Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interaction between a nano-formulation of atrazine and rhizosphere bacterial communities: atrazine degradation and bacterial community alterations†
Yujia
Zhai
a,
Fazel
Abdolahpur Monikh
 *a,
Juan
Wu
a,
Renato
Grillo
*a,
Juan
Wu
a,
Renato
Grillo
 b,
Daniel
Arenas-Lago
b,
Daniel
Arenas-Lago
 c,
Gopala Krishna
Darbha
c,
Gopala Krishna
Darbha
 d,
Martina G.
Vijver
d,
Martina G.
Vijver
 a and
Willie J. G. M.
Peijnenburg
a and
Willie J. G. M.
Peijnenburg
 ae
ae
aInstitute of Environmental Sciences (CML), Leiden University, P.O. Box 9518, 2300 RA, Leiden, The Netherlands. E-mail: f.a.monikh@cml.leidenuniv.nl
bDepartment of Physics and Chemistry, School of Engineering, São Paulo State University (UNESP), 15385-000, Ilha Solteira, SP, Brazil
cDepartment of Plant Biology and Soil Science, University of Vigo, As Lagoas, Marcosende, 36310, Vigo, Spain
dEnvironmental Nanoscience Laboratory, Department of Earth Sciences, Indian Institute of Science Education and Research Kolkata, Kolkata, Mohanpur, West Bengal 741246, India
eNational Institute of Public Health and the Environment (RIVM), P.O. Box 1, Bilthoven, The Netherlands
First published on 8th September 2020
Abstract
Nanotechnology can potentially revolutionize the agricultural industry by offering nano-formulations of pesticides, the so-called nano-pesticides, which can e.g. increase the efficacy and stability of the active ingredients of pesticides. However, it is unknown how a nano-formulation may modulate the interaction between the active ingredient and non-target soil (micro)organisms. Here, we show that long-term exposure to a high dosage of atrazine (ATZ) containing nano-pesticides (NPATZs), where ATZ is encapsulated in a biodegradable polymeric shell, significantly decreases the metabolic capacity of rhizosphere bacterial communities and alters their community structure and composition compared to rhizosphere bacterial communities exposed to the same amount of conventionally applied ATZ. In the rhizosphere, the NPATZs and ATZ were found to be initially degraded by Mycobacterium and Pseudomonas bacteria. As the exposure time increased, more bacterial consortia became involved in NPATZ degradation than in ATZ degradation, especially in metabolizing N-isopropylammelide to carboxybiuret catalyzed by the genes atzC and atzD. Our findings provide important insights into the time-resolved interactions between rhizosphere bacterial communities and nano-pesticides.
Environmental significanceNanotechnology can potentially revolutionize agriculture by offering opportunities e.g. to turn pesticides into nano-formulations of pesticides, the so-called nano-pesticides. Nano-formulations can significantly reduce the application rates of pesticides while increasing their efficiency. However, like the challenges associated with the application of other nanomaterials in the industry sector, the usage of nano-pesticides is also hampered by the unknown toxicity profile of the particles, especially since it has been realized that some nano-pesticides are designed to have enhanced mobility and bioavailability. In this study, we synthesized nano-pesticides using atrazine as the active ingredient and poly-ε-caprolactone as the biodegradable polymeric carrier, where ATZ is encapsulated in a nano-sized polymeric shell. We showed that nano-pesticides may have some distinct adverse effects on the non-target rhizosphere bacterial communities of plants after long-term exposure. However, this may be tackled by modifying the nano-pesticides, e.g. surface modification to increase the targetable delivery or lower release into non-target places, in the future considering the fact that nanotechnology is developing quickly. |
Introduction
Recently, nanotechnology has inspired the agricultural industry to develop nano-formulations of pesticides, the so-called nano-pesticides,1 to allow the targetable delivery of active ingredients to target organisms.2 Nano-pesticides are suggested to be more environmentally friendly compared to conventional formulations due to the targeted delivery and higher stability of the active ingredient. This allows one to significantly reduce application rates and minimizes the possible adverse effects on the environment and humans.1,3 Release and distribution can, for example, effectively be controlled and the active ingredient can be targeted and protected against degradation for a longer period of time.1 Research regarding the application of nano-pesticides is still in its infancy but some nano-fertilizers (e.g. N, P, K, Fe, Mn, Zn, Cu, and Mo developed in nano-forms and carbon nanotubes) and nano-pesticides (e.g. Ag, Cu, SiO2, ZnO and nano-formulations/encapsulations) have been commercialized in agriculture. Nano-pesticides may interact with plants by affecting the soil properties and symbiotic microbiota.4 For instance, copper(II) hydroxide [Cu(OH)2] nano-pesticides have been reported to affect the soil bacterial community, enzyme activity and microbial functioning (i.e. carbon and phosphorus cycling).5,6 The potential adverse effects of nano-pesticides on non-target (micro)organisms are however still largely unknown, especially since it has been realized that some nano-pesticides are designed to have enhanced mobility.7–9 As for any newly developed chemical or material, it is critical to understand the possible toxicity of nano-pesticides to non-target (micro)organisms.Atrazine (ATZ) is the second most widely used herbicide in the world, with for instance over 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tons applied yearly in the United States.10 Cost–benefit studies have reported that atrazine promoted corn yields up to 6%.11 Atrazine remains in soil for three months up to twelve years, depending on the soils.12 In soil, degradation of ATZ is primarily carried out by the action of soil bacteria.13,14 For example, some bacteria such as Pseudomonas sp. hydrolyze the C–Cl bond in ATZ (catalyzed by the hydrolase enzymes called AtzA, AtzB, and AtzC).15–17 Others such as Nocardioides, Arthrobacter, and Mycobacterium were found to encode atzB that is involved in dealkylation and dechlorination of ATZ.18 However, the presence of ATZ in the form of a nano-pesticide, where a shell protects the ATZ ingredient, might dramatically influence the physico-chemical properties of ATZ and its microbiological degradation.19 For instance, the encapsulation of atrazine in poly(epsilon-caprolactone) (PCL) nanocapsules was found to increase the colloidal stability and modified the ATZ release profile,19,20 where a greater pre-emergence herbicidal activity against plant seedlings was observed.21 Moreover, previous studies have been mostly focused on the degradation capacity of ATZ by individual bacterial species, such as Arthrobacter, Nocardioides, Pseudomonas, Mycobacterium or Rhodococcus.22–26 It is critical to also assess how the community of plant rhizosphere bacteria influence the degradation of the nano-formulation of ATZ. This may also influence the interactions between ATZ and the bacterial community and consequently affect the composition and diversity of rhizosphere microbial communities and change their metabolic activities.27 As a result, the soil crop production and soil sustainability may be affected over long periods of nano-pesticide application.
