Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mechanistic insights into toxicity pathways induced by nanomaterials in Daphnia magna from analysis of the composition of the acquired protein corona†
Laura-Jayne A.
Ellis
 and
Iseult
Lynch
and
Iseult
Lynch
 *
*
School of Geography, Earth and Environmental Sciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK. E-mail: i.lynch@bham.ac.uk
First published on 4th September 2020
Abstract
Identifying proteins present on the surface of nanomaterials (NMs) incubated with plasma, serum, and cell lysates can offer insights into the biological interactions of the NMs, including uptake and toxicity. Organisms such as Daphnia magna respond to the presence of toxicants by secreting proteins and other biomolecules; we demonstrate that the eco-corona acquired during exposure to NMs can provide similar important insights regarding the mechanistic pathways induced by the NM exposure as part of an ecotoxicity assessment. Using freshly dispersed (pristine) and ‘medium aged’ NMs (as environmental pollutants), differing in core material, size and surface properties, the influence of NM physicochemical characteristics and exposure environment on the protein corona composition and potential for in vivo uptake by the model test species Daphnia magna are investigated. Surface bound protein corona compositions changed between NM type, NM ‘age’ and the environmental medium in which the NMs were incubated, thus changing the NM biological identity and interactions. Proteins identified on the freshly dispersed NM surfaces were largely associated with metabolic damage, DNA damage, mitochondrial breakdown and energy processes, all of which are associated with cytotoxic damage. Significantly fewer proteins were bound to the aged NMs in all medium conditions, compared to the freshly dispersed NMs. Proteins bound only to the aged NMs were involved in calcium ion binding and cell redox homeostasis, indicative of significantly lower toxic responses to the aged NMs. Thus, NM protein corona composition can facilitate detection of organism responses to NM exposure and potentially identification of the molecular initiating events in adverse outcome pathways.
Environmental significanceUnderstanding nanomaterial–biological interactions, and organism responses to the presence of nanomaterials (NMs), is a crucial step in elucidating the environmental safety of NMs and facilitating their regulation. Model organisms were exposed to a variety of pristine and aged silver and titanium NMs in a standard culture medium and two natural water proxies. NMs recovered from the exposure media after 7 days of incubation with Daphnia magna (set-up as per OECD reproductive test) had surface-bound protein coronas which varied between NM type, ageing and medium composition. Compared to the aged NMs, the pristine NMs had the highest numbers of bound proteins relating to metabolic and cytotoxic damage, giving insights into the selectivity of the endogenous pathways triggered by the NMs. This study facilitates a better understanding of how exposure to pristine and aged NMs disrupts biological processes that would otherwise safeguard health, and provides key pointers regarding molecular initiating events for further study and establishment of adverse outcome pathways for NMs in the environment. |
1. Introduction
Understanding the interactions of nanomaterials (NMs) with biological matter, and the responses of organisms to the presence of NMs, is an essential step in elucidating the safety of the NMs and to facilitate their regulation.1 The application of NMs in a range of consumer applications undoubtedly arises due their unique sizes (1–100 nm), shape and surface physico-chemistry.2 As nano-applications evolve, for use in drug delivery, personal care items and engineering technologies to name a few, so do the requirements for careful assessment of the potential for unexpected toxicities and biological interactions,3 particularly when such materials enter the environment.4 Once released into the environment NMs have the ability to adsorb biomolecules to their surfaces.5,6 Biomolecules adsorbed onto the NMs surface are termed the corona, and they transform the NMs from their ‘pristine’ engineered identity providing them with a biological or environmental identity, enabling the NMs to interact with cellular surface receptors.7,8 Formation of the NM corona is known to be affected by the NMs physicochemical properties, medium conditions, and the nature/composition of the available biomolecules.9,10The entry of NMs into organisms and their translocation across biological barriers is particularly important for determining subsequent adverse outcomes. Due to their size and surface reactivity, NM-corona complexes can engage with a range of endogenous processes and potentially reach all tissues and organs.11 As a result, the range of affinities of biomolecules for NM surfaces,12 can change the corona composition as the NM moves from outside to inside cells. Additionally, the corona itself may provide insights into the uptake and translocation pathways utilised by the NMs,13–15 and give insights into the cellular responses following NM exposure.16 In mammalian systems, NMs associate with soluble serum compounds to form surface coronas, which affect the NM interactions in vivo. For example, common proteins identified in the coronas of NMs exposed to blood plasma were linked to lipid translocation, complement activation and ion transport.17 Connections between the NM properties and the corona composition and conformation of adsorbed proteins can provide insights into the biological impacts and interactions of the NMs. For example, protein unfolding related activation of cellular receptors and associated biological signalling pathways involved with the production of inflammation have been induced following fibrinogen binding to specific sizes of poly(acrylic acid)-coated gold NMs.18 Other studies have identified significant protein groups sharing 80% homology from exposure to two differently sized polystyrene particles, suggesting that the surface molecular properties are important when determining NM protein coronas.17 However, these studies all relate to the investigation of protein coronas acquired in human cell lines and blood plasma, with much less known about the role of the coronas acquired in the environment. Additionally, the range of biomolecules available in the environment is much broader, including the aforementioned proteins, as well as polysaccharides, hemicelluloses, lipids, small molecules and more, coming from a range of species.6
Indeed, protein and biomolecule corona relevance in the environment is at an early stage of investigation. For example, in earthworm coelomocytes, lysenin was identified as the major corona protein for silver (Ag) NMs in vitro (Hayashi et al., 2013 (ref. 73)). Gao, Lin19 identified specific proteins (vitellogenin and zona pellucida) in Ag NMs coronas that related to gender differences in the fish plasma in which they were exposed. The research raises the possibility of the Ag NMs being translocated into developing oocytes and the role of NM induced reproductive toxicity. Furthermore, analysis of the corona of titanium dioxide (TiO2) NMs, exposed to sea urchin (P. lividus) immune cells (and cell conditioned medium), found that negatively charged proteins bound preferentially to the NMs. The main proteins identified in the sea urchin cell-conditioned TiO2 NM protein coronas were involved in cellular adhesion and cytoskeletal organization (actin and tubulin).20 Differential secreted proteins related to cell-to-cell signalling were identified in polystyrene NM coronas exposed to Daphnia magna,21 and the acquired protein corona was shown to influence the gut retention, dissolution and toxicity of cerium dioxide and zinc oxide NMs.22
Daphnia magna are considered a perfect ecological test species as they have a complete sequenced genome, which is advantageous for monitoring stress/adaptive changes to their environments;23 they reproduce parthenogenetically, such that disturbing homeostasis leads to compensatory, responsive, and adaptive processes which can be measured by various omics techniques. There is a body of evidence that supports protein functions and their related pathways (expressed by the genome) being evolutionally conserved across species via lineage specific expansions identified by their sequenced genomes.24–27 Therefore, the biochemical properties of such proteins, including their mechanistic pathway involvements, interactions with receptors and cellular responses, have not changed with species evolution.28Daphnia have a complete set of protein sequences (UP000076858) which can easily be mapped to identify specific molecular pathways via their ‘GO’ (gene ontology) terms in the Uniprot database (https://www.uniprot.org/). Furthermore, Daphnia are a keystone species recognised by the Organization for Economic Cooperation and Development (OECD) for use in standard regulatory testing protocols29,30 although having been designed for molecular chemicals these tests require adaption for NMs,31 including the need to consider biomolecule corona formation from secreted biomolecules32 or from the natural organic matter present in typical freshwaters, and how this influences subsequent NM uptake and toxicity.
It was recently shown that the chronic (reproductive) ecotoxicity of Ag and TiO2 NMs to Daphnia magna is dramatically reduced by environmental ageing of the NMs in media of different ionic strengths and natural organic matter contents.33,34 Understanding the different proteins acquired by freshly dispersed (pristine engineered) and aged NMs in different environmentally relevant media, and their biological functions will provide important insights into their mechanisms of NM toxicity. Thus, as part of the overall assessment of the NMs chronic toxicity (Ellis et al., 202033), we characterised the secreted protein coronas that the NMs had acquired after 7 days in contact with Daphnia magna under each of the different medium conditions. The NMs tested were various surface coated Ag and TiO2 NMs in their pristine engineered (freshly dispersed) and 6-months medium-aged forms, to identify medium-specific and ageing-related effects on the composition of the protein coronas acquired in the presence of Daphnia magna. By choosing the 7-day time point, the hypothesis was that the organisms would have responded to the exposure of the NMs, secreting proteins into the medium, which are subsequently adsorbed by the NMs still in solution, thereby evolving their coronas. Additionally, as part of their filter feeding nature, we assume that at least some of the NMs in solution at day 7 will have passed though the daphnia gut, where their coronas would have further evolved due to contact with a different biological environment. By identifying the composition of the proteins adsorbed onto the various pristine and aged NMs coronas, in the different medium conditions, we can learn about the possible endogenous pathways activated by the specific NMs. Comparing the pristine and medium aged variants allows determination of protein binding selectivity and distinction of corona compositions between different NM core speciation and surface coatings. Additionally, we assessed if the water conditions had any influence on the preferential binding of proteins to the surface of the NMs. This was achieved by using 3 chemically differently waters, two of which were environmentally relevant in terms of their ionic strengths and natural organic matter contents, in comparison to the salt-only Daphnia culturing media recommended for use in the OECD standard test guidelines.
