Open Access Article
Open Access ArticleStructural characterization of a homopolysaccharide with hypoglycemic activity from the roots of Pueraria lobata†
Kai
Qian‡
ab,
Ting
Tan‡
 ab,
Hui
Ouyang
ab,
Shi-Lin
Yang
c,
Wei-Feng
Zhu
a,
Rong-Hua
Liu
a,
Quan
Wen
ab,
Hui
Ouyang
ab,
Shi-Lin
Yang
c,
Wei-Feng
Zhu
a,
Rong-Hua
Liu
a,
Quan
Wen
 *ab and
Yu-Lin
Feng
*abc
*ab and
Yu-Lin
Feng
*abc
aJiangxi University of Traditional Chinese Medicine, Nanchang 330006, Jiangxi, PR China. E-mail: qwen12@fudan.edu.cn
bThe National Pharmaceutical Engineering Center (NPEC) for Solid Preparation in Chinese Herbal Medicine, Nanchang 330006, Jiangxi, PR China. E-mail: fengyulin@126.com
cState Key Laboratory of Innovative Drug and Efficient Energy-Saving Pharmaceutical Equipment, Nanchang 330006, Jiangxi, PR China
First published on 3rd August 2020
Abstract
A water-soluble neutral homopolysaccharide (PLP-1) was obtained from the roots of Pueraria lobata by DEAE cellulose and Sephadex G-200 gel chromatography purification. The average molecular weight of PLP-1 was 16.2 kDa. Monosaccharide composition analysis showed that PLP-1 was composed of glucose as a glucan. The structure of PLP-1 was characterized on the basis of extensive physical and chemical analysis, which indicated that the backbone of PLP-1 was mainly composed of →3)-α-D-Glcp(1→ and →4)-β-D-Glcp(1→ with a molar ratio of 7.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0. Moreover, the hypoglycemic activity of PLP-1 was investigated by palmitic acid and high glucose induced insulin resistant HepG2 cells. The results elucidated that PLP-1 could decrease the glucose concentration by up-regulating the expression of PI3K and AKT, and down-regulating the expression of FoxO1, PCK2, and G6Pase in insulin resistant cells. Therefore, PLP-1 could serve as a dietary supplement to ameliorate insulin resistance for diabetic patients.
1.0. Moreover, the hypoglycemic activity of PLP-1 was investigated by palmitic acid and high glucose induced insulin resistant HepG2 cells. The results elucidated that PLP-1 could decrease the glucose concentration by up-regulating the expression of PI3K and AKT, and down-regulating the expression of FoxO1, PCK2, and G6Pase in insulin resistant cells. Therefore, PLP-1 could serve as a dietary supplement to ameliorate insulin resistance for diabetic patients.
1. Introduction
Diabetes mellitus, one of the most universal chronic diseases, has been emerging as a major health burden in the 21st century since its incidence has risen rapidly in the past few decades.1 415 million people worldwide were suffering from diabetes in 2014, and it is predicted that the number of patients will reach 642 million by 2040.2 More than 90% of the diabetic population are diagnosed as type II diabetes mellitus (T2DM), which is characterized by chronic hyperglycemia with disturbances of carbohydrate, fat and protein metabolism due to insulin resistance (IR) and/or inadequate insulin secretion.3 The mechanistic studies of IR have mainly drawn attention to improving IR due to its central process in the pathogenesis of T2DM.4 Currently, there are some oral hypoglycemic medications on the market, such as biguanides, sulfonylureas and thiazolidinediones, but these synthetic agents are associated with poor compliance and certain adverse effects.5 Therefore, natural edible plant products with low toxicity and hypoglycemic effect are attractive to many researchers.6,7Pueraria lobata (Willd.) Ohwi, belonging to the genus Pueraria of the family Leguminosae, is one of the earliest known and most important edible vine herbs in most parts of China.8 Biological studies have indicated that roots of P. lobata have potential effects, exhibiting antidiabetic,9 antihypertensive,10 antioxidative,11 antiosteoporosis12 and immunoregulatory13 activities. Aqueous extract of P. lobata has been used by local Chinese people and hospitals in folk medicine for the treatment of diabetes mellitus, or as an ingredient for many Chinese prescriptions for anti-diabetes formulations,14 but the active constituents of aqueous extract of P. lobata are ambiguous. Interestingly, it has been reported that the crude polysaccharides from roots of P. lobata have potential for treatment of T2DM in rats.15 However, which kind of homopolysaccharides with particular structures from such crude polysaccharides that exert pharmacological activity is still unclear.
Therefore, the aim of this study was to isolate and purify active hypoglycemic homopolysaccharides from water extract of roots of P. lobata, and to elucidate the structural characteristics. Furthermore, the potential mechanism of action was investigated using insulin resistant HepG2 cells.
2. Materials and methods
2.1. Materials and reagents
The roots of P. lobata were purchased from Zhangshu, Jiangxi province, China. The sample (P. lobata) was identified by Professor Fei Ge at School of Pharmacy, Jiangxi University of Traditional Chinese Medicine, Nanchang, China, where a voucher specimen (no. PL-190210-07) is deposited. Palmitic acid, metformin hydrochloride and glucose assay kit were purchased from Solarbio Science & Technology Co. Ltd (Beijing, China). DEAE-52 cellulose and Sephadex G-200 were purchased from Yuanye Bio-Technology Co. Ltd (Shanghai, China). Dextran standards and monosaccharide standards were purchased from National Institutes for Food and Drug Control. Dimethyl sulfoxide (DMSO) and iodomethane were purchased from Chemical Technology Co. Ltd (Shanghai, China). Sodium hydride and potassium bromide were purchased from Aladdin Reagent Co. Ltd (Shanghai, China). Deuterium oxide (D2O) was purchased from Qiaoyi Biotechnology Co. Ltd (Shanghai, China). Ultrapure water was prepared from a Milli-Q water purification system (Millipore, MA, USA). All other chemicals were of analytical grade. TRIzol was purchased from ThermoFisher Scientific Inc. (Invitrogen, USA). cDNA synthesis kit and TransStart Tip Green qPCR kit were purchased from TransGen Biotechnology Co. Ltd (Beijing, China). β-Actin, PI3K, GSK-3β, FoxO1, PCK2 primary antibody and lgG secondary antibody were purchased from Cell Signaling Technology (Boston, USA). AKT and G6Pase primary antibody were obtained from Abcam (Cambridge, UK).2.2. Extraction and purification of polysaccharides
The method of water extraction and alcohol precipitation in our laboratory was used to extract crude polysaccharides.16 Briefly, the dried roots of P. lobata (500 g) were crushed into coarse powder and extracted twice with 10 times the amount of deionized water for 3 h each time by a reflux device. All water extracts were combined and concentrated to about 1.0 L using a rotavapor vacuum concentrator (EYELA N-1300, Tokyo, Japan), which was then precipitated with anhydrous ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v, 3 L) at 4 °C for 12 h, followed by centrifugation at 4000g for 10 min using a centrifuge (Xiang li CTL550, Hunan, China). The precipitate dissolved in 400 mL of ultrapure water was then centrifuged at 4000g for 10 min. All supernatant liquids were collected and concentrated to about 200 mL, which was then lyophilized at −80 °C (FD-1C-80, Shanghai, China) to obtain crude polysaccharide PLP (10.2 g). PLP was dissolved in distilled water, then loaded on macroporous resin HP-20 (Mitsubishi, Japan) to remove proteins and pigments, which was eluted with deionized water by decompression filtration. Eluent was concentrated to about 100 mL, followed by centrifugation at 4000g for 10 min. All supernatant liquids were eluted by distilled water on DEAE-52 cellulose column until phenol–sulfuric acid reaction17 showed a colorless result. After that, the fraction was further purified by Sephadex G-200 chromatography,18 eluted with deionized water at a flow rate of 1.0 mL min−1 to obtain PLP-1 (1.2 g). The homogeneity of PLP-1 was determined by HPGPC-RID methods (Fig. S1†).
3, v/v, 3 L) at 4 °C for 12 h, followed by centrifugation at 4000g for 10 min using a centrifuge (Xiang li CTL550, Hunan, China). The precipitate dissolved in 400 mL of ultrapure water was then centrifuged at 4000g for 10 min. All supernatant liquids were collected and concentrated to about 200 mL, which was then lyophilized at −80 °C (FD-1C-80, Shanghai, China) to obtain crude polysaccharide PLP (10.2 g). PLP was dissolved in distilled water, then loaded on macroporous resin HP-20 (Mitsubishi, Japan) to remove proteins and pigments, which was eluted with deionized water by decompression filtration. Eluent was concentrated to about 100 mL, followed by centrifugation at 4000g for 10 min. All supernatant liquids were eluted by distilled water on DEAE-52 cellulose column until phenol–sulfuric acid reaction17 showed a colorless result. After that, the fraction was further purified by Sephadex G-200 chromatography,18 eluted with deionized water at a flow rate of 1.0 mL min−1 to obtain PLP-1 (1.2 g). The homogeneity of PLP-1 was determined by HPGPC-RID methods (Fig. S1†).