000 tons applied yearly in the United States.10 Cost–benefit studies have reported that atrazine promoted corn yields up to 6%.11 Atrazine remains in soil for three months up to twelve years, depending on the soils.12 In soil, degradation of ATZ is primarily carried out by the action of soil bacteria.13,14 For example, some bacteria such as Pseudomonas sp. hydrolyze the C–Cl bond in ATZ (catalyzed by the hydrolase enzymes called AtzA, AtzB, and AtzC).15–17 Others such as Nocardioides, Arthrobacter, and Mycobacterium were found to encode atzB that is involved in dealkylation and dechlorination of ATZ.18 However, the presence of ATZ in the form of a nano-pesticide, where a shell protects the ATZ ingredient, might dramatically influence the physico-chemical properties of ATZ and its microbiological degradation.19 For instance, the encapsulation of atrazine in poly(epsilon-caprolactone) (PCL) nanocapsules was found to increase the colloidal stability and modified the ATZ release profile,19,20 where a greater pre-emergence herbicidal activity against plant seedlings was observed.21 Moreover, previous studies have been mostly focused on the degradation capacity of ATZ by individual bacterial species, such as Arthrobacter, Nocardioides, Pseudomonas, Mycobacterium or Rhodococcus.22–26 It is critical to also assess how the community of plant rhizosphere bacteria influence the degradation of the nano-formulation of ATZ. This may also influence the interactions between ATZ and the bacterial community and consequently affect the composition and diversity of rhizosphere microbial communities and change their metabolic activities.27 As a result, the soil crop production and soil sustainability may be affected over long periods of nano-pesticide application.
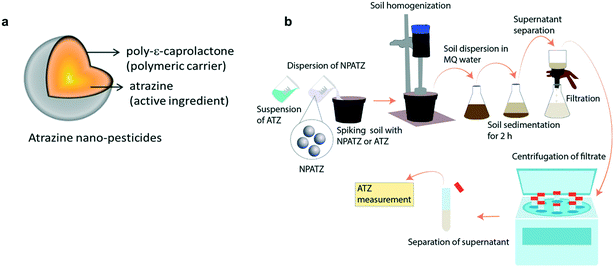
Herein, we synthesized nano-pesticides using ATZ as the active ingredient and poly-ε-caprolactone as the biodegradable polymeric carrier, where ATZ is encapsulated in a nano-sized polymeric shell (Fig. 1a). The rhizosphere microbial communities of Lactuca sativa were exposed to low, medium and high concentrations of ATZ nano-pesticides (NPATZs) for different exposure durations. We investigated (a) the influence of the NPATZs on the ecological functioning of L. sativa rhizosphere microbial communities, as non-target microorganisms, and (b) the influence of the bacteria on the degradation of the NPATZs. The metabolic capacity, functional composition, community structure and composition of the L. sativa associated rhizosphere microbial communities were monitored over 6 weeks of exposure. The bacterial consortia and functional genes associated with NPATZ degradation were also determined over time.
Materials and methods
Synthesis and characterization of atrazine nano-pesticides
The synthesis of the NPATZs was carried out following the method reported by Grillo et al. (2012), (see S1, ESI†).20 The morphology and size distribution of the NPATZs were measured using a scanning electron microscope (SEM, EVO-LS-15, Carls Zeiss), operated at high voltage of 20 kV with a spot size between 3.0 and 4.0 and a working distance (WD) of 9.5 mm. The relative frequency was measured using the ImageJ program (300 particles). The release of ATZ from the NPATZs was monitored over time in ultra-pure water (S2, ESI†) and in soil as described in Fig. 1b. The soil was spiked with the NPATZs or ATZ to reach a final concentration of 1 mg per kg soil, homogenized and incubated for 16 days. At days 1, 4, 8, and 16, an aliquot of the soil was dispersed in Milli-Q water and left to sediment. After 4 h of sedimentation, the supernatant was removed and filtered using Whatman® filter paper of 10 μm cut-off, followed by that with a 1 μm cut-off. The filtrates were centrifuged to separate the NPATZs from ATZ and measured using HPLC (Agilent 1100 series, S3, ESI†). In brief, 20 μl of sample was injected into a 4 by 250 mm Hypersil ODS column with acetonitrile/water as the mobile phase at a flow rate of 1 ml per minute.28 The hydrodynamic size and the zeta potential were measured using a Zetasizer Nano device (Malvern Panalytical, Netherlands).Plant material and growth conditions
Lactuca sativa was used as the plant model. The lettuce seeds were purchased from Floveg GmbH (Kall, Germany). After sterilizing and immersing in deionized water for 24 h, the seeds were germinated in Petri dishes filled with a rolled paper towel for 1 week (15 seeds per dish), and then pre-incubated hydroponically with a nutrient solution for 2 weeks as described previously.29 The composition of the culture medium is given in the ESI† (S4). Afterward, the uniformly pre-grown seedlings were transferred into plastic pots (9 cm long, 9 cm wide, 9.5 cm high) filled with 0.5 kg of soil collected from a clean area in The Netherlands for two weeks of further growth (no ATZ was detected in the soil). The soil was sandy loam with a pH at 7.5–8.5, containing 4 ± 0.6 mg kg−1 dissolved organic matter with the soil moisture at 18.4%. The physicochemical properties of the soil are reported in Table S1 (ESI†).Experimental design
An illustration of the experimental set-up is given in Fig. S1 (ESI†). Plants (L. sativa) were exposed to ATZ or NPATZs at a nominal concentration of 0.3, 1.5, or 3 mg ATZ per kg soil (nominal concentrations are expressed as ATZ content in the case of the nano-pesticide), representing a low concentration, medium concentration, and contaminated exposure scenario for ATZ.30 ATZ was dissolved in 5 ml acetone and further diluted to 50 ml using Milli-Q (MQ) water. To spike the soil, 15 ml of the ATZ or NPATZ suspensions were carefully and homogeneously dropped into the soil. Plants (L. sativa) were exposed to nominal concentrations of 0.3 (low concentration), 1.5 (medium concentration) and 3 (high concentration, which is representative of a contamination scenario) mg ATZ per kg soil by spiking the ATZ or NPATZs into the soil. The exposures were performed for three durations, including short-term (2 weeks), medium-term (4 weeks) and long-term (6 weeks) exposure. For the medium-term and long-term exposure, the plants were exposed at the beginning and after every two weeks of the treatment. The plants were also exposed to 1.5 mg of the polymeric carrier (PNC, without ATZ) per kg soil as a control. In another control experiment, the plants were not exposed to any chemicals.Substrate utilizations by rhizosphere bacterial communities
The collected rhizosphere soil samples were kept at 4 °C. The ability of the rhizosphere bacterial communities to utilize environmentally relevant substrates was determined following a previously reported methodology.31 Biolog EcoPlates (Biolog, Hayward, USA) were used, which contain environmentally-relevant carbon substrates. The substrate utilities offer a proxy of the metabolic potential of the rhizosphere bacterial communities in response to different treatments.32 For each rhizosphere soil sample, the soil was diluted 10 times with 10 mM BIS–TRIS buffer and centrifuged at 1500 rpm for 10 minutes (Sorvall RC5B Plus centrifuge, Fiberlite F21-8 × 50y rotor). The supernatant was then diluted 5 times using the same buffer to obtain the rhizosphere bacterial community extract.33,34 The extracts from different treatments, controls, ATZ and NPATZs, were incubated in EcoPlates separately for 96 h at 20 °C. Optical density was measured at 590 nm using a BioTek microplate reader at every 24 h during the incubation to ensure saturation of the substrate utilization. Average well color development (AWCD) was used to represent the average bacterial metabolic activity.31Soil DNA extraction and Illumina Miseq sequencing
A Qiagen DNeasy PowerSoil kit (Hilden, Germany) was used to extract the DNA from the rhizosphere soil samples in each treatment. The DNA concentration in each sample is listed in Table S2 (ESI†). Downstream sequencing was performed for quality control checking. PCR amplification was performed with a universal bacterial primer set (515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 909R: 5′-CCCGTCAATTCMTTTRAGT-3′) targeting the variable V4–V5 regions of bacterial 16S rRNA genes. Paired-end sequencing was done using a 2 × 300 bp Illumina Miseq platform (Illumina, Inc., San Diego, CA, USA) by BaseClear (Leiden, The Netherlands). The sequences have been deposited into the National Center for Biotechnology Information (NCBI) database (project number: PRJNA610041) with sample information provided in Table S3 (ESI†).Statistics and data analysis
We hypothesize that the ATZ toxicity to plant-associated rhizosphere bacterial communities is enhanced by the nano-encapsulation (NPATZ) over time compared with its chemical counterpart (ATZ). One-way ANOVA, principal coordinate analysis, and cluster and correlation analysis were used to test the hypothesis. The average well color development (AWCD) of the EcoPlates, which represents the average bacterial metabolic activity, was transformed using a natural logarithmic transformation according to our previous study.34 Statistically significant differences among the metabolic capacities of the rhizosphere bacterial communities of different treatments (controls, ATZ and NPATZ) within the same exposure duration were determined by means of one-way ANOVA and Tukey's honestly significant difference tests (with a significance level set as p < 0.05). The normality and equal variance were tested using the Shapiro–Wilk test and Bartlett's test. The significant differences between total ATZ in the NPATZ-spiked soil before and after centrifugation were determined using the t-test. The Quantitative Insights Into Microbial Ecology (QIIME version 1.8.0) pipeline (http://qiime.sourceforge.net) was used to process the 16S rRNA gene sequences (S8, ESI† for more information). After quality check, the generated OUT table was rarefied to remove sampling depth heterogeneity. Beta-diversity metrics assessing the differences among rhizosphere bacterial communities phylogenetically were computed based on the weighted UniFrac distance, and visualized with analyses such as principal coordinate analysis (PCoA). The core OTUs were selected based on the NCBI annotated ATZ-degrading bacterial genera, with presence in 100% of all the replicates for each treatment, clustered by UPGMA hierarchical clustering, and illustrated in a heatmap. The core functional genes were selected based on the KEGG pathway of ATZ degradation, with presence in 100% of all the replicates for each treatment. The correlation between the core OTUs and functional genes was assessed based on Pearson correlation analysis with a significance p < 0.05. Analyses were conducted using R v3.3.2, Paleontological Statistics (PAST, v3.14), and Statistical Analysis of Metagenomic Profiles (STAMP, v2.1.3).Results
Characterization of the nano-pesticides in different matrices
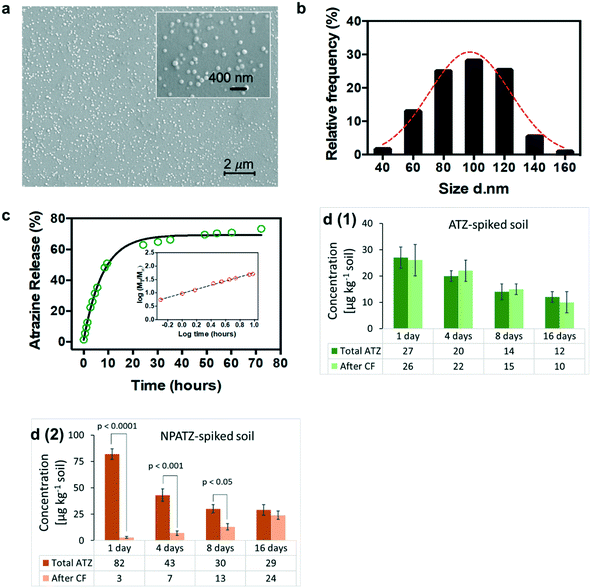
The SEM images (Fig. 2a) of the NPATZs show that the shape of the NPATZs is spherical and confirm that no particle agglomeration occurred. The median size distribution (Fig. 2b) of the NPATZs was around 100 nm as calculated from the SEM images. The hydrodynamic size of the NPATZs was 120 ± 10 nm and the zeta potential was −28 ± 4 mV. The negative charge of the particles could partially explain their stability against aggregation due to the repulsive forces between the particles.Due to the limitations in the existing analytical techniques, it is not possible to directly investigate the stability and behavior of the nano-pesticides in soil matrices. Thus, the in vitro release profile of ATZ from the NPATZs was, first, evaluated in water.35 The time to release 50% of ATZ (t50%) from the NPATZs was 11.5 hours (Fig. 2c) and the semi-empirical Korsmeyer–Peppas model (Fig. 2c – inset) indicated the transport kinetics of ATZ. The active ingredient is released through the rearrangement of polymeric chains of the carriers by a combination of solvent diffusion and polymeric relaxation processes.
We assumed that the release of ATZ from the NPATZs in the soil matrix is much lower compared to that in water due to the absence of diffusive media. To test this hypothesis, we spiked the soil with ATZ and NPATZs (1 mg per kg soil) and incubated it for two weeks (S9, ESI†). The total concentration of ATZ in the filtrate of the ATZ-spiked soil was significantly (P < 0.001) lower than the total ATZ concentration in the filtrate of the NPATZ-spiked soil (Fig. 2d). This can be explained by the low solubility of ATZ in water and confirms that the nano-formulation increased the solubility of ATZ in water. Furthermore, the filtrate samples were centrifuged and the concentration of ATZ in the supernatant was measured, assuming that the NPATZs will sediment while ATZ remains in the supernatant. After centrifugation, there was no significant variation in the total concentration of ATZ in the filtrate of the ATZ-spiked soil [Fig. 2d(1)]. However, a significantly lower concentration of ATZ was measured in the supernatant of the filtrate of the NPATZ-spiked soil, suggesting that ATZ is still present in the form of a nano-pesticide which sediments upon centrifugation [Fig. 2d(2)]. The ratio of the total ATZ after centrifugation to the total ATZ before centrifugation increased in the supernatant of the NPATZ-spiked soil over time, indicating that ATZ was released from the NPATZs in the soil matrix. However, the total concentration of ATZ in the NPATZ-spiked soil decreases over time, which could be due to the time-resolved degradation of ATZ.
The metabolic capacity of the rhizosphere bacterial communities
The measured concentrations (mean ± standard deviation) for ATZ were 0.27 ± 0.04, 1.69 ± 0.2 and 3.2 ± 0.3 mg kg−1 for low, medium and high concentrations, respectively. The measured concentrations of NPATZs were 0.25 ± 0.4, 1.38 ± 0.6 and 2.7 ± 0.5 mg kg−1 for low, medium and high concentrations, respectively. The metabolic capacity of the rhizosphere bacterial communities in response to ATZ and NPATZs after exposure for different durations is reported in Table 1. No changes in the metabolic capacity of the rhizosphere microbial communities were observed in the control samples. After short-term exposure, there were no differences in the metabolic activity of the bacterial communities exposed to ATZ or the NPATZs compared to the controls. After medium-term and long-term exposure, the metabolic capacity of the bacteria decreased as the exposure concentrations of ATZ and the NPATZs increased in comparison to the controls. A lower AWCD was observed upon exposure to a high concentration of the NPATZs compared to a higher concentration of ATZ. This indicates that an elevated concentration of the NPATZs decreased the metabolic capacity of the rhizosphere bacterial communities to a significantly higher extent as compared to the same concentration of the traditional formulation of ATZ.| Treatment | Short-term | Medium-term | Long-term |
|---|---|---|---|
| CK: control check, control plants without exposure to chemicals. ATZ-L: exposure to a low concentration of atrazine (ATZ). ATZ-M: exposure to a medium concentration of ATZ. ATZ-H: exposure to a high concentration of ATZ. PNC: exposure to the polymeric carrier without ATZ (control). NPATZ-L: exposure to a low concentration of ATZ nano-pesticides (NPATZs). NPATZ-M: exposure to a medium concentration of NPATZs. NPATZ-H: exposure to a high concentration of NPATZs. The different letters indicate significant differences among different treatments within the same exposure duration by one-way ANOVA and Tukey's honestly significant difference tests (with the significance level set as p < 0.05). | |||
| CK | 1.62 ± 0.14a | 1.33 ± 0.11a | 1.08 ± 0.09a |
| PNC | 1.55 ± 0.08a | 1.24 ± 0.12ab | 1.12 ± 0.06a |
| ATZ-L | 1.46 ± 0.22a | 1.23 ± 0.03a | 1.06 ± 0.08a |
| ATZ-M | 1.53 ± 0.12a | 1.13 ± 0.03ab | 0.94 ± 0.11ab |
| ATZ-H | 1.51 ± 0.07a | 1.14 ± 0.02ab | 0.82 ± 0.07b |
| NPATZ-L | 1.47 ± 0.11a | 1.12 ± 0.04bc | 0.89 ± 0.05b |
| NPATZ-M | 1.54 ± 0.12a | 1.10 ± 0.02bc | 0.76 ± 0.04bc |
| NPATZ-H | 1.51 ± 0.06a | 1.07 ± 0.02c | 0.65 ± 0.09c |
The structure and composition of the rhizosphere bacterial communities
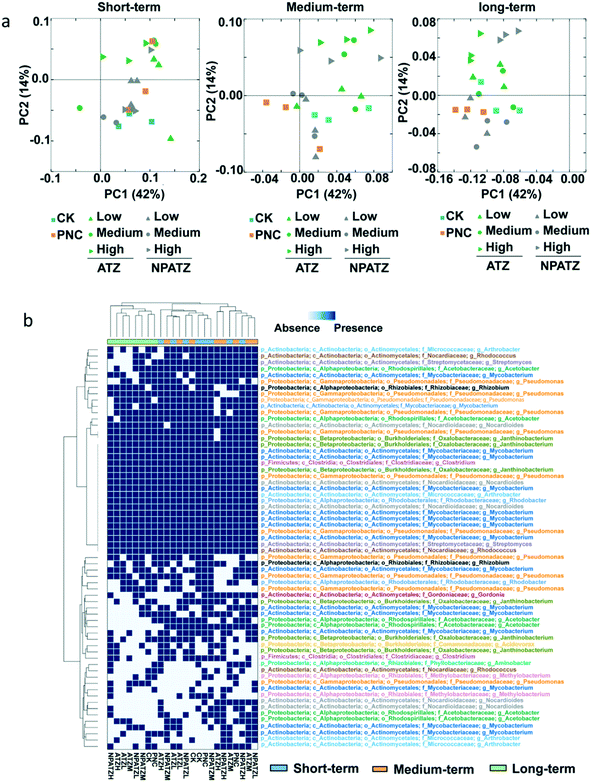
The effects of ATZ and NPATZs on the rhizosphere bacterial community structure were evaluated on the basis of sequencing data. The comparison between bacterial communities in different treatments (controls, ATZ and NPATZs) and after different exposure durations is provided in Fig. 3. The principal coordinate analysis results showed that all the bacterial communities in the different treatments clustered closely to each other after short-term exposure (Fig. 3a). By increasing the exposure duration to 4 weeks, the bacterial communities treated with a high concentration of ATZ and the NPATZs separated from the controls and from the communities treated with a lower concentration of ATZ and the NPATZs. However, there was no separation between the bacterial communities treated with high levels of ATZ and NPATZs. The differences between bacterial communities treated with the NPATZs and ATZ appeared after 4 weeks of exposure. For example, the bacterial communities exposed to a high concentration of ATZ clearly separated from the bacterial communities exposed to a high concentration of the NPATZs. This confirms that the same total amounts of ATZ and NPATZs at high concentrations induced different effects on the structure of the bacterial communities in the rhizosphere.Core operational taxonomic units (OTUs) were selected based on the ATZ-degrading bacterial genera annotated by the National Center for Biotechnology Information (NCBI) database, which were present in 100% of all the replicates for each treatment. In total, 64 core OTUs were selected and the presence/absence of these OTUs in each treatment is shown in Fig. 3b. The generated heatmap shows that, regardless of being exposed to ATZ or the NPATZs, the bacterial communities that were treated for a longer period of time (6 weeks) clustered together and separated from the bacterial communities that were treated for short-term and medium-term. After long-term exposure, the controls separated from all the ATZ and NPATZ treatments. Moreover, the highest concentration of the NPATZs induced a different mode of action on bacterial community structures compared to the highest concentration of ATZ. This was determined on the basis of the observed separation in the clusters of the bacteria in the heatmap, where bacterial communities exposed to the high concentration of the NPATZs were separated from the bacterial communities exposed to the high concentration of ATZ. After long-term exposure, the phyla of Proteobacteria (including Acetobacter, Pseudomonas, Rhizobium, Janthinobacterium, Aminobacter, Rhodobacter, Methylobacterium, Acidovorax), Actinobacteria (including Mycobacteriaceae, Gordonia, Rhodococcus, Nocardioides, Arthrobacter) and Firmicutes (including Clostridium) contributed most in inducing the observed differences between the high concentration of the NPATZs and the high concentration of ATZ.
The functional profiling of the rhizosphere bacterial community
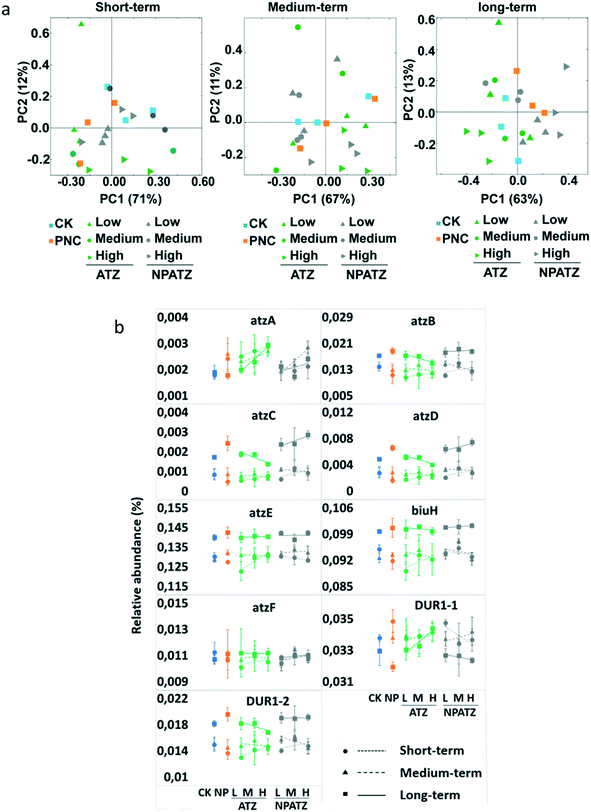
The functional composition of the metagenomes from the rhizosphere bacterial communities in different treatments and after different exposure durations was further analyzed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fig. 4). After the short-term exposure, there was no difference between all treatments (controls, ATZ and NPATZs) and the variation in the concentrations did not influence the bacterial functional composition (Fig. 4a). As the exposure time increased to medium-term, a separation was observed between the bacterial communities exposed to the high concentrations of ATZ and NPATZs and the communities treated with the low concentration of these chemicals. Interestingly, after long-term exposure, the functional composition of the bacterial communities exposed to the low concentration of the NPATZs was similar to the composition of the bacterial communities exposed to medium and high concentrations of the NPATZs. Similarly, the impact of long-term exposure to ATZ on bacterial communities was also concentration-independent, meaning that all ATZ concentrations showed a similar influence on the functional composition of the bacteria. However, the bacterial communities exposed to ATZ were separated from the communities exposed to NPATZs, confirming that conventional formulation of ATZ and the nano-formulation of ATZ induce distinct long-term adverse effects on the functional composition of rhizosphere bacterial communities.We further focused on the genes that are associated with ATZ degradation according to the KEGG pathway database. The genes with 100% occupation of all replicates for each treatment were defined as core functional genes. In total, nine core genes were selected, and the relative abundance of these genes in each treatment is presented in Fig. 4b. Different patterns were observed upon exposure to the NPATZs compared to ATZ. For example, after short-term exposure to ATZ, the relative abundance of all functional genes increased as the ATZ concentration increased. Increasing the exposure time to medium-term and long-term enhanced the number of suppressed functional genes, where the relative abundance of functional genes, i.e. atzB, atzC, atzD and DUR1-2, decreased with increasing ATZ concentration. We also observed that after short-term exposure to the NPATZs, the majority of the functional genes increased in relative abundance along with increasing NPATZ concentration, i.e. atzA, atzB, atzC, atzD, atzF and DUR1-2. As the exposure time increased to medium-term, the relative abundance of atzB, atzC, atzD and DUR1-2 decreased upon increasing the NPATZ concentration. However, after long-term exposure to NPATZ, the relative abundance of six out of nine functional genes, i.e. atzA, atzB, atzC, atzD, biuH and atzF, was increased. Only the relative abundance of DUR1-1 was found to be decreased as the exposure concentration of the NPATZs increased. This finding suggests that ATZ degradation might be promoted by the nano-formulation of ATZ upon long-term exposure.
Correlation between ATZ- or NPATZ-degrading bacterial genera and functional genes
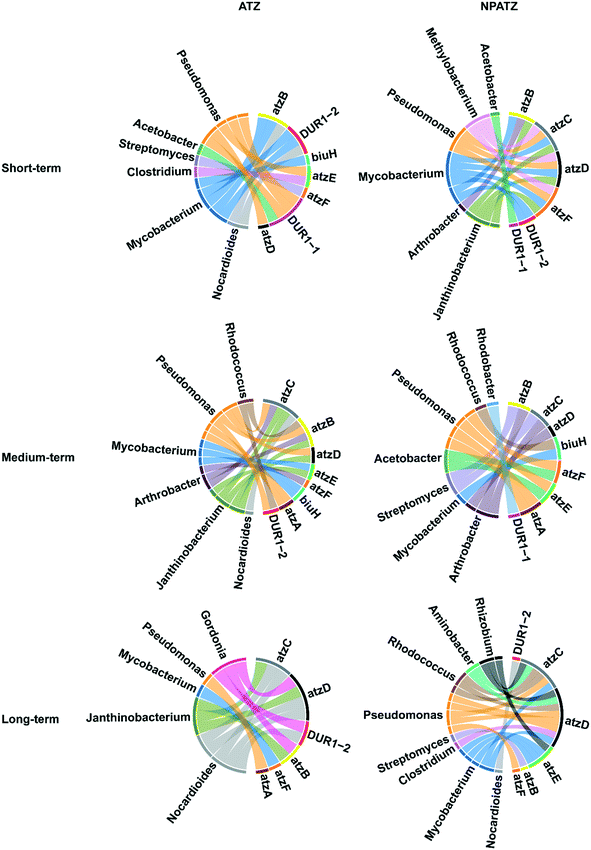
The correlation between the bacterial genera and functional genes with regard to ATZ and NPATZ degradation was investigated and the results are reported in Fig. 5. After short-term exposure, both ATZ degradation and NPATZ degradation were found to be dominated by Pseudomonas and Mycobacterium. These bacteria degraded ATZ and NPATZs from hydroxyatrazine to CO2 mostly using the atzB, atzD and DUR1 genes (Fig. S2, ESI†).As the exposure time increased to 4 weeks (medium-term), Arthrobacter became involved in the degradation of both ATZ and the NPATZs (Fig. 5). Arthrobacter and Pseudomonas encode the genes atzA, atzB, atzC and atzD, which contributed mainly to the degradation of ATZ and NPATZs to carboxybiuret. The further degradation of ATZ-related and NPATZ-related carboxybiuret products to CO2 was differentiated by bacterial species: Janthinobacterium, Mycobacterium and Arthrobacter for ATZ, and Pseudomonas and Acetobacter for NPATZs. This indicates that the ATZ and NPATZ degradation pathways became different as the exposure time increased, particularly with regard to the degradation process of carboxybiuret to CO2 (Fig. S2, ESI†).
After long-term exposure (6 weeks), the differences between the ATZ and NPATZ degradation pathways were even more pronounced. There were more genera involved in NPATZ degradation compared to ATZ degradation (Fig. 5). The bacterial genera involved in the NPATZ and ATZ degradation pathways were also different (Fig. S2, ESI†). For instance, the functional genes of atzC and atzD were encoded by Gordonia, Janthinobacterium, and Nocardioides for ATZ degradation, while they were encoded by Rhizobium, Aminobacter, Rhodococcus, Pseudomonas, Mycobacterium and Clostridium for NPATZ degradation. This clearly shows that the nano-pesticide formulation changed the rhizosphere bacterial consortia involved in the ATZ degradation pathway after long-term exposure.
Discussion
This is the first study revealing the interaction between plant rhizosphere bacterial communities and the nano-formulation of ATZ as a model of commercial pesticides to support the application of nanotechnology in the agricultural industry. There was no effect of ATZ and NPATZs on the bacterial community in short-term exposure, regardless of the exposure concentration applied for both formulations. However, the negative effects of high concentrations of ATZ and NPATZs started to appear after 4 weeks of exposure, as reflected by the reduction in the metabolic capacity of the rhizosphere bacterial community.Previous studies already showed the adverse effects of ATZ on the structure of soil microbial communities after long-term exposure.36–40 Here, we documented the chronic adverse effects of NPATZs on the rhizosphere bacterial communities of L. sativa. For instance, it was found that 6 months of soil treatment with ATZ decreased the diversity and evenness of the bacterial community of the soil compared to the control.38 We showed that medium-term to long-term exposure to NPATZs also induced adverse effects to non-target rhizosphere bacterial communities, which may challenge the future application of nano-pesticides for agricultural purposes. Nevertheless, we must emphasize that one of the main purposes of nano-pesticide applications is to reduce the concentration of the required active ingredient. A previous study showed that the application of NPATZs at a concentration of 10 times less than the concentration of ATZ induced the same impacts of the herbicide on target organisms.30 This suggests that the advantage of nano-pesticides compared to traditional pesticides is related to their efficacy at lower concentrations. This also indicates that short-term toxicity testing for nano-pesticides may be insufficient to reveal the adverse effects of nano-pesticides. For example, we observed that the exposure time significantly differentiated the observed effects on the metabolic capacities of bacterial communities between the high concentrations of ATZ and NPATZs. More inhibitions of bacterial metabolic capacity were detected after exposure to NPATZs compared to ATZ. Several recent studies have shown that nano-pesticides can enhance the biological activity of the active ingredients, e.g. herbicides loaded in solid lipid nanoparticles,41 chitosan/tripolyphosphate nanoparticles,35 and silver nanoparticles–chitosan.42 For instance, similar increased toxicity to microbial enzyme activity was observed when applying carboxymethyl-β-cyclodextrin–Fe3O4 magnetic nanoparticles–diuron as a nano-pesticide in soil.43 We further demonstrated that the NPATZs might also potentiate the activity of ATZ against the non-target rhizosphere bacterial community. The enhanced biological activity of NPATZs could be attributed to the delivery efficiency of ATZ to the rhizosphere microbial communities e.g. due to the higher mobility of the NPATZs in the soil matrix and the expected high bioavailability.21,44
Our findings revealed that the composition of rhizosphere bacterial communities was altered when the exposure concentration of and the exposure duration to the NPATZs increased. However, the alteration resulting from the NPATZs was not similar to the alteration caused by ATZ. Similar alterations in rhizosphere bacterial communities were also observed in long-term contamination by a metal-based nano-pesticide Cu(OH)2,6 and the toxicity of the Cu(OH)2 nano-pesticide was reported to be different from that of the copper compartment.5 In our results, after long-term exposure, Proteobacteria and Actinobacteria were found to play the most important roles in inducing the differences among rhizosphere bacterial communities in different treatments. Similar positive correlations between the frequencies of Actinobacteria and Proteobacteria and the concentrations of ATZ were also reported in soils containing agrochemicals.38 When comparing the changes in the community composition after exposure to the NPATZs and ATZ, we observed the presence of more bacterial genera involved in the NPATZ degradation compared to that in ATZ degradation. The number of OTUs in the NPATZ treated samples increased after long-term exposure, with some OTUs, e.g. Aminobacter, Clostridium and Nocardioides, occurring in the presence of the high concentration of the NPATZs but absent in the treatment with the high concentration of ATZ. These genera are well known for their diverse metabolic abilities and have been reported as ATZ degraders.18,45,46 The presence of these bacteria indicated that the nano-pesticide stimulated more bacterial genera in NPATZ degradation.
When focusing on the relative abundance of functional genes, the exposure to ATZ suppressed the functional genes as the exposure time increased, while the treatment with the NPATZs promoted the functional genes after long-term exposure. It is possible that due to the slow release of ATZ from the NPATZs over time, some bacteria could obtain the required genes for degradation of ATZ and further spread more rapidly into other bacteria.47 As we observed in our release experiment, the release of ATZ was reported to be postponed by the nano-pesticides, which depends partially on the polymer swelling and relaxation of the polymeric matrix.19,20,41 The long-term exposure by the gradual release of ATZ from the NPATZs might induce more bacterial genera in ATZ degradation.
The ATZ and NPATZ degradations were found to be initially driven by Mycobacterium and Pseudomonas that mainly harbor the functional genes atzB, atzC and atzD. It has been reported that Mycobacterium and Pseudomonas play important roles in metabolizing cyanuric acid derivatives of ATZ to carbon dioxide and ammonia catalyzed by atzB, atzD, atzE and atzF.18,48 After long-term exposure, the degradation of ATZ is likely to be taken over by Janthinobacterium and Nocardioides encoding atzB and atzC that could convert ATZ into cyanuric acid.46,49,50 However, the number of bacterial genera (i.e. Rhizobium, Aminobacter, Rhodococcus, Pseudomonas, Mycobacterium and Clostridium) that positively correlated with NPATZ degradation increased with exposure time compared to the number of bacteria correlated with ATZ degradation. This further indicated that as the exposure time increased, the nano-pesticides altered the bacterial consortia involved in ATZ degradation. The main differences between the ATZ and NPATZ degradation pathways were found in bacterial consortia associated with atzC and atzD that could metabolize N-isopropylammelide to carboxybiuret. Our data showed that the NPATZs decreased the community-level metabolic potential, while promoting the abundance of functional genes. We speculate that the NPATZs disrupted the bacterial community more than ATZ, by involving more bacterial species and functional genes in the degradation process, consequently decreasing the overall metabolic capacity of the community. The slow release of ATZ from the NPATZs might allow the rhizosphere bacterial community to adapt to the exposure and promoted the bacterial functioning in degrading the released ATZ. However, the long-term repeated application of ATZ in the form of a nano-pesticide with high herbicidal efficiency could inhibit the community's metabolic activity.
Conclusions
In this study, we found that ATZ and NPATZs have distinct adverse effects on the non-target rhizosphere bacterial communities of plants after long-term exposure. This finding is of paramount importance because nano-pesticides as a promising alternative for traditional pesticides may be applied for agricultural purposes but could also induce long-term adverse effects on the non-target microbiome. Long-term exposure to a high concentration of NPATZs was found to act more effectively and gave more microbial community impacts (i.e. decreased the community metabolic capacity and shifted the community structure and composition to a higher extent) as compared to the same amount of ATZ. This may be tackled by modifying the NPATZs, e.g. surface modification to increase the targetable delivery or lower release in non-target places, in the future considering the fact that nanotechnology is developing quickly. This potentially allows us to design safer nano-pesticides in the future, thus benefiting from other promising properties of these materials. Nevertheless, the nano-pesticides may have hidden long-term adverse effects that must be revealed before marketing. Although the application of the nano-formulation promoted the ATZ efficiency, the inhibited community-level metabolic capacity revealed the potential impact of NPATZs on soil fertility as mediated by microbial processes, especially on the non-target plant-associated rhizosphere bacterial communities. These results call for research on the understanding of long-term assessment of nano-pesticides in terrestrial agroecosystems.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The research was partially supported by the European Union's Horizon 2020 Research and Innovation Programme under grant agreement number 760813 “PATROLS”. R. G. would like to thank São Paulo Research Foundation (grant #2017/21004-5), the National Council for Scientific and Technological Development (grant #427498/2018-0) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001.References
- M. Kah, R. S. Kookana, A. Gogos and T. D. Bucheli, A critical evaluation of nanopesticides and nanofertilizers against their conventional analogues, Nat. Nanotechnol., 2018, 13, 677–684 Search PubMed.
- R. Grillo, N. F. de Melo, R. de Lima, R. W. Lourenço, A. H. Rosa and L. F. Fraceto, Characterization of atrazine-loaded biodegradable poly (hydroxybutyrate-co-hydroxyvalerate) microspheres, J. Polym. Environ., 2010, 18, 26–32 Search PubMed.
- A. Gogos, K. Knauer and T. D. Bucheli, Nanomaterials in plant protection and fertilization: current state, foreseen applications, and research priorities, J. Agric. Food Chem., 2012, 60, 9781–9792 Search PubMed.
- H. Chhipa, Nanofertilizers and nanopesticides for agriculture, Environ. Chem. Lett., 2017, 15, 15–22 Search PubMed.
- X. Zhang, Z. Xu, X. Qian, D. Lin, T. Zeng, J. Filser, L. Li and M. Kah, Assessing the Impacts of Cu(OH)2 nanopesticide and ionic copper on the soil enzyme activity and bacterial community, J. Agric. Food Chem., 2020, 68, 3372–3381 Search PubMed.
- M. Simonin, B. P. Colman, W. Tang, J. D. Judy, S. M. Anderson, C. M. Bergemann, J. D. Rocca, J. M. Unrine, N. Cassar and E. S. Bernhardt, Plant and microbial responses to repeated Cu(OH)2 nanopesticide exposures under different fertilization levels in an agro-ecosystem, Front. Microbiol., 2018, 9, 1769 Search PubMed.
- M. Kah, S. Beulke, K. Tiede and T. Hofmann, Nanopesticides: state of knowledge, environmental fate, and exposure modeling, Crit. Rev. Environ. Sci. Technol., 2013, 43, 1823–1867 Search PubMed.
- R. S. Kookana, A. B. Boxall, P. T. Reeves, R. Ashauer, S. Beulke, Q. Chaudhry, G. Cornelis, T. F. Fernandes, J. Gan and M. Kah, Nanopesticides: guiding principles for regulatory evaluation of environmental risks, J. Agric. Food Chem., 2014, 62, 4227–4240 Search PubMed.
- D. Fojtová, J. Vašíčková, R. Grillo, Z. Bílková, Z. Šimek, N. Neuwirthová, M. KahC and J. HofmanA, Supplementary Material Nanoformulations can significantly affect pesticide degradation and uptake by earthworms and plants, Environ. Chem., 2019, 16, 470–481 Search PubMed.
- M. Moore, R. Lizotte Jr, S. Knight, S. Smith Jr and C. Cooper, Assessment of pesticide contamination in three Mississippi Delta oxbow lakes using Hyalella azteca, Chemosphere, 2007, 67, 2184–2191 Search PubMed.
- F. Ackerman, The economics of atrazine, Int. J. Occup. Environ. Health, 2007, 13, 437–445 Search PubMed.
- A. Mudhoo and V. Garg, Sorption, transport and transformation of atrazine in soils, minerals and composts: a review, Pedosphere, 2011, 21, 11–25 Search PubMed.
- T. Anderson, E. Kruger and J. Coats, Enhanced degradation of a mixture of three herbicides in the rhizosphere of a herbicide-tolerant plant, Chemosphere, 1994, 28, 1551–1557 Search PubMed.
- A.-L. Marchand, S. Piutti, B. Lagacherie and G. Soulas, Atrazine mineralization in bulk soil and maize rhizosphere, Biol. Fertil. Soils, 2002, 35, 288–292 Search PubMed.
- B. Martinez, J. Tomkins, L. P. Wackett, R. Wing and M. J. Sadowsky, Complete nucleotide sequence and organization of the atrazine catabolic plasmid padp-1 from pseudomonassp. strain adp, J. Bacteriol., 2001, 183, 5684–5697 Search PubMed.
- M. L. De Souza, M. J. Sadowsky and L. P. Wackett, Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization, J. Bacteriol., 1996, 178, 4894–4900 Search PubMed.
- L. Wackett, M. Sadowsky, B. Martinez and N. Shapir, Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies, Appl. Microbiol. Biotechnol., 2002, 58, 39–45 Search PubMed.
- H. Fang, J. Lian, H. Wang, L. Cai and Y. Yu, Exploring bacterial community structure and function associated with atrazine biodegradation in repeatedly treated soils, J. Hazard. Mater., 2015, 286, 457–465 Search PubMed.
- A. E. Pereira, R. Grillo, N. F. Mello, A. H. Rosa and L. F. Fraceto, Application of poly (epsilon-caprolactone) nanoparticles containing atrazine herbicide as an alternative technique to control weeds and reduce damage to the environment, J. Hazard. Mater., 2014, 268, 207–215 Search PubMed.
- R. Grillo, N. Z. P. dos Santos, C. R. Maruyama, A. H. Rosa, R. de Lima and L. F. Fraceto, Poly (ε-caprolactone) nanocapsules as carrier systems for herbicides: Physico-chemical characterization and genotoxicity evaluation, J. Hazard. Mater., 2012, 231, 1–9 Search PubMed.
- H. C. Oliveira, R. Stolf-Moreira, C. B. R. Martinez, R. Grillo, M. B. de Jesus and L. F. Fraceto, Nanoencapsulation enhances the post-emergence herbicidal activity of atrazine against mustard plants, PLoS One, 2015, 10, e0132971 Search PubMed.
- K. Sajjaphan, P. Heepngoen, M. J. Sadowsky and N. Boonkerd, Arthrobacter sp. strain KU001 isolated from a Thai soil degrades atrazine in the presence of inorganic nitrogen sources, J. Microbiol. Biotechnol., 2010, 20, 602–608 Search PubMed.
- R. Behki, E. Topp, W. Dick and P. Germon, Metabolism of the herbicide atrazine by Rhodococcus strains, Appl. Environ. Microbiol., 1993, 59, 1955–1959 Search PubMed.
- K. L. Boundy-Mills, M. L. De Souza, R. T. Mandelbaum, L. P. Wackett and M. J. Sadowsky, The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway, Appl. Environ. Microbiol., 1997, 63, 916–923 Search PubMed.
- S. Piutti, E. Semon, D. Landry, A. Hartmann, S. Dousset, E. Lichtfouse, E. Topp, G. Soulas and F. Martin-Laurent, Isolation and characterisation of Nocardioides sp. SP12, an atrazine-degrading bacterial strain possessing the gene trzN from bulk-and maize rhizosphere soil, FEMS Microbiol. Lett., 2003, 221, 111–117 Search PubMed.
- M. M. Kandil, C. Trigo, W. C. Koskinen and M. J. Sadowsky, Isolation and characterization of a novel imidacloprid-degrading Mycobacterium sp. strain MK6 from an Egyptian soil, J. Agric. Food Chem., 2015, 63, 4721–4727 Search PubMed.
- G. Imfeld and S. Vuilleumier, Measuring the effects of pesticides on bacterial communities in soil: a critical review, Eur. J. Soil Biol., 2012, 49, 22–30 Search PubMed.
- T. Satapanajaru, P. Anurakpongsatorn, P. Pengthamkeerati and H. Boparai, Remediation of Atrazine-contaminated Soil and Water by Nano Zerovalent Iron, Water, Air, Soil Pollut., 2008, 192, 349–359 Search PubMed.
- J. Wu, G. Wang, M. G. Vijver, T. Bosker and W. J. Peijnenburg, Foliar versus root exposure of AgNPs to lettuce: Phytotoxicity, antioxidant responses and internal translocation, Environ. Pollut., 2020, 261, 114117 Search PubMed.
- H. C. Oliveira, R. Stolf-Moreira, C. B. R. Martinez, R. Grillo, M. B. de Jesus and L. F. Fraceto, Nanoencapsulation enhances the post-emergence herbicidal activity of atrazine against mustard plants, PLoS One, 2015, 10, e0132971 Search PubMed.
- J. L. Garland and A. L. Mills, Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization, Appl. Environ. Microbiol., 1991, 57, 2351–2359 Search PubMed.
- V. Echavarri-Bravo, L. Paterson, T. J. Aspray, J. S. Porter, M. K. Winson, B. Thornton and M. G. Hartl, Shifts in the metabolic function of a benthic estuarine microbial community following a single pulse exposure to silver nanoparticles, Environ. Pollut., 2015, 201, 91–99 Search PubMed.
- Y. Zhai, E. R. Hunting, M. Wouters, W. J. Peijnenburg and M. G. Vijver, Silver nanoparticles, ions, and shape governing soil microbial functional diversity: nano shapes micro, Front. Microbiol., 2016, 7, 1123 Search PubMed.
- Y. Zhai, E. R. Hunting, M. Wouterse, W. J. Peijnenburg and M. G. Vijver, Importance of exposure dynamics of metal-based nano-ZnO,-Cu and-Pb governing the metabolic potential of soil bacterial communities, Ecotoxicol. Environ. Saf., 2017, 145, 349–358 Search PubMed.
- R. Grillo, A. E. Pereira, C. S. Nishisaka, R. de Lima, K. Oehlke, R. Greiner and L. F. Fraceto, Chitosan/tripolyphosphate nanoparticles loaded with paraquat herbicide: an environmentally safer alternative for weed control, J. Hazard. Mater., 2014, 278, 163–171 Search PubMed.
- X. Liu, K. Chen, S. Chuang, X. Xu and J. Jiang, Shift in bacterial vommunity structure drives different atrazine-degrading efficiencies, Front. Microbiol., 2019, 10, 88 Search PubMed.
- J. Mahía, S. J. González-Prieto, A. Martín, E. Bååth and M. Díaz-Raviña, Biochemical properties and microbial community structure of five different soils after atrazine addition, Biol. Fertil. Soils, 2011, 47, 577–589 Search PubMed.
- I. Godoi, L. Sene and A. B. Caracciolo, Assessment of the bacterial community structure in a Brazilian clay soil treated with atrazine, Ann. Microbiol., 2014, 64, 307–311 Search PubMed.
- G. Briceño, M. Jorquera, R. Demanet, M. Mora, N. Durán and G. Palma, Effect of cow slurry amendment on atrazine dissipation and bacterial community structure in an agricultural Andisol, Sci. Total Environ., 2010, 408, 2833–2839 Search PubMed.
- X. Liu, C. Hui, L. Bi, M. Romantschuk, M. Kontro, R. Strömmer and N. Hui, Bacterial community structure in atrazine treated reforested farmland in Wuying China, Appl. Soil Ecol., 2016, 98, 39–46 Search PubMed.
- J. L. de Oliveira, E. N. V. R. Campos, C. M. G. da Silva, T. Pasquoto, R. Lima and L. F. Fraceto, Solid lipid nanoparticles co-loaded with simazine and atrazine: preparation, characterization, and evaluation of herbicidal activity, J. Agric. Food Chem., 2015, 63, 422–432 Search PubMed.
- S. K. R. Namasivayam and A. Aruna, Evaluation of silver nanoparticles-chitosan encapsulated synthetic herbicide paraquate (AgNp-CS-PQ) preparation for the controlled release and improved herbicidal activity against Eichhornia crassipes, Res. J. Biotechnol., 2014, 19–27 Search PubMed.
- W. Liu, J. Yao, M. Cai, H. Chai, C. Zhang, J. Sun, R. Chandankere and K. Masakorala, Synthesis of a novel nanopesticide and its potential toxic effect on soil microbial activity, J. Nanopart. Res., 2014, 16, 2677 Search PubMed.
- Y. Sun, J. Liang, L. Tang, H. Li, Y. Zhu, D. Jiang, B. Song, M. Chen and G. Zeng, Nano-pesticides: A great challenge for biodiversity?, Nano Today, 2019, 28, 100757 Search PubMed.
- I. R. McDonald, P. Kämpfer, E. Topp, K. L. Warner, M. J. Cox, T. L. C. Hancock, L. G. Miller, M. J. Larkin, V. Ducrocq and C. Coulter, Aminobacter ciceronei sp. nov. and Aminobacter lissarensis sp. nov., isolated from various terrestrial environments, Int. J. Syst. Evol. Microbiol., 2005, 55, 1827–1832 Search PubMed.
- K. Satsuma, M. Kameshiro, O. Hayashi, K. Sato and Y. Kato, Characterization of a Nocardioides-based, atrazine-mineralizing microbial colony isolated from Japanese riverbed sedimen, J. Pestic. Sci., 2006, 31, 420–423 Search PubMed.
- N. Udiković-Kolić, C. Scott and F. Martin-Laurent, Evolution of atrazine-degrading capabilities in the environment, Appl. Microbiol. Biotechnol., 2012, 96, 1175–1189 Search PubMed.
- I. Fruchey, N. Shapir, M. J. Sadowsky and L. P. Wackett, On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP, Appl. Environ. Microbiol., 2003, 69, 3653–3657 Search PubMed.
- A. E. Omotayo, M. O. Ilori, M. Radosevich and O. O. Amund, Metabolism of atrazine in liquid cultures and soil microcosms by Nocardioides strains isolated from a contaminated Nigerian agricultural soil, Soil Sediment Contam., 2013, 22, 365–375 Search PubMed.
- K. Satsuma, Complete biodegradation of atrazine by a microbial community isolated from a naturally derived river ecosystem (microcosm), Chemosphere, 2009, 77, 590–596 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0en00638f |
| This journal is © The Royal Society of Chemistry 2020 |