2. Methodology
2.1 Media and representative waters
Commercially available chemicals, solvents and humic acids (HA) were purchased from Sigma-Aldrich (Dorset, UK) of analytical reagent grade. Ultrapure water (UPW) with a maximum resistivity of 18.2 M Ω cm−1 was used throughout the experiments. Experiments were conducted in Daphnia high hardness combo media (HH combo), and class I and class V synthetic natural water standards,35 as described in Ellis et al.33 Briefly, the HH combo medium is a salt-only medium that matches the total hardness of water found in the environment,36 while class I and class V river waters have low alkalinity and dissolved organic carbon and high alkalinity and ionic strength, respectively.35 A description of the water compositions is presented in Tables S1 and S1A.†2.2 Nanomaterials and characterization
The NMs used in this study correspond to those used in Ellis et al.,33 which include PVP-coated Ag2S NMs (43.6 ± 14 nm) from AppNano, uncoated Ag (61 ± 36 nm) and PVP coated Ag (18.2 ± 11 nm) from Promethean Particles Ltd, bulk Ag (Sigma Aldrich, UK), uncoated (9 ± 2 nm) and PVP (9 ± 2 nm) TiO2 NMs from Promethean Particles Ltd. The range of NM sizes, surface coatings and core materials used were chosen to test the hypothesis that irrespective of starting characteristics, the physical and chemical transformation of NMs in the environmental causes a ‘convergence’, due to the high surface reactivity of the NMs with the surrounding environment, resulting in the “environmentally aged” NMs having similar chemical compositions which would be reflected in similar corona compositions and similar ecotoxicological effects. The choice of Ag and TiO2 NMs in the present study is based on their high production volumes and widespread use in a range of consumer and industrial products.37 For example, TiO2 NMs are commonly added to many healthcare products (particularly in suntan lotions) that are directly applied on skin. TiO2 is a known photocatalyst, which is reactive to produce hydroxyl radicals, superoxide anion radicals and other reactive oxygen species (ROS) as by-products leading to DNA damage, lipid peroxidation38 and genotoxicity.39 TiO2 is classified as a group 2B possible carcinogen to humans for inhalation exposure40 although this is currently under review. Ag NMs are added to medical products to exploit their antibacterial and anti-microbial properties41 and are known to be cytotoxic.42Dynamic light scattering (DLS), was used to measure hydrodynamic sizes of the pristine and aged NMs, using a Malvern Nanosizer 5000 (Table S2†). NM samples for transmission electron microscopy (TEM) were prepared by depositing a 20 μL drop of the NM suspension onto a 300 mesh carbon-coated copper TEM grid (Agar Scientific, UK) (Table S2†). A JEOL 1400EX 80 kV Max system was used to image the samples.
2.3 Nanomaterials chemical ageing
NMs stock solutions of 1000 mg L−1 were prepared in each of the HH combo, class I and class V water and aged at 4 °C (refrigerated) for at least 6 months prior to Daphnia exposures. The experimental design and NM ageing process are shown in Fig. 1.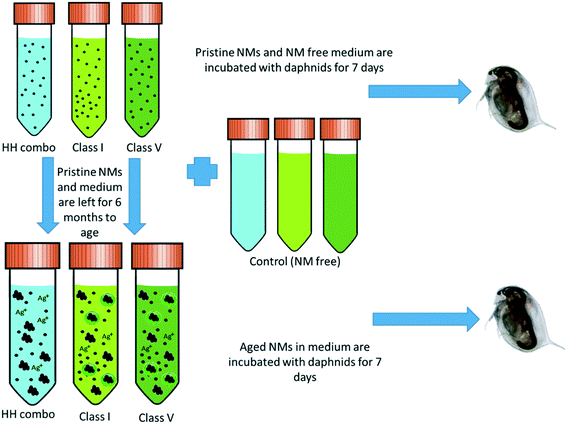 | ||
| Fig. 1 Experimental design showing how the three different water types are incubated for 7 days with the Daphnia (both NM containing and NM free) and the NM ageing process. | ||
2.4 Daphnia maintenance and culturing
Cultures of Daphnia magna Bham2 strain were maintained in HH combo medium, class I and class V water using pools of 3rd broods kept in a 20 °C temperature-controlled environment with 12-hour light and dark cycles for at least 3 generations before experimentation. Media were refreshed twice weekly, and daphnids were fed Chlorella vulgaris algae daily, 0.5 mg carbon for neonates to juveniles (days 0–5) and 0.75 mg carbon for juveniles to adulthood (from day 5).2.5 Experimental conditions
The Daphnia reproductive toxicity test was performed according to the OCED-211 test guidelines,30 using the EC30 (48-hour) values determined for the Ag NMs in a range finding study, and the EC5 value for the TiO2 NMs due to their significant chronic effects.33 Briefly, the exposures were set up as follows: for each NM type and medium condition, three beakers (250 mL) each with 10 neonates <24 hours from brood 3 were exposed to the NMs: 20 μg L−1 of uncoated an PVP-coated Ag NMs, 100 μg L−1 of Ag2S, and 5 mg L−1 for the TiO2 NMs. Media was either treated with NMs, or NM free for the controls which was left for the duration of the study (7 days). Daphnids were fed Chlorella vulgaris algae daily, 0.5 mg carbon for neonates to juveniles (days 0–5) and 0.75 mg carbon (days 5–7).2.6 Total bioaccumulated Ti and Ag concentrations
At day 7, pools of 10 F0 daphnids from each exposure condition were euthanized (using liquid nitrogen) and mechanically homogenised in 2% nitric acid (HNO3) using a Precellys 24 instrument (Bertin Technologies) with 2 cycles of 30s at a pulse speed of 6000 revolutions per second. Samples (in triplicate) were analysed for their total body burden metal concentration (Ag or Ti) using an inductively coupled plasma mass spectrometry (ICP-MS) (Nexion 300X instrument, Perkin Elmer). Samples were ran alongside an online germanium internal standard. Operating conditions were optimized to produce maximum Ag/Ti intensity using ionic standards. Instrument calibration was achieved by analysis of a blank and 3 dissolved Ag/Ti solutions ranging from 0 to 1 μg L−1. To ensure no deviations from instrumental drift over time, Ag/Ti dissolved calibration check standard was also ran in between sampling.2.7 NM distribution in the organisms
TEM cross sections of the daphnids after 7 days of exposure to the NMs were prepared by the Centre for Electron Microscopy at the University of Birmingham (UK). Briefly, whole Daphnia were euthanised and fixed immediately in a 2.5% glutaraldehyde 0.1 M phosphate buffer suspension. Daphnids were dehydrated in ethanol and embedded in epoxy resin before sectioning the gut areas using an ultramicrotome with a diamond knife to cut 1 μm thick sections. Sections were mounted on TEM grids and stained with toluidine blue. Image analysis was performed using a JEOL 1400EX 80 kV Max system.2.8 Hard-corona isolation and identification of proteins
To assess the protein coronas formed around the NMs during exposure to the daphnids, water samples were taken after 7 days of NM incubation with the daphnids. The NMs that had either not been taken up at all or had been excreted by the Daphnia were recovered (Fig. 2). For polyacrylamide gel electrophoresis (PAGE) the NM protein coronas were isolated using the methodology of Monopoli et al.43 Briefly, 25 mL water samples containing NMs were subjected to a series of centrifugation and washing steps to remove any contaminants and unbound proteins using phosphate buffered saline (PBS) and three replicates of centrifugation at 5200g at 4 °C for 20 minutes. In each replicate, the surface liquid was removed leaving the pellet containing the NM-protein complexes. The NM pellets were then incubated in the PAGE loading buffer (60 mM Tris-HCl (pH 6.8 at 25 °C), 2% (w/v) SDS, 0.1 M DTT, 10% glycerol, 0.01% (w/v) bromophenol blue) at 95 °C for 5 minutes to denature the proteins, which were then transferred to a 12.5% polyacrylamide gel. Using polyacrylamide gel electrophoresis (PAGE), a total of 20 μL of eluted protein was used per well and the gels were run at 100 V for approximately 2–3 hours. Bands were observed and developed using Coomassie blue (0.25%) and silver stain to confirm the presence of proteins isolated from the NMs hard corona. A protein standard ladder (p77125 bioLabs) 11–245 kDa was used to calculate to molecular weights of the identified protein bands.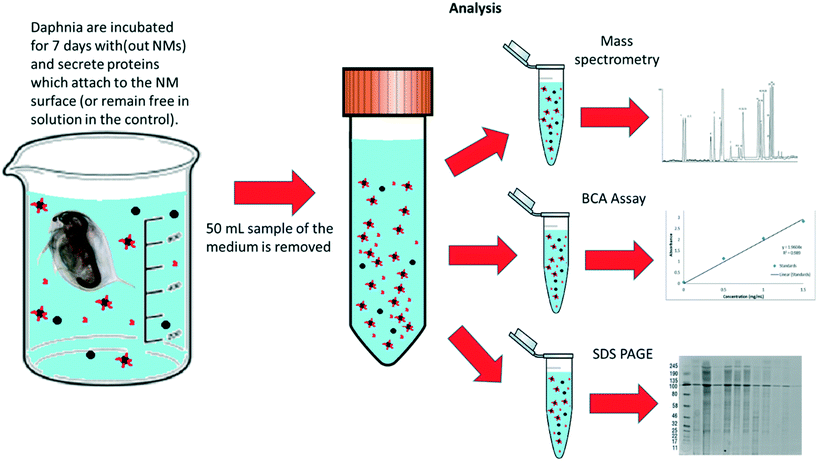 | ||
| Fig. 2 Schematic diagram of how the samples were prepared and split for protein and corona analysis after the daphnia were incubated with NMs (and NM free-controls) media for 7 days. | ||
A second set of 25 mL water samples containing NMs were also taken and sent for protein identification by mass spectrometry. Preparation for these samples also involved centrifugation and washing steps at 5200g at 20 °C for 15 minutes (three times) except ultrapure water was used (instead of PBS) keeping the NM pellet at the bottom each time. Importantly, there was no final suspension in the SDS-containing loading buffer used for the PAGE analysis, as SDS causes interference with the mass-spectrometer. The NM pellets were analysed using liquid chromatography tandem mass spectrometry (LC-MS/MS) using an ‘on particle’ digestion technique at the Functional Genomics and Proteomics Laboratories at the University of Birmingham (https://www.birmingham.ac.uk/facilities/genomics/index.aspx), as described in detail in the ESI† section 1.4. An UltiMate 3000 HPLC series (Dionex, Sunnyvale, CA USA) was used for peptide concentration and separation. The MS and MS/MS scans were searched against the Uniprot database (http://www.uniprot.org) using Proteome Discoverer 2.1 (ThermoFisher Scientific). Variable modifications are deamidation (N and Q), oxidation (M) and phosphorylation (S, T and Y). The precursor mass tolerance was 10 ppm and the MS/MS mass tolerance was 0.02 Da. Two missed cleavages were allowed and identified proteins are accepted as a real hit protein with at least two high confidence peptides. The corresponding molecular ‘Go’ functions were identified using http://www.uniprot.org.
