Enhanced curcumin solubility and antibacterial activity by encapsulation in PLGA oily core nanocapsules
Mengqian
Gao
,
Xue
Long
,
Jing
Du
,
Mengting
Teng
,
Weichen
Zhang
,
Yuting
Wang
,
Xingqi
Wang
,
Ziyuan
Wang
,
Peng
Zhang
 * and
Jun
Li
* and
Jun
Li
 *
*
School of Life Science, Jiangsu Normal University, Xuzhou 221116, Jiangsu, China. E-mail: zhangpeng@jsnu.edu.cn; lijun@jsnu.edu.cn; lijun2016@tsinghua.org.cn
First published on 2nd December 2019
Abstract
Infections caused by bacteria represent an emerging public health threat due to the development of antibiotic resistance in bacteria. Curcumin (CUR), a naturally derived substance, is found to be effective against several bacteria. However, its use is limited by its low water solubility and rapid degradation profile. Polymeric nanocapsules (NCs) represent an interesting drug delivery system with high incorporation rates due to their liquid core. The present study aimed to develop poly-(lactic-co-glycolic acid) (PLGA) NCs for the delivery of CUR for enhancing its solubility and antibacterial activity. The particle size, polydispersity index (PDI), zeta potential and drug entrapment efficiency of CUR NCs with optimal formulation were 158 nm, 0.156, −29.1 mV and 92.64%, respectively. The water solubility of CUR in NCs increased about 1500 fold compared to that of free CUR. TEM and AFM images proved the core–shell structure of PLGA NCs with narrow size distributions. The in vitro release profile of CUR from PLGA NCs showed a burst release in the initial 24 h followed by a sustained release of the interior CUR over 10 days. In vitro antibacterial experiments demonstrate that the minimum inhibitory concentrations (MICs) of CUR NCs were lower than those of free CUR for all different bacterial strains, especially for Gram-negative bacteria. CUR NCs exhibited broad-spectrum antibacterial effects compared with free CUR. These data suggest that these CUR-loaded PLGA NCs may provide a promising strategy as novel antibacterial agents.
Introduction
In recent years, bacterial infections are among the important infectious diseases. Traditional antibiotics are gradually losing their effectiveness due to bacterial resistance. Curcumin (CUR) is a natural polyphenol compound obtained from the rhizomes of Curcuma longa L. During the past few years, extensive study showed that this derivative exhibits antimicrobial activities at relatively nontoxic and safe doses.1,2 There have been several explanations of how CUR kills bacteria. Rai et al. reported that CUR interacts with FtsZ (a prokaryotic homologue of the eukaryotic cytoskeletal protein tubulin), thereby inhibiting the assembly of FtsZ protofilaments.3 Tyagi et al. demonstrated that CUR could inhibit the growth of bacteria by damaging the bacterial membrane.4 Lee et al. investigated the antibacterial mechanism of CUR due to an apoptosis-like response.5 However, the potential for therapeutic translation of CUR has been limited by its low water solubility, poor oral bioavailability and rapid degradation.6,7With the development of nanotechnology, nano-drug delivery systems have been designed for biomedical applications, including antitumor,8–11 antibacterial,12–14 anti-inflammatory15 and antioxidant applications.16 In the past few decades, polymeric nanocapsules (NCs) have attracted much interest for drug delivery.17–19 In contrast to nanospheres, nanocapsules (NCs) consist of a polymer shell layer and an inner liquid compartment. The main advantage of polymeric NCs as drug carriers is their high drug loading capacity based on optimized drug solubility in the liquid core.20 The shell composition and thickness can be modified to trigger the release of the drug and the degradation rate of the polymeric shell. Polymeric NCs based on biodegradable polymers are of interest with respect to their transition from basic research to clinical trials. Among these polymers, polylactic acid (PLA) and poly-(lactic-co-glycolic acid) (PLGA) have been widely investigated in nano drug delivery systems for both fundamental research and product development due to their safety and biofate.21–23 In recent years, PLGA-based nanoparticles have been used in extensive applications for drug delivery24 and tissue engineering.25,26 Di et al. reported ultrasound-triggered microgels integrated with insulin-loaded PLGA NCs that can provide pulsatile insulin release to achieve glycemic control.27
For this purpose, in an attempt to further improve CUR delivery, we developed PLGA NCs that incorporate CUR within their interior oily core. The particle size, size distribution, zeta potential, entrapment efficiency and morphology of these NCs were determined by dynamic light scattering (DLS), UV-Vis spectrophotometry, transmission electron microscopy (TEM) and atomic force microscopy (AFM). Also, the drug release behaviors of free CUR and CUR NCs were investigated in vitro. Furthermore, the minimum inhibitory concentration (MIC) study and antibacterial assay of free CUR and CUR NC formulations were performed against both Gram-negative and Gram-positive bacteria.
