photobleached the point-of-interest chosen from the x–y and y–z confocal scan images using the confocal two-foci. The data recorded with the rare occurrence of adsorbed molecules where we observed constant background were not taken into account.An exemplary single-molecule 2fFCS measurement performed at 220 V mm−1 is shown in Fig. 4a. Here, light-blue and red curves are the fits to two cross-correlation data points – forward and backward. The peak of the light-blue curve corresponds to the unidirectional (forward) flow. The other two are fits to the forward and backward autocorrelation data. The noisy nature of the intensity correlation (hν2/s2) at a fast timescale (which is not important here) is due to fewer photon counts of single molecules at a fast flow compared to at a slow flow where the noise is reduced due to high photon counts. Despite the noisy correlation data, the cross-correlations and auto-correlations can be used to obtain the velocity and diffusion coefficients, respectively. The velocity and diffusion coefficients from the curve fitting are D = 1.51 × 10–7 cm2 s−1 and v = −207 μm s−1. The negative value of the flow velocity infers that the direction of the flow is from focus 2 to focus 1, which can be altered by changing the applied polarity of the electric field. The linear relationship of electroosmotic flows at different applied electric fields is plotted in Fig. 4b. The linear fit has an r2 value of 0.992 with error bars hidden inside the markers of the data-points. This suggests that the 1D electroosmotic flow measurement is reproducible. However, a fluctuation of the diffusion coefficient is observed in the range of 1.1 × 10−7 to 9.6 × 10−7 (ESI† Fig. S11). Could this be only because diffusion coefficient values are not accurate under these conditions since the driven flow dominates the dynamics of the molecules? However, we also considered other factors as discussed later.
To further investigate the diffusion behaviour, we analysed the photon burst sizes of the molecular transits.61Fig. 4c shows the burst size distribution (BSD) of the time trace of flow measurement. The time trace is shown in the inset. The first peak with the lowest photon number is due to the single-molecule transits. The higher peaks are due to more than one molecule coinciding in the focus or molecules crawling at the wall or their transient unspecific bindings. If the molecules move independently as single entities, the BSD is given by the Poissonian distribution. Fig. 4d shows the first 600 ms of the inset 5 ms binned time-trace of Fig. 4c, where we burst single-molecule transits.
Interpretation of confined diffusion
For theoretical interpretation, we simulated similar confined nanofluidic molecular interactions at room temperature in the LAMMPS platform62 (Fig. 4e, also see the ESI† Movie S3), where the walls of the nanochannel and particles have the same charge (details in the ESI†). The simulation is performed in 2D only to understand the interaction of the similarly charged particles and walls. The main motivation of this simulation was not to simulate a complete 30 nm wide nanochannel. However, the coarse-grain model and the computational efficiency of LAMMPS enable much larger systems63–66 to be simulated in a parallel computer, which may seem impossible otherwise. We have simulated a nearly 10 nm wide nanochannel i.e. three times smaller than our experimental nanochannel where the wall interactions are expected to be similar to the actual size. We performed kymographic analysis in Movie S3† to show the motion of the particles along the three lines inside the nanochannel over time. Fig. 4f–h show kymographic analysis at three different regions inside the nanochannel with characteristic variations near the walls and the centre of the nanochannel. Along the centre line of the nanochannel, discrete single events are observable along with successive multiple-molecule-events (highlighted in green in Fig. 4f). Molecular interactions near the wall (Fig. 4g) are dominated by a large number of crawling events where molecules were moving along the wall for an extended duration – ranging from 10 to 130 frames. These events are highlighted in red. In Fig. 4h, at the wall–fluid–solid interface, both the red and green events are not dominant. Within the diffraction-limited confocal detection volumes of 2fFCS, besides the molecular shot noise, the crawling events are another potential cause behind the second less frequent peak in the BSD analysis and variation in diffusion