A Pierce BCA assay protein kit protein was used to quantify the total protein secreted by the Daphnia into the media.21 A range of dilutions (0–2 mg mL−1) of bovine serum albumin (BSA) of known concentration in triplicate were made using UPW water to create a standard curve (Fig. S1†) using a 96-well plate (Costar). Samples (100 μL each) of the test mediums were put into separate wells in the 96-well plate in triplicate. A 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of reagent A to reagent B was made ensuring enough volume to add 200 μL to each sample in the wells (total volume 300 μL). The 96-well plate was microwaved for 30 seconds alongside a beaker of water to act as a heat sink. The absorbance function set to 560 nm was used to obtain the absorbance of the different reaction wells. Samples of unknown concentration could then be found by inserting the absorbance provided by the plate reader for each of the samples to calculate their protein concentration. Further details are described in the ESI† section 1.4. A complete list of the NMs identified on each NM, pristine and aged, in each medium, including protein identifications, functions, % coverage and accession numbers are located in the Appendix; Workbooks 1–27.
1 mixture of reagent A to reagent B was made ensuring enough volume to add 200 μL to each sample in the wells (total volume 300 μL). The 96-well plate was microwaved for 30 seconds alongside a beaker of water to act as a heat sink. The absorbance function set to 560 nm was used to obtain the absorbance of the different reaction wells. Samples of unknown concentration could then be found by inserting the absorbance provided by the plate reader for each of the samples to calculate their protein concentration. Further details are described in the ESI† section 1.4. A complete list of the NMs identified on each NM, pristine and aged, in each medium, including protein identifications, functions, % coverage and accession numbers are located in the Appendix; Workbooks 1–27.
3. Results and discussion
3.1 NM characterization
To explore the molecular pathways induced by NM exposure, daphnids were exposed over 7 days to a library of different core and surface coated Ag and TiO2 NMs. The NMs used in this study were characterised (in all conditions) using TEM and DLS for both freshly dispersed and medium aged NM exposures (Table S2 and Fig. S2†). In all cases, when the NMs were suspended in the three different media to age for 6 months (to mimic an environmental pollutant), the particle sizes increased indicative of agglomeration. The increase in hydrodynamic diameter in solution for the aged NMs compared to their pristine counterparts, particularly for the NMs in the class I and class V water, may be due to NOM and surface protein adsorption to the NMs.3.2 Nanomaterial uptake
It is well known that Daphnia are filter feeders and take up food based on size.44 Uptake of the NMs was observed at the end of the study (day 7) by measuring the total bioaccumulated concentrations of Ag and Ti using ICP-MS (Table 1). In most conditions, daphnids internalised higher concentrations of the freshly dispersed NMs when compared to their aged counterparts. Interestingly, the NMs aged in the realistic waters (class I and V) were least bioaccumulated by the Daphnia after 7 days of incubation. There are several possible explanations for the reduced bioavailability of the aged NMs. Firstly, the NMs may have agglomerated and settled, making them less available for uptake in the aquatic phase. Secondly, the transformation of the NMs reduced the free NM surface area available for protein binding and therefore the daphnids may have been able to clear the NMs more readily following uptake.| Nanomaterial | Starting concentration (μg L−1) | HH combo | Class V | Class I | ||
|---|---|---|---|---|---|---|
| Freshly dispersed Ag NM exposure | Aged Ag NM exposure | Freshly dispersed Ag NM exposure | Aged Ag NM exposure | Aged Ag NM exposure | ||
| Measured Ag uptake per daphnid (μg L−1) | Measured Ag uptake per daphnid (μg L−1) | Measured Ag uptake per daphnid (μg L−1) | Measured Ag uptake per daphnid (μg L−1) | Measured Ag uptake per daphnid (μg L−1) | ||
| Uncoated Ag | 20 | 0.4 ± 0.09 | 0.7 ± 0.01 | 0.6 ± 0.01 | 0.3 ± 0.07 | 1.8 ± 0.04 |
| PVP Ag | 20 | 0.7 ± 0.00 | 0.5 ± 0.05 | 0.7 ± 0.01 | 0.3 ± 0.02 | 0.2 ± 0.00 |
| Ag2S | 100 | 0.7 ± 0.01 | 0.2 ± 0.03 | 1.3 ± 0.07 | 1 ± 0.03 | 1.2 ± 0.02 |
| TiO2 PVP | 5000 | 1.5 ± 0.09 | 1.2 ± 0.21 | 1.5 ± 0.07 | 1.2 ± 0.14 | 0.7 ± 0.03 |
| TiO2 uncoated | 5000 | 4.3 ± 0.11 | 1.2 ± 0.08 | 1.8 ±0.10 | 1.4 ± 0.09 | 1.1 ± 0.02 |
3.3 Protein corona composition: effects of medium and NM ageing
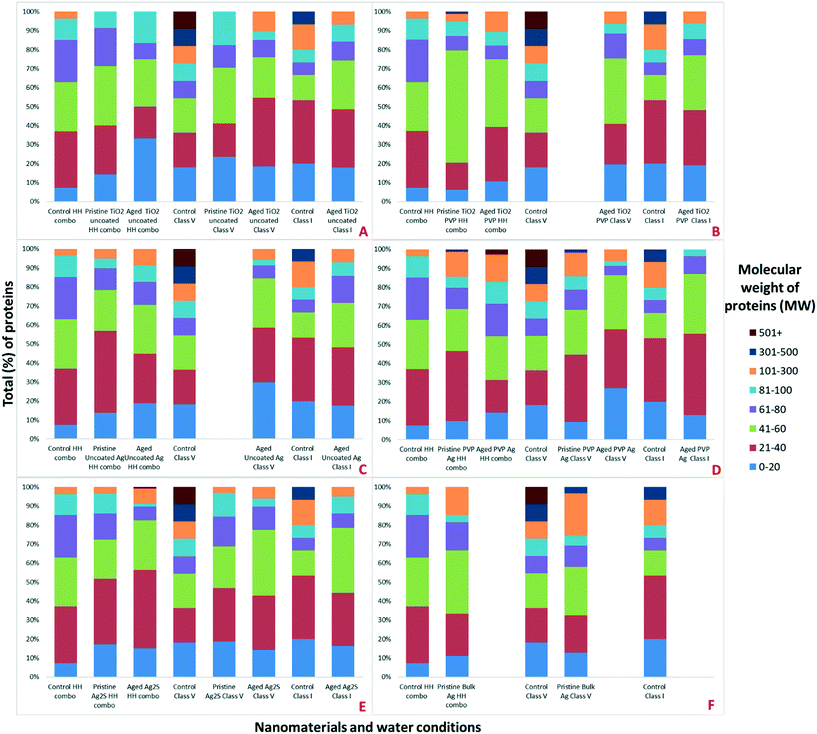
To further our understanding of the implications of ‘environmental ageing’ of NMs and their biological impacts, NMs from the culturing media were harvested after 7 days of exposure to the daphnids, as shown schematically in Fig. 2. Assessments were made to identify how the ‘freshly dispersed’ and ‘medium aged’ NMs differ in physicochemical characteristics, and how exposure medium influences the composition of their acquired protein coronas, which in turn reflects how the Daphnia responded to the NMs presence through secretion of proteins. Total protein concentrations in the media were quantified using BCA assays (Table S3 ESI†). The NM protein corona compositions were assessed using both SDS-PAGE (Fig. S3†) and shot-gun proteomics using LC-MS/MS (ESI† Appendix: Workbooks 1–27). The results for each technique were compared between each exposure and control population samples. Note, that the controls were conditioned medium samples from Daphnia cultured in NM-free media as described by Briffa et al.22 measuring only the proteins secreted by the Daphnia into the medium as part of their normal filtering processes. Thus, proteins associated with the NMs that were not present in the equivalent medium blank represent proteins that were secreted by the daphnids in response to their interaction with the NMs, and shed light on the mechanisms of toxicity induced by the different NMs, and the effect of medium composition and NMs ageing on the NM toxicity to daphnids.Preliminary assessments using PAGE qualitatively identified the presence of proteins on the surface of the NMs (Fig. S3†). Strong bands in the region of 20–100 (kDa), were also confirmed as the most abundant masses of proteins present when analysed by LC-MS (Fig. 3). Proteins identified in this mass region using LC-MS/MS were related to a range of molecular functions including, but not limited to, GTP-binding nuclear proteins, histone proteins, putative ATP synthase subunit alpha, mitochondrial, heat shock 70 kDa protein cognate, all of which are associated with DNA binding, APT energy binding mechanisms and metabolic processes. The complete protein ID datasets are provided in the ESI;† Workbooks 1–27.