Materials and methods
Materials
The copolymer PLGA 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 was purchased from Shandong Biomedicine Co. Poloxamer 188 (Lutrol® F68) and medium-chain triglycerides (MCTs) were purchased from BASF. Curcumin (CUR) was bought from Beijing solarbio Co., Ltd. Soy lecithin was obtained from Shanghai Advanced Viecle Technology Co., Ltd. All other chemicals were of reagent grade.
50 was purchased from Shandong Biomedicine Co. Poloxamer 188 (Lutrol® F68) and medium-chain triglycerides (MCTs) were purchased from BASF. Curcumin (CUR) was bought from Beijing solarbio Co., Ltd. Soy lecithin was obtained from Shanghai Advanced Viecle Technology Co., Ltd. All other chemicals were of reagent grade.
Microbial strains
Three Gram-negative bacteria, E. coli ATCC25922, Salmonella CGMCC1.1194 and P. aeruginosa CGMCC1.10274, and three Gram-positive bacteria, S. aureus ATCC 29213, B. sonorensis CGMCC1.10580 and B. licheniformis CGMCC1.10314, were used in this study.Methods
Preparation of the curcumin nanocapsules
Curcumin (CUR) nanocapsules (NCs) were prepared by the solvent displacement method with some modifications. Briefly, 25 mg of PLGA and 50 mg of lecithin were first dissolved in 5 ml solvent including 4.5 ml acetone and 0.5 ml ethanol. Then, 250 μl MCT oil containing CUR were added to the mixture solution. This organic solution was dropwise injected into water containing 10 mg poloxamer 188 under moderate magnetic stirring at room temperature. Afterwards, the solvent and part of water were evaporated under reduced pressure at 40 °C. Samples were centrifuged at 3000 rpm for 10 min to remove nonencapsulated CUR. The desired final volume of the CUR loaded nanocapsule suspension was 8 ml. The precipitated CUR was used to quantify the level of nonencapsulated CUR of NCs by UV-vis spectroscopy at 425 nm. The percentage of encapsulated CUR was calculated using eqn (1):| CUR encapsulation efficiency (EE%) = [(WT − WS)/WS] × 100%, | (1) |
Characterization of CUR NCs
The particle size, size distribution and zeta potential of the CUR NCs were measured by using a Zetasizer Nano ZS90, which is based on the principle of dynamic light scattering (DLS). The size and morphology of CUR NCs were determined by atomic force microscopy (AFM) and transmission electron microscopy (TEM).Freeze-drying
The CUR NC samples were frozen for at least 12 h at a temperature of −81 °C, and then the samples were lyophilised for 48 h at a temperature of −25 °C. The redispersibility of all the freeze-dried samples was tested by adding the same volume of water.In vitro drug release
Drug release study was performed by the dialysis bag method, where the dialysis bag was activated and soaked in release media (PBS; pH 7.4) overnight prior to the study. 4 ml of free CUR or CUR NC suspension was placed in the dialysis bag, and both ends of the dialysis bag were tightly tied and immersed in 100 ml of the release medium containing 0.5% (v/v) SDS to maintain sink conditions. While stirring the release medium using a horizontal rotary shaker at 150 rpm at 37 °C, samples (3 ml) were collected at different time points (0.5, 1, 2, 4, 6, 8, 10, 12, 24, 36, 48, 72, 96, 120, 144, 168, 192, 216 and 240 h) with replacement with an equal volume of fresh medium. Each study was carried out in triplicate, and data are presented as mean ± S.D (n = 3). Cumulative release data were fitted for evaluation of drug release kinetics. Regression coefficient (R2) was calculated for the best-fit model.Antibacterial activity study
Tested bacteria (Escherichia coli, Staphylococcus aureus, salmonella, P.aeruginosa, B.sonorensis, and Bacillus licheniformis) were cultured and activated in Luria–Bertani (LB) medium at 37 °C for 12 h. The activated bacteria were diluted with Mumbai High Bassein (MHB) medium. The fresh CUR NC suspension and CUR DMSO solution were diluted to diverse concentrations. Fifty microliters of them was added into a 96-well plate. The same volume of diluted bacteria was added into all the wells of the 96-well plate. The OD600 was determined with a micro-titer plate reader (Molecular Devices; Sunnyvale, CA, USA) and recorded as OD1. The 96-well plate was cultured at 37 °C. The OD600 of the plate was tested as OD2 after 18 h. OD3 is the value of OD2 minus OD1, and the OD600 of the negative control was OD4. The minimum inhibitory concentration (MIC) is the lowest concentration of antibacterial agents that can prevent bacterial growth. The bacterial growth inhibition effect was calculated using eqn (2):| Inhibition = (1 − OD3/OD4) × 100% | (2) |
Results and discussion
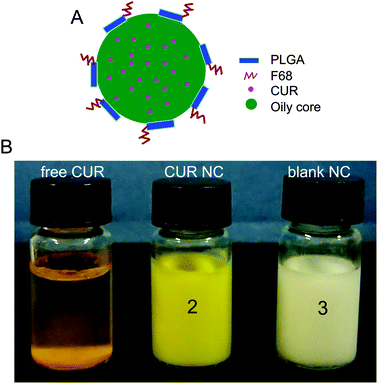
