Open Access Article
Open Access ArticleThe effect of quercetin on endothelial cells is modified by heterocellular interactions
Sarka
Tumova
 ab,
Michael J.
Houghton
ab,
Michael J.
Houghton
 ac and
Gary
Williamson
ac and
Gary
Williamson
 *ac
*ac
aUniversity of Leeds, Leeds, LS2 9JT, UK. E-mail: gary.williamson1@monash.edu
bBiology Center, Czech Academy of Sciences, Ceske Budejovice 37005, Czech Republic
cDepartment of Nutrition, Dietetics and Food, School of Clinical Sciences at Monash Health, Faculty of Medicine, Nursing and Health Sciences, Monash University, BASE Facility, 264 Ferntree Gully Road, Notting Hill, VIC 3168, Australia
First published on 4th May 2020
Abstract
Single cell-type models are useful for determining mechanisms, but in vivo, cell–cell interactions are important, and neighbouring cells can impact endothelial cell function. Quercetin can attenuate endothelial dysfunction by modulating vascular tone and reducing inflammation. We determined the effect of quercetin on a co-culture between Human Umbilical Vein Endothelial Cells (HUVEC) and human HepG2 hepatic cells or human LHCN-M2 muscle cells. Heme oxygenase-1 (HO-1) mRNA and protein were decreased, pyruvate dehydrogenase kinase (PDK) 4 and glucose transporter (GLUT) 3 mRNA increased, and GLUT1 protein decreased in HUVEC when cultured with HepG2. GLUT transporters, but not the other targets, were similarly regulated in co-culture with muscle cells. Some but not all of the effects were mediated by lactate and transforming growth factor β1. Quercetin added apically to the endothelial cells upregulated HO-1 and downregulated PDK4 both in monoculture and in co-culture, but the total PDK4 levels were higher in the presence of HepG2 cells. In the absence of general permeability changes, glucose transport across the endothelial monolayer was elevated in the presence of HepG2 cells, however this effect was moderated by quercetin applied on the apical side of the endothelial cells. At lower glucose concentration, apical quercetin also promoted glucose uptake in HepG2 cells. Co-culturing HUVEC with the HepG2 cells showed capacity to modulate quercetin-elicited changes in endothelial gene transcription and glucose transport.
1. Introduction
Endothelial cells fulfil a range of functions across the vascular beds in the body. They provide an interface between circulating blood and target tissue, provide access to nutrients, exchange gases, regulate blood flow and sustain local signalling to modulate physiological and pathophysiological responses, including inflammation or blood coagulation.1–3 Endothelial cell functionality is partly governed by the type of vessel, depending on whether they line artery, vein or capillary and, in particular for the latter, depends on the tissue microenvironment, which determines critical parameters such as mechanical forces and oxygen tension as well as hormonal and growth factor regulation. The properties of endothelial cells are finely tuned to their specific location4 and, starting from their emergence during development, they help to establish tissue-specific signalling and organ formation,5 promote regeneration6 and contribute to metabolic homeostasis in the tissues.7 Endothelium integrates signalling elicited by the circulating cells and molecules with the feedback from the adjacent tissues in order to secure adequate supply of nutrients and provide capacity for efficient organ maintenance and function.8 The context of the nascent environment can have a significant impact and is indispensable for endothelium-specific phenomena such as the nitric oxide (NO) mediated vasorelaxation, where the signalling between endothelial and smooth muscle cells in larger vessels is critical for the regulation of blood pressure.9 Similarly, the presence of astrocytes and pericytes is critical for blood-brain barrier integrity and maintenance.10 The communication between neighbouring cell types in capillaries can regulate endothelial cell proliferation and angiogenesis, and control access of growth factors and hormones to the tissues, as demonstrated for delivery of insulin to the skeletal muscle11,12 or modulation of hepatic regeneration and fibrosis.13 At the same time, interactions with cells in underlying tissues can modulate the effects of circulating molecules on the endothelial cells.Diet-derived small molecules such as polyphenols and their metabolites that circulate in the blood affect various aspects of endothelial function post-prandially. Intervention and epidemiological studies have found positive association of polyphenol-rich food with cardiovascular health benefits, including decrease in inflammatory markers and improved blood pressure.14–16 Further in vitro and animal studies attributed these benefits to pathways that improve redox balance and reduce inflammation,17 specifically via modulating endothelial nitric oxide synthase (eNOS) function, inhibiting NADPH oxidases (NOX) and other reactive oxygen species (ROS)-generating enzymes and stimulation of nuclear factor erythroid 2-related factor 2 (Nrf2) pathways.18,19 One of the best studied polyphenols is the flavonoid quercetin, with specific protective effects on the vasculature described in animal and in vitro studies. It can alleviate some consequences of atherogenic and inflammatory signalling, including endothelial damage induced by oxidized low-density lipoprotein (oxLDL) or angiotensin II, by activating SIRT1 and modulating NOX/eNOS activities,20 or by decreasing NOX subunit.21,22 In the ApoE−/− mouse model, quercetin attenuates atherosclerosis and improves NO bioavailability through heme oxygenase 1 (HO-1) regulation.23 It also improves redox balance by regulating oxidation, conjugation and export of glutathione in human aortic endothelial cells.24 Quercetin regulates blood pressure in rats and humans via local deconjugation of the plasma metabolite quercetin glucuronide.25,26 Quercetin can also downregulate adhesion molecules that attract monocytes during inflammation, such as vascular cell adhesion molecule 1 (VCAM1) or intercellular adhesion molecule 1 (ICAM1),27,28 and decrease attachment of monocytes to endothelial cells via Nrf2 activation.29 We have reported recently that quercetin can affect endothelial cell metabolism through hypoxia induced factor 1α (HIF1α) and the Nrf2 pathway.30 However, despite the fact that the endothelium in vivo functions as a part of a larger system, most of the in vitro models rely on single-type cell cultures.
In order to examine the impact of other cells on quercetin modulation of endothelial function to better mimic in vivo conditions, here we have used HUVEC as a model in non-contact co-cultures and studied the endothelial gene expression and glucose uptake and transport with and without quercetin. HUVEC were cultured in the presence of HepG2 cells or immortalized primary skeletal muscle cells to elucidate the impact on selected gene expression, and then the effects of plant-derived polyphenol quercetin on HUVEC were examined in mono-culture and co-culture with the carcinoma cells.