Strong bands in the polyacrylamide gel were identified around 100–120 kDa (Fig. S3†), which are associated with (but not limited to) the following proteins identified in specific NMs coronas using a shot-gun proteomic approach (Appendix; Workbooks 1–27); carbamoyl-phosphate synthase large chain (identified in the NM coronas of the pristine TiO2 PVP, pristine Ag2S NMs and pristine bulk Ag all in HH combo medium; aged TiO2 PVP, aged uncoated Ag and aged PVP Ag NMs all in HH combo medium; aged TiO2 PVP and aged PVP Ag NMs in class V water; and aged TiO2 uncoated, aged TiO2 PVP and aged uncoated Ag NMs in class I water), chorion peroxidase (identified in pristine uncoated Ag NMs in HH combo medium; aged Ag2S NMs in HH combo medium; and aged TiO2 uncoated NMs in class V water), ATP-dependent RNA helicase DDX23, (identified in pristine TiO2 PVP NMs in HH combo medium and aged TiO2 PVP NMs in HH combo), putative copper–zinc superoxide dismutase (identified in pristine PVP Ag NMs in HH combo medium; pristine bulk Ag and pristine PVP Ag NMs in class V water), putative ATP-binding cassette sub-family (identified in pristine bulk Ag and pristine Ag2S NMs in HH combo medium; aged uncoated Ag, aged PVP Ag and aged Ag2S NMs in HH combo medium), matrix metalloproteinase 1 (identified in aged TiO2 PVP and aged Ag2S NMs in HH combo medium; pristine bulk Ag in class V water; aged TiO2 uncoated, aged uncoated Ag, aged PVP Ag NMs in class V water and in the control in class V water).
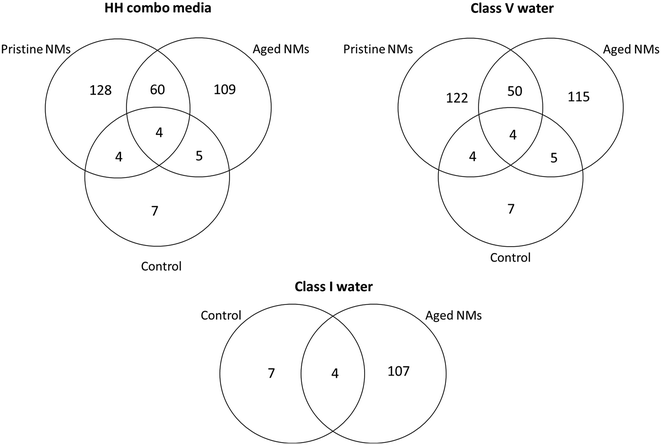
Interestingly, the number of individual identified proteins by LC-MS was always higher for the pristine NMs when compared the aged NMs (Fig. 4). It was also possible to identify differential protein binding patterns, with some proteins binding uniquely to the pristine NMs and some only to the aged NMs. There were also several proteins that were absent entirely from the control populations, indicating that they were secreted by the Daphnia in response to interaction with the NMs. For example (but not limited to), apoptosis-inducing factor 1, a mitochondrial protein was unique to the pristine PVP Ag NM corona in the class V water. The 2-domain haemoglobin protein (involved in heme and oxygen binding), 60S acidic ribosomal protein (ribosome biogenesis), 60S ribosomal L14 (ribosomal protein), creatine kinase U-type, (mitochondrial protein-energy metabolism) nucleoside diphosphate kinase and nucleosome assembly proteins (both ATP binding) were only identified in the coronas of pristine PVP Ag NMs in HH combo and class V water. Adenosine deaminase (catalytic and metal ion binding activity) proteins were identified in the coronas of both the aged PVP and uncoated Ag NMs in the class V water.
Additionally, there were proteins that were only identified once and were unique to the PVP Ag NMs. For example, aldehyde dehydrogenase was unique to the pristine PVP Ag NMs in HH combo media, alpha,alpha-trehalose-phosphate synthase and apoptosis-inducing factor 1 were uniquely associated with the pristine PVP Ag in class V water, and the 2-oxoglutarate dehydrogenase E1 protein (oxoglutarate dehydrogenase and tricarboxylic acid cycling) was unique to the corona of aged PVP Ag NMs in the class V water (Fig. S4†). Similarly, there were proteins unique to the corona of aged TiO2 PVP NMs in HH combo, including the anion exchange protein, and apolipoprotein D, highlighting that different proteins and interaction pathways are triggered by the different NMs, with both core and coating or surface species affecting corona composition. Fig. S5† identifies some of the ancestor pathways associated with the GO terms of these uniquely identified proteins.
Unique proteins were also associated the aged NMs across the different media, including (but not limited to); calmodulin, involved in calcium ion binding (aged Ag2S and aged uncoated Ag NMs in HH combo; pristine uncoated Ag NMs in class V; aged uncoated TiO2, aged TiO2 PVP, aged uncoated Ag, aged PVP Ag and aged Ag2S NMs in class I:), lethal(3)malignant blood neoplasm (aged uncoated TiO2 NMs in class V), lysine-specific histone demethylase 1A (aged uncoated TiO2 in class I), mitotic control dis3 (aged Ag2S in class V; pristine Ag2S and pristine PVP Ag NMs in HH combo), and boundary element associated factor proteins involved in peroxiredoxin activity and cell redox homeostasis (aged PVP Ag NMs in class V).
Common proteins that were secreted in response to NM exposure and identified in all NM coronas (except control secreted proteins) include (but are not limited to) the 40S ribosomal proteins, 60S ribosomal protein L13, actin-alpha skeletal muscle proteins, ADP-ribosylation factor (energy and GTP binding), ATP synthase subunit alpha/beta (ATP binding), copper–zinc superoxide dismutase (lipid transport, metal ion binding and superoxide metabolic processes), c analogous to peritrophins (chitin binding), elongation factor protein (GTP binding), and histone proteins (DNA binding) (Appendix Workbooks 1–27). Actin, a major component of the cytoskeleton (and muscle fibres), is one of the most abundant and highly conserved proteins in eukaryotes and is usually encoded in multiple genes.45,46 Actin was observed in the majority of the NM exposures (except controls) irrespective of medium or NM ageing. As evidenced by the differential and unique number of proteins on the NM surfaces, it is crucial to consider that the same NMs with different surface coatings, aged in different media will have different uptake potentials based on differences in their acquired coronas, the compositions of which are influenced by the ionic strength and NOM concentration in the medium (Table 2). This also demonstrates that NM transformation is a multi-parameter process driven by NM size, surface coating, age and the medium in which the NMs are dispersed prior to and during exposure.
| Material | HH combo ‘pristine’ NMs | HH combo ‘aged’ NMs | Class V ‘pristine’ NMs | Class V ‘aged’ NMs | Class I ‘aged’ NMs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of identified proteinsa | Number of identified molecular pathways | Number of molecular pathways unique to the control | Number of identified proteins | Number of identified molecular pathways | Number of molecular pathways unique to the control | Number of identified proteins | Number of identified molecular pathways | Number of molecular pathways unique to the control | Number of identified proteins | Number of identified molecular pathways | Number of molecular pathways unique to the control | Number of identified proteins | Number of identified molecular pathways | Number of molecular pathways unique to the control | |
| a Note: the number of proteins includes the total number of proteins identified in the NM coronas. This does not reflect the individual unique number of proteins, but reflects the collective total, including multiple identifications of proteins of the same origin. | |||||||||||||||
| Control | 27 | 10 | NA | 27 | 10 | NA | 11 | 8 | NA | 11 | 8 | NA | NA | NA | NA |
| Bulk Ag | 27 | 12 | 9 | NA | NA | NA | 21 | 13 | 7 | NA | NA | NA | NA | NA | NA |
| Uncoated Ag | 79 | 13 | 8 | 58 | 19 | 15 | 117 | 19 | 13 | 104 | 22 | 16 | 85 | 17 | 14 |
| PVP Ag | 223 | 32 | 27 | 35 | 17 | 14 | 246 | 43 | 37 | 81 | 23 | 18 | 54 | 13 | 11 |
| Ag2S | 58 | 18 | 14 | 126 | 25 | 19 | 32 | 11 | 6 | 49 | 20 | 15 | 79 | 21 | 16 |
| TiO2 uncoated | 35 | 15 | 12 | 12 | 8 | 6 | 15 | 9 | 8 | 108 | 23 | 17 | 101 | 26 | 20 |
| TiO2 PVP | 48 | 14 | 11 | 64 | 19 | 14 | NA | NA | NA | 61 | 22 | 16 | 83 | 19 | 17 |
There were also clear differences in the total number of combined surface bound proteins attached to the freshly dispersed and 6-month medium aged NM surfaces after exposure to the daphnids (Table 2). Previous studies have suggested that there are only a few tens of proteins that form a corona around a NM surface when introduced to complex media and environments.7 Interestingly, we were able to identify a total number of 223 surface bound proteins associated with the pristine PVP NMs in HH combo and 246 surface bound protein associated with freshly-dispersed PVP Ag NMs in class V water (Table 2). In most cases, there were higher total numbers of surface bound proteins identified on the freshly-dispersed NM surfaces in the HH combo media than on those that were aged under the same conditions, the exceptions being Ag2S and TiO2 PVP NMs. For the NMs in the class V water the trend was also that the freshly-dispersed NMs had the highest total of surface bound proteins in their corona, compared to their aged counterparts (exceptions; TiO2 uncoated). Explanations for the lower total surface bound proteins on the pristine Ag2S and TiO2 PVP NM in the HH combo media and TiO2 uncoated in the class V water, may be due to particle agglomeration, which is consistent with the size data (Table S2†). Thus, agglomeration reduces the free NM surface area available for protein binding and therefore, reduces the number of proteins identified. For the NMs in the class V water, NOM will competitively bind to the NM surface, also reducing the free surface available for protein binding.47 Agglomeration is driven by the ionic strength of the medium (HH combo) and by the presence of NOM (class V) interacting with the surface of the NMs changing their physicochemical properties.48
Although we did not analyse the NM coronas from NMs that were internalized in the daphnids, the higher number of total surface bound proteins on the pristine NMs corresponds with the increased total bioaccumulated metal concentrations recovered from the whole organisms (Table 1). In comparison, lower metal concentrations were identified in the whole organisms when exposed to the aged NMs. Several possibilities explain the higher internalized concentration of the freshly-dispersed NMs and their higher number of surface proteins. Firstly, freshly-dispersed NMs will have an increased binding affinity, due to higher surface reactivity49,50 particularly in the NOM free HH combo medium. Therefore, once taken up by the Daphnia, the NMs are more likely to concentrate and be retained in the gut by adhesion to the gut epithelium cells (Fig. 5). Moreover, the NMs will have a higher affinity to proteins and vesicles allowing their translocation into other cellular and tissue compartments. By contrast, the aged NMs are less surface reactive, and those aged in the artificial natural waters (class I and V) are likely to have NOM coronas that must be displaced by higher affinity proteins in order to form the protein corona, resulting in less surface bound proteins in the NM coronas.