2.3. Structural characterization of PLP-1
The total sugar was determined by the phenol-sulfuric acid method with D-glucose as standard.20 The content of protein was measured by the Bradford method with bovine serum albumin as standard.21
PLP-1 (10 mg) was dissolved in a reaction flask with anhydrous DMSO (2 mL), and placed on a magnetic agitator with nitrogen protection. Then the reaction solution (2.5 mL) was injected slowly and reacted at room temperature for 30 min. Afterwards, iodomethane was added to the reaction flask in an ice bath for 3 hours. After that, the resulting solution was dialyzed using 3500 Da dialysis membrane (MD 44, Solarbio, China) in flowing water for 48 h and deionized water for 24 h. The products of complete methylation were hydrolyzed, reduced and acetylated according to the above monosaccharide composition and analyzed by GC-MS. MS analysis conditions: injection temperature was 280 °C, electron energy was 70 eV, the ion source temperature was 230 °C, scanning mode was selective ion detection, solvent was removed during 2.5 min.
2.4. Assay of hypoglycemic activity in vitro
2.5. Statistical analysis
Each cell culture experiment was performed at least three separate times. All values were expressed as (![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s). The experimental data were statistically analyzed by ANOVA, where P < 0.05 was assumed to be statistically significant. SPSS 19.0 software was used for statistical analysis.
± s). The experimental data were statistically analyzed by ANOVA, where P < 0.05 was assumed to be statistically significant. SPSS 19.0 software was used for statistical analysis.
3. Results and discussion
3.1. Isolation and purification
The crude polysaccharide (PLP) was obtained from the roots of P. lobata by the water-extraction and alcohol-precipitation method, and the yield was about 2.0%. PLP was then separated by a DEAE-52 cellulose column to obtain water-eluted fraction. The fraction was further purified by a Sephadex G-200 column to obtain pure sub-fraction named PLP-1. The HPGPC trace showed a single symmetrical peak (Fig. 1), which indicated that PLP-1 was a homogeneous polysaccharide. The average molecular weight was 16.2 kDa based on the equation of dextran standard curve (lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Mw = −0.4232t + 9.5597, R2 = 0.997). Moreover, the total sugar and protein contents of PLP-1 were determined to be 98.7% and 0.4%, respectively (Table S2†).
Mw = −0.4232t + 9.5597, R2 = 0.997). Moreover, the total sugar and protein contents of PLP-1 were determined to be 98.7% and 0.4%, respectively (Table S2†).
3.2. Monosaccharide composition and FT-IR spectrum
PLP-1 was decomposed by acid hydrolysis, and its alditol acetates were subjected to GC-MS for analysis. Compared with monosaccharide standards, the results showed (Fig. 2A) that PLP-1 was composed of glucose as a glucan. Moreover, FT-IR spectral analysis is an effective method to study the structure of polysaccharides owing to the characteristic absorption of functional groups. As shown in Fig. 2B, a major broad peak at around 3305 cm−1 was due to O–H stretching vibrations of hydroxyl group. The small band at 2923 cm−1 was attributed to the C–H stretching and bending vibrations. The peak at 1642 cm−1 was caused by the bending vibration of –OH. The absorption peak at 1361 cm−1 was attributed to variable angle vibrations of C–H.23,32 Furthermore, the absorption peaks at 1149 cm−1, 1078 cm−1 and 1017 cm−1 were ascribed to the stretching vibrations of pyranose ring.333.3. Methylation and NMR spectroscopy analysis
Methylation analysis was applied to determine the linkage forms among monosaccharide units of PLP-1. Methylated alditol acetate (PMAA) derivatives were formed after PLP-1 being fully methylated, hydrolyzed and acetylated, followed by GC-MS measurement. It could be concluded that PLP-1 mainly contained two glycosidic linkages, namely →3)-D-Glcp-(1→ and →4)-D-Glcp-(1→, with a relative molar ratio of 7.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 (Table 1) based on analysis of PMAA peak retention time (Fig. S2†) and mass fragments described in the literature.34,35
1.0 (Table 1) based on analysis of PMAA peak retention time (Fig. S2†) and mass fragments described in the literature.34,35
| PMAA | Linkage type | Molar ratio | Mass fragments (m/z) |
|---|---|---|---|
| 2,4,6-Me3-Glcp | →3)-D-Glcp-(1→ | 7.0 | 43, 87, 101, 118, 129, 174, 202, 234, 277 |
| 2,3,6-Me3-Glcp | →4)-D-Glcp-(1→ | 1.0 | 43, 59, 99, 118, 129, 159, 173, 187, 233 |
The structure of PLP-1 was further determined by NMR spectra which are shown in Fig. 3. The chemical shift and coupling constant of the anomeric proton signals were used to determine the α/β configurations of residues.36,37 Two anomeric proton signals (δH 5.30 and δH 4.55 ppm) were identified in the 1H-NMR spectrum (Fig. 3A), which were ascribed to two different residue types of residue A [→3)-Glcp-(1→] and residue B [→4)-Glcp-(1→] according to integration. δH 5.30 ppm (A/H-1) is inferred as α-configuration owing to a small coupling constant J = 2.0 Hz. δH 4.55 ppm (B/H-1) is inferred as β-configuration due to a large coupling constant J = 8.0 Hz. The two corresponding anomeric carbon signals δC 101.5 and 97.7 ppm were determined in the 13C-NMR spectrum (Fig. 3B) by the correlations from δH 5.30 to δC 101.5 and δH 4.55 to δC 97.7 on the basis of the HSQC spectrum (Fig. 3C).