2. Methods
2.1 Cell culture and co-culture
HUVEC (Lonza Sales Ltd, Switzerland) were cultured in endothelial growth medium 2 (EGM-2) supplemented with EGM-2 BulletKit (Lonza) and used between passage 3–6. HepG2 cells (ATCC cat# HB-8065, LGC Promochem, Teddington, UK) were cultured in Eagle's Minimum Essential Medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 100 U ml−1 penicillin, 100 mg ml−1 streptomycin, 2% (v/v) non-essential amino acids, and 1% (v/v) sodium pyruvate (all from Sigma-Aldrich, Gillingham, UK). Muscle cells were immortalized skeletal muscle cells, LHCN-M2, a gift of Dr Vincent Mouly (Sorbonne Université, INSERM, Institute of Myology, Centre of Research in Myology, Unite Mixte de Recherche Scientifique 974, Paris, France).31 They were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Life Technologies, ThermoFisher, UK) supplemented with 5.5 mM glucose, 10% (v/v) FBS (heat-inactivated), 10% v/v heat-inactivated newborn calf serum (Gibco), hEGF (10 μg L−1; PeproTech, London, UK), hbFGF (1 μg L−1; Gibco) and dexamethasone (400 μg L−1) (Sigma), as described previously.32 Myocytes were differentiated upon confluency by switching to a differentiation medium (5.5 mM glucose in DMEM, 2% horse serum (Gibco)) for up to 4 days.32 For the co-cultures, HUVEC were seeded on semi-permeable membrane supports in 6-well (2 × 105 cells per well) Transwell plates (Corning, New York, USA). After 3 days, membrane inserts with the HUVEC monolayers were transferred on top of wells in 6-well plates with nearly confluent HepG2 cells or 3-day differentiated LHCN-M2 muscle cells, using 0.2% FBS in EGM-2 as the basal and apical medium, and the cells were co-cultured for a further 24 h. Alternatively, after 1 h incubation, the medium on apical side was replaced with fresh 0.2% FBS in EGM-2 with 5 μM quercetin or 0.01% DMSO and the cells were further incubated for 18 hours. At the end of the co-culture, monolayer trans-endothelial electrical resistance (TEER) was measured using a Millicell ERS voltohmmeter (Millipore (UK) Ltd, Watford, UK) according to the manufacturer's instructions. After that, the cells were harvested or the glucose uptake assays were performed as described below. The quercetin (Extrasynthese, Genay, France) stock was prepared at 50 mmol L−1 in DMSO and diluted to the final concentration of 5 μmol L−1 for the treatments. DMSO was used at 0.01% in the corresponding control. For TGFβ1 treatment, human recombinant TGFβ1 (Sigma) was applied to the cells in the EGM-2 medium at 50 ng ml−1.2.2 Glucose uptake and transport
Cells were washed with glucose-free buffer (2.5 mM MgCl2, 1 mM CaCl2, 0.14 M NaCl, 20 mM HEPES, 3.5% NaHCO3, pH 7.3), incubated for 10 min in glucose-free RPMI medium and then supplemented for 10 min with 5.5 mM D-[14C(U)]-glucose (0.1 μCi ml−1) (PerkinElmer, Seer Green, UK) on the apical side. Radioactive medium on the apical and basolateral side was collected and the radioactivity measured by scintillation counting. The cells were washed three times with cold PBS with Ca2+ and Mg2+, lysed in 1 M NaOH and radioactivity in the cell lysate aliquot, neutralized with 1 M HCl, was determined by scintillation counting. Measured radioactivity was converted to glucose concentration using standard calibration curves constructed using medium with known amounts of 14C-labelled glucose. Small aliquots of cell lysates were used to measure protein content by Coomassie Protein Assay (Pierce, Thermo Fisher Scientific, Altrincham, UK) and uptake corrected for protein content.2.3. mRNA quantification by droplet digital PCR (ddPCR)
The ddPCR was performed using cDNA obtained by reverse transcription (high capacity RNA-to-cDNA kit, Life Technologies) from mRNA, isolated from cell lysates (AurumTM-Total RNA Mini kit, Bio-Rad Laboratories Ltd, Herts, UK) as recommended by the standard manufacturer's protocols. Guidelines for digital PCR33 were followed as appropriate. FAM-labelled gene-specific probes were from Bio-Rad: glucose transporter (GLUT)1 (SLC2A1), dHsaCPE5035286; and Life Technologies: GLUT3 (SLC2A3), Hs00359840_m1; GLUT6 (SLC2A6), Hs01115485_m1; HO-1, Hs01110250_m1; ATP binding cassette subfamily B member 1 (ABCB1) Hs00184500_m1; ATP binding cassette subfamily C member 2 (ABCC2), Hs00166123_m1; ATP binding cassette subfamily G member 2 (ABCG2), Hs01053790_m1; monocarboxylate transporter (MCT)1 (SLC16A1), Hs00161826_m1; MCT4 (SLC16A3), Hs00358829_m1; MCT5 (SLC16A4), Hs00190794_m1; pyruvate dehydrogenase kinase (PDK) 2, Hs00176865_m1; PDK4, Hs01037712_m1. TATA binding protein (TBP) was used as a reference for mRNA quantification using VIC-labelled probe (TBP, Hs00427620_m1, Life Technologies). Samples corresponding to 10–15 ng mRNA were analysed for specific gene expression using QX100 Droplet Digital PCR system (Bio-Rad) as described previously.34 Briefly, the PCR reaction mix with the gene-specific fluorescent probes was dispersed into oil droplets using QX-100 Droplet Generator. After 40 cycles on the X1000 Touch, droplets were analysed by QX100 Droplet Reader. QuantaSoft software (Kosice, Slovakia) was used to analyse the data and determine the number of copies present in the mix, which were then expressed as the relevant mRNA/TBP ratio, or converted to copies per ng of mRNA, assuming 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 RT PCR efficiency.