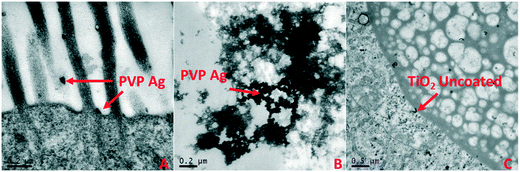
To confirm cellular translocation, pristine PVP Ag and pristine PVP TiO2 (HH combo medium exposures) were visualised in the gut and in cellular material when imaged under TEM (Fig. 5). There was also a high abundance of proteins related to the cell surface, cytoskeleton, cytosol and lipid transport identified in the corona of the pristine PVP Ag NMs in the HH combo medium exposures, including endocuticle structural glycoprotein SgAbd-2, putative Ccp84Ae and copper–zinc superoxide dismutase proteins. Similarly, the pristine uncoated Ag (HH combo medium exposures), Ag2S (class V exposures), aged TiO2 uncoated TiO2 PVP (class V) NMs also contained these and similar pathway related proteins, suggesting transport of the respective NMs into the cells of the gut epithelium. Additionally, the importin subunit alpha-4 protein, which is responsible for importing proteins into the nucleus was identified in the corona of the pristine PVP Ag NMs in the class V water exposure, further evidencing NM translocation via protein binding in the corona.
Protein coronas: pathway association
Collectively, the freshly-dispersed and 6-months medium aged NMs had proteins in their coronas relating to 110 GO terms compared to the 8 GO terms for the secreted proteins in the control (medium only) populations (Fig. 6). The GO terms describe ancestral genes that are annotated to describe the gene products, that are involved in the molecular and biological pathways and protein synthesis.51 The pathways are defined as the interacting elements (genes) of genomes that perform a specific molecular function or a biological process.24 Thus, identifying the proteins that are in the NM coronas (Fig. 7 and 8) gives insight into what functions these proteins have in complex molecular and biological processes, or as cellular components, and sheds light on pathways activated in response to exposure to NMs generally or to a specific NM.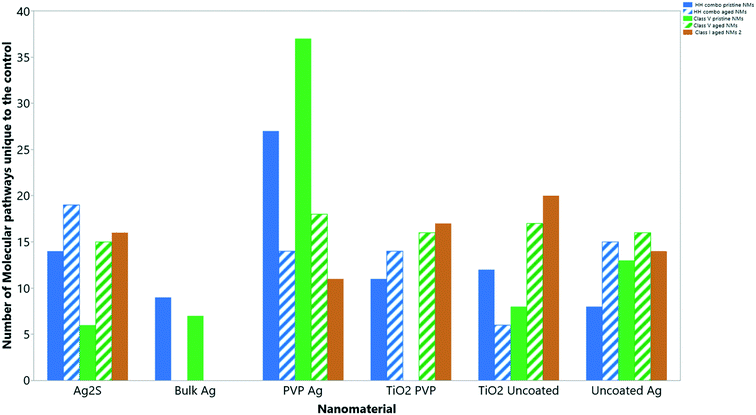 | ||
| Fig. 6 The total number of ‘GO’ molecular pathways uniquely identified compared to the controls for each NM condition. | ||
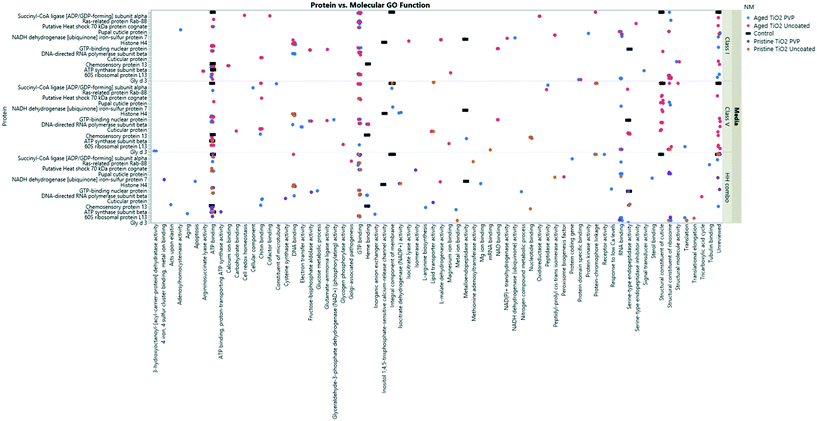 | ||
| Fig. 7 TiO2 NM coronas for each medium condition, showing the associated GO terms relating to biological and molecular pathways for each protein ID. Note, not all the protein IDs are not listed on the Y axis due to character spacing, the figure is intended to give an insight into the most popular GO terms and associated pathways. Some proteins are involved in multiple pathways, and therefore are listed only as their primary GO terms for comparison as described at http://www.uniprot.org. A full list of the protein IDs are located in the ESI† Table S4. | ||
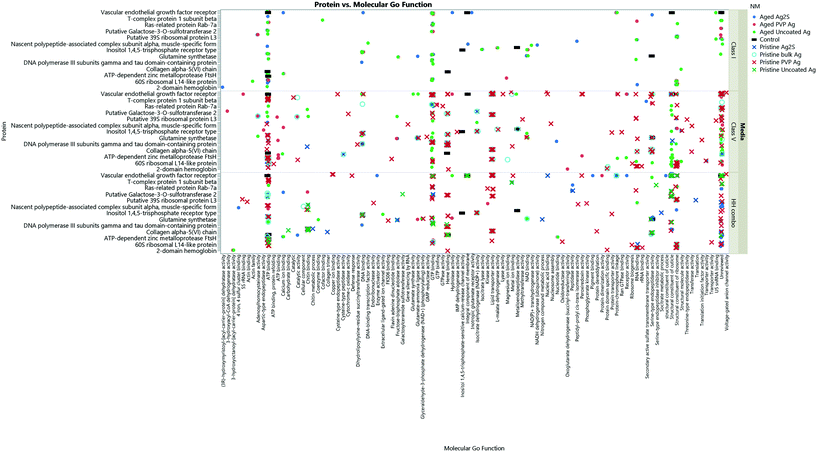 | ||
| Fig. 8 The collective Ag NM coronas for each medium condition, showing the associated GO terms relating to biological and molecular pathways for each protein ID. Note, not all the protein IDs are not listed on the Y axis due to character spacing; the figure is intended to give an insight into the most popular GO terms and associated pathways. Some proteins are involved in multiple pathways, and therefore are listed only as their primary GO terms for comparison as described at http://www.uniprot.org. A full list of the protein IDs are located in the ESI† Table S4. Individual Ag NMs and associated GO terms are identified in Fig. S9–S11.† | ||
The proteins secreted by the unexposed control organisms were mainly associated with pathways involved in general cellular homeostasis, biological and metabolic processes, catalytic activity, molecule function, ion and small molecule binding, all of which are normal organism functions (Table S4 ESI†). By contrast, the unique pathways associated with the proteins secreted by the daphnids in response to all NM exposures and subsequently incorporated into the NM coronas were mainly associated with metabolic activity, DNA damage, mitochondrial break down and energy processes associated with cytotoxic damage.
Functions of the most commonly identified proteins in the NMs coronas
As shown in Fig. 7 and 8, three of the most common GO term pathways that were associated with all NMs were ATP binding, GTP binding and DNA binding. ATP binding is involved in protein function and energy consumption.52 This pathway was identified by various proteins including actin, ATP synthase subunit, heat shock 70 kDa protein cognate, S-adenosylmethionine synthase, chaperone protein ClpB to name a few. GTP binding is responsible for the G protein family activation, effectively the ‘switching on’ of signalling transduction pathways.53 The common proteins that were identified in GTP binding were elongation factor G, ras-related protein Rab-7a, GTP-binding protein SAR1a and tubulin alpha and beta chain proteins.Proteins involved in DNA binding including DNA-directed RNA polymerase subunit beta, and various histone proteins which were identified in the NM coronas. Histones are central for transcription regulation, DNA repair, DNA replication and chromosomal stability.54 DNA accessibility is regulated via a complex set of post-translational modifications of histones,55 such as histone H1.3, histone H2A, histone H2B, histone H3.2, histone H2B, and histone H4, all of which were identified in high abundance in the all freshly dispersed and aged NM coronas, irrespective of media. Therefore, each of these pathways were mediated by both the pristine and aged NM exposures, since they were not identified in the control media in the absence of NMs (Daphnia secreted proteins only).