coefficients. Surface chemistry – a function of surface roughness – and ionic contaminants play an important role in electrokinetic transport at the nanometre length scale as observed by Golestanian and co-workers.67 The observation of molecular shot noise at the nanometric level using photon statistics of BSD analysis is a fundamental achievement. The reliability of observing molecular shot noise could not be possible if the electrodynamics of the molecules were unpredictable. The uniform electrodynamic interaction of the molecules with all-silica nanochannels led to this level of precision. Fig. 3 shows that controlling the number of the single molecules in the focal volume is dependent on the concentration of single molecules in the milli-reservoirs. At an ultra-low concentration, we still observe molecular shot noise, which is also supported by this MD simulation.Conclusion
In summary, this report presents a dynamic single-molecule nanofluidic detection method at the length scale where molecular interactions can be influenced by surface charge and dielectric confinement for photophysical artefacts. The 1D flow of single organic fluorophores, sub-2 nm GQDs, and 11 nm DNA molecules was achieved using electroosmosis inside the nanochannels at a single-molecule level due to the nanofabrication method and 2fFCS. A broad range of velocities up to 300 μm s−1 was achieved by varying the applied electric field. BSD analysis confirms that the observed transits were mainly due to single emitters, and such analysis at the nanometer length scale has not been reported earlier. The ability to resolve molecular shot noise is justified with BSD analysis and MD simulation, which were also compared earlier. Diffusion inside nanofluidic channels requires further investigation and opens another path of fundamental research. In the future, trapping nanoscale objects of less than 2 nm in size for a large residence time should be also feasible using these nanochannels. Biomolecular interactions with DNA, protein aggregation, and structural biology of molecules under physiological conditions can be also studied at the single-molecule level using the SAEBD based nanofluidic devices. In this work, we have presented single-molecule nanofluidics using fluorescent single molecules. However, fluorescent labelling does not limit the method. Several non-dissipative detection methods has potential possibilities inside these single-molecule nanofluidic device. Among them, we are currently working on resonance scattering and long-range interaction.68 The simple approach to nanofabrication, i.e. suitable for high-throughput and large scale production of the nanofluidic chip without restricting target materials, not only paves the way towards fundamentals of single-molecule nanofluidics but also opens up the possibility of detecting early onsets of diseases. Single-molecule handpicking techniques at room temperature will be a paramount advancement of nanoscience and engineering, and we envision that this work is a step towards that.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are thankful to the International Max Planck Research School for Physics of Biological and Complex Systems and the Ministry of Science and Culture (Lower Saxony) for funding SG and NK. The authors thank the internal funding from the University of Göttingen associated with the Third Institute of Physics for this project. Professor Jörg Enderlein has provided the original BSD algorithm and given countless important advice. The experiments were also held in Enderlein's laboratory. We cannot thank Prof Enderlein enough for his generosity and interest towards moving the field ahead. Dr Mitja Platen, Ditter Hille, and Falk Schneider have supported us in AFM, building the SAEBD stages, and buffer, respectively. The authors also thank Xuxing Lu (Single-Molecule Optics Group, Leiden) for the suggestions on the simulations.References
- A. E. Cohen, Phys. Rev. Lett., 2003, 91, 235506 CrossRef.
- A. E. Cohen and W. Moerner, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4362–4365 CrossRef CAS.
- N. J. Dovichi, J. C. Martin, J. H. Jett, M. Trkula and R. A. Keller, Anal. Chem., 1984, 56, 348–354 CrossRef CAS.
- D. C. Nguyen, R. A. Keller, J. H. Jett and J. C. Martin, Anal. Chem., 1987, 59, 2158–2161 CrossRef CAS.
- W. E. Moerner and L. Kador, Phys. Rev. Lett., 1989, 62, 2535 CrossRef CAS.
- P. M. Goodwin, M. E. Johnson, J. C. Martin, W. P. Ambrose, B. L. Marrone, J. H. Jett and R. A. Keller, Nucleic Acids Res., 1993, 21, 803–806 CrossRef CAS.
- P. V. Jones, G. L. Salmon and A. Ros, Anal. Chem., 2017, 89, 1531–1539 CrossRef CAS.
- M. J. Skaug, C. Schwemmer, S. Fringes, C. D. Rawlings and A. W. Knoll, Science, 2018, 359, 1505–1508 CrossRef CAS.
- J. C. Eijkel and A. Van Den Berg, Microfluid. Nanofluid., 2005, 1, 249–267 CrossRef CAS.
- J. K. Holt, H. G. Park, Y. Wang, M. Stadermann, A. B. Artyukhin, C. P. Grigoropoulos, A. Noy and O. Bakajin, Science, 2006, 312, 1034–1037 CrossRef CAS.
- P. Goswami and S. Chakraborty, Langmuir, 2009, 26, 581–590 CrossRef.
- R. H. Tunuguntla, R. Y. Henley, Y.-C. Yao, T. A. Pham, M. Wanunu and A. Noy, Science, 2017, 357, 792–796 CrossRef CAS.
- F. Persson and J. O. Tegenfeldt, Chem. Soc. Rev., 2010, 39, 985–999 RSC.
- S. K. Min, W. Y. Kim, Y. Cho and K. S. Kim, Nat. Nanotechnol., 2011, 6, 162 CrossRef CAS.
- B. M. Venkatesan and R. Bashir, Nat. Nanotechnol., 2011, 6, 615 CrossRef CAS.
- C. Wang, R. L. Bruce, E. A. Duch, J. V. Patel, J. T. Smith, Y. Astier, B. H. Wunsch, S. Meshram, A. Galan and C. Scerbo, et al. , ACS Nano, 2015, 9, 1206–1218 CrossRef CAS.
- A. Mitra, B. Deutsch, F. Ignatovich, C. Dykes and L. Novotny, ACS Nano, 2010, 4, 1305–1312 CrossRef CAS.
- M. N. Hamblin, J. Xuan, D. Maynes, H. D. Tolley, D. M. Belnap, A. T. Woolley, M. L. Lee and A. R. Hawkins, Lab Chip, 2010, 10, 173–178 RSC.
- J.-L. Fraikin, T. Teesalu, C. M. McKenney, E. Ruoslahti and A. N. Cleland, Nat. Nanotechnol., 2011, 6, 308 CrossRef CAS.
- A. de la Escosura-Muñiz and A. Merkoçi, Small, 2011, 7, 675–682 CrossRef.
- S. Faez, Y. Lahini, S. Weidlich, R. F. Garmann, K. Wondraczek, M. Zeisberger, M. A. Schmidt, M. Orrit and V. N. Manoharan, ACS Nano, 2015, 9, 12349–12357 CrossRef CAS.
- B. H. Wunsch, J. T. Smith, S. M. Gifford, C. Wang, M. Brink, R. L. Bruce, R. H. Austin, G. Stolovitzky and Y. Astier, Nat. Nanotechnol., 2016, 11, 936 CrossRef CAS.
- N. Kavokine, S. Marbach, A. Siria and L. Bocquet, Nat. Nanotechnol., 2019, 1 Search PubMed.
- A. Siria, M.-L. Bocquet and L. Bocquet, Nat. Rev. Chem., 2017, 1, 0091 CrossRef CAS.
- F. Persson, L. H. Thamdrup, M. B. L. Mikkelsen, S. Jaarlgard, P. Skafte-Pedersen, H. Bruus and A. Kristensen, Nanotechnology, 2007, 18, 245301 CrossRef.
- D. C. Bien, H. W. Lee and R. M. Saman, ECS Solid State Lett., 2012, 1, P45–P47 CrossRef CAS.
- S. H. Kim, Y. Cui, M. J. Lee, S.-W. Nam, D. Oh, S. H. Kang, Y. S. Kim and S. Park, Lab Chip, 2011, 11, 348–353 RSC.
- F. Westerlund, F. Persson, A. Kristensen and J. O. Tegenfeldt, Lab Chip, 2010, 10, 2049–2051 RSC.
- P. Mao and J. Han, Lab Chip, 2005, 5, 837–844 RSC.
- R. Hildner, D. Brinks, J. B. Nieder, R. J. Cogdell and N. F. van Hulst, Science, 2013, 340, 1448–1451 CrossRef CAS.
- E. Wientjes, J. Renger, A. G. Curto, R. Cogdell and N. F. Van Hulst, Nat. Commun., 2014, 5, 4236 CrossRef CAS.
- S. Ghosh, M. Kumbhakar, M. Platen, I. Gregor and J. Enderlein, Single Molecule Spectroscopy and Superresolution Imaging VII, 2014, p. 895008 Search PubMed.
- H. Cheng, PhD. thesis, Georg-August-Universität Göttingen, 2017 Search PubMed.
- N. Karedla, S. C. Stein, D. Hähnel, I. Gregor, A. Chizhik and J. Enderlein, Phys. Rev. Lett., 2015, 115, 173002 CrossRef.
- S. Ghosh, M. Ghosh, M. Seibt and G. M. Rao, Nanoscale, 2016, 8, 2632–2638 RSC.
- A. S. Backer, M. Y. Lee and W. Moerner, Optica, 2016, 3, 659–666 CrossRef CAS.
- J. Z. Low, G. Kladnik, L. L. Patera, S. Sokolov, G. Lovat, E. Kumarasamy, J. Repp, L. M. Campos, D. Cvetko and A. Morgante, et al. , Nano Lett., 2019, 19, 2543–2548 CrossRef CAS.
- S. Kühn and V. Sandoghdar, Appl. Phys. B: Lasers Opt., 2006, 84, 211–217 CrossRef.
- S. Kühn, G. Mori, M. Agio and V. Sandoghdar, Mol. Phys., 2008, 106, 893–908 CrossRef.
- J.-W. Liaw, J.-H. Chen, C.-S. Chen and M.-K. Kuo, Opt. Express, 2009, 17, 13532–13540 CrossRef CAS.
- J. S. Meisner, S. Ahn, S. V. Aradhya, M. Krikorian, R. Parameswaran, M. Steigerwald, L. Venkataraman and C. Nuckolls, J. Am. Chem. Soc., 2012, 134, 20440–20445 CrossRef CAS.
- J. F. Lesoine, P. A. Venkataraman, P. C. Maloney, M. E. Dumont and L. Novotny, Nano Lett., 2012, 12, 3273–3278 CrossRef CAS.
- F. Ruggeri, F. Zosel, N. Mutter, M. Różycka, M. Wojtas, A. Ożyhar, B. Schuler and M. Krishnan, Nat. Nanotechnol., 2017, 12, 488 CrossRef CAS.
- M. Krishnan, N. Mojarad, P. Kukura and V. Sandoghdar, Nature, 2010, 467, 692–695 CrossRef CAS.
- S. Ghosh, A. M. Chizhik, N. Karedla, M. O. Dekaliuk, I. Gregor, H. Schuhmann, M. Seibt, K. Bodensiek, I. A. Schaap and O. Schulz, et al. , Nano Lett., 2014, 14, 5656–5661 CrossRef CAS.
- M. Brinkmeier, K. Dörre, J. Stephan and M. Eigen, Anal. Chem., 1999, 71, 609–616 CrossRef CAS.
- T. Dertinger, V. Pacheco, I. von der Hocht, R. Hartmann, I. Gregor and J. Enderlein, ChemPhysChem, 2007, 8, 433–443 CrossRef CAS.
- T. Dertinger, A. Loman, B. Ewers, C. B. Müller, B. Krämer and J. Enderlein, Opt. Express, 2008, 16, 14353–14368 CrossRef.
- T. J. Arbour and J. Enderlein, Lab Chip, 2010, 10, 1286–1292 RSC.
- J. Merkel, T. Sontheimer, B. Rech and C. Becker, J. Cryst. Growth, 2013, 367, 126–130 CrossRef CAS.
- S. Egger, A. Ilie, S. Machida and T. Nakayama, Nano Lett., 2007, 7, 3399–3404 CrossRef CAS.
- D. M. Mattox, Handbook of physical vapor deposition (PVD) processing, William Andrew, 2010 Search PubMed.
- X. Huang, M. J. Gordon and R. N. Zare, Anal. Chem., 1988, 60, 1837–1838 CrossRef CAS.
- N. A. Patankar and H. H. Hu, Anal. Chem., 1998, 70, 1870–1881 CrossRef CAS.
- N. J. Dovichi and J. Zhang, Angew. Chem., Int. Ed., 2000, 39, 4463–4468 CrossRef CAS.
- S. Chakraborty and A. K. Srivastava, Langmuir, 2007, 23, 12421–12428 CrossRef CAS.
- M. Kosmulski, Chemical properties of material surfaces, CRC press, 2001 Search PubMed.
- M. Keswani, S. Raghavan, P. Deymier and S. Verhaverbeke, Microelectron. Eng., 2009, 86, 132–139 CrossRef CAS.
- S. Ghosh, M. Awasthi, M. Ghosh, M. Seibt and T. A. Niehaus, 2D Materials, 2016, vol. 3, p. 041008 Search PubMed.
- H. Risken, in The Fokker-Planck Equation, Springer, 1996, pp. 63–95 Search PubMed.
- J. Enderlein, D. L. Robbins, W. P. Ambrose and R. A. Keller, J. Phys. Chem. A, 1998, 102, 6089–6094 CrossRef CAS.
- S. Plimpton, J. Comput. Phys., 1995, 117, 1–19 CrossRef CAS.
- H. Liu and G. Cao, J. Phys. Chem. C, 2014, 118, 25223–25233 CrossRef CAS.
- A. Loya, J. L. Stair and G. Ren, Int. Nano Lett., 2015, 5, 1–7 CrossRef CAS.
- F. Vargas-Lara, S. M. Stavis, E. A. Strychalski, B. J. Nablo, J. Geist, F. W. Starr and J. F. Douglas, Soft Matter, 2015, 11, 8273–8284 RSC.
- M. Ghosh, S. Ghosh, H. Attariani, K. Momeni, M. Seibt and G. Mohan Rao, Nano Lett., 2016, 16, 5969–5974 CrossRef CAS.
- S. Ebbens, D. Gregory, G. Dunderdale, J. Howse, Y. Ibrahim, T. Liverpool and R. Golestanian, EPL, 2014, 106, 58003 CrossRef.
- S. Ghosh, Nanofluidic and Nanophotonic Study of Biomolecular Interactions, Deutsche Forschungsgemeinschaft Projekt number 405479535, 2018, DFG Research Proposal Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Contains the methods of nanofabrication, integrating the nanofluidic device to electrodes, CND/GQD interaction behaviour, emCCD-based measurements, description of the 2fFCS setup and fitting function, controlled 1D diffusion of single Atto 488 fluorophores, electro-osmotic flow and diffusion of single DNA molecules, and LAMMPS simulation. See DOI: 10.1039/d0lc00398k |
| ‡ Present address: Maxwell Centre, Cavendish Laboratory and Centre for Misfolding Diseases, Department of Chemistry, St Johns College, University of Cambridge, Cambridge, UK. Leiden Institute of Physics, Leiden University, The Netherlands. Open Academic Research Group, Cambridge, UK, email: E-mail: sg915@cam.ac.uk. |
| § Present address: Physical and Theoretical Chemistry, University of Oxford, Oxford, UK. |
| This journal is © The Royal Society of Chemistry 2020 |