1 RT PCR efficiency.
2.4. Protein detection by simple western
GLUT1, GLUT3, HO-1 and Galectin1 protein levels were detected by automated capillary western blotting on WES (ProteinSimple, Bio-Techne, San Jose, CA, USA). At the end of the co-culture, HUVEC on filters were washed in cold PBS and lysed in 60 mM octyl-glucoside, 150 mM NaCl, 20 mM Tris, pH 7.4 with protease inhibitors. Standard conditions for the western protocol were followed as recommended by the manufacturer, except the incubation with the primary antibodies was extended to 60 min. Rabbit antibodies against GLUT1 (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50; Abcam, cat# ab115730, RRID:AB_10903230) and GLUT3 (dilution 1
50; Abcam, cat# ab115730, RRID:AB_10903230) and GLUT3 (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; Abcam, cat# ab191071, RRID:AB_2736916) were used with rabbit anti-Na,K-ATPase (dilution 1
100; Abcam, cat# ab191071, RRID:AB_2736916) were used with rabbit anti-Na,K-ATPase (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50; Cell Signaling Technology, New England Biolabs (UK) Ltd, Hitchin, UK, cat# 3010 RRID:AB_2060983) for a loading control, with denaturation performed for 20 min at 37 °C to prevent aggregation. Goat anti-HO-1 (20 μg ml−1, R&D Systems, Bio-Techne, cat# AF3776, RRID:AB_2295362) was used for detection with anti-calnexin (8 μg ml−1, Santa Cruz Biotechnology, Dallas, TX, USA, cat# sc-6465, RRID:AB_2069146) as the loading control and the standard denaturation protocol for 5 min at 95 °C was followed for the sample preparation. Galectin-1 was detected with mouse monoclonal antibody (dilution 1
50; Cell Signaling Technology, New England Biolabs (UK) Ltd, Hitchin, UK, cat# 3010 RRID:AB_2060983) for a loading control, with denaturation performed for 20 min at 37 °C to prevent aggregation. Goat anti-HO-1 (20 μg ml−1, R&D Systems, Bio-Techne, cat# AF3776, RRID:AB_2295362) was used for detection with anti-calnexin (8 μg ml−1, Santa Cruz Biotechnology, Dallas, TX, USA, cat# sc-6465, RRID:AB_2069146) as the loading control and the standard denaturation protocol for 5 min at 95 °C was followed for the sample preparation. Galectin-1 was detected with mouse monoclonal antibody (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; Novus Biologicals, Bio-Techne, cat# MAB1152, RRID:AB_2811716) using rabbit anti-α-actinin D6F6 antibody (1
20; Novus Biologicals, Bio-Techne, cat# MAB1152, RRID:AB_2811716) using rabbit anti-α-actinin D6F6 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50; Cell Signaling Technology, NEB, cat# 6487, RRID:AB_11179206) as a control. Incubation with anti-goat-IgG-HRP (dilution 1
50; Cell Signaling Technology, NEB, cat# 6487, RRID:AB_11179206) as a control. Incubation with anti-goat-IgG-HRP (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25; R&D Systems), anti-rabbit-IgG-HRP (ProteinSimple, Bio-Techne), or anti-mouse-IgG-HRP with 20× anti-rabbit-IgG-HRP (ProteinSimple, Bio-Techne) followed by luminol/peroxide was used to generate the chemiluminescent signal for quantification. All sets of antibodies gave a linear response in the range between 0.1–0.4 mg protein per ml and 0.2 mg ml−1 was selected for the GLUT and Galectin-1 analysis, while 0.4 mg ml−1 was used for HO-1.
25; R&D Systems), anti-rabbit-IgG-HRP (ProteinSimple, Bio-Techne), or anti-mouse-IgG-HRP with 20× anti-rabbit-IgG-HRP (ProteinSimple, Bio-Techne) followed by luminol/peroxide was used to generate the chemiluminescent signal for quantification. All sets of antibodies gave a linear response in the range between 0.1–0.4 mg protein per ml and 0.2 mg ml−1 was selected for the GLUT and Galectin-1 analysis, while 0.4 mg ml−1 was used for HO-1.
2.5. TGFβ1 ELISA
Nearly confluent HepG2 cells or 3-days differentiated LHCN-M2 cells were incubated with 0.2% FBS in EGM-2 medium for 19 h. Conditioned medium was centrifuged for 10 min at 600g and aliquots were stored frozen at −80 °C until use. Total secreted TGFβ1 was quantified after activation of the latent form, using Dual TGFβ1 Quantikine ELISA (R&D Systems, Bio-Techne) according to the manufacturer's instructions. The EGM-2 medium with 0.2% FBS, which contained less than 31 pg ml−1 TGFβ1, was used as a blank. The measured values were within the linear range of the calibration curve (31–1000 pg ml−1) for the recombinant TGFβ1 provided by the manufacturer.2.6. Lactate quantification
Lactate in the cell medium was detected and quantified by High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAE-PAD) using the Dionex ICS-4000 Ion Chromatography System (Thermo Scientific). After treatments, medium was removed from the cell monolayers and centrifuged at 1000g to remove cellular debris. Cell-free medium was deproteinized by adding an equal volume of ACN, incubating on ice for 5 min and centrifuging for 10 min at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C. The sample was diluted 50-fold in MilliQ water to fall within the linear range of lactate detection (1–20 μg ml−1) and applied on the Dionex IonPac AS11 (Thermo Scientific) anion exchange column, equilibrated in 1 mM KOH, and a 1–60 mM KOH gradient was used for elution. A standard calibration curve was obtained from linear range of concentrations of ICE standard lactate (Sigma).
000g at 4 °C. The sample was diluted 50-fold in MilliQ water to fall within the linear range of lactate detection (1–20 μg ml−1) and applied on the Dionex IonPac AS11 (Thermo Scientific) anion exchange column, equilibrated in 1 mM KOH, and a 1–60 mM KOH gradient was used for elution. A standard calibration curve was obtained from linear range of concentrations of ICE standard lactate (Sigma).