We suggest that the Daphnia try to counteract the cytotoxic responses from exposure to the NMs by increasing basic energy metabolism, as evidenced by the amount of secreted proteins incorporated in the NM coronas that are associated with ATP binding pathways compared to the controls (Fig. 7 and 8). As NMs come into contact with the cells, the cell treats the NM as a foreign body by coding for certain proteins involved in the cellular metabolism and defence mechanisms including: chymotrypsin BI, aldehyde dehydrogenase, copper–zinc superoxide dismutase, chorion peroxidase, and putative actin, alpha skeletal muscle proteins,56,57 which were identified in a range of the NM coronas (Appendix; Workbooks 1–27). To protect against cytotoxic damage the daphnids increased protein degradation via carboxypeptidase activity, and increased the regulation of cellular cycles via the activity of cyclin dependant kinase, as observed in the coronas of the NMs associated with the pristine Ag2S in the HH combo medium, aged PVP Ag NMs in the HH combo medium, aged TiO2 PVP in both the HH combo and class I media as examples. Proteins associated with cell cycle regulation were observed in all media irrespective of NM ageing, suggesting that the effects on the daphnids are a general response to NM exposure, since these proteins were not secreted in the NM-free controls.
Furthermore, cysteine synthase (identified in the pristine PVP-coated TiO2 NM coronas in the HH combo) are proteins involved in glutathione synthesis,58 whose expression may have increased as a result of inhibition of homeostasis by the affected Daphnia when exposed to the NMs. Other pathways that were indicative of cytotoxic damage include GTP binging processes (Ras-related protein Rab-11B), and isocitrate dehydrogenase (NADP+) activity, lipid transporter activity, nucleotide binding and oxidoreductase activity, which were all identified in the coronas of the pristine and aged NMs irrespective of the medium (Fig. 7 and 8).
Further evidence of induced stress caused by exposure to the NMs was via the presence of heat-shock proteins which were not observed in the control populations. The intracellular build-up of damaged proteins can lead to proteotoxic stress, resulting in ageing phenotypes due to increased stress.59 Heat-shock proteins provide a molecular defence mechanism against proteotoxic stress, facilitating protein folding and unfolding, which essentially renatures or degrades damaged proteins, to restore normal cellular homoeostasis.45,60 Heat shock proteins were present in high abundance in the NM coronas of both pristine and aged Ag2S, pristine and aged PVP Ag, aged uncoated Ag NMs and pristine bulk Ag in HH combo medium; aged uncoated TiO2, aged TiO2 PVP, aged uncoated Ag, and aged Ag2S NMs in class I water; aged uncoated TiO2, aged TiO2 PVP, aged uncoated Ag and aged PVP Ag NMs, and pristine PVP Ag NMs in class V water, confirming cytotoxic stress. Moreover, this pathway is also involved in the ageing processes as the presence of apolipoprotein D (identified specifically in the corona of aged TiO2 PVP in HH combo medium) was also identified alongside the heat shock proteins and matrix metalloproteinase 1, which is responsible for apoptosis. As it is widely understood, if any living cell is unable to break down/remove the foreign body (in this case the NM) then the cell switches to apoptosis to kill the cell.61 Both the aged and pristine PVP Ag NMs (HH combo exposure) had evidence of lysosomal proteins in their NM coronas, evidencing that signalling pathways for DNA damage and apoptosis were triggered. Further evidence of apoptosis driven by cytotoxic responses to the NMs included the presence of (among others) cytochrome c-2, matrix metalloproteinase 1 proteins and the presence of apolipoprotein D.
Apolipoprotein D (observed in the corona of aged PVP-coated TiO2 NM in the HH combo medium) is also linked to ageing, lipid transport and other developmental processes (Fig. S5†).62 Apolipoprotein D, is part of the lipocalin family which has a high degree of sequence conservation from insects to mammals.62 It is a lipid binding protein whose expression is strongly induced in the mammalian brain during ageing62 and age-dependent neurodegenerative diseases63 such as Alzheimer's disease (AD), where it can play an important function as a neuroprotective and antioxidant protein. Increasing evidence suggests that the gradual increase in free radicals and oxidative stress with age is the primary determinant of ageing.64 The presence of ageing related proteins in the NMs coronas is consistent with the morphological evidence presented previously34,65 that exposure to Ag NMs results in dramatic tail shortening and increased lipid droplets, two phenotypical traits of ageing, suggesting that the NMs induce accelerated ageing in the developing daphnids. Other proteins which have a high degree of species conservation that were linked to lipid transport, antioxidant processes and developmental processes are copper–zinc superoxide dismutase, vascular endothelial growth factor receptor, and S-adenosylmethionine synthase. Copper–zinc superoxide dismutase were found in most of the NM coronas irrespective of NM ageing and media type, and is a highly conserved protein that provides antioxidant processes that counteract ROS. If ROS remains elevated in the cells, it can cause lipid peroxidation, inactivation of enzymes and DNA/protein oxidation.57,66 Fig. S6† describes the main copper–zinc superoxide dismutase molecular and biological pathways. Vascular endothelial growth factor receptor (found in the coronas of aged PVP Ag and aged TiO2 PVP NMs in HH combo) in crustaceans plays a pivotal role in growth, development, cell division and the differentiation of endothelial cells.67 In mammalian cells, inhibition of the vascular endothelial growth factor receptors have been linked to hypotension and cardiovascular disease and such inhibitors have uses in anticancer and antitumor activity therapies.68,69 Fig. S7† shows the binding processes, signalling pathways and receptor activation pathways. Lastly, S-adenosylmethionine synthase is the main methyl donor for DNA, RNA and histone methylation reactions in one-carbon metabolism.70 Metal contaminants have been shown to interrupt the regulation of DNA methylation of the one-carbon cycle, having implications for the fitness of Daphnia.71 Fig. S8† shows the S-adenosylmethionine synthase cycle related biological processes.
Orthologous proteins have the same structure and evolutionary origin (homologous) and the genes that code for these proteins are therefore conserved between species. By identifying the domain architecture in the sequential arrangement along a protein sequence, we can be identify the functional annotation transfer between species.72 Using orthologs of species that have complete mapped genomes it is possible to identify the pathways that have been conserved across species deep in the evolutionary origins in the animal phylogeny.24 When checked against the ancestry ortholog (via OrthoDB at https://www.orthodb.org/) it was possible to find common ancestral genes that are conserved between Daphnia magna and Homo sapiens by identifying the overlapping homologous superfamilies.
For apolipoprotein D, both Homo sapiens and Daphnia magna had matches at InterPro domain at IPR022271, 12674, 02969 and 00566. Similarly, between Daphnia magna and Homo sapiens for copper–zinc superoxide dismutase there were 35% overlaps between the InterPro domain IPR036163 06121 24134 36423 01424 18152 and 30% at InterPro domain IPR036423 24134 01424 18152. For the vascular endothelial growth factor receptor, there were overlaps with the InterPro domain at IPR029034 00072, and for S-adenosylmethionine synthase there were overlaps at InterPro domain at IPR002133 22636 22628 22629 22631 22630 between the two species.
The overall interpretation of this data highlights that the biological and molecular pathways induced are not unique to Daphnia and are in fact shared between a host of species. Although this evidence is based primarily on the identification of specific proteins via linked pathways, the research is still fundamental for identifying the provoked pathways in model non-mammalian species shared between common ancestral phylogeny. This information strengthens the findings from eco-toxicological studies, towards a more general understanding of how exposure to NM pollutants disrupts biological processes that otherwise ensure animal (including human) health. With the increasing move away from animal testing, extraction of increased mechanistic understanding of the toxicity of NMs, and subsequent building of adverse outcome pathways becomes possible. The identification of proteins related to mechanisms of toxicity in the acquired protein coronas on NMs recovered from the exposure waters, especially in the pristine NMs, indicates that NMs may be usefully applied as probes of modes of action in studies with a range of pollutant types, wherein the NMs could be utilised to concentrate low abundance proteins following removal of the test organisms from the solution to ensure no contribution of organism response to the presence of the NMs. Even more importantly, future work should also focus on exploring organism responses at more environmentally realistic concentrations – for example at predicted exposure concentrations, as the data presented here suggest that the NM corona can concentrate and support identification of proteins involved in normal organism function and repair functions.
Conclusion
Using a variety of ‘pristine’ and ‘aged’ NMs that differed in core material, size and surface properties, in environmentally relevant exposure media, the mechanism of toxicity induced by the NMs was explored through investigation of the compositions of the secreted coronas that had been acquired during 7 days of exposure to Daphnia. The surface bound proteins identified varied between NM type, NM ageing and medium composition. Proteins identified on both of the pristine and aged NMs were largely associated with metabolic damage, DNA damage, mitochondria breakdown, and energy processes, which are all associated with cytotoxic damage, gaving insights into the selectivity in the endogenous pathways. Results were also medium specific, with the highest number of proteins, and numbers of proteins being strongly associated with damage and toxicity, identified in the coronas of the freshly dispersed (pristine) NMs in the HH combo medium (particularly the pristine PVP Ag NMs). Thus, the salt only medium and pristine particles (which had the highest surface energies) were the most toxic to the daphnids, resulting in the highest protein secretions and the most proteins bound to the NMs. The higher degree of toxicity-related proteins in the coronas of the pristine NMs was likely a consequence of insufficiently quenched surface reactivity casing increased protein secretion, as shown by comparison with the corona composition on the environmentally aged NMs. The environmental water samples, with their varying concentrations of natural organic matter, had lower amounts of proteins adsorbed, likely due to pre-coating by the NOM reducing the available surface for protein binding, as well as the lower numbers of total proteins secreted under these conditions. The aged NMs also had lower uptake of the NMs as evidenced by the lower Ti/Ag concentrations detected in the daphnids. These realistic water conditions (i.e. the model waters with various NOM contents) are the most likely exposure scenarios for aquatic organisms when naïve NMs are first released directly into the environment. The likelihood of direct contact between the pristine NMs with the aquatic organisms would be negligible due to rapid (essentially instantaneous) transformations that would occur due to the high surface reactivity of the NMs with the surrounding environment.The proteins secreted by the unexposed control organisms were mainly associated with pathways involved in general cellular homeostasis, biological and metabolic processes, catalytic activity, molecule function, ion and small molecule binding, all of which are normal organism functioning. By contrast, although the aged NMs had significantly less proteins identified in their coronas, the unique pathways associated with the proteins secreted by the daphnids in response to all NM exposures (pristine and aged NMs) were mainly associated with metabolic activity, DNA damage, mitochondrial break down and energy processes associated with cytotoxic damage.