In the HMBC spectrum (Fig. 3D), a cross-peak signal from δH 5.30 (A/H-1) to δC 78.6 (A/C-3) was assigned, which suggested a repeat unit →3)-α-D-Glcp(1→3)-α-D-Glcp(1→ as backbone. In addition, the correlation between δH 3.56 (B/H-4) and δC 101.5 (A/C-1), δH 5.30 (A/H-1) and δC 78.1 (B/C-4) indicated that C-1 of residue A was linked to C-4 of residue B. Furthermore, in the DEPT-135° spectrum (Fig. S5†), PLP-1 had negative methylene signals at δC 62.3 ppm without downfield shift, indicating that there was an absence of linkage at C-6 position of residues A and B, which were consistent with results of methylation. Thus, the possible structure of PLP-1 was deduced (Fig. S8†). Additionally, the signals for carbon and hydrogen for PLP-1 are fully shown in Table 2 according to the HMBC spectrum and the literature.36–40
| Sugar residue | Chemical shift (ppm) | |||||
|---|---|---|---|---|---|---|
| C1/H1 | C2/H2 | C3/H3 | C4/H4 | C5/H5 | C6/H6 | |
| →3)-α-D-Glcp(1→(A) | 101.5 | 73.1 | 78.5 | 71.2 | 73.5 | 62.3 |
| 5.30 | 3.86 | 3.87 | 3.73 | 3.75 | 3.84/3.52 | |
| →4)-β-D-Glcp(1→(B) | 97.7 | 75.3 | 75.9 | 78.1 | 74.6 | 62.3 |
| 4.55 | 3.24 | 3.67 | 3.56 | 3.31 | 3.82/3.53 | |
3.4. AFM analysis
AFM is a powerful technique to characterize the microstructure of polysaccharides.39,41 Therefore, AFM was utilized to observe the molecular morphology of PLP-1. As shown in Fig. 4, the 2D images demonstrated that the polysaccharide rings of PLP-1 were closely arranged due to intermolecular van der Waals forces, and the polysaccharide chains were also closely twisted together to form a structure of equally distributed clusters. The surface roughness of PLP-1 ranged from approximately −1.3 to 1.2 nm.3.5. Effect of PLP-1 on glucose content in the IR cell models
When cells were treated with 0.5 mM PA and 30 mM Glu for 8 h, the glucose content was increased significantly compared with the normal group (P < 0.01), indicating that establishment of insulin resistance model was due to the accumulation of PA. MTT assay results of PLP-1 (Fig. 5A) showed that there was no cytotoxicity at a concentration of 1.925–30.8 μM. To assess the effect of PLP-1 on reversing insulin resistance, the IR cells were incubated with PLP-1 (30.8 μM). In Fig. 5B, compared with model group, PLP-1 obviously improved glucose content in the IR HepG2 cells. At a concentration of 30.8 μM, PLP-1 showed an obvious glucose content decrease which was nearly comparable to the normal group. Therefore, PLP-1 (30.8 μM) was chosen for the following mechanism study.3.6. PLP-1 activated the PI3K/AKT signal pathway in IR HepG2 cells
It is generally acknowledged that the PI3K/AKT signaling pathway plays a crucial role in the regulation of glucose metabolism, including glucose transport, glycogen synthesis and gluconeogenesis suppression.42 PI3K is considered as a second messenger, playing an important role in cellular chemical signal transfer. Activated PI3K can phosphorylate AKT, thus activating AKT.43 GSK-3β is a negative regulatory gene in glycogen synthesis, which is regulated by AKT. In the state of insulin resistance, AKT phosphorylation is weakened, the activity of GSK-3β is strong, which inhibits the activity of glycogen synthase, thus inhibiting glycogen synthesis. On the contrary, activated AKT can reduce the expression of GSK-3β and promote glycogen synthesis.44,45 Besides, AKT stimulation can restrain gluconeogenesis by inhibiting PCK2 and G6Pase expressions and phosphorylating FoxO1.46 Therefore, the PI3K/AKT signaling pathway plays an exceedingly significant role in mediating insulin metabolism and is closely correlated with insulin resistance.In recent years, IR HepG2 cell models have been used to study the hypoglycemic activities and mechanisms of plant polysaccharides. For instance, the polysaccharide EFSP-1 from Euryale ferox increased the expression of IRS-1, PI3K, AKT and GLUT4 in mRNA and protein levels in IR HepG2 cells.32 In addition, previous research reported that polysaccharides LSP, OJP and LMP from herbs Maidong upregulated the levels of PI3K, AKT, InsR, and PPARγ and downregulated the levels of PTP1B in mRNA and protein expression in IR HepG2 cells.23
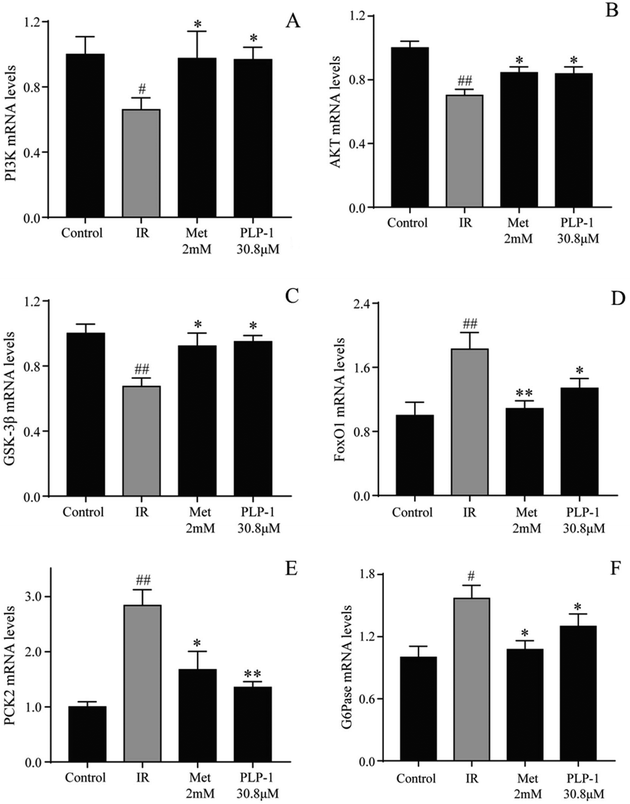
To determine whether the PI3K/AKT signal pathway was affected by PLP-1 treatment in IR HepG2 cells, the expressions of main protein and mRNA in glucose transport pathway including PI3K, AKT, GSK-3β, FoxO1, PCK2 and G6Pase were investigated. As shown in Fig. 6, the downregulation of mRNA expression of PI3K, AKT, and GSK-3β was observed in the model group, while the upregulation of mRNA expression of FoxO1, PCK2 and G6Pase was observed in the model group, indicating that there was insulin resistance in HepG2 cells. When treated with PLP-1 and Met, both groups obviously increased the expression of PI3K, AKT, and GSK-3β and decreased the expression of FoxO1, PCK2, and G6Pase in mRNA levels as compared to the model group. Meanwhile, both groups also significantly increased the expression of PI3K, AKT, and GSK-3β and decreased the expression of FoxO1, PCK2, and G6Pase in protein levels as compared to the IR control (Fig. 7). Furthermore, to confirm the role of FoxO1 in gluconeogenesis stimulated by PLP-1 and Met, the expression of FoxO1 was demonstrated by immunofluorescence staining. As shown in Fig. 8, PLP-1 and Met obviously increased FoxO1 expression and enhanced nuclear-cytoplasmic translocation in HepG2 cells compared with the model group. These results indicated that IR HepG2 cells could be mitigated by treatment with PLP-1, the mechanism of which may be associated with the regulation of PI3K/AKT/FoxO1 signaling pathway to reduce the content of glucose.
 | ||
| Fig. 8 PLP-1 enhanced FoxO1 protein expression in IR HepG2 cells (control: normal group; IR: model group; Met: 2 mM; PLP-1: 30.8 μM). | ||
4. Conclusions
In this study, a novel water-soluble polysaccharide, PLP-1, was purified from the roots of P. lobata. PLP-1 is a glucan with a molecular weight of 16.2 kDa. The backbone of PLP-1 was mainly composed of →3)-α-D-Glcp-(1→ and →4)-β-D-Glcp-(1→ with a relative ratio of 7.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 and special spatial structure. PLP-1 could ameliorate adverse effects of glucose metabolism caused by PA and high glucose via down-regulating the expression of FoxO1, which might occur by activating the PI3K/AKT/FoxO1 signal pathway in IR HepG2 cells. These findings provide evidence that PLP-1 may serve as an alternative functional food or clinical adjuvant therapy to mitigate metabolic disorders with T2DM. However, the hypoglycemic effect and underlying mechanisms of PLP-1 require further verification in vivo.
1.0 and special spatial structure. PLP-1 could ameliorate adverse effects of glucose metabolism caused by PA and high glucose via down-regulating the expression of FoxO1, which might occur by activating the PI3K/AKT/FoxO1 signal pathway in IR HepG2 cells. These findings provide evidence that PLP-1 may serve as an alternative functional food or clinical adjuvant therapy to mitigate metabolic disorders with T2DM. However, the hypoglycemic effect and underlying mechanisms of PLP-1 require further verification in vivo.
Funding
This work was supported by the National Natural Science Foundations of China under grant number 81860684 and the National Key R&D Program of China (numbers 2019YFC1712300 and 2017YFC1702902).Conflicts of interest
No potential conflict of interest was reported by the authors.References
- K. Ogurtsova, J. D. da Rocha Fernandes, Y. Huang, U. Linnenkamp, L. Guariguata, N. H. Cho, D. Cavan, J. E. Shaw and L. E. Makaroff, IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040, Diabetes Res. Clin. Pract., 2017, 128, 40–50 CrossRef CAS PubMed.
- L. M. Jaacks, K. R. Siegel, U. P. Gujral and K. M. V. Narayan, Type 2 diabetes: A 21st century epidemic, Best Pract. Res., Clin. Gastroenterol., 2016, 30, 331–343 CrossRef PubMed.
- W. Kerner and J. Brückel, Definition, classification and diagnosis of diabetes mellitus, Exp. Clin. Endocrinol. Diabetes, 2014, 122, 384–386 CrossRef CAS PubMed.
- K. P. Wang, Z. H. Tang, Z. M. Zheng, P. Cao, W. Z. Shui, Q. Li and Y. Zhang, Protective effects of Angelica sinensis polysaccharide against hyperglycemia and liver injury in multiple low-dose streptozotocin-induced type 2 diabetic BALB/c mice, Food Funct., 2016, 7, 4889–4897 RSC.
- C. Chen, Q. Huang, C. Li and X. Fu, Hypoglycemic effects of a Fructus Mori polysaccharide in vitro and in vivo, Food Funct., 2017, 8, 2523–2535 RSC.
- M. I. Kazeem and T. C. Davies, Anti-diabetic functional foods as sources of insulin secreting, insulin sensitizing and insulin mimetic agents, J. Funct. Foods, 2016, 20, 122–138 CrossRef CAS.
- J. Martel, D. M. Ojcius, C. J. Chang, C.-S. Lin, C. C. Lu, Y. F. Ko, S. F. Tseng, H. C. Lai and J. D. Young, Anti-obesogenic and antidiabetic effects of plants and mushrooms, Nat. Rev. Endocrinol., 2017, 13, 149 CrossRef CAS PubMed.
- K. H. Wong, G. Q. Li, K. M. Li, V. Razmovski-Naumovski and K. Chan, Optimisation of Pueraria isoflavonoids by response surface methodology using ultrasonic-assisted extraction, Food Chem., 2017, 231, 231–237 CrossRef CAS PubMed.
- J. K. Prasain, N. Peng, R. Rajbhandari and J. M. Wyss, The Chinese Pueraria root extract (Pueraria lobata) ameliorates impaired glucose and lipid metabolism in obese mice, Phytomedicine, 2012, 20, 17–23 CrossRef CAS PubMed.
- C. F. Ng, C. M. Koon, D. W. S. Cheung, M. Y. Lam, P. C. Leung, C. B. S. Lau and K. P. Fung, The anti-hypertensive effect of Danshen (Salvia miltiorrhiza) and Gegen (Pueraria lobata) formula in rats and its underlying mechanisms of vasorelaxation, J. Ethnopharmacol., 2011, 137, 1366–1372 CrossRef CAS PubMed.
- L. Bebrevska, K. Foubert, N. Hermans, S. Chatterjee, E. Van Marck, G. De Meyer, A. Vlietinck, L. Pieters and S. Apers, In vivo antioxidative activity of a quantified Pueraria lobata root extract, J. Ethnopharmacol., 2010, 127, 112–117 CrossRef PubMed.
- T. Tanaka, H. Tang, F. Yu, S. Michihara, Y. Uzawa, N. Zaima, T. Moriyama and Y. Kawamura, Kudzu (Pueraria lobata) vine ethanol extracts improve ovariectomy-induced bone loss in female mice, J. Agric. Food Chem., 2011, 59, 13230–13237 CrossRef CAS PubMed.
- Z. Dong, M. Zhang, H. Li, Q. Zhan, F. Lai and H. Wu, Structural characterization and immunomodulatory activity of a novel polysaccharide from Pueraria lobata (Willd.) Ohwi root, Int. J. Biol. Macromol., 2020, 154, 1556–1564 CrossRef PubMed.
- Y. Yu, M. Y. Shen, Q. Q. Song and J. H. Xie, Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review, Carbohydr. Polym., 2018, 183, 91–101 CrossRef CAS PubMed.
- C. C. Cai, X. U. Yan-Ying, H. X. Wang, S. Wang and D. O. Endocrinology, The study of therapeutic effect and mechanism of Puerariae Radix polysaccharides on type 2 diabetes mellitus rats, Tianjin J. Tradit. Chin. Med., 2014, 94–97, DOI:10.11656/j.issn.1672-1519.2014.02.09.
- J. J. Wen, H. Gao, J. L. Hu, Q. X. Nie, H. H. Chen, T. Xiong, S. P. Nie and M. Y. Xie, Polysaccharides from fermented Momordica charantia ameliorate obesity in high-fat induced obese rats, Food Funct., 2019, 10, 448–457 RSC.
- X. H. Yu, Y. Liu, X. L. Wu, L. Z. Liu and D. D. Song, Isolation, purification, characterization and immunostimulatory activity of polysaccharides derived from American ginseng, Carbohydr. Polym., 2016, 156, 9–18 CrossRef PubMed.
- Q. X. Yuan, L. Y. Zhao, Q. Q. Cha, Y. Sun, H. Ye and X. X. Zeng, Structural Characterization and Immunostimulatory Activity of a Homogeneous Polysaccharide From Sinonovacula Constricta, J. Agric. Food Chem., 2015, 63, 7986–7994 CrossRef CAS PubMed.
- M. A. M. Ghoneim, A. I. Hassan, M. G. Mahmoud and M. S. Asker, Effect of polysaccharide from Bacillus subtilis sp. on cardiovascular diseases and atherogenic indices in diabetic rats, BMC Complementary Altern. Med., 2016, 16, 112 CrossRef PubMed.
- N. Blumenkrantz and A. H. Gustav, New method for quantitative determination of uronic acids, Anal. Chem., 1973, 54, 484–489 CAS.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- J. Lehrfeld, Simultaneous gas-liquid chromatographic determination of aldonic acids and aldoses, Anal. Chem., 1985, 57, 346–348 CrossRef CAS.
- Y. J. Gong, J. Zhang, F. Gao, J. W. Zhou, Z. N. Xiang, C. G. Zhou, L. S. Wan and J. C. Chen, Structure features and in vitro hypoglycemic activities of polysaccharides from different species of Maidong, Carbohydr. Polym., 2017, 173, 215–222 CrossRef CAS PubMed.
- I. M. Sims, S. M. Carnachan, T. J. Bell and S. F. R. Hinkley, Methylation analysis of polysaccharides: Technical advice, Carbohydr. Polym., 2018, 188, 1–7 CrossRef CAS PubMed.
- C. L. Cao, C. Li, Q. Chen, Q. Huang, M. E. M. Pérez and X. Fu, Physicochemical characterization, potential antioxidant and hypoglycemic activity of polysaccharide from Sargassum pallidum, Int. J. Biol. Macromol., 2019, 139, 1009–1017 CrossRef CAS PubMed.
- Y. Yao, Y. Y. Zhu, Y. Gao, Z. X. Shi, Y. B. Hu and G. X. Ren, Suppressive effects of saponin-enriched extracts from quinoa on 3 T3-L1 adipocyte differentiation, Food Funct., 2015, 6, 3282–3290 RSC.
- F. J. Yan, G. H. Dai and X. D. Zheng, Mulberry anthocyanin extract ameliorates insulin resistance by regulating PI3K/AKT pathway in HepG2 cells and db/db mice, J. Nutr. Biochem., 2016, 36, 68–80 CrossRef CAS PubMed.
- Q. Zhang, X. J. Kong, H. Yuan, H. J. Guan, Y. Li and Y. C. Niu, Mangiferin Improved Palmitate-Induced-Insulin Resistance by Promoting Free Fatty Acid Metabolism in HepG2 and C2C12 Cells via PPARα: Mangiferin Improved Insulin Resistance, J. Diabetes Res., 2019, 2019, 2052675 Search PubMed.
- M. Nácher-Vázquez, N. Ballesteros, Á. Canales, S. Rodríguez Saint-Jean, S. I. Pérez-Prieto, A. Prieto, R. Aznar and P. López, Dextrans produced by lactic acid bacteria exhibit antiviral and immunomodulatory activity against salmonid viruses, Carbohydr. Polym., 2015, 124, 292–301 CrossRef PubMed.
- L. H. Chen, X. H. Sun, H. M. Xiao, F. T. Xu, Y. Yang, Z. Y. Lin, Z. Q. Chen, S. J. Quan and H. Q. Huang, PAQR3 regulates phosphorylation of FoxO1 in insulin-resistant HepG2 cells via NF-κB signaling pathway, Exp. Cell Res., 2019, 381, 301–310 CrossRef CAS PubMed.
- H. Matsuzaki, H. Daitoku, M. Hatta, K. Tanaka and A. Fukamizu, Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 11285–11290 CrossRef CAS PubMed.
- W. N. Zhang, R. N. Su, L. L. Gong, W. W. Yang, J. Chen, R. Yang, Y. Wang, W. J. Pan, Y. M. Lu and Y. Chen, Structural characterization and in vitro hypoglycemic activity of a glucan from Euryale ferox Salisb. seeds, Carbohydr. Polym., 2019, 209, 363–371 CrossRef CAS PubMed.
- Y. F. Gao, Y. B. Zhou, Q. Zhang, K. Zhang, P. Peng, L. Chen and B. Xiao, Hydrothermal extraction, structural characterization, and inhibition HeLa cells proliferation of functional polysaccharides from Chinese tea Zhongcha 108, J. Funct. Foods, 2017, 39, 1–8 CrossRef CAS.
- J. Y. Jiang, F. S. Kong, N. S. Li, D. Z. Zhang, C. Y. Yan and H. C. Lv, Purification, structural characterization and in vitro antioxidant activity of a novel polysaccharide from Boshuzhi, Carbohydr. Polym., 2016, 147, 365–371 CrossRef CAS PubMed.
- C. Xu, N. B. Qin, C. Y. Yan and S. M. Wang, Isolation, purification, characterization and bioactivities of a glucan from the root of Pueraria lobata, Food Funct., 2018, 9, 2644–2652 RSC.
- E. I. Nep, S. M. Carnachan, N. C. Ngwuluka, V. Kontogiorgos, G. A. Morris, I. M. Sims and A. M. Smith, Structural characterisation and rheological properties of a polysaccharide from sesame leaves (Sesamum radiatum Schumach. & Thonn.), Carbohydr. Polym., 2016, 152, 541–547 CrossRef CAS PubMed.
- K. J. Murphy, N. McLay and D. A. Pye, Structural Studies of Heparan Sulfate Hexasaccharides: New Insights into Iduronate Conformational Behavior, J. Am. Chem. Soc., 2008, 130, 12435–12444 CrossRef CAS PubMed.
- X. M. Wang, R. G. Sun, J. Zhang, Y. Y. Chen and N. N. Liu, Structure and antioxidant activity of polysaccharide POJ-U1a extracted by ultrasound from Ophiopogon japonicus, Fitoterapia, 2012, 83, 1576–1584 CrossRef CAS PubMed.
- J. K. Yan, Y. Y. Wang, Z. B. Wang, H. L. Ma, J. J. Pei and J. Y. Wu, Structure and antioxidative property of a polysaccharide from an ammonium oxalate extract of Phellinus linteus, Int. J. Biol. Macromol., 2016, 91, 92–99 CrossRef CAS PubMed.
- T. Zhao, G. H. Mao, W. W. Feng, R. W. Mao, X. Y. Gu, T. Li, Q. Li, Y. T. Bao, L. Q. Yang and X. Y. Wu, Isolation, characterization and antioxidant activity of polysaccharide from Schisandra sphenanthera, Carbohydr. Polym., 2014, 105, 26–33 CrossRef CAS PubMed.
- N. B. Liao, S. G. Chen, X. Q. Ye, J. J. Zhong, X. Ye, X. Z. Yin, J. Tian and D. H. Liu, Structural characterization of a novel glucan from Achatina fulica and its antioxidant activity, J. Agric. Food Chem., 2014, 62, 2344–2352 CrossRef CAS PubMed.
- E. L. Whiteman, H. Cho and M. J. Birnbaum, Role of Akt/protein kinase B in metabolism, Trends Endocrinol. Metab., 2002, 13, 444–451 CrossRef CAS PubMed.
- X. T. Feng, T. Z. Wang, J. Leng, Y. Chen, J. B. Liu, Y. Liu and W. J. Wang, Palmitate Contributes to Insulin Resistance through Downregulation of the Src-Mediated Phosphorylation of Akt in C2C12 Myotubes, Biosci., Biotechnol., Biochem., 2012, 76, 1356–1361 CrossRef CAS PubMed.
- M. Wan, K. F. Leavens, R. W. Hunter, S. Koren, A. von Wilamowitz-Moellendorff, M. Lu, S. Satapati, Q. Chu, K. Sakamoto, S. C. Burgess and M. J. Birnbaum, A noncanonical, GSK3-independent pathway controls postprandial hepatic glycogen deposition, Cell Metab., 2013, 18, 99–105 CrossRef CAS PubMed.
- C. Lipina, X. Huang, D. Finlay, E. J. McManus, D. R. Alessi and C. Sutherland, Analysis of hepatic gene transcription in mice expressing insulin-insensitive GSK3, Biochem. J., 2005, 392, 633–639 CrossRef CAS PubMed.
- P. J. Klover and R. A. Mooney, Hepatocytes: critical for glucose homeostasis, Int. J. Biochem. Cell Biol., 2004, 36, 753–758 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0fo01234c |
| ‡ Co-first author. |
| This journal is © The Royal Society of Chemistry 2020 |