2.7. Data analysis
Test and control data obtained in pairs were compared using independent Student t-test. ANOVA with Bonferroni post-hoc analysis was used for sets with multiple samples. Differences with p < 0.05 (*) were considered significant. All results were obtained from at least 2–3 independent experiments. Origin software (OriginLab Corporation, Northampton, MA, USA) was used for data analysis and the data are presented as mean ± SEM.3. Results
3.1 Co-culturing HUVEC with HepG2 or muscle cells affects differently endothelial gene expression
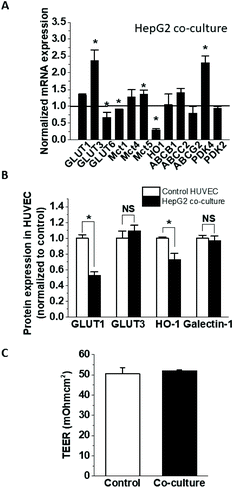
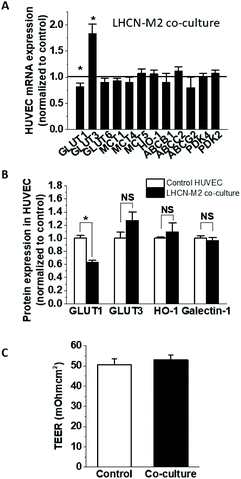
To establish the impact of HepG2 cells on endothelial cell expression of glucose, anion and drug transporters and metabolism-related genes, confluent HUVEC on Transwell inserts were cultured for 24 h with or without HepG2 cells on their basolateral side. This affected the mRNA expression of several endothelial genes (Fig. 1A), including significant increases in GLUT3 (236 ± 32% of the control) and PDK4 (229 ± 21% of the control), and decreases in GLUT6, MCT1 and HO-1 (66.7 ± 15.7%, 91.0 ± 1.5% and 29.7 ± 4.5% of the control, respectively), while the expression of the other tested genes remained unchanged. The downregulation of HO-1 was confirmed at the protein level (72.9 ± 7.7% of the control) (Fig. 1B), but the upregulation of GLUT3 mRNA did not translate to a significant increase in protein (Fig. 1B). On the other hand, protein levels of endothelial GLUT1 were almost halved (52.5 ± 4.8% of the control) in the co-culture even in the absence of mRNA changes (Fig. 1B), indicating involvement of posttranscriptional regulation. Galectin-1 expression has been previously reported to change in endothelial co-cultures, but galectin-1 protein levels were similar between the HUVEC monoculture and co-culture (Fig. 1B). Some of the observed effects were specific for HepG2 cells, as HUVEC co-culture with differentiated skeletal muscle cells affected endothelial mRNA and protein levels differently (Fig. 2A and B). GLUT3 mRNA was still significantly increased in the co-culture (183 ± 18.5% of the control), but in contrast to the HepG2 co-cultures, GLUT6, MCT1, PDK4 and HO-1 mRNA remained unaffected, while GLUT1 mRNA was downregulated by about 20% (80.9 ± 7.9% of the control). At the protein level, similar to the effect on HepG2 cells, the co-incubation with muscle cells triggered GLUT1 downregulation (63.1 ± 3.0% of the control), while GLUT3 and galectin-1 were not affected. However, in contrast to the HepG2, muscle cells did not affect the endothelial HO-1 protein levels (Fig. 2B).3.2. The effects of HepG2-conditioned media on HUVEC gene expression
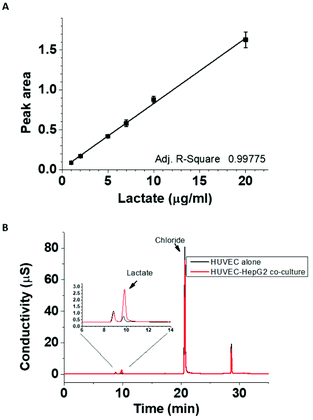
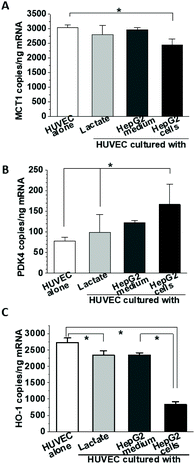
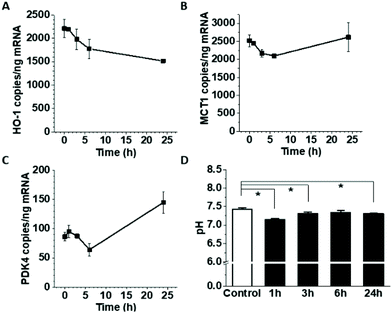
One of the most prominent features of HepG2 metabolism is lactate production. In the current setting, the lactate concentration in the media was measured by HPAE-PAD chromatography and was estimated to reach 8.3 ± 0.3 mM at the end of the 24 h co-culture (Fig. 3), a significant increase compared to 1.1 ± 0.1 mM lactate measured in corresponding HUVEC monoculture. To test whether the additional lactate or other factors secreted by HepG2 cells into the basolateral medium could be responsible for the observed gene expression changes, HUVEC were cultured for 18 h in the presence of 8 mM lactate, HepG2-conditioned medium, or HepG2 cells on the basolateral side and the mRNA for MCT1, PDK4 and HO-1 was detected by ddPCR (Fig. 4A, B and C). A small effect of lactate or conditioned media was observed on PDK4 and HO-1 mRNA expression (Fig. 4B and C), but did not reach the magnitude of the changes triggered by HepG2 cell co-culturing. The time-course of the mRNA expression for MCT1, PDK4 and HO-1 was further followed in HUVEC incubated with 8 mM lactate for 1, 3, 6 and 24 h (Fig. 5). Lactate transiently decreased PDK4 and MCT1 mRNA levels while HO-1 mRNA declined steadily with maximum decrease of ∼30% after 24 h. This indicates that time-dependent changes in lactate concentration may contribute to MCT1 and HO-1 regulation, but are not solely responsible for the full effect.3.3. The role of TGFβ1 secreted by HepG2 cells in the HO-1 downregulation
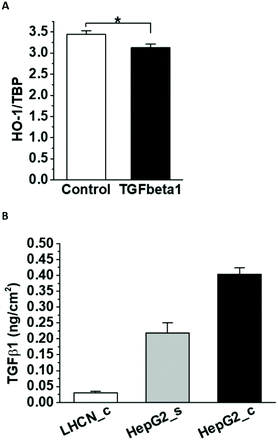
One of the few known negative regulators of HO-1 expression is TGFβ1.35 In agreement, incubation with human recombinant TGFβ1 decreased HO-1 mRNA expression in HUVEC (Fig. 6A). HepG2 cells secreted a significant amount of TGFβ1 in the media (Fig. 6B), which could contribute to HO-1 downregulation. Considerably lower amounts of TGFβ1 were detected in the muscle-conditioned media, consistent with their lack of effect on HO-1 expression in HUVEC.3.4. Effects of quercetin on HO-1 and PDK4 expression in HUVEC in the presence and absence of HepG2 cells
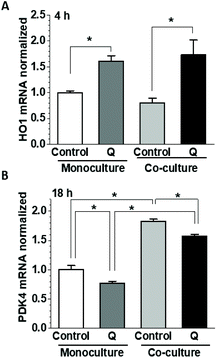
We have shown previously in HUVEC that incubation with quercetin dramatically upregulated HO-1 mRNA at 4 h while decreasing PDK4 mRNA expression at 18 h.30 As both genes were significantly affected by co-culturing with HepG2 cells, we have tested the impact of quercetin in the co-culture. Quercetin was applied to the apical side of HUVEC during the culture in the presence and absence of HepG2 cells and the mRNA for HO-1 and PDK4 was analysed after 4 or 18 h. Compared to untreated cultures, incubation with quercetin for 4 h significantly upregulated HO-1 mRNA in both monoculture (160 ± 11%) and co-culture (214 ± 35%) (Fig. 7A), consistent with the previous work.30 In contrast to the longer, 24 h incubation (Fig. 1A and 4C), there was no significant effect of co-culturing on the HO-1 mRNA level at 4 h. At 18 h, the HUVEC PDK4 mRNA was significantly downregulated by quercetin in monoculture as well as co-culture (77.4 ± 2.8 and 85.9 ± 2.8%, of the respective untreated controls), but because of the PDK4 upregulation in the co-culture (182.8 ± 3.3%), the absolute PDK4 levels in the quercetin-treated co-culture were significantly higher (∼2-fold) than in monoculture (Fig. 7B). Thus, the effects elicited by co-culturing did not reverse, but rather modulated quercetin-induced changes in PDK4 expression, and did not significantly affect the short-term HO-1 upregulation.3.5. Glucose transport in HUVEC co-cultures
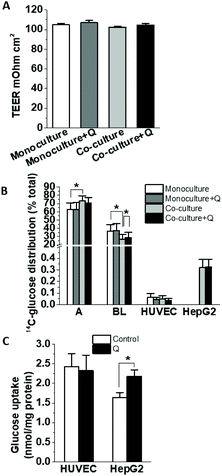