The information presented provides a snapshot in time; the chronic studies themselves continued to day 24 (and beyond in cases where the NMs resulted in reproductive delays), and as such, further studies should include characterisation of the corona at later time points, wherein the effects from the NMs are more diversified. The distinct differences in acquired coronas, and the convergence of pathways activated even with the low-toxic aged NMs, provides important new insights that will be helpful in the establishment of adverse outcome pathways for NMs. Furthermore, the results also demonstrate the risk of over estimating the toxicity of pristine NMs using the current regulatory tests, as in real environments NOM and secreted biomolecules with essentially instantaneously bind to the NMs reducing their surface reactivity and consequent toxicity to aquatic organisms.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded via a NERC highlight topic grant (NE/N006569/1). The authors acknowledge use of the Functional Genomics and Proteomics Laboratories at the University of Birmingham. Additional support from the H2020 project NanoSolveIT (Grant Agreement No. 814572) is acknowledged, and the data are deposited in the NanoCommons (Grant Agreement No. 731032) Knowledge Base.References
- M. A. Gatoo, S. Naseem, M. Y. Arfat, A. Mahmood Dar, K. Qasim and S. Zubair, Physicochemical properties of nanomaterials: implication in associated toxic manifestations, BioMed Res. Int., 2014, 498420 Search PubMed.
- P.-C. Lin, S. Lin, P. C. Wang and R. Sridhar, Techniques for physicochemical characterization of nanomaterials, Biotechnol. Adv., 2014, 32(4), 711–726 Search PubMed.
- K. L. Aillon, Y. Xie, N. El-Gendy, C. J. Berkland and M. L. Forrest, Effects of nanomaterial physicochemical properties on in vivo toxicity, Adv. Drug Delivery Rev., 2009, 61(6), 457–466 Search PubMed.
- A. R. Petosa, D. P. Jaisi, I. R. Quevedo, M. Elimelech and N. Tufenkji, Aggregation and deposition of engineered nanomaterials in aquatic environments: role of physicochemical interactions, Environ. Sci. Technol., 2010, 44(17), 6532–6549 Search PubMed.
- M. Markiewicz, J. Kumirska, I. Lynch, M. Matzke, J. Köser and S. Bemowsky, et al. Changing environments and biomolecule coronas: consequences and challenges for the design of environmentally acceptable engineered nanoparticles, Green Chem., 2018, 20(18), 4133–4168 Search PubMed.
- F. Nasser, J. Constantinou and I. Lynch, Nanomaterials in the Environment Acquire an ‘Eco-Corona’Impacting their Toxicity to Daphnia Magna–a Call for Updating Toxicity Testing Policies, Proteomics, 2019, 20, 1800412 Search PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz and R. Hecht, et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology, Nat. Nanotechnol., 2013, 8(10), 772–781 Search PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, What the cell “sees” in bionanoscience, J. Am. Chem. Soc., 2010, 132(16), 5761–5768 Search PubMed.
- G. Pulido-Reyes, F. Leganes, F. Fernández-Piñas and R. Rosal, Bio-nano interface and environment: A critical review, Environ. Toxicol. Chem., 2017, 36(12), 3181–3193 Search PubMed.
- I. Lynch, K. A. Dawson, J. R. Lead and E. Valsami-Jones, Macromolecular coronas and their importance in nanotoxicology and nanoecotoxicology, Frontiers of Nanoscience, Elsevier, 2014, vol. 7, pp. 127–156 Search PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells, ACS Nano, 2012, 6(7), 5845–5857 Search PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin and H. Nilsson, et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2050–2055 Search PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7(12), 779–786 Search PubMed.
- M. Lundqvist, J. Stigler, T. Cedervall, T. Berggård, M. B. Flanagan and I. Lynch, et al. The evolution of the protein corona around nanoparticles: a test study, ACS Nano, 2011, 5(9), 7503–7509 Search PubMed.
- F. Bertoli, D. Garry, M. P. Monopoli, A. Salvati and K. A. Dawson, The intracellular destiny of the protein corona: a study on its cellular internalization and evolution, ACS Nano, 2016, 10(11), 10471–10479 Search PubMed.
- S. Juling, A. Niedzwiecka, L. Böhmert, D. Lichtenstein, S. Selve Sr and A. Braeuning, et al. Protein Corona Analysis of Silver Nanoparticles Links to Their Cellular Effects, J. Proteome Res., 2017, 16(11), 4020–4034 Search PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(38), 14265–14270 Search PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation, Nat. Nanotechnol., 2011, 6(1), 39 Search PubMed.
- J. Gao, L. Lin, A. Wei and M. S. Sepúlveda, Protein corona analysis of silver nanoparticles exposed to fish plasma, Environ. Sci. Technol. Lett., 2017, 4(5), 174–179 Search PubMed.
- A. Alijagic, O. Benada, O. Kofroňová, D. Cigna and A. Pinsino, Sea urchin extracellular proteins design a complex protein corona on titanium dioxide nanoparticle surface influencing immune cell behaviour, Front. Immunol., 2019, 10, 2261 Search PubMed.
- F. Nasser and I. Lynch, Secreted protein eco-corona mediates uptake and impacts of polystyrene nanoparticles on Daphnia magna, J. Proteomics, 2016, 137, 45–51 Search PubMed.
- S. Briffa, F. Nasser, E. Valsami-Jones and I. Lynch, Uptake and impacts of polyvinylpyrrolidone (PVP) capped metal oxide nanoparticles on Daphnia magna: role of core composition and acquired corona, Environ. Sci.: Nano, 2018, 5(7), 1745–1756 Search PubMed.
- T. Vandenbrouck, O. A. Jones, N. Dom, J. L. Griffin and W. De Coen, Mixtures of similarly acting compounds in Daphnia magna: from gene to metabolite and beyond, Environ. Int., 2010, 36(3), 254–268 Search PubMed.
- I. B. Rogozin, D. Managadze, S. A. Shabalina and E. V. Koonin, Gene family level comparative analysis of gene expression in mammals validates the ortholog conjecture, Genome Biol. Evol., 2014, 6(4), 754–762 Search PubMed.
- A. Prachumwat and W.-H. Li, Protein function, connectivity, and duplicability in yeast, Mol. Biol. Evol., 2005, 23(1), 30–39 Search PubMed.
- J. Yang, R. Lusk and W.-H. Li, Organismal complexity, protein complexity, and gene duplicability, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(26), 15661–15665 Search PubMed.
- W. Qian and J. Zhang, Genomic evidence for adaptation by gene duplication, Genome Res., 2014, 24(8), 1356–1362 Search PubMed.
- S. B. Carroll, Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution, Cell, 2008, 134(1), 25–36 Search PubMed.
- OECD, OECD Guidline for the testing of chemicals, Test No. 211, Daphnia Magna Reproduction Test, OECD Publishing, 2012.
- OECD Guideline for testing of chemicals. Daphnia sp., Acute Immobilisation Test 202, Adpoted April 2004, ( 2004).
- K. Hund-Rinke, A. Baun, D. Cupi, T. F. Fernandes, R. Handy and J. H. Kinross, et al. Regulatory ecotoxicity testing of nanomaterials–proposed modifications of OECD test guidelines based on laboratory experience with silver and titanium dioxide nanoparticles, Nanotoxicology, 2016, 10(10), 1442–1447 Search PubMed.
- F. Nasser and I. Lynch, Updating traditional regulatory tests for use with novel materials: Nanomaterial toxicity testing with Daphnia magna, Saf. Sci., 2019, 118, 497–504 Search PubMed.
- L.-J. A. Ellis, E. Valsami-Jones and I. Lynch, Exposure medium and particle ageing moderate the toxicological effects of nanomaterials to Daphnia magna over multiple generations: a case for standard test review?, Environ. Sci.: Nano, 2020, 7, 1136–1149 Search PubMed.
- L. J. A. Ellis, S. Kissane, E. Hoffman, J. B. Brown, E. Valsami-Jones and J. Colbourne, et al. Multigenerational Exposures of Daphnia Magna to Pristine and Aged Silver Nanoparticles: Epigenetic Changes and Phenotypical Ageing Related Effects, Small, 2020, 2000301 Search PubMed.
- J. Hammes, J. A. Gallego-Urrea and M. Hassellöv, Geographically distributed classification of surface water chemical parameters influencing fate and behavior of nanoparticles and colloid facilitated contaminant transport, Water Res., 2013, 47(14), 5350–5361 Search PubMed.
- S. S. Kilham, D. A. Kreeger, S. G. Lynn, C. E. Goulden and L. Herrera, COMBO: a defined freshwater culture medium for algae and zooplankton, Hydrobiologia, 1998, 377(1–3), 147–159 Search PubMed.
- C. O. Hendren, X. Mesnard, J. Dröge and M. R. Wiesner, Estimating Production Data for Five Engineered Nanomaterials As a Basis for Exposure Assessment, Environ. Sci. Technol., 2011, 45(7), 2562–2569 Search PubMed.
- J. Virkutyte, S. R. Al-Abed and D. D. Dionysiou, Depletion of the protective aluminum hydroxide coating in TiO2-based sunscreens by swimming pool water ingredients, Chem. Eng. J., 2012, 191, 95–103 Search PubMed.
- L. Armand, A. Tarantini, D. Beal, M. Biola-Clier, L. Bobyk and S. Sorieul, et al. Long-term exposure of A549 cells to titanium dioxide nanoparticles induces DNA damage and sensitizes cells towards genotoxic agents, Nanotoxicology, 2016, 10(7), 913–923 Search PubMed.
- IARCA, International Agency for Research on Cancer: Agents classified by the IARC, IARC, Lyon, France, 2011.
- I. De Leersnyder, L. De Gelder, I. Van Driessche and P. Vermeir, Influence of growth media components on the antibacterial effect of silver ions on Bacillus subtilis in a liquid growth medium, Sci. Rep., 2018, 8(1), 9325 Search PubMed.
- J. W. Han, S. Gurunathan, J.-K. Jeong, Y.-J. Choi, D.-N. Kwon and J.-K. Park, et al. Oxidative stress mediated cytotoxicity of biologically synthesized silver nanoparticles in human lung epithelial adenocarcinoma cell line, Nanoscale Res. Lett., 2014, 9(1), 459 Search PubMed.
- in Nanomaterial Interfaces in Biology: Methods and Protocols, ed. P. Bergese and K. Hamad-Schifferli, Methods in Molecular Biology, Springer Science+Business Media New York, 2013, vol. 1025, DOI:10.1007/978-1-62703-462-3_11.
- S. Humphries, Filter feeders and plankton increase particle encounter rates through flow regime control, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(19), 7882–7887 Search PubMed.
- K. A. Otte, T. Fröhlich, G. J. Arnold and C. Laforsch, Proteomic analysis of Daphnia magna hints at molecular pathways involved in defensive plastic responses, BMC Genomics, 2014, 15(1), 306 Search PubMed.
- S. Yuan, H. Li, Y. Dang and C. Liu, Effects of triphenyl phosphate on growth, reproduction and transcription of genes of Daphnia magna, Aquat. Toxicol., 2018, 195, 58–66 Search PubMed.
- M. Baalousha, K. Afshinnia and L. Guo, Natural organic matter composition determines the molecular nature of silver nanomaterial-NOM corona, Environ. Sci.: Nano, 2018, 5(4), 868–881 Search PubMed.
- F. Seitz, S. Lüderwald, R. R. Rosenfeldt, R. Schulz and M. Bundschuh, Aging of TiO2 nanoparticles transiently increases their toxicity to the pelagic microcrustacean Daphnia magna, PLoS One, 2015, 10(5), e0126021 Search PubMed.
- R. Duffin, L. Tran, D. Brown, V. Stone and K. Donaldson, Proinflammogenic effects of low-toxicity and metal nanoparticles in vivo and in vitro: highlighting the role of particle surface area and surface reactivity, Inhalation Toxicol., 2007, 19(10), 849–856 Search PubMed.
- S. R. Saptarshi, A. Duschl and A. L. Lopata, Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle, J. Nanobiotechnol., 2013, 11(1), 26 Search PubMed.
- H. Mi, Q. Dong, A. Muruganujan, P. Gaudet, S. Lewis and P. D. Thomas, PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium, Nucleic Acids Res., 2010, 38(suppl_1), D204–D210 Search PubMed.
- A. Sturm, P. Cunningham and M. Dean, The ABC transporter gene family of Daphnia pulex, BMC Genomics, 2009, 10(1), 170 Search PubMed.
- Y. Takai, T. Sasaki and T. Matozaki, Small GTP-binding proteins, Physiol. Rev., 2001, 81(1), 153–208 Search PubMed.
- R. J. Sims III, K. Nishioka and D. Reinberg, Histone lysine methylation: a signature for chromatin function, Trends Genet., 2003, 19(11), 629–639 Search PubMed.
- C. M. Hammond, C. B. Strømme, H. Huang, D. J. Patel and A. Groth, Histone chaperone networks shaping chromatin function, Nat. Rev. Mol. Cell Biol., 2017, 18(3), 141 Search PubMed.
- H. Liu, Y. Liu, C. Song and Z. Cui, A chymotrypsin-like serine protease from Portunus trituberculatus involved in pathogen recognition and AMP synthesis but not required for prophenoloxidase activation, Fish Shellfish Immunol., 2017, 66, 307–316 Search PubMed.
- K. Lyu, X. Zhu, Q. Wang, Y. Chen and Z. Yang, Copper/zinc superoxide dismutase from the cladoceran Daphnia magna: Molecular cloning and expression in response to different acute environmental stressors, Environ. Sci. Technol., 2013, 47(15), 8887–8893 Search PubMed.
- G. Wu, Y.-Z. Fang, S. Yang, J. R. Lupton and N. D. Turner, Glutathione metabolism and its implications for health, J. Nutr., 2004, 134(3), 489–492 Search PubMed.
- D. Clancy and J. Birdsall, Flies, worms and the free radical theory of ageing, Ageing Res. Rev., 2013, 12(1), 404–412 Search PubMed.
- K. Pauwels, R. Stoks and L. De Meester, Coping with predator stress: interclonal differences in induction of heat-shock proteins in the water flea Daphnia magna, J. Evol. Biol., 2005, 18(4), 867–872 Search PubMed.
- M. Redza-Dutordoir and D. A. Averill-Bates, Activation of apoptosis signalling pathways by reactive oxygen species. Biochimica et Biophysica Acta (BBA)-Molecular, Cell Res., 2016, 1863(12), 2977–2992 Search PubMed.
- S. Dassati, A. Waldner and R. Schweigreiter, Apolipoprotein D takes center stage in the stress response of the aging and degenerative brain, Neurobiol. Aging, 2014, 35(7), 1632–1642 Search PubMed.
- D. Sanchez, R. Bajo-Grañeras, M. Del Caño-Espinel, R. Garcia-Centeno, N. Garcia-Mateo and R. Pascua-Maestro, et al. Aging without Apolipoprotein D: Molecular and cellular modifications in the hippocampus and cortex, Exp. Gerontol., 2015, 67, 19–47 Search PubMed.
- E. Martínez, A. Navarro, C. Ordónez, E. del Valle and J. Tolivia, Oxidative stress induces apolipoprotein D overexpression in hippocampus during aging and Alzheimer's disease, J. Alzheimer's Dis., 2013, 36(1), 129–144 Search PubMed.
- P. Karatzas, G. Melagraki, L.-J. A. Ellis, I. Lynch, D.-D. Varsou, A. Afantitis, A. Tsoumanis, P. Doganis and H. Sarimveis, Development of deep learning models for predicting the effects of exposure to engineered nanomaterials on Daphnia Magna, Small, 2020, 16, 2001080 Search PubMed.
- H. Wu, R. Li, Y. Liu, X. Zhang, J. Zhang and E. Ma, A second intracellular copper/zinc superoxide dismutase and a manganese superoxide dismutase in Oxya chinensis: Molecular and biochemical characteristics and roles in chlorpyrifos stress, Ecotoxicol. Environ. Saf., 2020, 187, 109830 Search PubMed.
- S.-c. Zheng, J.-y. Xu and L. H-p, Cellular entry of white spot syndrome virus and antiviral immunity mediated by cellular receptors in crustaceans, Fish Shellfish Immunol., 2019, 93, 580–588 Search PubMed.
- K. B. Neves, F. J. Rios, L. Van Der Mey, R. Alves-Lopes, A. C. Cameron and M. Volpe, et al. VEGFR (Vascular Endothelial Growth Factor Receptor) inhibition induces cardiovascular damage via redox-sensitive processes, Hypertension, 2018, 71(4), 638–647 Search PubMed.
- J.-M. Lee, A. Cimino-Mathews, C. J. Peer, A. Zimmer, S. Lipkowitz and C. M. Annunziata, et al. Safety and clinical activity of the programmed death-ligand 1 inhibitor durvalumab in combination with poly (ADP-ribose) polymerase inhibitor olaparib or vascular endothelial growth factor receptor 1-3 inhibitor cediranib in women's cancers: a dose-escalation, phase I study, J. Clin. Oncol., 2017, 35(19), 2193 Search PubMed.
- F. Kusari, A. M. O'Doherty, N. J. Hodges and M. W. Wojewodzic, Bi-directional effects of vitamin B 12 and methotrexate on Daphnia magna fitness and genomic methylation, Sci. Rep., 2017, 7(1), 1–9 Search PubMed.
- C. G. Athanasio, U. Sommer, M. R. Viant, J. K. Chipman and L. Mirbahai, Use of 5-azacytidine in a proof-of-concept study to evaluate the impact of pre-natal and post-natal exposures, as well as within generation persistent DNA methylation changes in Daphnia, Ecotoxicology, 2018, 27(5), 556–568 Search PubMed.
- K. Forslund, I. Pekkari and E. L. Sonnhammer, Domain architecture conservation in orthologs, BMC Bioinf., 2011, 12(1), 326 Search PubMed.
- Y. Hayashi, T. Miclaus, C. Scavenius, K. Kwiatkowska, A. Sobota, P. Engelmann, J. J. Scott-Fordsmand, J. J. Enghild and D. S. Sutherland, Species Differences Take Shape at Nanoparticles: Protein Corona Made of the Native Repertoire Assists Cellular Interaction, Environ. Sci. Technol., 2013, 47, 24 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0en00625d |
| This journal is © The Royal Society of Chemistry 2020 |