The solvent displacement method, also called the nanoprecipitation method, is a direct, rapid and easy technique to prepare polymeric NCs.28 In general, the polymeric NC synthesis needs both organic and aqueous phases. The organic phase essentially consists of a solution of a polymer, drug and oil in a water-miscible solvent. On the other hand, the aqueous phase is mainly water, supplemented with surfactants. In this study, after the organic solution is injected into Pluronic F68 (poloxamer 188) water solution, the resulting mixed phase immediately turned milky as a result of the formation of NCs due to the diffusion of the organic solvent towards the aqueous phase. Fig. 1A shows the schematic representation of the copolymer PLGA NCs incorporated with CUR. The CUR NCs demonstrate a reservoir system composed of core–shell structures with a MCT oily core, in which the CUR is dispersed, surrounded by an outer polymer shell with a hydrophobic PLGA block and hydrophilic Pluronic F68 block. Since CUR is lipophilic with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of about 3.7, most of this drug remains in the oily MCT phase. Furthermore, the hydrophobic core composed of MCT oil is effective in providing an ideal microenvironment to keep CUR more stable and protection after administration.29
P values of about 3.7, most of this drug remains in the oily MCT phase. Furthermore, the hydrophobic core composed of MCT oil is effective in providing an ideal microenvironment to keep CUR more stable and protection after administration.29
CUR shows very low water solubility and has poor bioavailability.30 The solubility of CUR in water was determined to be about 0.6 μg ml−1.31 In order to improve its water solubility, CUR NC formulations were prepared using the modified solvent displacement method. As shown in Fig. 1B (tube 1), most of the CUR particles were precipitated and could not dissolve in water. Owing to its water solubility, pharmacological potential of CUR is limited.32 In this study, using PLGA NCs as nanoformulations could overcome the water insolubility of CUR, as seen in Fig. 1B (tube 2).
Table 1 shows the particle size, size distribution, zeta potential and EE% of the different CUR NC formulations. When the volume ratio of the organic phase to the water phase (O/W ratio) was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the CUR NCs had mean diameters between 132 and 185 nm with a narrow PDI between 0.146 and 0.185. The water-solubility of CUR in the NC suspension was obtained at the highest value of 926 μg ml−1, which increased to about 1500-fold compared to free CUR. When the O/W ratio was 1
2, the CUR NCs had mean diameters between 132 and 185 nm with a narrow PDI between 0.146 and 0.185. The water-solubility of CUR in the NC suspension was obtained at the highest value of 926 μg ml−1, which increased to about 1500-fold compared to free CUR. When the O/W ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, the NCs had larger particle sizes between 229 nm and 304 nm and increased PDI values between 0.216 and 0.295. The water-solubility of CUR in the NC suspension can reach 755 μg ml−1.
4, the NCs had larger particle sizes between 229 nm and 304 nm and increased PDI values between 0.216 and 0.295. The water-solubility of CUR in the NC suspension can reach 755 μg ml−1.
| O/W | CUR (mg) | Z-Ave (nm) | PDI | Zeta potentials | EE (%) | CUR solubility in NC (μg ml−1) |
|---|---|---|---|---|---|---|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
0 | 132 ± 11 | 0.146 | −33.1 ± 0.32 | — | — |
| 5 | 141 ± 5 | 0.182 | −32.5 ± 1.26 | 94.56 | 591 | |
| 8 | 158 ± 19 | 0.156 | −29.1 ± 0.44 | 92.64 | 926 | |
| 10 | 185 ± 16 | 0.165 | −30.1 ± 1.58 | 65.54 | 819 | |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
0 | 229 ± 12 | 0.216 | −31.6 ± 1.23 | — | — |
| 5 | 243 ± 13 | 0.223 | −29.2 ± 1.88 | 83.34 | 521 | |
| 8 | 231 ± 7 | 0.258 | −29.5 ± 1.34 | 75.46 | 755 | |
| 10 | 304 ± 17 | 0.295 | −30.1 ± 2.33 | 51.25 | 641 |
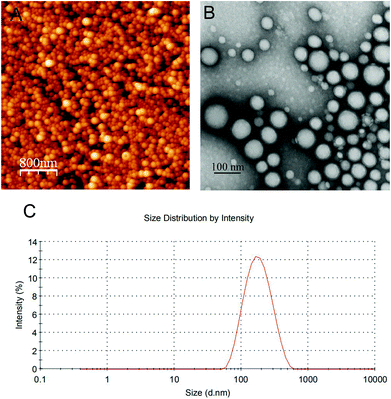
Electron microscopy can provide direct information on the particle shape and morphology. Fig. 2 shows the AFM and TEM images of NCs. AFM provides the possibility for visualizing the particle shape and structure in a natural environment.33 The CUR NCs showed homogeneous particles with a spherical shape in the AFM images (Fig. 2A). It could also be seen that the NC surfaces were smooth. TEM structural and morphological study of polymeric NCs can be made using the negative staining method. In the TEM image, the negatively stained formulation revealed spherical NCs with a narrow size distribution (Fig. 2B). The NCs also exhibited a core–shell structure with homogeneous shells. And, the particle size of CUR NCs from DLS (Fig. 2C) was in agreement with that determined by TEM and AFM.