To determine whether co-culture with HepG2 cells affects glucose uptake and transport by HUVEC, cells seeded on Transwells were cultured alone or with HepG2 cells in the presence or absence of 5 μM quercetin on the apical side, and then D-[14C(U)]-glucose transport was measured. There were no changes in the monolayer integrity as measured by the TEER (Fig. 8A). Compared to the monoculture, HUVEC in co-culture incubated with 5 mM glucose transported less D-[14C(U)]-glucose across the monolayer, and this effect was slightly alleviated in the presence of quercetin (Fig. 8B). No statistically significant effect was observed for the glucose uptake in the HUVEC or HepG2 cells. Depending on the role of different glucose transporters in the uptake, the effects could be modulated by the total glucose levels in the uptake assay and so we tested lower glucose concentration in the transport/uptake solution. When 0.5 mM glucose was used in the HUVEC-HepG2 co-culture, quercetin pre-incubation significantly increased the glucose uptake in the HepG2 cells, but not in the HUVEC (Fig. 8C). At this concentration, the transport across the monolayer was not significantly affected (data not shown).4. Discussion
Food-derived polyphenols have been studied as candidates for interventions to lower the risk of type 2 diabetes, based on their reported ability to regulate glucose absorption and metabolism and alleviate endothelial dysfunction. The mechanism of these activities has been explored through numerous in vitro, animal and human studies. In recent years, a limited number of co-culture cell models have been used to investigate some aspects of polyphenol metabolism and signalling.36 For example, co-cultures of Caco2 enterocytes and HT29-MTX goblet cells were used to study polyphenol transport and conjugation.37 Protective effects of polyphenols during inflammation have been elucidated in models combining macrophages and gingival fibroblasts38 or adipocytes,39 or monocytes and Caco-2 cells.40 Other studies explored anti-inflammatory potential of anthocyanins in co-cultures of gingival epithelial cells and fibroblasts41 or epithelial and endothelial cells.42,43 Recently, quercetin was shown to attenuate apoptosis of pancreatic β-cells through NO signalling in endothelial cells.44 However, our comprehension of the interplay between different cells in similar systems is still limited. Therefore, here we aimed to extend our previous study on the impact of quercetin on endothelial cell metabolism30 by including liver cells in the model, in order to examine the potential role of microenvironment and paracrine messengers. Different aspects of the interaction between endothelial and liver cells have been studied previously in organoids and systems combining endothelial cells with hepatocytes or HepG2 cells.45–47 HepG2 cells were shown to regulate lymphocyte transmigration across sinusoidal endothelial cells.48 Several studies addressed the communication via soluble factors including vesicles,49 although physical contact between HUVEC and HepG2 cells was required for angiogenic stimulation.50In vivo, liver endothelial cells form a discontinuous, fenestrated layer, and are separated from the other liver cells by a thin Disse space, allowing for free exchange of soluble and other factors. Therefore, we have investigated the impact of liver cells on intact mature endothelial monolayers by utilizing a contact-free co-culture of endothelial and HepG2 cells and further studied the changes in the endothelial gene expression under the combined impact of co-culture and quercetin exposure. Because of the challenges associated with culturing primary hepatocytes, carcinoma-derived HepG2 cells were used as they are well characterized and afford better reproducibility. However, since there are metabolic differences between normal and transformed cells, it is possible that our results are more pertinent to tumour environment than normal tissue. We found that in the presence of HepG2 cells, HUVEC expressed increased levels of PDK4 mRNA and lower levels of GLUT1 and HO-1 protein. The HepG2 cells secreted TGFβ1 and dramatically increased lactate concentration in the medium compared to the HUVEC cultured alone. These and other soluble factors possibly contributed to the observed changes in gene expression, but continuous feedback from HepG2 cells was required for the maximum impact as HepG2-conditioned medium had only limited effect. These were specific to the HepG2 cells as the only detected change in endothelial cells co-cultured with LHCN-M2 immortalized skeletal muscle cells was downregulation of GLUT1 protein. Neither cell line affected the tightness of the endothelial monolayer. Since endothelial HO-1 and PDK4 gene expression has been shown to be significantly affected by quercetin exposure after 4 and 18 h respectively,30 we looked at whether this is altered in the co-culture. HO-1 mRNA was upregulated by 4 h-exposure to quercetin both in monoculture and co-culture. At the same time there were no significant differences between the HO-1 levels in the two settings in the absence of quercetin, most likely because the 4 h-incubation was too short for the full effect of the co-culture to take place. PDK4 mRNA was upregulated in the co-culture after 18 h, but quercetin decreased the mRNA levels in co-cultured HUVEC to the levels comparable to the untreated cells in monoculture. Thus, although the general impact of quercetin on endothelial expression of PDK4 and HO-1 was similar in endothelial monoculture and co-culture, the presence of HepG2 cells moderated the quercetin effects on PDK4, but not HO-1 expression.Co-culturing with HepG2 cells decreased the amount of glucose transported across the HUVEC monolayer, possibly related to the GLUT1 downregulation. This was slightly alleviated by quercetin, consistent with our previous finding that quercetin could increase glucose transport in HUVEC. In addition, quercetin application to the apical side of endothelial cells stimulated glucose uptake in HepG2 cells on the basolateral side, but only for low glucose concentration, possibly as a downstream effect of quercetin-triggered signalling. At this quercetin concentration, the amount of the polyphenol transported to the basolateral side of the endothelial monolayer would be quite small, so an indirect effect through HUVEC is more plausible than quercetin acting on the HepG2 directly. Altered expression of glucose or drug transporters as well as changes in ROS and extracellular signalling in the endothelial cells could have a downstream effect on the adjacent HepG2 cells. Thus, in addition to direct effects, polyphenols such as quercetin could act on the target tissues through shifting the redox and metabolic balance in the endothelium and modifying the associated endothelial signalling to the underlying tissues.
5. Conclusions
In summary, we have used endothelial cells in co-culture with HepG2 cells as a more physiologically relevant configuration to study the impact of diet-derived polyphenol quercetin on their physiology. We have shown that the co-culture affects expression of various genes involved in metabolism (GLUT1 and PDK4) or redox balance (HO-1) in HUVEC, which can then contribute to regulating changes triggered by quercetin application, supporting the notion that the local environment in tissues has potential to modulate the impact of circulating molecules.Abbreviations
| ABCB1 | ATP binding cassette subfamily B member 1 |
| ABCC2 | ATP binding cassette subfamily C member 2 |
| ABCG2 | ATP binding cassette subfamily G member 2 |
| ApoE | Apolipoprotein E |
| ddPCR | Droplet digital PCR |
| DMEM | Dulbecco's modified Eagle medium |
| EGM-2 | Endothelial cell growth medium 2 |
| eNOS | Endothelial nitric oxide synthase |
| GLUT | Glucose transporter |
| HIF1α | Hypoxia induced factor 1α |
| HO-1 | Heme oxygenase-1 |
| HPAE-PAD | High performance anion exchange chromatography with pulsed amperometric detection |
| HUVEC | Human umbilical vein endothelial cells |
| ICAM1 | Intercellular adhesion molecule 1 |
| MCT | Monocarboxylate transporter |
| NO | Nitric oxide |
| NOX | NADPH oxidase |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| oxLDL | Oxidized low density lipoprotein |
| PDK | Pyruvate dehydrogenase kinase |
| ROS | Reactive oxygen species |
| SIRT1 | Sirtuin 1 |
| TGFβ | Transforming growth factor beta |
| VCAM1 | Vascular cell adhesion molecule 1 |
| TBP | TATA binding protein |
| TEER | Transendothelial electrical resistance |
Author contributions
ST and GW conceived and planned the project. ST carried out the experiments and analysed the data with MJH providing assistance with the muscle cell cultures. ST and GW wrote the manuscript and all authors provided feedback and contributed to the final version of the manuscript.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
The presented research has been supported by funding from the European Research Council advanced grant number 322467 (‘POLYTRUE?’). We thank Dr Vincent Mouly (Sorbonne Université, INSERM, Paris, France) for providing the immortalized skeletal muscle cells LHCN-M2.References
- V. Bauer and R. Sotnikova, Nitric oxide – the endothelium-derived relaxing factor and its role in endothelial functions, Gen. Physiol. Biophys., 2010, 29, 319–340 CrossRef CAS PubMed.
- M. J. Durand and D. D. Gutterman, Diversity in mechanisms of endothelium-dependent vasodilation in health and disease, Microcirculation, 2013, 20, 239–247 CrossRef CAS PubMed.
- H. F. Galley and N. R. Webster, Physiology of the endothelium, Br. J. Anaesth., 2004, 93, 105–113 CrossRef CAS PubMed.
- S. Rafii, J. M. Butler and B. S. Ding, Angiocrine functions of organ-specific endothelial cells, Nature, 2016, 529, 316–325 CrossRef CAS PubMed.
- J. G. Camp, K. Sekine, T. Gerber, H. Loeffler-Wirth, H. Binder, M. Gac, S. Kanton, J. Kageyama, G. Damm, D. Seehofer, L. Belicova, M. Bickle, R. Barsacchi, R. Okuda, E. Yoshizawa, M. Kimura, H. Ayabe, H. Taniguchi, T. Takebe and B. Treutlein, Multilineage communication regulates human liver bud development from pluripotency, Nature, 2017, 546, 533–538 CrossRef CAS PubMed.
- B. S. Ding, D. J. Nolan, J. M. Butler, D. James, A. O. Babazadeh, Z. Rosenwaks, V. Mittal, H. Kobayashi, K. Shido, D. Lyden, T. N. Sato, S. Y. Rabbany and S. Rafii, Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration, Nature, 2010, 468, 310–315 CrossRef CAS PubMed.
- X. Pi, L. Xie and C. Patterson, Emerging Roles of Vascular Endothelium in Metabolic Homeostasis, Circ. Res., 2018, 123, 477–494 CrossRef CAS PubMed.
- A. Godecke and J. Haendeler, Intra- and Interorgan Communication in the Cardiovascular System: A Special View on Redox Regulation, Antioxid. Redox Signaling, 2017, 26, 613–615 CrossRef PubMed.
- J. K. Freed and D. D. Gutterman, Communication Is Key: Mechanisms of Intercellular Signaling in Vasodilation, J. Cardiovasc. Pharmacol., 2017, 69, 264–272 CrossRef CAS PubMed.
- J. Keaney and M. Campbell, The dynamic blood-brain barrier, FEBS J., 2015, 282, 4067–4079 CrossRef CAS PubMed.
- T. Kubota, N. Kubota, H. Kumagai, S. Yamaguchi, H. Kozono, T. Takahashi, M. Inoue, S. Itoh, I. Takamoto, T. Sasako, K. Kumagai, T. Kawai, S. Hashimoto, T. Kobayashi, M. Sato, K. Tokuyama, S. Nishimura, M. Tsunoda, T. Ide, K. Murakami, T. Yamazaki, O. Ezaki, K. Kawamura, H. Masuda, M. Moroi, K. Sugi, Y. Oike, H. Shimokawa, N. Yanagihara, M. Tsutsui, Y. Terauchi, K. Tobe, R. Nagai, K. Kamata, K. Inoue, T. Kodama, K. Ueki and T. Kadowaki, Impaired insulin signaling in endothelial cells reduces insulin-induced glucose uptake by skeletal muscle, Cell Metab., 2011, 13, 294–307 CrossRef CAS PubMed.
- M. Konishi, M. Sakaguchi, S. M. Lockhart, W. Cai, M. E. Li, E. P. Homan, C. Rask-Madsen and C. R. Kahn, Endothelial insulin receptors differentially control insulin signaling kinetics in peripheral tissues and brain of mice, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E8478–E8487 CrossRef CAS PubMed.
- B. S. Ding, Z. Cao, R. Lis, D. J. Nolan, P. Guo, M. Simons, M. E. Penfold, K. Shido, S. Y. Rabbany and S. Rafii, Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis, Nature, 2014, 505, 97–102 CrossRef PubMed.
- T. Schewe, Y. Steffen and H. Sies, How do dietary flavanols improve vascular function? A position paper, Arch. Biochem. Biophys., 2008, 476, 102–106 CrossRef CAS PubMed.
- K. Yamagata, M. Tagami and Y. Yamori, Dietary polyphenols regulate endothelial function and prevent cardiovascular disease, Nutrition, 2015, 31, 28–37 CrossRef CAS PubMed.
- N. Suganya, E. Bhakkiyalakshmi, D. V. Sarada and K. M. Ramkumar, Reversibility of endothelial dysfunction in diabetes: role of polyphenols, Br. J. Nutr., 2016, 116, 223–246 CrossRef CAS PubMed.
- T. Hussain, B. Tan, Y. Yin, F. Blachier, M. C. Tossou and N. Rahu, Oxidative Stress and Inflammation: What Polyphenols Can Do for Us?, Oxid. Med. Cell. Longevity, 2016, 2016, 7432797 Search PubMed.
- G. E. Mann, B. Bonacasa, T. Ishii and R. C. Siow, Targeting the redox sensitive Nrf2-Keap1 defense pathway in cardiovascular disease: protection afforded by dietary isoflavones, Curr. Opin. Pharmacol., 2009, 9, 139–145 CrossRef CAS PubMed.
- J. H. Kim, C. Auger and V. B. Schini-Kerth, Activation of eNOS by polyphenol-rich products and polyphenolic compounds, Curr. Pharm. Des., 2014, 20, 3521–3529 CrossRef CAS PubMed.
- C. H. Hung, S. H. Chan, P. M. Chu and K. L. Tsai, Quercetin is a potent anti-atherosclerotic compound by activation of SIRT1 signaling under oxLDL stimulation, Mol. Nutr. Food Res., 2015, 59, 1905–1917 CrossRef CAS PubMed.
- M. Sanchez, M. Galisteo, R. Vera, I. C. Villar, A. Zarzuelo, J. Tamargo, F. Perez-Vizcaino and J. Duarte, Quercetin downregulates NADPH oxidase, increases eNOS activity and prevents endothelial dysfunction in spontaneously hypertensive rats, J. Hypertens., 2006, 24, 75–84 CrossRef CAS PubMed.
- H. S. Jones, A. Gordon, S. G. Magwenzi, K. Naseem, S. L. Atkin and F. L. Courts, The dietary flavonol quercetin ameliorates angiotensin II-induced redox signaling imbalance in a human umbilical vein endothelial cell model of endothelial dysfunction via ablation of p47phox expression, Mol. Nutr. Food Res., 2016, 60, 787–797 CrossRef CAS PubMed.
- Y. Shen, N. C. Ward, J. M. Hodgson, I. B. Puddey, Y. Wang, D. Zhang, G. J. Maghzal, R. Stocker and K. D. Croft, Dietary quercetin attenuates oxidant-induced endothelial dysfunction and atherosclerosis in apolipoprotein E knockout mice fed a high-fat diet: a critical role for heme oxygenase-1, Free Radical Biol. Med., 2013, 65, 908–915 CrossRef CAS PubMed.
- C. Li, W. J. Zhang, J. Choi and B. Frei, Quercetin affects glutathione levels and redox ratio in human aortic endothelial cells not through oxidation but formation and cellular export of quercetin-glutathione conjugates and upregulation of glutamate-cysteine ligase, Redox Biol., 2016, 9, 220–228 CrossRef CAS PubMed.
- P. Galindo, I. Rodriguez-Gomez, S. Gonzalez-Manzano, M. Duenas, R. Jimenez, C. Menendez, F. Vargas, J. Tamargo, C. Santos-Buelga, F. Perez-Vizcaino and J. Duarte, Glucuronidated quercetin lowers blood pressure in spontaneously hypertensive rats via deconjugation, PLoS One, 2012, 7, e32673 CrossRef CAS PubMed.
- A. Perez, S. Gonzalez-Manzano, R. Jimenez, R. Perez-Abud, J. M. Haro, A. Osuna, C. Santos-Buelga, J. Duarte and F. Perez-Vizcaino, The flavonoid quercetin induces acute vasodilator effects in healthy volunteers: correlation with beta-glucuronidase activity, Pharmacol. Res., 2014, 89, 11–18 CrossRef CAS PubMed.
- S. Tribolo, F. Lodi, C. Connor, S. Suri, V. G. Wilson, M. A. Taylor, P. W. Needs, P. A. Kroon and D. A. Hughes, Comparative effects of quercetin and its predominant human metabolites on adhesion molecule expression in activated human vascular endothelial cells, Atherosclerosis, 2008, 197, 50–56 CrossRef CAS PubMed.
- C. Li, W. J. Zhang and B. Frei, Quercetin inhibits LPS-induced adhesion molecule expression and oxidant production in human aortic endothelial cells by p38-mediated Nrf2 activation and antioxidant enzyme induction, Redox Biol., 2016, 9, 104–113 CrossRef CAS PubMed.
- T. Koga and M. Meydani, Effect of plasma metabolites of (+)-catechin and quercetin on monocyte adhesion to human aortic endothelial cells, Am. J. Clin. Nutr., 2001, 73, 941–948 CrossRef CAS PubMed.
- S. Tumova, A. Kerimi and G. Williamson, Long term treatment with quercetin in contrast to the sulfate and glucuronide conjugates affects HIF1alpha stability and Nrf2 signaling in endothelial cells and leads to changes in glucose metabolism, Free Radical Biol. Med., 2019, 137, 158–168 CrossRef CAS PubMed.
- C. H. Zhu, V. Mouly, R. N. Cooper, K. Mamchaoui, A. Bigot, J. W. Shay, J. P. Di Santo, G. S. Butler-Browne and W. E. Wright, Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies, Aging Cell, 2007, 6, 515–523 CrossRef CAS PubMed.
- M. J. Houghton, A. Kerimi, V. Mouly, S. Tumova and G. Williamson, Gut microbiome catabolites as novel modulators of muscle cell glucose metabolism, FASEB J., 2019, 33, 1887–1898 CrossRef CAS PubMed.
- J. F. Huggett, C. A. Foy, V. Benes, K. Emslie, J. A. Garson, R. Haynes, J. Hellemans, M. Kubista, R. D. Mueller and T. Nolan, et al., The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments, Clin. Chem., 2013, 59, 892–902 CrossRef CAS PubMed.
- S. Tumova, A. Kerimi, K. E. Porter and G. Williamson, Transendothelial glucose transport is not restricted by extracellular hyperglycaemia, Vasc. Pharmacol., 2016, 87, 219–229 CrossRef CAS PubMed.
- Y. Okita, A. Kamoshida, H. Suzuki, K. Itoh, H. Motohashi, K. Igarashi, M. Yamamoto, T. Ogami, D. Koinuma and M. Kato, Transforming growth factor-beta induces transcription factors MafK and Bach1 to suppress expression of the heme oxygenase-1 gene, J. Biol. Chem., 2013, 288, 20658–20667 CrossRef CAS PubMed.
- C. Grootaert, S. Kamiloglu, E. Capanoglu and J. Van Camp, Cell Systems to Investigate the Impact of Polyphenols on Cardiovascular Health, Nutrients, 2015, 7, 9229–9255 CrossRef CAS PubMed.
- F. Jailani and G. Williamson, Effect of edible oils on quercetin, kaempferol and galangin transport and conjugation in the intestinal Caco-2/HT29-MTX co-culture model, Food Funct., 2014, 5, 653–662 RSC.
- M. P. Morin and D. Grenier, Regulation of matrix metalloproteinase secretion by green tea catechins in a three-dimensional co-culture model of macrophages and gingival fibroblasts, Arch. Oral Biol., 2017, 75, 89–99 CrossRef CAS PubMed.
- Y. Sakamoto, J. Kanatsu, M. Toh, A. Naka, K. Kondo and K. Iida, The Dietary Isoflavone Daidzein Reduces Expression of Pro-Inflammatory Genes through PPARalpha/gamma and JNK Pathways in Adipocyte and Macrophage Co-Cultures, PLoS One, 2016, 11, e0149676 CrossRef PubMed.
- H. Zhang, Y. I. Hassan, J. Renaud, R. Liu, C. Yang, Y. Sun and R. Tsao, Bioaccessibility, bioavailability, and anti-inflammatory effects of anthocyanins from purple root vegetables using mono- and co-culture cell models, Mol. Nutr. Food Res., 2017, 61, 1600928 CrossRef PubMed.
- T. B. Lombardo Bedran, D. Palomari Spolidorio and D. Grenier, Green tea polyphenol epigallocatechin-3-gallate and cranberry proanthocyanidins act in synergy with cathelicidin (LL-37) to reduce the LPS-induced inflammatory response in a three-dimensional co-culture model of gingival epithelial cells and fibroblasts, Arch. Oral Biol., 2015, 60, 845–853 CrossRef CAS PubMed.
- S. Kuntz, H. Asseburg, S. Dold, A. Rompp, B. Frohling, C. Kunz and S. Rudloff, Inhibition of low-grade inflammation by anthocyanins from grape extract in an in vitro epithelial-endothelial co-culture model, Food Funct., 2015, 6, 1136–1149 RSC.
- S. Kamiloglu, C. Grootaert, E. Capanoglu, C. Ozkan, G. Smagghe, K. Raes and J. Van Camp, Anti-inflammatory potential of black carrot (Daucus carota L.) polyphenols in a co-culture model of intestinal Caco-2 and endothelial EA.hy926 cells, Mol. Nutr. Food Res., 2017, 61, 1600455 CrossRef PubMed.
- N. Suganya, K. P. Mani, D. Sireesh, P. Rajaguru, M. Vairamani, T. Suresh, T. Suzuki, S. Chatterjee and K. M. Ramkumar, Establishment of pancreatic microenvironment model of ER stress: Quercetin attenuates beta-cell apoptosis by invoking nitric oxide-cGMP signaling in endothelial cells, J. Nutr. Biochem., 2018, 55, 142–156 CrossRef CAS PubMed.
- Y. B. Kang, S. Rawat, J. Cirillo, M. Bouchard and H. M. Noh, Layered long-term co-culture of hepatocytes and endothelial cells on a transwell membrane: toward engineering the liver sinusoid, Biofabrication, 2013, 5, 045008 CrossRef PubMed.
- M. A. Guzzardi, F. Vozzi and A. D. Ahluwalia, Study of the crosstalk between hepatocytes and endothelial cells using a novel multicompartmental bioreactor: a comparison between connected cultures and cocultures, Tissue Eng., Part A, 2009, 15, 3635–3644 CrossRef CAS PubMed.
- Y. Nahmias, M. Casali, L. Barbe, F. Berthiaume and M. L. Yarmush, Liver endothelial cells promote LDL-R expression and the uptake of HCV-like particles in primary rat and human hepatocytes, Hepatology, 2006, 43, 257–265 CrossRef CAS PubMed.
- S. Edwards, P. F. Lalor, G. B. Nash, G. E. Rainger and D. H. Adams, Lymphocyte traffic through sinusoidal endothelial cells is regulated by hepatocytes, Hepatology, 2005, 41, 451–459 CrossRef PubMed.
- F. Royo, L. Moreno, J. Mleczko, L. Palomo, E. Gonzalez, D. Cabrera, A. Cogolludo, F. P. Vizcaino, S. van-Liempd and J. M. Falcon-Perez, Hepatocyte-secreted extracellular vesicles modify blood metabolome and endothelial function by an arginase-dependent mechanism, Sci. Rep., 2017, 7, 42798 CrossRef CAS PubMed.
- G. G. Y. Chiew, A. Fu, K. Perng Low and K. Qian Luo, Physical supports from liver cancer cells are essential for differentiation and remodeling of endothelial cells in a HepG2-HUVEC co-culture model, Sci. Rep., 2015, 5, 10801 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |