Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microbiological water quality in a decentralized Arctic drinking water system†
Stephanie L.
Gora
 *a,
Tim
Anaviapik Soucie
b,
Nicole E.
McCormick
*a,
Tim
Anaviapik Soucie
b,
Nicole E.
McCormick
 a,
C. Carolina
Ontiveros
a,
C. Carolina
Ontiveros
 a,
Vincent
L'Hérault
c,
Megan
Gavin
d,
Benjamin F.
Trueman
a,
Vincent
L'Hérault
c,
Megan
Gavin
d,
Benjamin F.
Trueman
 a,
Jessica
Campbell
a,
Jessica
Campbell
 ae,
Amina K.
Stoddart
ae,
Amina K.
Stoddart
 a and
Graham A.
Gagnon
a and
Graham A.
Gagnon
 *a
*a
aDepartment of Civil and Resource Engineering, Dalhousie University, Halifax, NS B3H 4R2, Canada. E-mail: stephanie.gora@dal.ca; graham.gagnon@dal.ca
bCommunity of Pond Inlet, Pond Inlet, NU X0A 0S0, Canada
cARCTIConnexion, Québec, QC G1L 1Y8, Canada
dNunavut Arctic College, Iqaluit, NU X0A 0H0, Canada
eHalifax Water, 450 Cowie Hill Rd., Halifax, NS B3P 2V3, Canada
First published on 28th May 2020
Abstract
Drinking water samples were collected from the water source, water delivery truck, domestic water storage tanks, and at the point of use in a decentralized drinking water system in Pond Inlet, Nunavut, a predominantly Inuit community in Canada's Arctic region. The samples were analyzed for standard water quality parameters (turbidity, colour, pH, conductivity), biological water quality parameters (coliforms, adenosine triphosphate), and free and total chlorine. The microbial communities in a subset of water and biofilm samples were characterized with DNA analysis. Physiochemical characteristics at the tap were influenced by source water quality and building-specific conditions. Multiple aesthetic water quality parameters were above recommended values including turbidity and colour. Turbidity and biological activity (measured as cellular ATP) varied temporally in some locations. DNA analysis at the phylum, family, and genus level demonstrated that microbial ecology evolved from source water to tap and that individual storage tanks and taps were influenced by distinct microbial communities.
Water impactThis study demonstrated that drinking water quality in a decentralized trucked drinking water system in an Arctic community was influenced by both system-wide and building-specific factors and that the system is potentially vulnerable to microbial water quality hazards. This study is the first to report 16S gene information at the genus level for a decentralized trucked drinking water system in Canada. |
Introduction
Drinking water quantity and safety are perennial concerns in Arctic communities. Most homes in Nunavut have access to in-home drinking water (>95%) but a high proportion of homes report “water that is not safe to drink at least sometimes” (>80%) based on surveys conducted between 2001 and 2006.1 More recent research has demonstrated that Inuit are more likely to experience acute gastrointestinal (AGI) infections than people in Southern Canada but less likely to seek healthcare.2 In-person interviews with residents in Northern communities indicate that some people associate GI symptoms with tap water.3 Many people also consume water from secondary sources such as brooks, lakes, or icebergs or purchase bottled water. The use of gathered or purchased water is related to traditional lifestyle and/or personal preferences associated with aesthetic water quality3–7 but it is also used as a coping mechanism in communities that experience frequent water shortages.5 Challenges such as incomplete monitoring, lack of trained personnel, and difficulty maintaining chlorine residual after disinfection further complicate the provision of clean and safe drinking water in Arctic communities.5,8,9 Additionally, climate change has resulted increased temperatures, decreasing permafrost, and, depending on local soil conditions and lake characteristics, increased inputs of phosphorus and other nutrients10,11 in Arctic regions. These changes have been linked to potentially negative impacts on environmental and social determinants of Inuit health including food and water security12 as well as on water and sanitation systems in Arctic regions.13Water storage tanks have been specifically identified as potential sources of risk and microbial contamination in trucked drinking water systems in Nunavut,14 including by residents in Pond Inlet.8 A study by Martin et al.3 in Nunavik found that a small number of domestic water storage tanks contained total coliforms, Escherichia coli, (E. coli) and/or Enterococcus and secondary storage containers (5 L) used to store gathered water in the home were much more likely to be contaminated with bacteria than the raw water sources. These concerns are echoed by water quality results from studies conducted in other small and/or Indigenous communities in Canada that rely on trucked drinking water, which have demonstrated that storage tanks and secondary storage containers can act as sinks for microbial contaminants.3,7,15,16 Farenhorst et al.15 examined water quality at various points within a trucked water system in a fly-in First Nation community in Northern Manitoba, Canada. This community has a water treatment plant and two thirds of the homes are served by a piped distribution system. The remaining homes have domestic water storage tanks or no running water. The researchers found that chlorine levels were consistently below guidelines at all locations and high levels of total coliforms (including E. coli) were detected throughout the system but especially in secondary storage containers and at the community standpipe. Levels in the source and in domestic water storage tanks were higher than at the outlet of the water treatment plant and in the trucks. Another study with a focus on water quality in trucked water systems that used a mixed method approach that included monthly water quality sampling (chlorine, turbidity, temperature, total coliforms, E. coli) detected total coliforms in the trucks and most cisterns and taps and E. coli in a small number of cisterns and taps.16 Microbiological contamination was more likely to occur in August than at other times of year, suggesting seasonal/temperature effects.16
The majority of the biological water quality studies that have been conducted in Canada's Northern communities have relied on rapid onsite microbial analysis methods (e.g. Colilert™) by necessity; as laboratory facilities are scarce in Northern Canada and shipping times from the communities to laboratories in the south often exceed acceptable sample hold times. A small number of studies have also gathered DNA samples and to identify potential pathogens or to characterize the overall microbial community. Farenhorst et al.15 analyzed DNA in water samples gathered at locations through the water treatment and distribution infrastructure in a partially trucked drinking water system and their results indicated that the bacterial communities present at different points in the system were distinct from one another. Daley et al.17 analyzed samples from different points in the drinking water systems of four communities in Nunavut, including Pond Inlet, for the presence of microbial pathogens using qPCR and did not detect any species of concern in samples collected from the Pond Inlet drinking water supply system. Anaviapik-Soucie et al.8 reported detected Listeria monocytogenes in the drinking water source used in Pond Inlet at levels below the safety threshold.
The main objective of this study was to examine how source water quality, treatment (chlorination), distribution, and building-specific conditions influenced the general and microbiological quality of point of use drinking water in a decentralized drinking water system in Pond Inlet. Specific research questions included:
1. Do water samples collected from different parts of the Pond Inlet drinking water supply system achieve territorial regulations and/or Health Canada guidelines for drinking water quality?
2. What are the impacts of treatment and domestic water storage on general water quality parameters and biological activity as measured by ATP?
3. How do the microbial communities in the source water (ahead of chlorination), truck (after chlorination), domestic water storage tanks, and at the tap compare to one another?
4. Are any microbial contaminants of concern present in the drinking water supply system in Pond Inlet?
Experimental
Description of system
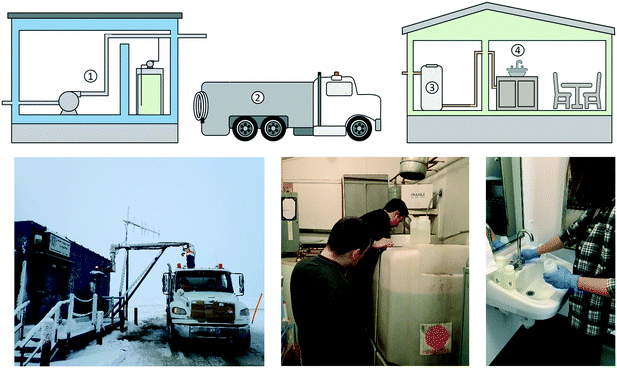
We studied the microbiological quality of the water in a decentralized drinking water system located in the community of Pond Inlet, Nunavut, located at 72°42N, 77°59W, the northernmost tip of Baffin Island in the Canadian Arctic. The community is populated by approximately 1600 people, represented by a majority of Inuit residents. Fig. 1 shows the main components of the drinking water system in Pond Inlet, NU.The community draws its water from a reservoir (Water Lake) located southwest of the hamlet. The pumphouse contains a raw water pump, a chemical pump and a sodium hypochlorite tank. Water is pumped from the reservoir, dosed with liquid sodium hypochlorite (“chlorine”) and transported directly to the community in water trucks. A minimum specific chlorine contact time (20 minutes) is satisfied in the truck before delivery to the first customer, which is in line with current recommended practices in the territory.18,19 Community buildings are supplied with chlorinated trucked water, which is stored in an onsite water holding tank until it is required by users.
The community has a longstanding tradition of freshwater gathering from nearby sources including lakes, streams and rivers, underground water, ice, and iceberg melt water.4 Some residents of the community prefer such sources to tap water, which many of them consider to be of low quality.8 However, source water uses can greatly vary in the community according to demographic and social factors (gender, age, etc.). Tap water still remains the main water type consumed by residents, some homes will use some form of in-home treatment system or device to improve the aesthetic quality of tap water. Residents can also purchase bottled water or pay a fee to fill secondary storage containers (e.g. water jugs) using reverse osmosis systems available at local stores.
Collection of water samples
Water samples were gathered from multiple locations in the Pond Inlet drinking water system between February 2018 and January 2019. The majority of the samples discussed in this paper were taken in May 2018, though some of the results presented here are also related to sampling events in February 2018 and July 2018, and two longer-term sampling programs that ran between mid-May 2018 and mid-July 2018 and from mid-October 2018 until the end of January 2019.Duplicate grab samples were collected from the pumphouse, truck, and a selection of domestic water storage tanks in sterile 250 mL HDPE sample bottles. Pumphouse samples were gathered from a tap located after the raw water pump and ahead of the chlorine dosing point. Water samples were collected from the water truck at the first delivery location after the truck had been filled at the pumphouse. The sample bottles were filled directly from the brass nozzles attached to the flexible hose used to fill the tanks. Tank water samples were only collected in February 2018, May 2018, and July 2018. The cover was removed from the top of the tank and a sterile 250 mL HDPE sample bottle was dipped in the tank and filled with water near the surface of the tank. Tap water samples were collected in sterile 250 mL HDPE bottles with the exception of those intended for DNA analysis, which were collected in sterile 1 L HDPE bottles. Water samples intended for biological and DNA analysis were collected after one minute of flushing while those intended for chemical/physical analysis were collected immediately after opening the tap (“first flush”). DNA water samples were collected in May 2018 and filtered through sterile 0.45 μm filters using sterilized instruments onsite in Pond Inlet. The filters were kept frozen until extraction at Dalhousie University in Halifax, NS.
Collection of biofilm samples
Biofilm swabs were collected from 5 storage tanks that were identified by the Nunavut Housing Authority in May 2018 for sampling. In each case, an approximately 5 cm (2 inch) square area of the inside tank wall near the surface of the water was swabbed using a sterile swab and placed in a sterile container to be transported to the lab.Chemical/physical water quality parameters
Water samples were analyzed for conductivity and pH with a YSI multiparameter sonde and turbidity was measured on a portable turbidimeter (HACH 2100Q). Free and total chlorine were measured according to the DPD colorimetric method on a handheld chlorine analyzer (HACH). Turbidity, pH, conductivity, and free and total chlorine were analyzed onsite in Pond Inlet using field instruments. Water treated with reverse osmosis was purchased at the local co-op and used to rinse probes and sample vials between samples. The calibration of the turbidimeter was checked using the manufacturer's turbidity standards before any samples were analyzed. An analytical blank consisting of reverse osmosis treated water purchased at the local co-op was run on the turbidimeter once a day and this value was subtracted from the turbidity measurements of the actual samples. The sonde that was used to measure pH and conductivity was calibrated before each use.A 15 mL aliquot of each sample was dispensed into a clean HDPE tube and shipped to Dalhousie University in Halifax, Canada, where it was analyzed for colour on a UV Vis spectrometer (HACH DR5000). Samples were also analyzed for metals (copper, iron, manganese, and lead) as described in Gora et al.20 Analytical blanks were included for all colour and metals analyses. For colour, a blank sample consisting of milliQ purified water was analyzed at the beginning of each set of samples. The absorbance of this blank sample was subtracted from that of the samples. The collection and transport of these samples and the analysis of iron, manganese, lead, and copper are described in detail in Gora et al.20
Analysis of microbiological parameters
Water samples from pumphouse and truck were analyzed for total and faecal coliforms using the Colilert Quanti-Tray method (IDEXX Laboratories, Inc., Westbrook, ME, USA), according to manufacturers' instructions. 100 mL of water sample was mixed with Colilert reagent, added to Quanti-Tray, and sealed. Quanti-Trays were incubated at 35 °C for 24 hours. Following the incubation period, positive wells were counted (yellow cells for total coliforms and fluorescent for faecal coliforms) and compared to the MPN table provided by manufacturer.Adenosine triphosphate (ATP) is an indicator of microbial activity that has been broadly used in drinking water research studies and has been shown to be correlated with standard heterotrophic plate counts.21–23 Water samples were collected from the pumphouse, truck, and domestic water storage tanks, along with the biological water samples gathered from drinking water taps after one minute of flushing, were analyzed for cellular ATP (cATP) using the QGA™ (Quench-Gone Aqueous, LuminUltra®) kit according to manufacturer's instructions. This method measures ATP from intact, viable cells. The water sample is filtered through a membrane (0.7 μm pore size), which traps intact cells but allows smaller compounds and lysed or damaged cells to pass through. At this stage the filtrate, which contains damaged cells and ATP that has been released from damaged cells, is discarded. A lysing reagent is then applied to the filter membrane that lyses the intact cells and releases intracellular ATP. The filter lysate is collected and the ATP is quantified using a luminometer. In the current study, 50 mL of sample was filtered and resuspended in the 9 mL UltraLute tubes, using 1 mL of UltraLyse 7. Then, 0.1 mL of prep sample was mixed with 0.1 mL of luminase enzyme and measured RLUs using a PhotonMaster. The chemiluminescent signal of the luminase used in the QGA ATP method was checked before each set of samples was analyzed and this value was used along with the signal from each sample to calculate the concentration of cATP in each sample according to the manufacturer's instructions.
It was not always possible to analyze blank samples for the cATP and Colilert™ tests for reasons including a lack of consistent access to sterile purified water.
DNA extraction and analysis
All materials used for DNA sample filtration were pre-sterilized at Dalhousie University before being transported to Pond Inlet in May 2018. Water samples were collected in sterile containers and filtered through sterile nitrocellulose membrane filters, (0.45 μm pore size and 25 mm diameter, Millipore®). Afterwards, filters were placed in a sterile 15 mL centrifuge tube (Fisher Scientific) and kept frozen until the moment of extraction. Biofilm samples were collected using a sterile cotton swab and preserved frozen until the moment of extraction. Frozen filters and swabs were transported to Dalhousie University in Halifax, Nova Scotia to the Centre of Water Research for DNA extraction. DNA was extracted in a sterile biosafety cabinet using the Qiagen DNeasy Powersoil kit. The filter or swab was added to the PowerBead Tube (provided by the kit) and processed according to the kit's instructions. The extracted DNA samples were sequenced at the Centre for Comparative Genomics and Evolutionary Bioinformatics at Dalhousie University in Halifax, Nova Scotia, Canada using primers to amplify the V4V 5 region of the 16S rRNA gene24 and the V4 region of the 18S rRNA gene.25 Results from the microbial community sequencing were processed using QIIME 2-v 2019 (ref. 26) following the microbiome helper workflow27 and visualized in R.28 Briefly, FastQC29 was used to inspect read quality, the cutadapt QIIME2 plugin was used to trim primers,30 sequences were denoised with DADA2,31 and taxonomy was assigned from the SILVA database v132.32 QIIME2 was used to generate alpha rarefaction curves to confirm sufficient sampling depth and alpha diversity metrics including richness, Shannon's diversity index, and Pielou's evenness index. The Shannon diversity index measures species diversity in a given community and takes into account the number of species present (richness) and the number of individuals present (abundance). Pielou's evenness index indicates how even the distribution of species is within a community. The Pielou index ranges from 0 and 1, and the more dominant species are present within a community (less evenness) the closer to 0 the index will be. Alpha diversity group significance was calculated using the Kruskal–Wallis test both methods used the Benjamini–Hochberg correction for false discovery rate (FDR). Individual ASV nucleotide sequences were compared against the NCBI database using BLASTn.Statistical analysis and graphics
All statistical analyses were conducted in R.28,33 Photographs were taken by Stephanie Gora and Jessica Campbell. Data visualizations were prepared using the ggplot2 package in R.34 The Kruskal–Wallis test was used to assess whether significant differences existed between turbidity, colour, pH, conductivity, free chlorine, and total chlorine measured in samples from the pumphouse, truck, tanks, and at the taps in May 2018 and July 2018 because these datasets deviated from normality according to the Shapiro–Wilk test. Standard ANOVA analysis with Tukey's HSD posthoc test was used to assess the impacts of sampling location and sampling date on cellular ATP levels because this dataset met the requirements of the general linear model (normality, etc.). All statistical tests were conducted at the 95% confidence level.Research license
This research was conducted under license # 02 038 18N-M issued by Nunavummi Qaujisaqtulirijikkut/Nunavut Research Institute (Iqaluit, NU).Community engagement
This study was initiated based on concerns expressed by community members related to potential microbial regrowth inside of their domestic water storage tanks. A community member was responsible for collecting and analyzing many of the water samples discussed in this paper. Preliminary results from the study were presented to community members at a public forum and discussed with Hamlet Council. The results were also communicated to the Health Department at the Government of Nunavut in the form of regular update reports. The abstract of each update report was translated into Inuktitut.Results
General water quality
Fig. 2 summarizes the turbidity, colour, pH, and free and total chlorine measured in samples collected from different points in the Pond Inlet drinking water system during an intensive sampling program executed between May 18, 2018 and May 22, 2018.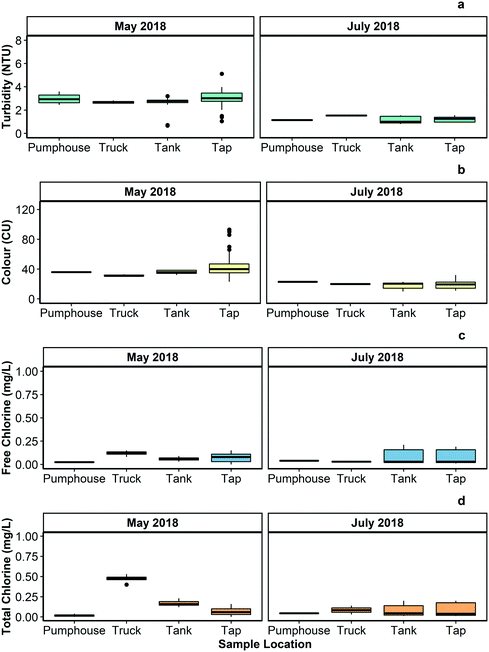 | ||
| Fig. 2 Boxplots showing (a) turbidity, (b) colour, (c) free chlorine, and (d) total chlorine, measured in water samples gathered from the pumphouse, water truck, 7 water storage tanks, and 19 taps in Pond Inlet in May 2018 and July 2018. Median, maximum, and minimum values and the number of samples analyzed for each parameter from each location in drinking water supply system are provided Tables S1 and S2 of the ESI.† | ||
Total coliforms
A total of 72 samples collected from different locations within the drinking water system were tested for coliforms using the Colilert™ test between February 2018 and January 2019. Coliform bacteria are widespread in the environment, and a total coliforms detection is not necessarily an indicator of the presence of pathogenic bacteria. Only seven samples tested positive for total coliforms. All but one of these positive samples was from the pumphouse (before chlorine addition). Additional details about the coliform results are provided in the ESI.†Cellular ATP (cATP)
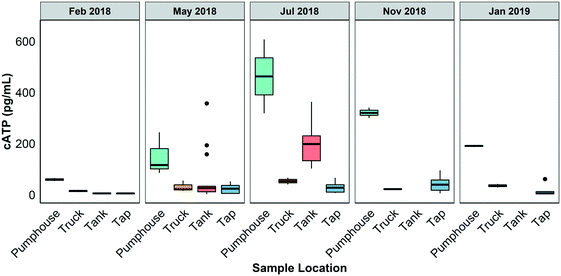
Fig. 3 shows the results of cATP measurements made on water samples gathered from different points in the Pond Inlet drinking water system in February 2018, May 2018, July 2018, November 2018, and January 2019. A two way ANOVA test conducted on the dataset indicated that there were significant differences between the sample events (May 2018 and July 2018) and sample locations and that there were interactions between these two factors at the 95% confidence level. In general, cATP was highest in the pumphouse samples and lower in the truck, in the storage tanks, and at the taps, though Tukey's posthoc testing indicated these differences were only statistically significant during the July 2018 sampling event. At this time, the pumphouse samples had cATP levels ranging from 318 to 607 pg mL−1 (median = 463 pg mL−1, n = 2) those of the truck samples ranged from 40 to 66 pg mL−1 (median = 53 pg mL−1, n = 2), the tank samples ranged from 102 to 363 pg mL−1 (median = 198 pg mL−1, n = 10), and the tap samples ranged from 6 to 65 pg mL−1 (median = 10 pg mL−1, n = 10). cATP measured in the pumphouse samples, which represent unchlorinated surface water, were significantly higher in May and July 2018 than at colder times of the year.16S microbial community analysis for bacterial DNA
The biofilm sample from building 2 had insufficient sampling depth and was not included in further analysis. The remaining samples (n = 10) were analyzed for alpha diversity and taxonomic profiles.Richness and two alpha diversity indexes (Shannon diversity index and Pielou's evenness index) were calculated for every sequenced sample (Fig. 4). The pumphouse and truck samples had the highest richness (ASVs of 254 and 247, respectively). Species richness was lower in all of the water samples (ASVs average 136 ± 55.3) and biofilm samples (ASVs average = 99.7 ± 77.7) gathered from buildings. The pumphouse and truck samples had similar alpha diversity indices (Shannon diversity index = 6.81 and 6.76 respectively, Pielou's evenness index = 0.85 in both samples) while the water and biofilm samples from the community buildings were more variable. All of the community samples had a Pielou's evenness index above 0.5 (average = 0.74 ± 0.11), which indicates species within the bacterial communities in these samples were relatively evenly distributed.
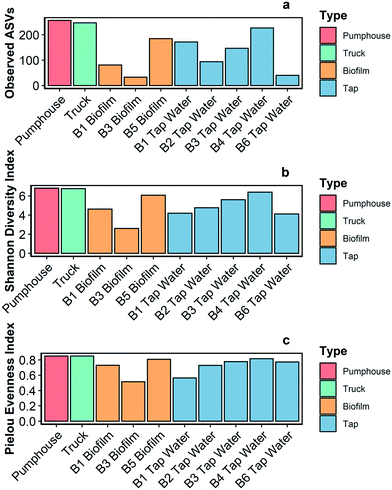 | ||
| Fig. 4 Richness (a), Shannon diversity index (b), and Pielou evenness index (c) in 10 samples gathered from various points in the Pond Inlet drinking water system in May 2018. | ||
The taxonomic profiles of the ten samples are summarized in Fig. 5 (genus level) and in Fig. S2 (phylum level) and Fig. S3 (family level) in the ESI.† More detailed information is provided in Tables S3–S5 in the ESI.†
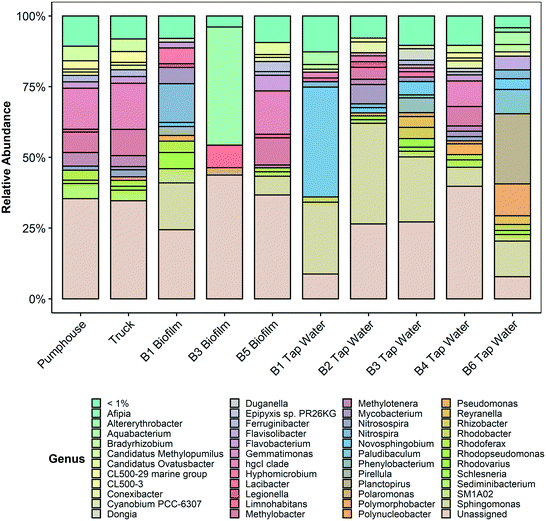 | ||
| Fig. 5 Microbial communities at the genus level (>1% abundance) in 10 samples gathered from various points in the Pond Inlet drinking water system in May 2018 and sequenced for prokaryotic DNA (16S V4–V5). Additional identification and abundance details are provided in Table S5 in the ESI.† | ||
Analysis of the taxa present in each sample found that all samples were composed of the phyla Proteobacteria, Planctomycetes, Bacteriodetes, and Actinobacteria, however, the relative abundances of these phyla in each sample were variable (see ESI†). The phylum Cyanobacteria was also present at low abundances in samples from the pumphouse, truck, and several community buildings.
Legionella, a genus that includes the pathogenic species Legionella pneumophila that is responsible for Legionnaires' disease, was found in the biofilm sample from building 1 (1.2%) and the water sample from building 2 (1.9%). Legionella was also present at less than 1% abundance in water samples from buildings 1, 3, 4, the pumphouse, the truck, and the biofilm samples from building 3 and building 5. Mycobacterium, a genus that includes pathogenic species such as Mycobacterium tuberculosis, non-tuberculous Mycobacteria opportunistic pathogens, and various non-pathogenic Mycobacterium species, was detected in the biofilm sample from building 1 (5.8%) and the water samples from building 2 (6.8%), building 3 (1.6%), building 4 (1.9%), the pumphouse (1%), and the truck (1.2%). Mycobacterium was also present at less than 1% abundance in the water samples from building 1 and building 6 and the biofilm samples from building 3 and building 5. All of the water samples and most of the biofilm samples contained ASVs from the family Sphingomonadceae, though the abundance varied from 7.1% in the biofilm sample from building 5 to 65% in the tap water sample from building 1. Bacteria from this family are ubiquitous in the environment, including in tap water and health-care settings, and have been associated with opportunistic infections.35
Lead was detected at taps in community buildings in Pond Inlet in two previous studies.17,20 Genera that contain species associated with lead corrosion, including Sediminibacterium,36Hyphomicrobium,36Duganella,37Sphinogomonas,37Novosphingobium,37Afipia,37 and Flavobacterium37 were present in all samples but, in aggregate, their abundance was greater in water from building 1 (66.4%), building 2 (40%), building 3 (31.9%), and building 6 (18.9%), and in the biofilm samples from building 1 (25.7%). These genera were present in the pumphouse and truck water samples as well but in lower abundances (7.5% and 6.1%, respectively). The abundance of each lead corrosion genus is presented in Table 1.
| Genus | Sediminibacterium | Hyphomicrobium | Duganella | Sphingomonas | Novosphingobium | Afipia | Flavobacterium |
|---|---|---|---|---|---|---|---|
| n.d. = not detected. | |||||||
| Pumphouse | 5.3% | n.d. | n.d. | n.d. | n.d. | n.d. | 2.3% |
| Truck | 3.7% | n.d. | n.d. | n.d. | n.d. | n.d. | 2.4% |
| B1 water | <1.0% | <1.0% | <1.0% | 25.0% | 38.9% | <1.0% | <1.0% |
| B2 water | 1.3% | <1.0% | n.d. | 35.6% | 1.9% | n.d. | 1.1% |
| B3 water | <1.0% | 1.8% | n.d. | 23.0% | 4.7% | <1.0% | 1.2% |
| B4 water | 2.6% | <1.0% | 1.2% | 6.7% | <1.0% | <1.0% | 2.2% |
| B6 water | <1.0% | <1.0% | n.d. | 12.6% | 3.8% | 1.6% | n.d. |
| B1 biofilm | n.d. | 5.6% | <1.0% | 16.6% | 1.4% | n.d. | 2.0% |
| B3 biofilm | n.d. | <1.0% | n.d. | <1.0% | n.d. | <1.0% | <1% |
| B5 biofilm | 1.6% | <1.0% | <1.0% | 6.6% | <1.0% | n.d. | 5.5% |
Other genera of potential concern in drinking water systems such as Campylobacter, Salmonella, Shigella, and Vibrio, and those that encompass coliform bacteria such as Citrobacter, Enterobacter, Escherichia, Hafnia, Klebsiella, Serratia, and Yersinia were not present in any of the biofilm or water samples.38
18S microbial community analysis for eukaryotic DNA
The extracted DNA was also sequenced for eukaryotes, and, to the authors' knowledge, this is the first study to report non-targeted 18S gene information for a decentralized trucked drinking water system in North America. The water samples from building 1, building 2, building 3, and building 6, and the biofilm sample from building 1 had insufficient sampling depth and were not included in further analysis. No sequences were amplified from the biofilm sample taken in building 3. The remaining samples (n = 5) were analyzed for alpha diversity (Fig. S4†) and taxonomic profiles (Table S6†).Richness was greatest in the biofilm sample from building 5 (ASVs of 121) and lowest in the water sample from the pumphouse (ASVs of 29). The Shannon diversity index was lowest at the pumphouse and truck samples (2.21 and 2.55, respectively) and highest in the biofilm sample from building 5 (4.98). Pielou's evenness index was also lowest in the pumphouse and truck water samples (0.46 and 0.41, respectively) and highest in the biofilm sample from building 5 (0.72). There were no significant differences in diversity or evenness when comparing water and biofilm samples or when comparing samples taken from the community buildings and the samples taken from the water source (pumphouse and truck samples).
The taxonomic profiles of eukaryotes in the samples are summarized in Table S6.† The communities in the pumphouse and truck samples were similar to one another and mainly composed of the order Cyclopoida (64.9% and 67.1%, respectively) and the genus Biecheleria (22.9% and 14%, respectively). The taxonomy of the water and biofilm samples taken from the community buildings was different from that in the pumphouse and truck samples and included several microorganisms of potential interest. For example, the biofilm samples from building 2 and building 5 contained the dinoflagellate Symbiodinium (83.1% and 28.6%, respectively). The waterborne pathogenic genera Cryptosporidium and Giardia were not found in any samples.
Discussion
General water quality and compliance with territorial and federal water quality guidelines
Elevated turbidity can negatively impact the effectiveness of disinfection processes, including chlorination, by consuming disinfectant chemicals and/or shielding microorganisms. In May 2018, the turbidity of the water samples collected from the Pond Inlet drinking water system was above Health Canada's recommended level for distributed water quality (1.0 NTU) but within the current acceptable turbidity levels as established by the Government of Nunavut (5.0 NTU). Turbidity was lower in July 2018, but still occasionally above the Health Canada guideline.In the context of water quality, colour refers to the presence of dissolved organic matter, metals (e.g., iron; manganese), and other substances that absorb, disperse, or otherwise interact with visible light. The Government of Nunavut recommends that drinking water have colour less than 15 CU, which is in line with Health Canada's recommendations for this parameter.39 The colour measurements in the samples gathered from the Pond Inlet drinking water system were consistently above 15 CU. Colour was higher during the May 2018 sampling event than the July 2018 sampling event, suggesting that there may be temporal differences in colour levels in the community's drinking water at different times of the year.
Conductivity was higher in the samples collected from the taps in the community buildings than those collected at other points in the drinking water supply system, suggesting that ionic species were leaching into the water from the premise plumbing leading into and out of the storage tanks or from the walls of the tanks. This is in line with the results reported in Gora et al.,20 which indicated that lead and copper were released from premises plumbing in community buildings. The pH of the water was generally below Health Canada's recommended operational range of 7.0 to 10.5. This range is recommended because it helps to limit the corrosion of distribution system and premise plumbing components.40
The Government of Nunavut's Guidelines for Drinking Water Delivery specifies that delivered water should contain 0.20 mg L−1 of free chlorine.19 Studies in trucked drinking water systems in Arctic communities17,41 and elsewhere in Canada15 have reported free chlorine levels below regulatory values. This trend was also observed in this study – all of the free chlorine measurements made in May 2018 and July 2018 were below 0.20 mg L−1. Total chlorine was higher than free chlorine in many locations during May 2018 sampling event, indicating that most of the chlorine that was added to the water was converted to less powerful chlorine-based compounds before it was delivered to individual buildings.
Impact of sample location on biological activity as measured by cellular ATP (cATP)
cATP is created by cells during metabolic processes and has become popular as a measure of the number of viable cells in water. Although cATP is not a regulated parameter in Nunavut or any other Canadian jurisdiction, it can provide useful information about the amount of living biomass present in water.The concentration of cATP at the pumphouse increased in warmer months and decreased in cooler months. Van der Wielen et al.42 observed similar temporal changes in ATP levels in an unchlorinated southern water distribution system. Samples gathered from the truck (post-chlorination) had lower cATP concentrations than samples gathered at the pumphouse (pre-chlorination), indicating that the chlorination process reduced the overall biological activity in the water (Fig. 3). When chlorine is added to water it damages and permeabilizes bacterial cell membranes, which releases intracellular compounds (including ATP) and results in cell death.43,44 In this study, samples from the truck were collected and ATP was measured after 20 minutes of exposure to chlorine. This is sufficient time for cell membrane damage and lysis to occur. The cATP method employed in this study only measures ATP from intact cells, so the reduction in ATP between the pumphouse and the truck observed in Fig. 3 is due to a reduction in biological activity. Additional sampling over multiple years would be required to confirm that biological activity consistently increases in the storage tanks in warmer months.
Microbial composition of samples collected from different parts of the drinking water supply system
Bacterial richness, diversity, and evenness were similar for the pumphouse and truck samples, but lower and more variable among the community buildings (Fig. 4). This pattern was also observed in the eukaryotic results, however, in this case the richness, diversity, and evenness were lowest in the pumphouse and truck samples, and greatest in the biofilm from building 5 (Fig. S4†). DNA sequencing and microbial community analysis indicated that samples gathered from different parts of the drinking water system in Pond Inlet had overlapping but nonetheless distinct microbial communities.Bacterial genera that encompass pathogenic species
Several of the samples collected from the Pond Inlet drinking water system in May 2018 contained the genera Mycobacterium and Legionella. These genera encompass pathogenic species like Mycobacterium tuberculosis, opportunistic pathogens like non-tuberculous Mycobacteria and Legionella pneumophila, as well as non-pathogenic species. No data was available at the species level, so it was not possible to confirm whether this DNA was from pathogenic or non-pathogenic species.Mycobacterium was present in the source water (i.e. pumphouse and truck samples) at low abundance (1% and 1.2%, respectively), but was present in higher abundance in the water sample from building 2 and the biofilm from building 1 (6.8% and 5.8% respectively). Mycobacterium species are ubiquitous in the environment45 and have been detected in water and biofilms in piped drinking water systems.46,47 The genus Mycobacterium includes the pathogen Mycobacterium tuberculosis, which is the causative agent of tuberculosis, as well as non-tuberculous Mycobacteria opportunistic pathogens. Although Canada has one of the lowest incidence rates in the world, tuberculosis infections continue to be a concern in the Canadian Arctic.48
Legionella was more abundant in the water sample from building 2 (1.9%) and the biofilm sample from building 1 (1.2%), than in water and biofilm samples from the pumphouse and the truck (<1%). Legionella, including the opportunistic pathogen Legionella pneumophila, is associated with Legionnaire's disease. Legionella enters the human body via the inhalation of aerosols produced when infected water is used for activities such as showering.49 Various species of Legionella have been observed in biofilms in water distribution systems and premises plumbing,50 including showerheads and hot water tanks.51Legionella is more likely to be detected when the temperature of the water is above 18 °C and the free chlorine residual is low (<1 mg L−1).52 Water storage tanks in Pond Inlet are located indoors in heated rooms and evidence from multiple studies, including this one, suggests that the drinking water often has low free chlorine residuals. Elevated concentrations of iron (>50 μg L−1) have also been linked to Legionella occurrence.53 Iron concentrations above Health Canada's recommended aesthetic objective of 300 μg L−1 (ref. 54) have been detected in drinking water in Pond Inlet in past studies.17,20
Although these results are limited to the genus level, and as such the identity of pathogenic Mycobacterium and Legionella species cannot be confirmed, these data indicate that potential health risks are present in the current drinking water system. Future DNA sampling in the community should include methods for targeted identification of pathogenic Mycobacterium and Legionella species.
Bacteria associated with lead corrosion
Several genera that include species associated with lead corrosion were found at a low abundance in the source water and were enriched in the community buildings (Table 1). Water samples from community buildings, in particular building 1 and building 6, were previously found to have elevated lead levels relative to the source water samples and other buildings in the community.20 It has been previously suggested that bacteria and biofilms may actively contribute to lead corrosion.37Sediminibacterium and Hyphomicrobium36 have been observed in biofilms in bench scale experiments examining the effect of orthophosphate on lead and copper in premises plumbing, and Duganella, Sphingomonas, Novosphingobium, Afipia, and Flavobacterium were found in biofilm on a corroded lead service line collected from a distributed drinking water system in a previous study.37 The lead corrosion associated genera found in this study may be associated with the elevated lead levels found in some Pond Inlet community buildings.Dinoflagellates as a potential source of taste and odour in drinking water
There was a large abundance of dinoflagellates present in all samples, including the genera Symbiodinium and Biecheleria. These two genera accounted for 83.1% and 44.7% of the total eukaryotic population in the biofilms of building 2 and building 5 respectively. The residents of these buildings had expressed concerns about the quality of their point of use drinking water due in part to a constant and pervasive fishy odor. Dinoflagellates have previously been reported to cause taste and odor problems in drinking water,55,56 specifically noted as a fishy odor, and these genera may be responsible for the undesirable smell associated with the water in buildings 2 and 5. Given the unexpected identification of Symbiodinium in premises plumbing sourced from a fresh water lake in the Canadian high Arctic, the individual ASV DNA sequences were compared to the NCBI database using BLASTn to confirm the taxonomic classification assigned by QIIME2 and the SILVA database. To the authors' knowledge this is the first reporting of the genus Symbiodinium in the biofilm within drinking water premises plumbing, and the first reporting of the genus in the Canadian high Arctic. Symbiodinium has previously been reported to produce several toxins,57,58 though no instances of toxicity in humans have been reported.Cyanobacteria
The phylum Cyanobacteria was detected in samples from the pumphouse, truck, and several community buildings, albeit at low abundances. Cyanobacteria species are ubiquitous in the global environment and have previously been detected in lakes,59,60 benthic mats,61 and snow62 in the Arctic. Some Cyanobacteria species can release toxins with negative human health effects, but the data collected in this study was insufficient to confirm the presence of toxin-releasing species or any direct human health impacts related to Cyanobacteria species. Instead, the detection of organisms in the Cyanobacteria phylum in the drinking water supply in Pond Inlet is potentially associated with changing climatic conditions in Arctic regions, including increased temperatures and, in some areas, increased inputs of phosphorus and other nutrients into surface water bodies.10,11Conclusions
The results of this study suggest that point of use drinking water quality in Pond Inlet was out of compliance with territorial and federal water quality guidelines, influenced by both system-wide and building-specific factors; and vulnerable to microbial water quality hazards. Turbidity and colour exceeded Health Canada and Nunavut territorial water quality recommended guidelines while free chlorine was below territorial recommendations for drinking water. In general, physical parameters such as pH, turbidity, and colour were constant throughout the drinking water system. This was not surprising as the only treatment applied to the water is chlorination, which would not be expected have any substantial impact on these parameters at the levels applied in Pond Inlet. Other parameters such as conductivity and free and total chlorine varied from one sample location to another.cATP levels were higher in samples collected from the pumphouse (ahead of chlorination) than at other points in the drinking water supply system. Elevated cATP levels were observed in some storage tanks during the warmer months. The cATP dataset for this study was limited and the cATP measurement does not differentiate between pathogenic and non-pathogenic biomass. Nonetheless, the results of this study do suggest that conditions in some parts of the Pond Inlet drinking water system, including the storage tanks, are conducive to the growth of microorganisms.
Ten non-replicated DNA samples were collected from the Pond Inlet drinking water supply system in May 2018 and analyzed at the phylum, family, and genus level. Although the DNA data presented here should not be assumed to be representative of the drinking water system in Pond Inlet at all times of the year or of Arctic drinking water systems more generally, they do provide some insight into the disparate microbial communities present in the source water (pumphouse and truck) and Pond Inlet community buildings. Collectively, these results suggest that the microbial composition of the drinking water system in Pond Inlet shifted from the source water, which had high bacterial species diversity and low abundances of the genera Mycobacterium and Legionella, lead corrosion associated bacteria, and dinoflagellates, to the community buildings, which had lower bacterial diversity and were enriched with these microorganisms. These results justify future investigation into the presence of potentially pathogenic organisms in drinking water infrastructure in the community.
Conflicts of interest
The authors do not have any conflicts of interest to declare.Acknowledgements
This work was funded through a research agreement with the Nunavut Arctic College (Project #3592), as supported through a Value Proposition Agreement with Irving Shipbuilding Inc. As well, the authors acknowledge partial funding support for this work through NSERC/Halifax Water Industrial Research Chair program (IRCPJ: 349838-16). The authors would like to acknowledge the contributions of members of the community of Pond Inlet, including homeowners and business operators who gave researchers access to their taps and water storage tanks. The authors would also like to acknowledge the technical assistance of Heather Daurie, Javier Locsin, Meaghan MacGillivray, and Dallys Serracin at the Centre for Water Resources Studies at Dalhousie University, and Dr. Claudio Slamovits at the Centre for Comparative Genomics and Evolutionary Bioinformatics at Dalhousie University for his expertise in Symbiodinium.References
- T. W. Hennessy and J. M. Bressler, Improving health in the Arctic region through safe and affordable access to household running water and sewer services: an Arctic Council initiative, Int. J. Circumpolar Health, 2016, 75(1), 31149 CrossRef PubMed.
- S. L. Harper, V. L. Edge, J. Ford, M. K. Thomas, D. Pearl and J. Shirley, et al., Healthcare use for acute gastrointestinal illness in two Inuit communities: Rigolet and Iqaluit, Canada, Int. J. Circumpolar Health, 2015, 74(1), 26290 CrossRef PubMed.
- D. Martin, D. Bélanger, P. Gosselin, J. Brazeau, C. Furgal and S. Déry, Drinking Water and Potential Threats to Human Health in Nunavik: Adaptation Strategies under Climate Change Conditions, Arctic, 2007, 60(2), 195–202 Search PubMed , Available from: https://journalhosting.ucalgary.ca/index.php/arctic/article/view/63306.
- T. Anaviapik-Soucie, T. Arreak and V. L'Hérault, Building Capacity to Monitor Fresh Water Quality in Pond Inlet, Elders of Pond Inlet. Historical and cultural perspective on water uses - risks to human health, climate, and environmental changes - recommendations for adaptations, Health Canada, 2015, p. 53 Search PubMed.
- K. Daley, H. Castleden, R. Jamieson, C. Furgal and L. Ell, Water systems, sanitation, and public health risks in remote communities: Inuit resident perspectives from the Canadian Arctic, Soc. Sci. Med., 2015, 135, 124–132 CrossRef PubMed.
- RICG, IHACC Research Team, C. J. Wright, J. M. Sargeant, V. L. Edge and J. D. Ford, et al., Water quality and health in northern Canada: stored drinking water and acute gastrointestinal illness in Labrador Inuit, Environ. Sci. Pollut. Res., 2018, 25(33), 32975–32987 CrossRef PubMed.
- C. J. Wright, J. M. Sargeant, V. L. Edge, J. D. Ford, K. Farahbakhsh and I. Shiwak, et al., How are perceptions associated with water consumption in Canadian Inuit? A cross-sectional survey in Rigolet, Labrador, Sci. Total Environ., 2018, 618, 369–378 CrossRef CAS PubMed.
- T. Anaviapik-Soucie, V. L'Hérault, T. Arreak, E. Maktar, M. Milton, J. Pitseolak and et al., Final Report on Research Activity 2015-16. 2016, p. 96 Search PubMed.
- A. S. Medeiros, P. Wood, S. D. Wesche, M. Bakaic and J. F. Peters, Water security for northern peoples: review of threats to Arctic freshwater systems in Nunavut, Canada, Reg. Environ. Change, 2017, 17(3), 635–647 CrossRef.
- W. B. Lyons and J. C. Finlay, Biogeochemical processes in high-latitude lakes and rivers in Polar Lakes and Rivers - Limnology of Arctic and Antarctic Aquatic Ecosystems, Oxford University Press, Oxford, UK, 2008 Search PubMed.
- J. E. Vonk, S. E. Tank, W. B. Bowden, I. Laurion, W. F. Vincent and P. Alekseychik, et al., Reviews and syntheses: Effects of permafrost thaw on Arctic aquatic ecosystems, Biogeosciences, 2015, 12(23), 7129–7167 CrossRef CAS.
- S. L. Harper, V. L. Edge, J. Ford, A. C. Willox, M. Wood, IHACC Research Team, RICG and S. A. McEwen, Climate-sensitive health priorities in Nunatsiavut, Canada, BMC Public Health, 2015, 15, 605 CrossRef PubMed.
- J. A. Warren, J. E. Berner and T. Curtis, Climate change and human health: infrastructure impacts to small remote communities in the north, Int. J. Circumpolar Health, 2005, 64(5), 487–497 CrossRef PubMed.
- K. Lane, A. K. Stoddart and G. A. Gagnon, Water safety plans as a tool for drinking water regulatory frameworks in Arctic communities, Environ. Sci. Pollut. Res., 2018, 25(33), 32988–33000 CrossRef PubMed.
- A. Farenhorst, R. Li, M. Jahan, H. M. Tun, R. Mi and I. Amarakoon, et al., Bacteria in drinking water sources of a First Nation reserve in Canada, Sci. Total Environ., 2017, 575, 813–819 CrossRef CAS PubMed.
- L. Bradford, C. Waldner, K. McLaughlin, R. Zagozewski and L. Bharadwaj, A mixed-method examination of risk factors in the truck-to-cistern drinking water system on the Beardy's and Okemasis First Nation Reserve, Saskatchewan, Canadian Water Resources Journal/Revue canadienne des ressources hydriques, 2018, 43(4), 383–400 CrossRef.
- K. Daley, L. Truelstrup Hansen, R. C. Jamieson, J. L. Hayward, G. S. Piorkowski and W. Krkosek, et al., Chemical and microbial characteristics of municipal drinking water supply systems in the Canadian Arctic, Environ. Sci. Pollut. Res., 2018, 25(33), 32926–32937 CrossRef CAS PubMed.
- Government of Nunavut, Environmental Health Manual, 2018, Available from: https://www.gov.nu.ca/sites/default/files/ehm-oct_1_2018-final.pdf Search PubMed.
- Government of Nunavut and Department of Health, Guidelines for Drinking Water Delivery Search PubMed.
- S. L. Gora, B. F. Trueman, T. Anaviapik-Soucie, M. K. Gavin, C. C. Ontiveros, J. Campbell, V. L’Hérault, A. K. Stoddart and G. A. Gagnon, Source Water Characteristics and Building-specific Factors Influence Corrosion and Point of Use Water Quality in a Decentralized Arctic Drinking Water System, Environ. Sci. Technol., 2020, 54(4), 2192–2201 CrossRef CAS PubMed.
- E. Delahaye, B. Welté, Y. Levi, G. Leblon and A. Montiel, An ATP-based method for monitoring the microbiological drinking water quality in a distribution network, Water Res., 2003, 37(15), 3689–3696 CrossRef CAS PubMed.
- R. A. Deininger and J. Lee, Rapid determination of bacteria in drinking water using an ATP assay, Field Anal. Chem. Technol., 2001, 5(4), 185–189 CrossRef.
- I. Douterelo, J. B. Boxall, P. Deines, R. Sekar, K. E. Fish and C. A. Biggs, Methodological approaches for studying the microbial ecology of drinking water distribution systems, Water Res., 2014, 65, 134–156 CrossRef CAS PubMed.
- W. Walters, E. R. Hyde, D. Berg-Lyons, G. Ackermann, G. Humphrey, A. Parada and H. Bik, et al., Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys, mSystems, 2016, 1(1), e00009-15 CrossRef PubMed.
- A. M. Comeau, W. K. W. Li, J.-É. Tremblay, E. C. Carmack, C. Lovejoy and J. A. Gilbert, Arctic Ocean Microbial Community Structure before and after the 2007 Record Sea Ice Minimum, PLoS One, 2011, 6(11), e27492 CrossRef CAS PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet and G. A. Al-Ghalith, et al., Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 852–857 CrossRef CAS PubMed . Available from: http://www.nature.com/articles/s41587-019-0209-9.
- A. M. Comeau, G. M. Douglas, M. G. I. Langille and J. Eisen, Microbiome Helper: a Custom and Streamlined Workflow for Microbiome Research, mSystems, 2017, 2(1), e00127-16 CrossRef PubMed.
- R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2018, Available from: https://www.R-project.org/ Search PubMed.
- S. Andrews, FastQC: A quality control tool for high throughput sequence data, Babraham Bioinformatics, Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Search PubMed.
- M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet J., 2011, 17(1), 10–12 CrossRef.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13(7), 581–583 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer and P. Yarza, et al., The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2012, 41(D1), D590–D596 CrossRef PubMed.
- H. Wickham and J. Bryan, readxl: Read Excel Files. 2018, Available from: https://CRAN.R-project.org/package=readxl.
- H. Wickham, Tidyverse: Easily install and load the “Tidyverse” R Package, 2017, Available from: https://CRAN.R-project.org/package=tidyverse Search PubMed.
- I. Vaz-Moreira, O. C. Nunes and C. M. Manaia, Diversity and Antibiotic Resistance Patterns of Sphingomonadaceae Isolates from Drinking Water, Appl. Environ. Microbiol., 2011, 77(16), 5697–5706 CrossRef CAS PubMed.
- S. J. Payne, G. S. Piorkowski, L. T. Hansen and G. A. Gagnon, Impact of Zinc Orthophosphate on Simulated Drinking Water Biofilms Influenced by Lead and Copper, J. Environ. Eng., 2016, 142(2), 04015067 CrossRef.
- C. White, M. Tancos and D. A. Lytle, Microbial Community Profile of a Lead Service Line Removed from a Drinking Water Distribution System, Appl. Environ. Microbiol., 2011, 77(15), 5557–5561 CrossRef CAS PubMed.
- Standing Committee of Analysts, Microbiology of Drinking Water - Part 1 - Water Quality and Public Health in Methods for the Examination of Waters and Associated Materials, United Kingdom: Environment Agency, 2002, Available from: http://www.standingcommitteeofanalysts.co.uk/Methods/Microbiology/drinkw.html Search PubMed.
- Health Canada, Guidelines for Canadian Drinking Water Quality - Summary Table, Health Canada, 2019 Search PubMed.
- Federal-Provincial-Territorial Committee on Drinking Water (Canada), Canada, Health Canada and Canada, Water and Air Quality Bureau, Guidelines for Canadian drinking water quality: guideline technical document - pH of drinking water, 2016, Available from: http://oaresource.library.carleton.ca/wcl/2016/20160519/H144-28-2016-eng.pdf Search PubMed.
- S. L. Harper, V. L. Edge, C. J. Schuster-Wallace, O. Berke and S. A. McEwen, Weather, Water Quality and Infectious Gastrointestinal Illness in Two Inuit Communities in Nunatsiavut, Canada: Potential Implications for Climate Change, Ecohealth, 2011, 8(1), 93–108 CrossRef PubMed.
- P. W. J. J. van der Wielen and D. van der Kooij, Effect of water composition, distance and season on the adenosine triphosphate concentration in unchlorinated drinking water in the Netherlands, Water Res., 2010, 44(17), 4860–4867 CrossRef CAS PubMed.
- C. Venkobachar, L. Iyengar and A. V. S. Prabhakara Rao, Mechanism of disinfection: Effect of chlorine on cell membrane functions, Water Res., 1977, 11, 727–729 CrossRef CAS.
- M. K. Ramseier, U. von Gunten, P. Freihofer and P. Hammes, Kinetics of membrane damage to high (HNA) and low (LNA) nucleic acid bacterial clusters in drinking water by ozone, chlorine, chlorine dioxide, monochloramine, ferrate(VI), and permanganate, Water Res., 2011, 45(3), 1490–1500 CrossRef CAS PubMed.
- M. Tebruegge and N. Curtis, Mycobacterium Nontuberculosis Species, in Principles and Practice of Pediatric Infectious Diseases, Elsevier, 2018, pp. 806.e4–812.e4, Available from: https://linkinghub.elsevier.com/retrieve/pii/B9780323401814001353 Search PubMed.
- T. C. Covert, M. R. Rodgers, A. L. Reyes and G. N. Stelma, Occurrence of Nontuberculous Mycobacteria in Environmental Samples, Appl. Environ. Microbiol., 1999, 65, 5 CrossRef.
- G. Liu, Y. Zhang, E. van der Mark, A. Magic-Knezev, A. Pinto and B. van den Bogert, et al., Assessing the origin of bacteria in tap water and distribution system in an unchlorinated drinking water system by SourceTracker using microbial community fingerprints, Water Res., 2018, 138, 86–96 CrossRef CAS PubMed.
- J. Vachon, V. Gallant and W. Siu, Tuberculosis in Canada, 2016, Can. Commun. Dis. Rep., 2018, 44(3/4), 75–81 CrossRef CAS PubMed.
- R. F. Breiman, B. S. Fields, G. N. Sanden, V. LaJean, A. Meier and J. S. Spika, Association of shower use with Legionnaires' disease. Possible role of amoebae, JAMA, J. Am. Med. Assoc., 1990, 263(21), 2924–2926 CrossRef CAS PubMed.
- H. Y. Lau and N. J. Ashbolt, The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water, J. Appl. Microbiol., 2009, 107(2), 368–378 CrossRef CAS PubMed.
- M. Pryor, S. Springthorpe, S. Riffard, T. Brooks and Y. Huo, Investigation of opportunistic pathogens in municipal drinking water under different supply and treatment regimes, Water Sci. Technol., 2004, 83–90 CrossRef CAS.
- M. W. LeChevallier, Occurrence of culturable Legionella pneumophila in drinking water distribution systems, AWWA Water Sci., 2019, 1(3), e1139 CrossRef CAS.
- W. J. Rhoads, E. Garner, P. Ji, N. Zhu, J. Parks and D. O. Schwake, et al., Distribution System Operational Deficiencies Coincide with Reported Legionnaires' Disease Clusters in Flint, Michigan, Environ. Sci. Technol., 2017, 51(20), 11986–11995 CrossRef CAS PubMed.
- Health Canada, Guidelines for Canadian Drinking Water Quality: Guideline Technical Document – Iron, 1987 Search PubMed.
- I. H. M. Suffet, D. Khiari and A. Bruchet, The drinking water taste and odor wheel for the millenium: Beyond geosmin and 2-methylisoborneol, Water Sci. Technol., 1999, 40(6), 1–13 CrossRef CAS.
- Algae detection and removal strategies for drinking water treatment plants, ed. D. R. U. Knappe, AWWA Research Foundation and Water Resources Research Institute of the University of North Carolina, AWWA Research Foundation: American Water Works Association, Denver, CO, 2004, p. 466 Search PubMed.
- T. Fukatsu, K. Onodera, Y. Ohta, Y. Oba, H. Nakamura and T. Shintani, et al., Zooxanthellamide D, a Polyhydroxy Polyene Amide from a Marine Dinoflagellate, and Chemotaxonomic Perspective of the Symbiodinium Polyols, J. Nat. Prod., 2007, 70(3), 407–411 CrossRef CAS PubMed.
- M. Kita, N. Ohishi, K. Konishi, M. Kondo, T. Koyama and M. Kitamura, et al., Symbiodinolide, a novel polyol macrolide that activates N-type Ca2+ channel, from the symbiotic marine dinoflagellate Symbiodinium sp, Tetrahedron, 2007, 63(27), 6241–6251 CrossRef CAS.
- A. M. Comeau, T. Harding, P. E. Galand, W. F. Vincent and C. Lovejoy, Vertical distribution of microbial communities in a perennially stratified Arctic lake with saline, anoxic bottom waters, Sci. Rep., 2012, 2(1), 604 CrossRef PubMed , Available from: http://www.nature.com/articles/srep00604.
- M. P. Lizotte, Phytoplankton and preimary production, in Polar Lakes and Rivers - Limnology of Arctic and Antarctic Aquatic Ecosystems, Oxford University Press, Oxford, UK, 2008 Search PubMed.
- A. Quesada, E. Fernandez-Valiente, I. Hawes and C. Howard-Williams, Benthic primary production in polar lakes and rivers, in Polar Lakes and Rivers - Limnology of Arctic and Antarctic Aquatic Ecosystems, Oxford University Press, Oxford, UK, 2008 Search PubMed.
- A. K. Møller, D. A. Søborg, W. Abu Al-Soud, S. J. Sørensen and N. Kroer, Bacterial community structure in High-Arctic snow and freshwater as revealed by pyrosequencing of 16S rRNA genes and cultivation, Polar Res., 2013, 32(1), 17390 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ew00019a |
| This journal is © The Royal Society of Chemistry 2020 |