The CUR-loaded NCs are more stable without any precipitation for at least 3 weeks. Fig. 3 shows the particle size and PDI of CUR-loaded NCs in 3 weeks. After 3-week storage of CUR NCs at 4 °C, no appreciable size change of nanoparticles with a very slight decrease in the PDI value was detected by DLS, and no aggregation of CUR NCs was observed. However, many physical and chemical factors may destabilize the NC suspensions to have poor long-term stability.34
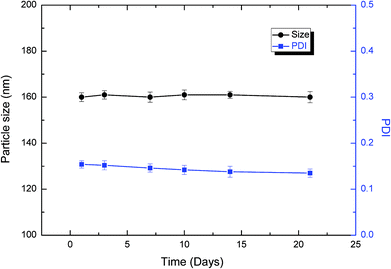 | ||
| Fig. 3 Particle size and PDI of CUR NCs determined by DLS in 3 weeks. Data are presented as mean ± standard deviation, N = 3. | ||
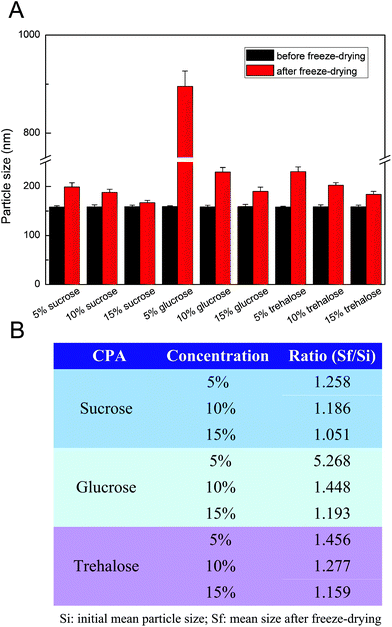
Freeze-drying is the widely used drying method of nanoparticle suspensions in pharmaceutical applications.35,36 During the freeze-drying process, cryoprotective agents (CPAs) may be added to the nanoparticle suspensions to increase their stability. In this study, redispersion cannot be possible and macroscopic aggregation was observed when the freeze-drying process of NC formulations was performed without CPAs. Thus, the CUR NCs were lyophilised in the presence of CPAs (sucrose, glucose, and trehalose) with increasing concentrations. Fig. 4A shows the most suitable CPAs with different concentrations for the freeze-drying process of CUR NCs. All the lyophilised NC samples could be easy and rapid to reconstitute. However, in the presence of 5% sucrose, the particle size of NCs showed a large change after freeze-drying and reconstitution, which could be due to the formation of aggregated particles. Fig. 4B shows the Sf/Si ratio before and after freeze-drying in the presence of different CPAs. Compared with glucose and trehalose, sucrose with a smaller Sf/Si ratio was identified as being the most suitable CPA for freeze-drying NCs.
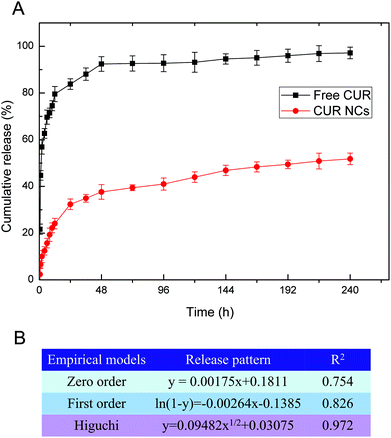
In drug release study in vitro, PBS containing 0.5% (v/v) SDS was used to maintain the sink conditions of CUR. And, 5% (w/v) Pluronic F68 was used during CUR NC formulation, which could be effective solubilization enhancers to improve the solubility and dissolution of water-insoluble drugs.37 From Fig. 5A, CUR release from CUR-NCs showed a burst release in the first 24 h to reach 31.92% release because of dissolution of CUR accumulated onto the NC surface followed by the sustained release of the interior CUR in the MCT oily core over 10 days to reach 48.5% release. This result suggests that these CUR NCs could stay therapeutically active for a minimum of 10 days. This drug release result indicated that CUR NCs for antibacterial administration can be reduced once every 10 days, which can improve patient compliance and reduce medical costs.
The drug release from polymeric nanocarriers is a complex procedure, which may be affected by many factors such as containing drug diffusion, drug distribution, polymer chain relaxation and polymer erosion.38 To clarify the mechanism of CUR release from PLGA NCs, data were fitted to zero order, first order and Higuchi models. Fig. 5B shows the correlation coefficient values (R2) as the indicators of the best fitting model for CUR NCs. The R2 value (0.972) for the Higuchi model was the greatest, which indicates that CUR release is controlled by diffusion mechanisms. A probable reason for this behavior could be due to the most amount of CUR remaining in the interior oily phase of PLGA NCs.
In this study, bacterial strains are used for the determination and examination of the antibacterial activity of free CUR and CUR NCs, by evaluating the minimum inhibitory concentration (MIC) against six bacterial strains, including three Gram negative (Escherichia coli ATCC25922, Salmonella typhimurium CGMCC1.1194 and Pseudomonas aeruginosa CGMCC1.10274) and three Gram positive (Staphylococcus aureus ATCC 29213, Bacillus sonorensis CGMCC1.10580 and Bacillus licheniformis CGMCC1.10314) bacteria. The CUR NC solution was prepared with distilled water, while the free CUR solution was prepared with distilled water and DMSO was added to the solution. The results were obtained after treating different bacterial strains over 18 h. The MIC value was defined as the lowest concentration of free CUR/CUR NCs that can prevent bacterial growth.
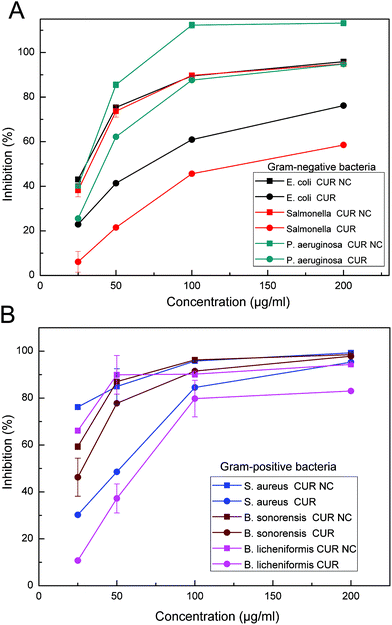
Table 2 shows the MICs of free CUR and CUR NCs against different bacteria. The higher bacterial sensitivity was found in Gram-positive bacteria of S. aureus (MIC = 100 μg ml−1), B. sonorensis (MIC = 100 μg ml−1), and B. licheniformis (MIC = 100 μg ml−1) when treated with free CUR, compared with CUR NC treatment for S. aureus (MIC = 75 μg ml−1), B. sonorensis (MIC = 75 μg ml−1), and B. licheniformis (MIC = 75 μg ml−1). And, the lower sensitivity was found in Gram-negative bacteria of E. coli (MIC = 300 μg ml−1), Salmonella (MIC = 300 μg ml−1), and P. aeruginosa (MIC = 200 μg ml−1) when treated with free CUR, compared with CUR NC treatment for E. coli (MIC = 100 μg ml−1), Salmonella (MIC = 100 μg ml−1), and P. aeruginosa (MIC = 100 μg ml−1). This means that selected Gram-positive bacteria (S. aureus, B. sonorensis, and B. licheniformis) had higher sensitivity than selected Gram-negative bacteria (E. coli, Salmonella, and P. aeruginosa) for both free CUR and CUR NCs. This result may be explained by the fact that Gram-negative bacteria have a double membrane envelope that prevents many antibiotics from accessing their targets.39,40 More interestingly, MICs of CUR NCs in this study were lower than those of free CUR for all different bacterial strains, especially for Gram-negative bacteria. These results showed that CUR NC formulations had enhanced antibacterial activity than free CUR in DMSO, which revealed that the therapeutic efficiency of CUR improved upon NC formation. The higher effectiveness of CUR NCs on Gram-negative bacteria is most important for the treatment of MDR bacterial infections.41
| Organism | MIC (μg ml−1) | ||
|---|---|---|---|
| Curcumin (DMSO) | Curcumin NC (water) | ||
| Gram-negative bacteria | E. coli | 300 | 100 |
| Salmonella | 300 | 100 | |
| P. aeruginosa | 250 | 100 | |
| Gram-positive bacteria | S. aureus | 100 | 75 |
| B. sonorensis | 100 | 75 | |
| B. licheniformis | 100 | 75 | |
To further investigate the antibacterial efficacy, the inhibition of all tested bacterial strains was determined with increasing the concentration from 25 μg ml−1 to 200 μg ml−1 in a dose-dependent manner (Fig. 6). In the selected Gram-negative bacteria, P. aeruginosa had a highest inhibition % when treated with CUR NCs. In the selected Gram-positive bacteria, S. aureus had a highest inhibition % when treated with CUR NCs. Moreover, the results clearly show that CUR NCs had more ability to inhibit all tested bacterial strains compared with free CUR. The enhanced antibacterial activity of CUR NCs could be mainly due to the reduced particle sizes. The particle size of CUR NCs (158 nm) in this study is much smaller than the size of CUR particles dissolved in DMSO (500–800 nm), which is responsible for higher cellular uptake,42 leading to more damage in the cell membrane of the bacteria. These results are in agreement with the earlier reports on CUR nanoparticles with high antibacterial activity.43,44 Basniwal et al. showed that CUR nanoparticles could bind to the bacterial cell walls, break the peptidoglycan layer and disrupt the structure of cell organelles.44
Conclusion
In summary, we have successfully developed CUR PLGA NCs composed of an MCT oily core with mean particle sizes of around 158 nm and a narrow size distribution (PDI < 0.2). Our work shows that CUR NCs can achieve higher aqueous solubility of CUR, which could enhance its use in pharmacological formulations. CUR release was evaluated in vitro for 10 days, and only 50% of the encapsulated CUR had been released. Moreover, the results also indicated that encapsulating CUR in PLGA NCs improved the antibacterial activity of CUR against all tested bacterial strains, which displayed broad-spectrum antimicrobial activity against both Gram-positive and Gram-negative bacteria. Therefore, CUR-loaded PLGA oily core NCs may be a promising candidate as antibacterial agents.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Undergraduate Training Programs for Innovation and Entrepreneurship (201810320034Z), the Natural Science Foundation of Jiangsu Normal University (16XLR046), the Natural Science Foundation of Jiangsu Province (BK20170244), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).References
- S. Z. Moghadamtousi, H. A. Kadir, P. Hassandarvish, H. Tajik, S. Abubakar and K. Zandi, A review on antibacterial, antiviral, and antifungal activity of curcumin, BioMed. Res. Int., 2014, 2014, 186864 Search PubMed.
- S. Shome, A. D. Talukdar, M. D. Choudhury, M. K. Bhattacharya and H. Upadhyaya, Curcumin as potential therapeutic natural product: a nanobiotechnological perspective, J. Pharm. Pharmacol., 2016, 68, 1481–1500 CrossRef CAS.
- D. Rai, J. K. Singh, N. Roy and D. Panda, Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity, Biochem. J., 2008, 410, 147–155 CrossRef CAS PubMed.
- P. Tyagi, M. Singh, H. Kumari, A. Kumari and K. Mukhopadhyay, Bactericidal activity of curcumin I is associated with damaging of bacterial membrane, PLoS One, 2015, 10, e0121313 CrossRef.
- D. G. Yun and D. G. Lee, Antibacterial activity of curcumin via apoptosis-like response in Escherichia coli, Appl. Microbiol. Biotechnol., 2016, 100, 5505–5514 CrossRef CAS.
- K. Mahmood, K. M. Zia, M. Zuber, M. Salman and M. N. Anjum, Recent developments in curcumin and curcumin based polymeric materials for biomedical applications: A review, Int. J. Biol. Macromol., 2015, 81, 877–890 CrossRef CAS.
- M. M. Yallapu, P. K. Nagesh, M. Jaggi and S. C. Chauhan, Therapeutic Applications of Curcumin Nanoformulations, AAPS J., 2015, 17, 1341–1356 CrossRef CAS.
- J. Li, H. Liang, J. Liu and Z. Wang, Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy, Int. J. Pharm., 2018, 546, 215–225 CrossRef CAS.
- O. Guo, X. Li, Y. Yang, J. Wei, Q. Zhao, F. Luo and Z. Qian, Enhanced 4 T1 breast carcinoma anticancer activity by co-delivery of doxorubicin and curcumin with core-shell drug-carrier based on heparin modified poly(L-lactide) grafted polyethylenimine cationic nanoparticles, J. Biomed. Nanotechnol., 2014, 10, 227–237 CrossRef.
- H. Batra, S. Pawar and D. Bahl, Curcumin in combination with anti-cancer drugs: A nanomedicine review, Pharmacol. Res., 2018, 139, 91–105 CrossRef PubMed.
- U. Shimanovich, G. J. Bernardes, T. P. Knowles and A. Cavaco-Paulo, Protein micro- and nano-capsules for biomedical applications, Chem. Soc. Rev., 2014, 43, 1361–1371 RSC.
- A. Kaur, S. Preet, V. Kumar, R. Kumar and R. Kumar, Synergetic effect of vancomycin loaded silver nanoparticles for enhanced antibacterial activity, Colloids Surf., B, 2018, 176, 62–69 CrossRef PubMed.
- K. Zhou, C. Li, D. Chen, Y. Pan, Y. Tao, W. Qu, Z. Liu, X. Wang and S. Xie, A review on nanosystems as an effective approach against infections of Staphylococcus aureus, Int. J. Nanomed., 2018, 13, 7333–7347 CrossRef CAS PubMed.
- P. Walvekar, R. Gannimani and T. Govender, Combination drug therapy via nanocarriers against infectious diseases, Eur. J. Pharm. Sci., 2019, 127, 121–141 CrossRef CAS.
- M. E. El-Naggar, F. Al-Joufi, M. Anwar, M. F. Attia and M. A. El-Bana, Curcumin-loaded PLA-PEG copolymer nanoparticles for treatment of liver inflammation in streptozotocin-induced diabetic rats, Colloids Surf., B, 2019, 177, 389–398 CrossRef CAS.
- J. Sheorain, M. Mehra, R. Thakur, S. Grewal and S. Kumari, In vitro anti-inflammatory and antioxidant potential of thymol loaded bipolymeric (tragacanth gum/chitosan) nanocarrier, Int. J. Biol. Macromol., 2019, 125, 1069–1074 CrossRef CAS.
- N. Erdoğar, S. Akkın and E. Bilensoy, Nanocapsules for Drug Delivery: An Updated Review of the Last Decade, Recent Pat. Drug Delivery Formulation, 2018, 12, 252–266 CrossRef PubMed.
- M. Peleteiro, E. Presas, J. V. González-Aramundiz, B. Sánchez-Correa, R. Simón-Vázquez, N. Csaba, M. J. Alonso and Á. González-Fernández, Polymeric Nanocapsules for Vaccine Delivery: Influence of the Polymeric Shell on the Interaction With the Immune System, Front. Immunol., 2018, 9, 791 CrossRef PubMed.
- S. Nicolas, M. A. Bolzinger, L. P. Jordheim, Y. Chevalier, H. Fessi and E. Almouazen, Polymeric nanocapsules as drug carriers for sustained anticancer activity of calcitriol in breast cancer cells, Int. J. Pharm., 2018, 550, 170–179 CrossRef CAS PubMed.
- L. A. Frank, R. V. Contri, R. C. Beck, A. R. Pohlmann and S. S. Guterres, Improving drug biological effects by encapsulation into polymeric nanocapsules, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 623–639 CAS.
- Z. Davoudi, N. Peroutka-Bigus, B. Bellaire, M. Wannemuehler, T. A. Barrett, B. Narasimhan and Q. Wang, Intestinal organoids containing poly(lactic-co-glycolic acid) nanoparticles for the treatment of inflammatory bowel diseases, J. Biomed. Mater. Res., Part A, 2018, 106, 876–886 CrossRef CAS PubMed.
- M. Mir, N. Ahmed and A. U. Rehman, Recent applications of PLGA based nanostructures in drug delivery, Colloids Surf., B, 2017, 159, 217–231 CrossRef CAS PubMed.
- K. Wang, Y. Feng, S. Li, W. Li, X. Chen, R. Yi, H. Zhang and Z. Hong, Oral Delivery of Bavachinin-Loaded PEG-PLGA Nanoparticles for Asthma Treatment in a Murine Model, J. Biomed. Nanotechnol., 2018, 14, 1806–1815 CrossRef CAS PubMed.
- Z. Davoudi, N. Peroutka-Bigus, B. Bellaire, M. Wannemuehler, T. A. Barrett, B. Narasimhan and Q. Wang, Intestinal Organoids Containing PLGA Nanoparticles for the Treatment of Inflammatory Bowel Diseases, J. Biomed. Mater. Res., Part A, 2018, 106, 876–886 CrossRef CAS PubMed.
- Q. Wang, S. Jamal, M. S. Detamore and C. Berkland, PLGA-chitosan/PLGA-alginate Nanoparticles Blends as Biodegradable Colloidal Gels for Seeding Human Umbilical Cord Mesenchymal Stem Cells, J. Biomed. Mater. Res., Part A, 2011, 96, 520–527 CrossRef PubMed.
- Q. Wang, Z. Gu, S. Jamal, M. S. Detamore and C. Berkland, Hybrid Hydroxyapatite Nanoparticle Colloidal Gels are Injectable Fillers for Bone Tissue Engineering, Tissue Eng., Part A., 2013, 19, 2586–2593 CrossRef CAS.
- J. Di, J. Yu, Q. Wang, S. Yao, D. Suo, Y. Ye, M. Pless, Y. Zhu, Y. Jing and Z. Gu, Ultrasound-triggered Noninvasive Regulation of Blood Glucose Levels Using Microgels Integrated with Insulin Nanocapsules, Nano Res., 2017, 10, 1393–1402 CrossRef CAS.
- C. E. Mora-Huertas, H. Fessi and A. Elaissari, Polymer-based nanocapsules for drug delivery, Int. J. Pharm., 2010, 385, 113–142 CrossRef CAS.
- K. H. Bae, Y. Lee and T. G. Park, Oil-encapsulating PEO-PPO-PEO/PEG shell cross-linked nanocapsules for target-specific delivery of paclitaxel, Biomacromolecules, 2007, 8, 650–656 CrossRef CAS.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Bioavailability of curcumin: problems and promises, Mol. Pharm., 2007, 4, 807–818 CrossRef CAS.
- B. T. Kurien, A. Singh, H. Matsumoto and R. H. Scofieldm, Improving the solubility and pharmacological efficacy of curcumin by heat treatment, Assay Drug Dev. Technol., 2007, 5, 567–576 CrossRef CAS.
- B. Salehi, Z. Stojanović-Radić, J. Matejić, M. Sharifi-Rad, N. V. A. Kumar, N. Martins and J. Sharifi-Rad, The therapeutic potential of curcumin: A review of clinical trials, Eur. J. Med. Chem., 2019, 163, 527–545 CrossRef CAS.
- D. Alsteens, V. Dupres, G. Andre and Y. F. Dufrêne, Frontiers in microbial nanoscopy, Nanomedicine, 2011, 6, 395–403 CrossRef PubMed.
- L. Wu, J. Zhang and W. Watanabe, Physical and chemical stability of drug nanoparticles, Adv. Drug Delivery Rev., 2011, 63, 456–469 CrossRef CAS.
- A. Umerska, K. J. Paluch, M. J. Santos-Martinez, O. I. Corrigan, C. Medina and L. Tajber, Freeze drying of polyelectrolyte complex nanoparticles: Effect of nanoparticle composition and cryoprotectant selection, Int. J. Pharm., 2018, 552, 27–38 CrossRef CAS.
- P. Fonte, S. Reis and B. Sarmento, Facts and evidences on the lyophilization of polymeric nanoparticles for drug delivery, J. Controlled Release, 2016, 225, 75–86 CrossRef CAS PubMed.
- F. Yan, C. Zhang, Y. Zheng, L. Mei, L. Tang, C. Song, H. Sun and L. Huang, The effect of poloxamer 188 on nanoparticle morphology, size, cancer cell uptake, and cytotoxicity, Nanomedicine, 2010, 6, 170–178 CrossRef CAS PubMed.
- C. Janas, Z. Mostaphaoui, L. Schmiederer, J. Bauer and M. G. Wacker, Novel polymeric micelles for drug delivery: Material characterization and formulation screening, Int. J. Pharm., 2016, 509, 197–207 CrossRef CAS.
- R. Kudva, K. Denks, P. Kuhn, A. Vogt, M. Müller and H. G. Koch, Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways, Res. Microbiol., 2013, 164, 505–534 CrossRef CAS.
- M. T. Cabeen and C. Jacobs-Wagner, Bacterial cell shape, Nat. Rev. Microbiol., 2005, 3, 601–610 CrossRef CAS PubMed.
- P. A. Smith, M. F. T. Koehler, H. S. Girgis, D. Yan, Y. Chen, Y. Chen, J. J. Crawford, M. R. Durk, R. I. Higuchi, J. Kang, J. Murray, P. Paraselli, S. Park, W. Phung, J. G. Quinn, T. C. Roberts, L. Rougé, J. B. Schwarz, E. Skippington, J. Wai, M. Xu, Z. Yu, H. Zhang and M. W. Tan, C.E Heise, Optimized arylomycins are a new class of Gram-negative antibiotics, Nature, 2018, 561, 189–194 CrossRef CAS.
- M. Wu, H. Guo, L. Liu, Y. Liu and L. Xie, Size-dependent cellular uptake and localization profiles of silver nanoparticles, Int. J. Nanomed., 2019, 14, 4247–4259 CrossRef.
- M. A. Adahoun, M. H. Al-Akhras, M. S. Jaafar and M. Bououdina, Enhanced anti-cancer and antimicrobial activities of curcumin nanoparticles, Artif. Cells, Nanomed., Biotechnol., 2017, 45, 98–107 CrossRef CAS.
- Bhawana, R. K. Basniwal, H. S. Buttar, V. K. Jain and N. Jain, Curcumin nanoparticles: preparation, characterization, and antimicrobial study, J. Agric. Food Chem., 2011, 59, 2056–2061 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |