Open Access Article
Open Access ArticlePolystyrene nanoplastics accumulate in ZFL cell lysosomes and in zebrafish larvae after acute exposure, inducing a synergistic immune response in vitro without affecting larval survival in vivo†
Irene
Brandts
ab,
Marlid
Garcia-Ordoñez
ab,
Lluis
Tort
b,
Mariana
Teles
*ab and
Nerea
Roher
 *ab
*ab
aInstitute of Biotechnology and Biomedicine (IBB), Universitat Autònoma de Barcelona, 08193 Barcelona, Spain. E-mail: nerea.roher@uab.cat; mariana.teles@uab.cat
bDepartment of Cell Biology, Animal Physiology and Immunology, Universitat Autònoma de Barcelona, 08193 Barcelona, Spain
First published on 22nd July 2020
Abstract
The presence of small-sized plastic particles in marine and freshwater environments is a global problem but their long-term impact on ecosystems and human health is still far from being understood. Nanoplastics (<1000 nm) could pose a real and uncontrolled ecological challenge due to their smaller size and sharp ability to penetrate living organisms at any trophic level. Few studies evaluate the impact of nanoplastics in vivo on the immune system of aquatic organisms, while most of them assessed the impact on indirect markers of immune response such as regulation of gene expression, ROS production or DNA genotoxicity, among others. Moreover, the study of the effects of nanoplastics on aquatic vertebrate species in vivo is still scarce. In this context, we seek to shed light on the underlying effects of polystyrene nanoplastics (PS-NPs) on the immune response in a model fish species (Danio rerio, zebrafish) after an acute exposure, with a combination of in vitro and in vivo experiments. Our results show that PS-NPs (65 nm) are efficiently taken up by zebrafish liver cells, accumulating mainly in lysosomes. Furthermore, the expression of immune genes presents a synergy when cells were simultaneously exposed to PS-NPs, at a low dose and early time point (12 h) and challenged with a viral stimulus (poly(I:C)). Moreover, zebrafish larvae also internalize PS-NPs, accumulating them in the gut and pancreas. However, at concentrations of up to 50 mg l−1 in an acute exposure (48 h), PS-NPs do not interfere with the survival of the larvae after a lethal bacterial challenge (Aeromonas hydrophila). This study addresses the relevant environmental question of whether a living organism exposed to PS-NPs can cope with a real immune threat. We show that, although PS-NPs can induce an immune response, the survival of zebrafish larvae challenged with a bacterial infection after an acute exposure to PS-NP is not decimated with respect to unexposed larvae.
Environmental significanceSmall sized plastic particles have been reported around the planet in all kinds of environments and it is generally accepted that microplastics are able to cause adverse effects on organisms. However, the understanding of the effects and possible implications of nanoplastics on vertebrate species is still scarce. The impact of polystyrene nanoplastics (PS-NPs) on indirect markers of immune response (such as regulation of gene expression, ROS production or DNA genotoxicity) has been to some extent previously assessed. Nevertheless, a key question remains unexplored: can an organism exposed to nanoplastics successfully cope with an additional immune challenge? This is the first study to provide data on how the immune system of a vertebrate species might respond to a combined challenge of nanoplastics and a pathogen in the surrounding environment. We show that an acute exposure of zebrafish larvae to PS-NPs at high concentrations has no effect on the survival rate of the larvae after a bacterial infection. Thus, this study provides novel insight beyond the current understanding of the potential interplay between pathogen and nanoplastic contamination. |
1. Introduction
After more than 50 years of plastic production, with 360 million tonnes manufactured solely in 2018,1 and the subsequent accumulation in the natural environment, the impact of plastics in ecosystems worldwide is just starting to be understood. Although nanoplastics, defined as particles ranging from 1 nm to 1000 nm resulting from degradation of plastic objects,2,3 have already been detected in the environment,4 their precise quantification in natural aquatic systems and the establishment of environmentally relevant concentration are still under debate. There is a general consensus on how microplastics are able to cause adverse effects on marine and freshwater species (see Chae et al.5 for a review). However, there are still many unanswered questions in nanoplastic research, regarding for example their final destiny once absorbed/ingested, their long-term effects or their interaction with microbial communities. One of these relevant knowledge gaps is whether nanoplastic pollution can impair the capacity of organisms to fight against pathogens. This publication aims to start the unravelling of this last question, combining an in vitro and in vivo approach.Zebrafish (Danio rerio) is considered a good model for eco-toxicological studies as well as a tool to understand eco-physiological adaptations in vertebrates. The versatility of this model, together with the availability of molecular and cellular tools, has popularized the use of zebrafish. Previous studies exploring the effects of polystyrene nanoplastics (PS-NPs) on zebrafish have described a broad physiological response, not particularly specific to a given tissue or cell type, but more typical of a general stress response (e.g. oxidative stress, complement system or glucose metabolism). Alterations in behaviour, measured by a decrease6 or increase7 in locomotor activity, changes in glucose metabolism and cortisol levels,7 intestinal and skin damage triggering a pro-inflammatory response8 and production of oxidative stress and lipid accumulation,9 have been documented in zebrafish after waterborne exposure to PS-NPs. Other studies have evaluated the effects of direct injection of PS-NPs on zebrafish embryos, describing changes in the transcriptome10 or production of reactive oxygen species (ROS) and apoptosis.11
Moreover, PS-NPs have been previously found to interfere with the immune system of zebrafish8,10,12 as well as other aquatic species.13,14 In D. rerio, PS-NPs have been shown to upregulate the expression of inflammatory cytokines,8 activate the complement system and oxidative stress mechanisms10 and provoke DNA damage, induced by ROS and apoptosis.11 In this context, we seek to address the question of whether their interaction with the immune system can affect the survival of populations in their natural environments, where organisms are frequently exposed to pathogens. To our knowledge, barely any studies on the combination of PS-NPs and an immune challenge have been performed. In a recent study, Sendra et al.14 evaluated the response of Mytilus galloprovincialis haemocytes in vitro when infected with Vibrio splendidus, following an exposure to PS-NPs. They reported that the invertebrate's haemocytes showed resilience when infected with the bacteria, being able to recover their phagocytic capacity. In the present work, we focus the attention in this direction, to determine whether the combination of an acute exposure to PS-NPs and a subsequent lethal bacterial challenge can shed light on the underlying toxic effects of PS-NPs on aquatic organisms.
At the present time, assessment of environmental concentrations of nanoplastics is largely speculative, primarily due to the lack of efficient analytical methods.15 In one relevant study by Gallego-Urrea et al.16 measured 107–109 particles per ml (100–250 nm size) in solution, using nanoparticle tracking analysis (NTA), in different sample sites of Scandinavian waters.16 The exposure concentrations used in our study fall within this particle range and were chosen taking into account the concentrations used in relevant studies with PS-NPs published in the recent literature6,8,14,17 and our own initial in vitro results, aimed at assessing the interaction of the zebrafish liver (ZFL) cell line with PS-NPs. Even though the tested concentrations could be in the higher range of estimated environmental nanoplastic concentrations, they allow an approximation to the potential response of zebrafish to a double challenge with PS-NPs and a pathogen.
Our results present evidence that PS-NPs are toxic to ZFL cells at doses higher than 75 mg l−1 and can accumulate rapidly inside liver cells, specifically in lysosomes. We show that PS-NPs at low doses (5 mg l−1) and early stimulation times (12 h) synergistically activated the expression of anti-viral genes, while low doses and a longer exposure time did not have any effect. Moreover, we did not observe this synergistic interaction at a high dose (50 mg l−1) and early stimulation time. Similarly, in the in vivo experiment, PS-NPs accumulated in the zebrafish larvae intestine and pancreas, but this accumulation did not impair the larvae's defence mechanisms against a bacterial lethal challenge with Aeromonas hydrophila. Thus, zebrafish larvae pre-exposed to PS-NPs had the same levels of survival as unexposed larvae.
2. Materials and methods
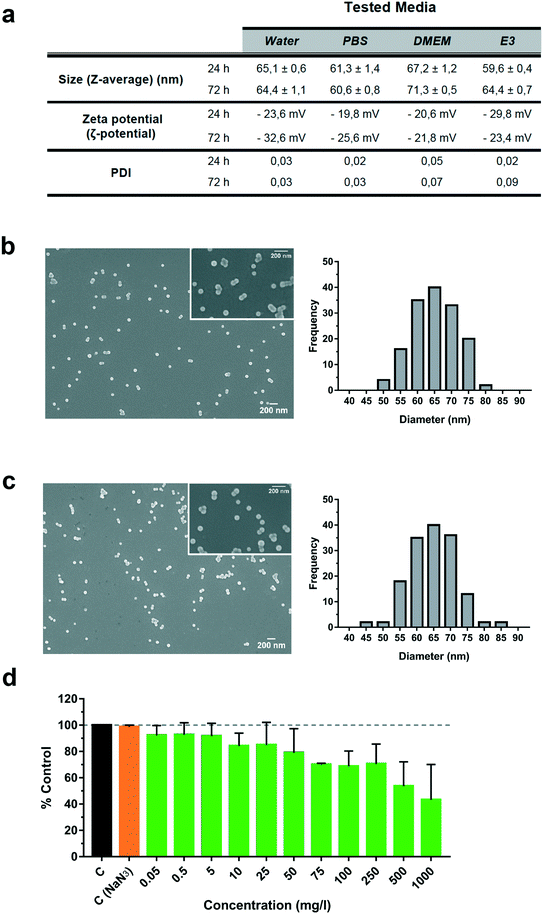
Characterization of polystyrene nanoplastics (PS-NPs) by DLS and electronic microscopy
Fluorescently labelled PS-NPs (FSDG001 Dragon Green, Bangs Ltd.) were purchased from Bang Laboratories (Fisher, IN, USA). PS-NPs were provided as a 1% (w/w) suspension in water with 2 mM NaN3. The particle size distribution and zeta potential were measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments, UK), under different incubation conditions: pure water (Sigma), PBS (Sigma), cell culture medium (DMEM, Gibco) and E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 and 0.1% methylene blue). Emission scanning electron microscopy (FESEM, Zeiss Merlin) was used to determine the external morphology and physical dimensions of the PS-NPs in pure water and cell culture medium. Exposure solutions were prepared by resuspending the stock solution of PS-NPs in PBS and further diluting in each medium, at 100 μg ml−1. For electron microscopy, 20 μl of each solution were pipetted onto silicon chips and air-dried O/N. A palladium–gold coating was applied prior to observation. Images were analysed using the Fiji open source image processing package,18 measuring the dimensions of a minimum of 150 particles for each condition. Size distribution histograms were generated using Prism 7.01 (GraphPad software).Decay of PS-NP fluorescence in vitro and in cell culture
Fluorescently-labelled nanoplastics could leach fluorophores, since they are not covalently linked to the PS-NPs.19 Catarino et al.20 suggested careful testing of the fluorescent particle uptake in order to avoid wrong conclusions. To establish the true internal accumulation of fluorescent PS-NPs, the following experiment was set up: 1 ml of PS-NP suspension (1000, 100 and 50 mg l−1) was centrifuged at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 60 min, then the supernatant (SN) was transferred to a new Eppendorf tube and the pellet (P) resuspended in 1 ml of PBS. The fluorescence intensity in the different fractions (P, SN and stock solution) was evaluated in solution using a Cary Eclipse fluorescence spectrophotometer (Agilent). At the lowest tested concentration, approximately 25% of the fluorescence remained in SN and we can therefore assume that it leached from the PS-NPs. To further confirm that the fluorescence inside the cells was not due to leached fluorophore, ZFL cells were treated with the PS-NP original suspension, SN and P and the uptake was monitored by cytometry as explained below.
000 × g for 60 min, then the supernatant (SN) was transferred to a new Eppendorf tube and the pellet (P) resuspended in 1 ml of PBS. The fluorescence intensity in the different fractions (P, SN and stock solution) was evaluated in solution using a Cary Eclipse fluorescence spectrophotometer (Agilent). At the lowest tested concentration, approximately 25% of the fluorescence remained in SN and we can therefore assume that it leached from the PS-NPs. To further confirm that the fluorescence inside the cells was not due to leached fluorophore, ZFL cells were treated with the PS-NP original suspension, SN and P and the uptake was monitored by cytometry as explained below.
Zebrafish liver (ZFL) and RTGut cell culture
ZFL cells (CRL-2643, ATCC) were cultured at 28 °C, 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) 4.5 g l−1 glucose, supplemented with 0.01 mg ml−1 insulin, 50 ng ml−1 EGF, 5% (v/v) antibiotic/antimycotic solution, 10% (v/v) heat inactivated fetal bovine serum (FBS) and 0.5% (v/v) heat-inactivated trout serum (TS) as described in Torrealba et al.21 RTGut cells were obtained from Dr. Carolina Tafalla's laboratory and cultured as described by Kawano et al.22 Briefly, cells were kept at 20 °C in Leibovitz's L-15 Medium GlutaMAX™ (Gibco) #31415029), supplemented with heat-inactivated FBS 10% (v/v) and 1% antibiotic/antimycotic.PS-NP cytotoxicity studies in ZFL
Cytotoxic and cytostatic effects of PS-NPs on ZFL were assessed using the MTT assay. After 2.5 h in minimal medium (0–0.5% FBS; 2% antibiotic/antimycotic), cultures were incubated with PS-NPs at 0, 0.05, 0.5, 5, 10, 25, 50, 100, 250, 500 and 1000 mg l−1 for 20 h at 28 °C. A 0.2 mM sodium azide control was also included. Cells were then washed in PBS and the MTT substrate (Sigma-Aldrich) was added to 10% of the total volume and further incubated at 28 °C for 1–2 h. The solution was removed, the cells were solubilized in DMSO and the lysate was read on a Victor 3 (PerkinElmer) at 550 nm. The experiment was repeated three times. Data were normalized using Prism 7.01 (GraphPad) such that the control readings were set at 100%. One-way ANOVA was performed with Dunnett's multiple comparison test, comparing treatment and control means.Uptake of PS-NPs by ZFL assessed using flow cytometry and confocal microscopy and in RTGut by microscopy
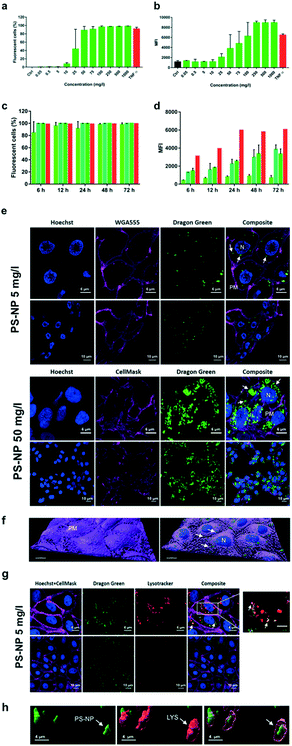
To test cellular uptake, fluorescently labelled PS-NPs were added to ZFL cultures at 70% confluence after 2 h of incubation in minimal medium, at the doses and times indicated below. Representative cytometry plots with the gating strategy and the threshold of fluorescence are shown in ESI† Fig. S1.For dose–response assays, cultures were incubated for 20 h with PS-NPs at 0.05, 0.5, 5, 10, 25, 50, 100, 250, 500 and 1000 mg l−1, at 28 °C. For time-course assays, cultures were incubated with PS-NPs at 25, 50 and 75 mg l−1 for 6, 12, 24, 48 and 72 h, at 28 °C. For positive control of uptake, Atto-488 conjugated TNFα was used, as its uptake has been previously characterized by the research group in this cell line.21 Both dose–response and time-course experiments were performed in triplicate. Post treatment, cells were washed in PBS and incubated at 28 °C with 1 mg ml−1 trypsin (Gibco) for 15 min. This strong trypsinization step aimed to remove PS-NPs attached to the cell surface.21 Then, two volumes of complete medium were added, and cells were retrieved by centrifugation at 300 × g for 5 min. Pellets were resuspended in PBS for flow cytometry (FACSCalibur BD), and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were counted. Data were analysed using Flowing Software 2.5.1 (University of Turku, Finland) and plotted with Prism 7.01 (GraphPad). One-way ANOVA followed by Dunnett's multiple comparison test was performed, comparing treatment and control means. To confirm that the fluorescent PS-NPs were inside the cells, confocal microscopy was performed (Leica SP5). ZFL cells were seeded on ibidi 35 mm glass bottom dishes (Ibidi GmbH, Germany). The next day, cells at approximately 60% confluence were placed in minimal medium. PS-NPs were added 2–3 h later at a concentration of 5 or 50 mg l−1 and cells were incubated for 24 h at 28 °C. Cells were then washed and the medium replaced with new minimal medium. RTGut cells were treated in the same way for confocal microscopy but incubated with PS-NPs at 10 and 25 mg l−1, at 20 °C. The cells were stained with Hoechst (nuclei), either WGA Alexa Fluor555 or Cell Mask Deep Red (membrane) and LysoTracker Red (lysosomes). Images were analysed using Imaris software v9.3 (Bitplane) and Image J.
000 events were counted. Data were analysed using Flowing Software 2.5.1 (University of Turku, Finland) and plotted with Prism 7.01 (GraphPad). One-way ANOVA followed by Dunnett's multiple comparison test was performed, comparing treatment and control means. To confirm that the fluorescent PS-NPs were inside the cells, confocal microscopy was performed (Leica SP5). ZFL cells were seeded on ibidi 35 mm glass bottom dishes (Ibidi GmbH, Germany). The next day, cells at approximately 60% confluence were placed in minimal medium. PS-NPs were added 2–3 h later at a concentration of 5 or 50 mg l−1 and cells were incubated for 24 h at 28 °C. Cells were then washed and the medium replaced with new minimal medium. RTGut cells were treated in the same way for confocal microscopy but incubated with PS-NPs at 10 and 25 mg l−1, at 20 °C. The cells were stained with Hoechst (nuclei), either WGA Alexa Fluor555 or Cell Mask Deep Red (membrane) and LysoTracker Red (lysosomes). Images were analysed using Imaris software v9.3 (Bitplane) and Image J.
Lipid peroxidation
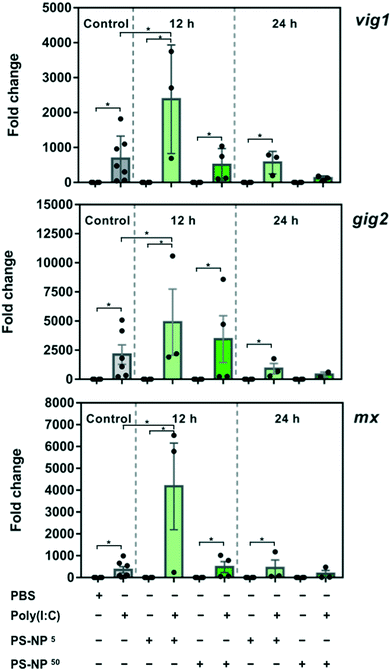
ZFL cells at 60% confluence were cultured in minimal medium (0.5% FBS; 2% A/A) for 2–3 h and then exposed to PS-NPs at 5 mg l−1 and 50 mg l−1 for 24 h. After the exposure, cells were stimulated with poly(I:C) (25 μg ml−1), a synthetic analog of dsRNA virus, for 16 h. Controls were: poly(I:C) 25 μg ml−1 (Sigma-Aldrich) and control cells with neither viral stimulus nor PS-NP exposure. Moreover, each PS-NP exposure time and concentration had a respective control group, with no viral stimulus. After completing the exposure, ZFL cells were washed with PBS and total lipid peroxidation was assessed using the lipid peroxidation (MDA) assay kit (Sigma) following the manufacturer's instructions. The experiment was repeated twice.RNA extraction and gene expression analysis of ZFL cells treated with PS-NPs
ZFL cells were plated and grown to 60% confluence, cultured in minimal medium for 2–3 h and then exposed to PS-NPs ± poly I:C as described above, for the lipid peroxidation assay. For gene expression analysis, an additional exposure time to PS-NPs of 12 h was added, together with the 24 h exposure previously described. Total RNA was extracted using TriReagent (Sigma-Aldrich) following the manufacturer's instructions. RNA was quantified using a nanodrop ND-1000 (Thermo Fisher Scientific) and integrity was checked on an Agilent 2100 Bioanalyser using the RNA 6000 Nano Lab-Chip kit (Agilent Technologies). The experiment was repeated, and four complete sets of high-quality RNA from two independent experiments were selected for cDNA synthesis using 1 μg of total RNA and the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was performed in a CFX384 touch real-time PCR detection system (Bio-Rad) using the iTaq universal SYBR green supermix kit (Bio-Rad) following the manufacturer's instructions. In brief, each PCR mixture consisted of 5 μl SYBR green supermix, 0.4 μM specific primers (ESI† Table S1), 2 μl diluted cDNA and 2.6 μl water (Sigma-Aldrich) in a final volume of 10 μl. Primers for the assessed genes had been previously designed and tested by the research team in the ZFL cell line.23 A reference gene (elongation factor 1alpha (ef1-α)) and three gene markers of the innate immune response to viral infection (IFN-induced protein Mx (mx), viperin (vig1) and grass-carp-reovirus-induced gene 2 (gig2)) were used. All the samples were run in triplicate, and data were analysed using the Livak method.24 For statistical analysis used, a one-way ANOVA test, followed by Dunnett's multiple comparisons for each treatment versus control, was conducted; p < 0.05 was considered statistically significant in all analyses (GraphPad Prism v7.0).Zebrafish husbandry and breeding
Wild type zebrafish (D. rerio) were kept in a re-circulating aquarium with water temperature maintained between 26 and 28 °C. The lighting conditions were 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 h (light
10 h (light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark) and adult fish were fed twice a day at a rate of 2% bodyweight. Ammonia, nitrite, pH and nitrate levels were measured once a week. Ammonia and nitrite levels were kept below the detection level and pH maintained between 6.8 and 7.5. The nitrate levels were maintained to be <100 mg l−1. For in-tank breeding, one female and three males were transferred to a breeding tank in the late afternoon. The divider was removed on the next morning after the onset of light. Embryos were collected after 1–2 h and cultured in embryo medium (E3 medium) in a Petri dish (Deltalab). Fertilized eggs were separated from unfertilized eggs using a plastic pipette (Deltalab). All experiments involving zebrafish (D. rerio) were performed following International Guiding Principles for Research Involving Animals (EU 2010/63) and previously authorized by the Ethics Committee of the Universitat Autònoma de Barcelona (UAB, CEEH number 1582).
dark) and adult fish were fed twice a day at a rate of 2% bodyweight. Ammonia, nitrite, pH and nitrate levels were measured once a week. Ammonia and nitrite levels were kept below the detection level and pH maintained between 6.8 and 7.5. The nitrate levels were maintained to be <100 mg l−1. For in-tank breeding, one female and three males were transferred to a breeding tank in the late afternoon. The divider was removed on the next morning after the onset of light. Embryos were collected after 1–2 h and cultured in embryo medium (E3 medium) in a Petri dish (Deltalab). Fertilized eggs were separated from unfertilized eggs using a plastic pipette (Deltalab). All experiments involving zebrafish (D. rerio) were performed following International Guiding Principles for Research Involving Animals (EU 2010/63) and previously authorized by the Ethics Committee of the Universitat Autònoma de Barcelona (UAB, CEEH number 1582).
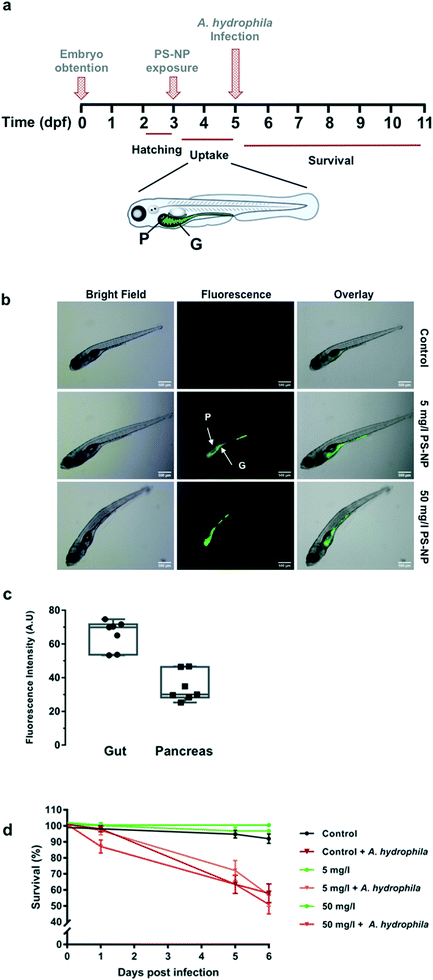
Uptake of PS-NPs by zebrafish larvae assessed by fluorescence microscopy
Groups of 14 larvae (n = 14 per condition) were distributed on 96-well plates (ThermoFisher) with one larva per well containing 200 μl E3 medium or fluorescent PS-NPs at 5, 50 and 100 mg l−1. Mortality was recorded for 96 h and zebrafish larvae were observed using a fluorescence stereomicroscope (Nikon SMZ800) coupled with a camera (Nikon DS-Fi2).A. hydrophila culture and zebrafish larvae infection
Experimental infections were performed as previously described.25 Briefly, bacteria were grown on LB agar plates overnight at 28 °C, collected from the plates in an Eppendorf tube, washed with PBS and finally resuspended with E3 to obtain a stock solution containing approximately 1010 colony-forming units (CFUs) per ml (OD620nm = 1.3). Dilutions at the desired concentration were all prepared from the stock solution. A. hydrophila infection was carried out by bath immersion according to Ji et al.25 Groups of 72 larvae (n = 72 per condition) were distributed on 96-well plates (ThermoFisher) with one larva per well containing 200 μl E3 medium or PS-NPs. For infection, the bacteria were diluted from the stock solution using E3 medium in serial 100-fold dilutions from 10−1 to 10−9. The bacterial dilutions of 10−7, 10−8 and 10−9 (100 μl of each) from the stock solution were inoculated on LB plates and incubated overnight at 28 °C to calculate the real CFUs during the infection. The survival curves were analysed, and statistical differences were assessed using the log-rank test (GraphPad Prism v7.0).3. Results and discussion
Characterization of PS-NPs in biological relevant buffers
The size and shape of fluorescently labelled PS-NP particles were evaluated under different conditions by scanning electronic microscopy and DLS (Fig. 1a–c). We performed incubations at 24 and 72 h in different biological media: phosphate buffer saline (PBS), cell culture medium (DMEM) and larvae water (E3) (Fig. 1a). DLS results showed a particle size of 65.1 ± 0.6 nm in water, 61.3 ± 1.4 nm in PBS, 67.2 ± 1.2 nm in DMEM and 59.6 ± 0.4 nm in E3 after 24 h and 72 h. No major changes were observed using DLS under any conditions with respect to pure sterile water. No aggregation of the particles over time was found (PDI less than 0.09) and the zeta potential was stable and did not change significantly over time, ranging between −20 and −32 mV and thus indicating a stable dispersion of the PS-NPs in suspension. Further confirmation of their characteristics was performed by direct sizing of FESEM images, showing values of 65.2 ± 6.5 nm (water) and 64.9 ± 7 nm (DMEM) (Fig. 1b and c). Moreover, the expected spherical morphology was observed by FESEM under both conditions (Fig. 1b and c). As shown in Fig. 1, the mean size, aggregation and morphology did not change significantly after 24 and 72 h of incubation in DMEM nor E3. Overall, common biological buffers and media did not change the physico-chemical properties of PS-NPs. The same commercially available PS-NPs were used by Cui et al.,26 who reported a similar zeta potential and mean size in a slightly different zebrafish incubation medium. Pitt et al.6 reported a similar zeta potential but different mean size (34.8 ± 10 nm). This difference is probably due to the different compositions of the zebrafish incubation media.To further study PS-NPs' interaction dynamics with living cells, we used PS-NPs labelled with a green fluorophore (Dragon Green). This fluorophore is not covalently linked to the PS-NPs and could be drained from the NPs. In order to discard uncontrolled loss of fluorescence, we evaluated the fluorophore both by fluorimetry (ESI† Fig. S2a) and cytometry (ESI† Fig. S2b and c). The fluorescence signal at three different concentrations (1000, 100 and 50 mg l−1) was stable under the assayed conditions and the amount of leached fluorophore reached a maximum of 25% after dilution and drastic manipulation (ESI† Fig. S2a). Although there is an apparent increase of fluorescence loss as the PS-NP concentration decreases, this can be explained by the inner filter effect, a well-known phenomenon that causes a decrease of fluorescence intensity at high doses and a better yield at low doses. Therefore, the fluorescence measured at 50 mg l−1 is more precise to obtain real percentages of fluorescence loss. When the distinct fractions (PS-NPs; P, pellet; SN, supernatant) were used to evaluate the internalization of PS-NPs by ZFL cells (ESI† Fig. S2b and c), we observed that a remnant of 24.7 ± 2.4% of cells were fluorescent when incubated with SN, while when incubated with the P fraction positive fluorescent cells were 78.2 ± 1.2%. According to Catarino et al., fluorescent nanoplastics could have a significant loss of fluorescence and this draining has to be characterised using appropriate controls.20 In the present study, most of the observed fluorescence is stably associated with PS-NPs and the fluorescence signal computed as a positive fluorescence signal by cytometry and confocal microscopy is truly bound to PS-NPs.
In vitro toxicity, uptake and accumulation of PS-NPs in ZFL cells
Liver is the main detoxification organ in vertebrates and accumulation of PS-NPs in zebrafish liver has previously been reported.6,9 Understanding the interaction of PS-NPs with liver cells would help to better understand how aquatic vertebrates cope with the presence of nanoplastics in their natural environment. Therefore, in order to evaluate the endocytosis and potential toxic effects of PS-NPs, we choose ZFL cells as an in vitro model. ZFL cells exposed to PS-NPs showed a significant decrease in viability (68.8 ± 11.6%) after 20 h at concentrations of 100 mg l−1 and higher (Fig. 1d), although at 75 mg l−1 we already observed a non-significant decrease trend in cell viability. The calculated LD50 of PS-NPs in ZFL cells was 188,8 mg l−1 (ESI† Fig. S3), extremely high compared to other toxics or genotoxics such as KBrO3 or MMS.27 Other studies using different fish cell lines showed that toxicity is highly dependent on the cell line28,29 but no data was available on any zebrafish cell line. In a recent study by Cortés et al.,27 no toxicity was reported at concentrations of up to 200 mg l−1 PS-NPs, after 24 h of treatment. It is worth mentioning that the LD50 calculation methodology is originally designed to calculate lethal doses of drugs or chemicals. Due to this, and since no specific adaptation to nanoplastic toxicity has been made, we sustain that these LD50 calculations should be interpreted with caution. Considering the results of the viability and uptake assays as well as the previous literature, we decided to use 5 and 50 mg l−1 as a representative non-toxic low and high PS-NP dose. In all the cytometry experiments, a positive control of fluorescent nanoparticles with a well-characterised internalization behaviour was included.21Results for the PS-NPs endocytosis showed that 100% of ZFL cells took up the fluorescent PS-NPs after 6 h of incubation (Fig. 2a and c). Johnston et al.30 showed that 20 nm fluorescent PS-NPs were rapidly internalized in mammalian hepatocytes, as after a 30 min incubation the fluorescence was located inside the cells. The ZFL internalization dynamics follows a dose–response pattern, with cells incubated at higher doses and longer times presenting higher fluorescence (Fig. 2b–d). Internalization was confirmed by confocal microscopy at 5 and 50 mg l−1 PS-NPs (Fig. 2e; see ESI† Fig. S4 for the control). PS-NPs were internalized and found in the cytosol as quite large clusters (Fig. 2e and f, white arrows). At 50 mg l−1, there was a massive accumulation in the cytosol (Fig. 2e) even though the cell viability was barely affected (Fig. 1d). Co-staining with Cell Mask and 3D reconstruction using Imaris software confirmed that PS-NPs were totally embedded inside the cytosol (Fig. 2f). Confocal image analysis indicated that ≈9 PS-NP agglomerates of different sizes were observed per cell (9.23 ± 3.4 agglomerates per cell at 5 mg l−1). Moreover, co-staining with Cell Mask (magenta), Hoechst (blue) and LysoTracker (red), a lysosome specific fluorophore, suggested that PS-NPs (green) accumulated in acidic lysosomes (Fig. 2g, white arrows). Lysosomes are dense spherical organelles, but they can display considerable variation in size and shape as a result of differences in the materials that have been taken up31. The 3D image reconstruction showed that PS-NPs and lysosomes co-localize in quite large lysosomal structures (Fig. 2h). To our knowledge, few studies have studied the internalization dynamics of nanoplastics in cells of aquatic vertebrates. Different authors have shown that microplastics and larger nanoplastics (larger than 200 nm) are not easily internalised but particles of 100 nm or less are readily taken up by the endocytic cell machinery.32,33 Johnston et al. described that 20 nm PS-NPs did not accumulate in lysosomes but suggested that in human and rat cell lines NPs ended up in the mitochondria.30 Recently, it has been demonstrated that granulocytes from the mussel M. galloprovincialis internalized 50 nm PS-NPs and accumulated them in lysosomes.14 In our work, we showed for the first time the internalization dynamics of PS-NPs in aquatic vertebrate hepatocytes and we demonstrated that they reached the lysosomal compartment (Fig. 2f). The final intracellular fate of nanoplastics could be different depending on the species, size, charge and plastic polymer composition, pointing out the need for more studies focused on nanoplastic internalization dynamics in vertebrates.
Transcriptional changes in zebrafish liver cells associated with PS-NP and poly(I:C) treatment
To gain insight into the effects of PS-NPs alone and in combination with an immune stimulus, we used poly(I:C), a synthetic dsRNA compound, to mimic a viral infection. The assessed target genes (mx, vig1 and gig2) are canonical genes related to the antiviral response that codify for interferon induced proteins34 and their upregulation in ZFL cells in response to poly(I:C), mounting a typical and strong antiviral response, has been previously characterised.23,35,36 As shown in Fig. 3, ZFL cells respond to poly(I:C) by upregulating antiviral genes such as mx, vig1 and gig2, at similar levels to those reported previously.21,23,35 When ZFL cells were pre-exposed for 12 h to PS-NPs and then treated with poly(I:C), a clear synergistic effect was observed (Fig. 3). Interestingly, when the cells were pre-exposed to PS-NPs for 24 h instead of 12 h, this synergistic effect was not observed, showing a typical poly(I:C) stimulation pattern. To further assess whether cells remained in a normal state after 24 h, we assessed lipid peroxidation levels after the PS-NP exposure and subsequent poly(I:C) treatment. No alterations in cell peroxidation status due to PS-NP exposure were observed (ESI† Fig. S5). Lipid peroxidation is a direct consequence of ROS production and we saw that after a 24 h exposure cells can deal with stress and maintain homeostasis. Altogether, data suggest that PS-NPs are internalized, accumulating in lysosomes, and initially provoke a strong and general stress state, thus potentiating the anti-viral response system. It could be speculated that PS-NP aggregation at higher doses could play a role in the uptake dynamics and, in turn, lead to differences in gene expression. However, we discarded this hypothesis, since the PS-NP stability data shown in Fig. 1 indicate very low aggregation in DMEM after 72 h of incubation (PDI value of 0.07 versus 0.03 in water). A second hypothesis is that interferences in the endocytosis and associated mechanisms could be affected by the PS-NP dose and exposure time. TLR3, the main receptor for poly(I:C) responsible for the initial activation of the immune response, is located in the endosomal membrane,34 the same subcellular compartment where the PS-NPs traffic and accumulate. We hypothesise that at a low PS-NP concentration the antiviral response signals through TLR3 and synergises with a cell stress response. At a higher dose or longer exposure (leading to an internalization of a higher number of particles), the endosomal architecture might be overloaded and the TLR3 pathway and cell stress response disturbed. Nevertheless, the antiviral response can signal alternatively through RIG-1, LGP-2 or MDA5,37,38 thus maintaining the poly(I:C) response at high dose or long exposure. To further explore this hypothesis, we analysed the average size of the PS-NP clusters inside the ZFL cells after exposure to 5 and 50 mg l−1 PS-NPs. We found a significant difference in cluster size between the two concentrations (0.2691 ± 0.01395 – low dose versus 0.3873 ± 0.01387 – high dose), which could sustain the possibility of a lysosomal overload (ESI† Fig. S6c). In line with this, we also tested RTGut cells, a different fish cell line derived from intestinal epithelia, which could be more conceptually relevant for waterborne absorption. In this cell line, the morphology and size of the endosomal–lysosomal system are massively changed at low versus high dose (ESI† Fig. S6a and b). We observed that at high dose: the number of clusters inside the cell increased (n = 464 at high dose versus n = 950 at low dose) and their size almost tripled (mean 0.1549 ± 0.0058 – high dose versus mean 0.05902 ± 0.001895 – low dose). Coincidentally, Canesi et al.39 found a perturbation on the endolysosomal system of M. galloprovincialis haemocytes after exposure to PS-NPs. They observed a decrease in lysosomal membrane stability (LMS) after exposures to 5 and 50 mg l−1 PS-NPs.39 The endosomal–lysosomal system is known to participate in several aspects of cell physiology, besides its classical role in protein degradation. As Neefjes and collaborators40 say: “endosomal system constitutes a key negotiator between the environment of a cell and its internal affairs”. For example, lysosome overload has an impact on the mRNA levels of several lysosomal proteases41 and is extremely important for a correct regulation of antigen receptor and pattern recognition receptor signalling in the innate immune system.42On the other hand, a cell stress response in which low doses cause a greater impact than high doses has been previously documented in nanoparticle toxicology. For example, Teles et al.43 observed changes in TOS (total oxidative status) and in mRNA levels of antioxidant-related genes after exposure to gold nanoparticles in Sparus aurata. Changes only occurred at the tested low and intermediate concentrations, revealing a non-monotonic dose–response curve, a pattern of response frequently found for endocrine disrupting chemicals. Moreover, Brandts et al.44 found an increased mRNA expression of genes involved in biotransformation (cypp11, pgp) and immune function (cathepsin) and a decrease in ChE in M. galloprovincialis only at lower PS-NP doses (0.05 and 0.5 mg l−1). In the same species, Canesi et al.39 found a greater increase in ROS production in haemocytes exposed to PS-NPs at low concentration (1 mg l−1). This suggests that, under some circumstances, low doses of nanoplastics could cause a greater impact than higher doses. Maybe a combination of scenarios (disturbance of the endolysosomal system and/or cell stress response) could explain the gene expression synergy observed at low dose. However, further work would be necessary to understand the precise role of intracellular PS-NP accumulation and how this affects the immune response.
In vivo uptake and accumulation of PS-NPs in zebrafish larvae and effects on survival after bacterial infection
Zebrafish larvae 5 dpf were imaged after 48 h of in vivo exposure to fluorescent PS-NPs (5 and 50 mg l−1) (see Fig. 4a), to confirm uptake. PS-NPs were taken up by the larvae and accumulated in the intestine and pancreas (Fig. 4b). The distribution of the fluorescence intensity normalized by the organ area was 65 ± 8.7% in the gut and 34 ± 8.7% in the pancreas (Fig. 4c). The zebrafish pancreas reaches its final position and shape by 6 dpf, the intestine is open to the surrounding environment between 3 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dpf,45,46 and the larva has a fully functional digestive system at day 5,47 therefore imaging at 5 dpf is an optimal window of time. The pancreas is located asymmetrically on the right side of the body and has an elongated shape and characteristic head–neck–tail morphology, where the anterior part contains islet tissue surrounded by exocrine tissue, while the neck and tail consist of exocrine tissue.45 We hypothesise that the larvae immersed in the solution of fluorescent PS-NPs could have ingested the PS-NPs and absorbed them through the intestinal epithelia, distributing them systemically and accumulating also in the pancreas. Nevertheless, we do not have definite proof of this uptake route. We do not discard that PS-NPs may accumulate in other organs such as the liver, spleen or brain at later life stages. We actually expected to find fluorescence in the liver, but even at continuous PS-NP exposure and saturating doses (60 h, 100 mg l−1) we could not detect PS-NPs' fluorescence in the zebrafish liver. This pattern of accumulation has been previously reported although with some differences, likely due to the specific exposure time, dose, size and larvae age. For example, Brun et al. observed accumulation of 25 nm PS-NPs (20 mg l−1)7,8 in the intestine, exocrine pancreas and gall bladder after 48 h of exposure at 120 hpf (5 dpf). It is worth mentioning that there is a considerable variation in between studies, with PS-NPs being located in the intestine and neuromast,7 in the heart region,10 in the digestive track and eyes,48 in the gut and gills region49 and in the gastrointestinal tract, gallbladder, liver, pancreas, heart and brain.6 Differences with published data could also be attributed to the use of other ZF strains (e.g. AB/TL wild type as in van Pomeren et al.,48 Veneman et al.,10 Brun et al.7 or Sökmen et al.11), instead of wild type larvae as in our case. We decided to use wild type zebrafish (no strain) to approach the individual variability present in a natural population, in order to better emulate an environmental situation.
dpf,45,46 and the larva has a fully functional digestive system at day 5,47 therefore imaging at 5 dpf is an optimal window of time. The pancreas is located asymmetrically on the right side of the body and has an elongated shape and characteristic head–neck–tail morphology, where the anterior part contains islet tissue surrounded by exocrine tissue, while the neck and tail consist of exocrine tissue.45 We hypothesise that the larvae immersed in the solution of fluorescent PS-NPs could have ingested the PS-NPs and absorbed them through the intestinal epithelia, distributing them systemically and accumulating also in the pancreas. Nevertheless, we do not have definite proof of this uptake route. We do not discard that PS-NPs may accumulate in other organs such as the liver, spleen or brain at later life stages. We actually expected to find fluorescence in the liver, but even at continuous PS-NP exposure and saturating doses (60 h, 100 mg l−1) we could not detect PS-NPs' fluorescence in the zebrafish liver. This pattern of accumulation has been previously reported although with some differences, likely due to the specific exposure time, dose, size and larvae age. For example, Brun et al. observed accumulation of 25 nm PS-NPs (20 mg l−1)7,8 in the intestine, exocrine pancreas and gall bladder after 48 h of exposure at 120 hpf (5 dpf). It is worth mentioning that there is a considerable variation in between studies, with PS-NPs being located in the intestine and neuromast,7 in the heart region,10 in the digestive track and eyes,48 in the gut and gills region49 and in the gastrointestinal tract, gallbladder, liver, pancreas, heart and brain.6 Differences with published data could also be attributed to the use of other ZF strains (e.g. AB/TL wild type as in van Pomeren et al.,48 Veneman et al.,10 Brun et al.7 or Sökmen et al.11), instead of wild type larvae as in our case. We decided to use wild type zebrafish (no strain) to approach the individual variability present in a natural population, in order to better emulate an environmental situation.
In aquatic invertebrates, the scenario is similar, and the accumulation pattern of nanoplastics depends on age, dose, material or size. For example, oyster (Crassostrea gigas) larvae have been found to ingest both nanoplastics and microplastics at different ages, being the younger larvae the ones with the highest capacity to ingest 160 nm PS-NPs.50 Sea urchin (Paracentrotus lividus) embryos accumulated PS–COOH-NPs inside the embryo's digestive tract without any toxic effects, while PS–NH2-NPs induced strong toxic effects.51 Finally, mussels (M. galloprovincialis) can accumulate 50 nm PS-NPs after 24 h of waterborne exposure, in the digestive system, muscle and gills.14 A recent review by Kögel et al.52 pointed out how different parameters such as charge, size or concentration could affect the uptake and toxicity of nanoplastics in aquatic organisms.
In our study, the highest PS-NP concentration tested (100 mg l−1) in a continuous 96 h exposure provoked 100% mortality, while continuous exposure to 50 mg l−1 provoked around 35% mortality (ESI† Table S2). However, concentrations of 5 and 50 mg l−1 for 48 h did not provoke any mortality different from control larvae (Fig. 4c). Low dose (5 mg l−1) and high dose (50 mg l−1) exposures were tested for a subsequent infection with A. hydrophila. As shown in Fig. 4d, the PS-NP exposure did not affect the survival after infection, as both treated and non-treated larvae had the same survival curves. Different authors have described how exposure to nanoplastics may affect the immune system by interfering in immune related gene expression, detoxifying enzyme activity (catalase etc.), or in ROS production, but this is the first study investigating the effects of nanoplastic exposure on the capacity to fight a lethal infection in zebrafish. At both tested concentrations, zebrafish larvae could cope with an acute PS-NP exposure and the physiological machinery was able to maintain the homeostasis. Only one previous work studied the interaction between nanoplastic exposure and pathogenic infection. In the said study, an in vitro infection study using isolated mussel haemocytes,14 the authors showed that the hemocytes' phagocytic capacity decreased when exposed to PS-NPs but not when the hemocytes were exposed to a combination of PS-NPs and V. splendidus. Nevertheless, a chronic or repeated acute exposure could perhaps lead to a failure of the pathogen response mechanisms. To further understand the effects of PS-NPs on fish, chronic or repeated acute exposure should be investigated, in order to determine if PS-NPs could affect the immune system in the long-term.
Conclusions
Our study shows that PS-NPs are efficiently taken up by ZFL cells in vitro, tending to accumulate in lysosomes, possibly in an attempt of the cell machinery to degrade PS-NPs. The expression of immune genes was synergistically affected by a viral stimulus (poly(I:C)) at low doses and early time points in ZFL cells. Even though we see this synergistic effect of PS-NPs and poly(I:C) at 12 h and 5 mg l−1, this effect disappears at a longer exposure time (24 h) as well as a higher dose (50 mg l−1). Therefore, it suggests that ZFL cells can regain homeostasis and normal function after the PS-NP challenge, presenting the expected response to a viral stimulus. Furthermore, zebrafish larvae also incorporated PS-NPs, potentially through ingestion, and accumulated them in the gut and pancreas. Nevertheless, on exposure to PS-NPs at concentrations of up to 50 mg l−1, PS-NPs did not show toxicity nor interference with a normal immune response against A. hydrophila. Altogether, although PS-NPs can induce an immune response, the survival of zebrafish larvae challenged with a bacterial infection after an acute exposure to PS-NPs is not decimated with respect to unexposed larvae.Authorship
IB and NR designed the study. IB and NR designed and performed the experiments. IB, MT, MGO and NR did the data analysis. NR wrote the manuscript and all authors contributed to the preparation of the final manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by grants from the Spanish Ministry of Science, European Commission and AGAUR funds to NR (RTI2018-096957-B-C21, MINECO/FEDER and 2014SGR-345 AGAUR). IB was funded by a pre-doctoral scholarship from UAB (2018FI_B_00711). We thank the LLEB (UAB), Servei de Citometria (SCAC-UAB) and Servei de Miscroscopia (UAB) for technical support and Dr. C. Tafalla for the generous gift of RTGut cells.References
- An analysis of European plastics production, demand and waste data, Plast. – Facts 2018, PlasticsEurope, 2018, p. 38 Search PubMed.
- J. Gigault, A. ter Halle, M. Baudrimont, P. Y. Pascal, F. Gauffre, T. L. Phi, H. El Hadri, B. Grassl and S. Reynaud, Current opinion: What is a nanoplastic?, Environ. Pollut., 2018, 235, 1030–1034 CrossRef CAS PubMed.
- N. B. Hartmann, T. Hüffer, R. C. Thompson, M. Hassellöv, A. Verschoor, A. E. Daugaard, S. Rist, T. Karlsson, N. Brennholt, M. Cole, M. P. Herrling, M. C. Hess, N. P. Ivleva, A. L. Lusher and M. Wagner, Are We Speaking the Same Language? Recommendations for a Definition and Categorization Framework for Plastic Debris, Environ. Sci. Technol., 2019, 53, 1039–1047 CrossRef CAS PubMed.
- A. Ter Halle, L. Jeanneau, M. Martignac, E. Jarde, B. Pedrono, L. Brach and J. Gigault, Nanoplastic in the North Atlantic Subtropical Gyre, Huanjing Kexue Yu Jishu, 2017, 51, 13689–13697 CrossRef CAS PubMed.
- Y. Chae and Y. J. An, Effects of micro- and nanoplastics on aquatic ecosystems: Current research trends and perspectives, Mar. Pollut. Bull., 2017, 124, 624–632 CrossRef CAS PubMed.
- J. A. Pitt, J. S. Kozal, N. Jayasundara, A. Massarsky, R. Trevisan, N. Geitner, M. Wiesner, E. D. Levin and R. T. Di Giulio, Uptake, tissue distribution, and toxicity of polystyrene nanoparticles in developing zebra fish (Danio rerio), Aquat. Toxicol., 2018, 194, 185–194 CrossRef CAS PubMed.
- N. R. Brun, P. van Hage, E. R. Hunting, A. P. G. Haramis, S. C. Vink, M. G. Vijver, M. J. M. Schaaf and C. Tudorache, Polystyrene nanoplastics disrupt glucose metabolism and cortisol levels with a possible link to behavioural changes in larval zebrafish, Commun. Biol., 2019, 2, 382 CrossRef PubMed.
- N. R. Brun, B. E. V. Koch, M. Varela, W. J. G. M. Peijnenburg, H. P. Spaink and M. G. Vijver, Nanoparticles induce dermal and intestinal innate immune system responses in zebrafish embryos, Environ. Sci.: Nano, 2018, 5, 904–916 RSC.
- Y. Lu, Y. Zhang, Y. Deng, W. Jiang, Y. Zhao, J. Geng, L. Ding and H. Ren, Uptake and accumulation of polystyrene microplastics in zebrafish (Danio rerio) and toxic effects in liver, Environ. Sci. Technol., 2016, 50, 4054–4060 CrossRef CAS PubMed.
- W. J. Veneman, H. P. Spaink, N. R. Brun, T. Bosker and M. G. Vijver, Pathway analysis of systemic transcriptome responses to injected polystyrene particles in zebrafish larvae, Aquat. Toxicol., 2017, 190, 112–120 CrossRef CAS PubMed.
- T. Ö. Sökmen, E. Sulukan, M. Türkoğlu, A. Baran, M. Özkaraca and S. B. Ceyhun, Polystyrene nanoplastics (20 nm) are able to bioaccumulate and cause oxidative DNA damages in the brain tissue of zebrafish embryo (Danio rerio), Neurotoxicology, 2020, 77, 51–59 CrossRef PubMed.
- Y. Jin, J. Xia, Z. Pan, J. Yang, W. Wang and Z. Fu, Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish, Environ. Pollut., 2018, 235, 322–329 CrossRef CAS PubMed.
- E. Bergami, A. Krupinski Emerenciano, M. González-Aravena, C. A. Cárdenas, P. Hernández, J. R. M. C. Silva and I. Corsi, Polystyrene nanoparticles affect the innate immune system of the Antarctic sea urchin Sterechinus neumayeri, Polar Biol., 2019, 42, 743–757 CrossRef.
- M. Sendra, A. Saco, M. P. Yeste, A. Romero, B. Novoa and A. Figueras, Nanoplastics: From tissue accumulation to cell translocation into Mytilus galloprovincialis hemocytes. resilience of immune cells exposed to nanoplastics and nanoplastics plus Vibrio splendidus combination, J. Hazard. Mater., 2020, 388, 121788 CrossRef CAS PubMed.
- S. Wagner and T. Reemtsma, Things we know and don't know about nanoplastic in the environment, Nat. Nanotechnol., 2019, 14, 300–301 CrossRef CAS PubMed.
- J. A. Gallego-Urrea, J. Tuoriniemi, T. Pallander and M. Hassellöv, Measurements of nanoparticle number concentrations and size distributions in contrasting aquatic environments using nanoparticle tracking analysis, Environ. Chem., 2010, 7, 67–81 CrossRef CAS.
- E. Kelpsiene, O. Torstensson, M. T. Ekvall, L. A. Hansson and T. Cedervall, Long-term exposure to nanoplastics reduces life-time in Daphnia magna, Sci. Rep., 2020, 10, 5979 CrossRef CAS PubMed.
- T. Schindelin, J. Arganda-Carreras, I. Frise, E. Kaynig, V. Longair, M. Pietzsch, D. J. Preibisch, S. Rueden, C. Saalfeld, S. Schmid, B. Tinevez, J.-Y. White, A. Hartenstein, V. Eliceiri, K. Tomancak and P. Cardona, Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef PubMed.
- T. Fishers, TechNote 103: Fluorescent/Dyed Microspheres, Bangs Lab. Inc., 2013, pp. 1–5 Search PubMed.
- A. I. Catarino, A. Frutos and T. B. Henry, Use of fluorescent-labelled nanoplastics (NPs) to demonstrate NP absorption is inconclusive without adequate controls, Sci. Total Environ., 2019, 670, 915–920 CrossRef CAS PubMed.
- D. Torrealba, D. Parra, J. Seras-franzoso, E. Vallejos-vidal, D. Yero, I. Gibert, A. Villaverde, E. Garcia-Fruitós and N. Roher, Nanostructured recombinant cytokines: A highly stable alternative to short-lived prophylactics, Biomaterials, 2016, 107, 102–114 CrossRef CAS PubMed.
- A. Kawano, C. Haiduk, K. Schirmer, R. Hanner, L. E. J. Lee, B. Dixon and N. C. Bols, Development of a rainbow trout intestinal epithelial cell line and its response to lipopolysaccharide, Aquacult. Nutr., 2011, 17, e241–e252 CrossRef.
- R. Thwaite, J. Ji, D. Torrealba, J. Coll, M. Sabés, A. Villaverde and N. Roher, Protein nanoparticles made of recombinant viral antigens: A promising biomaterial for oral delivery of fish prophylactics, Front. Immunol., 2018, 9, 1652 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of relative gene expression data using Real-Time quantitative PCR and the 2 -ΔΔCT method, Methods, 2001, 408, 402–408 CrossRef PubMed.
- J. Ji, S. Merino, J. M. Tomás and N. Roher, Nanoliposomes encapsulating immunostimulants modulate the innate immune system and elicit protection in zebrafish larvae, Fish Shellfish Immunol., 2019, 92, 421–429 CrossRef CAS PubMed.
- R. Cui, S. W. Kim and Y. J. An, Polystyrene nanoplastics inhibit reproduction and induce abnormal embryonic development in the freshwater crustacean Daphnia galeata, Sci. Rep., 2017, 7, 1–10 CrossRef CAS PubMed.
- C. Cortés, J. Domenech, M. Salazar, S. Pastor, R. Marcos and A. Hernández, Nanoplastics as a potential environmental health factor: Effects of polystyrene nanoparticles on human intestinal epithelial Caco-2 cells, Environ. Sci.: Nano, 2020, 7, 272–285 RSC.
- M. Almeida, M. A. Martins, A. M. V. Soares, A. Cuesta and M. Oliveira, Polystyrene nanoplastics alter the cytotoxicity of human pharmaceuticals on marine fish cell lines, Environ. Toxicol. Pharmacol., 2019, 69, 57–65 CrossRef CAS PubMed.
- M. Ruiz-Palacios, M. Almeida, M. A. Martins, M. Oliveira, M. Á. Esteban and A. Cuesta, Establishment of a brain cell line (FuB-1) from mummichog (Fundulus heteroclitus) and its application to fish virology, immunity and nanoplastics toxicology, Sci. Total Environ., 2020, 708, 134821 CrossRef CAS PubMed.
- H. J. Johnston, M. Semmler-Behnke, D. M. Brown, W. Kreyling, L. Tran and V. Stone, Evaluating the uptake and intracellular fate of polystyrene nanoparticles by primary and hepatocyte cell lines in vitro, Toxicol. Appl. Pharmacol., 2010, 242, 66–78 CrossRef CAS PubMed.
- G. Cooper, The Cell: A Molecular Approach, Sinauer Associates, Sunderland, MA, 2nd edn, 2000 Search PubMed.
- T. dos Santos, J. Varela, I. Lynch, A. Salvati and K. A. Dawson, Effects of transport inhibitors on the cellular uptake of carboxylated polystyrene nanoparticles in different cell lines, PLoS One, 2011, 6, e24438 CrossRef CAS PubMed.
- M. Forte, G. Iachetta, M. Tussellino, R. Carotenuto, M. Prisco, M. De Falco, V. Laforgia and S. Valiante, Polystyrene nanoparticles internalization in human gastric adenocarcinoma cells, Toxicol. In Vitro, 2016, 31, 126–136 CrossRef CAS PubMed.
- O. Takeuchi and S. Akira, Pattern recognition receptors and inflammation, Cell, 2010, 140, 805–820 CrossRef CAS PubMed.
- A. Ruyra, M. Cano-Sarabia, S. A. MacKenzie, D. Maspoch and N. Roher, A novel liposome-based nanocarrier loaded with an LPS-dsRNA cocktail for fish innate immune system stimulation, PLoS One, 2013, 8, 1–13 CrossRef PubMed.
- A. Ruyra, D. Torrealba, D. Morera, L. Tort, S. MacKenzie and N. Roher, Zebrafish liver (ZFL) cells are able to mount an anti-viral response after stimulation with Poly (I:C), Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2015, 182, 55–63 CrossRef CAS PubMed.
- M. F. Mian, A. N. Ahmed, M. Rad, A. Babaian, D. Bowdish and A. A. Ashkar, Length of dsRNA (poly I:C) drives distinct innate immune responses, depending on the cell type, J. Leukocyte Biol., 2013, 94, 1025–1036 CrossRef CAS PubMed.
- B. Wang, Y. B. Zhang, T. K. Liu, J. Shi, F. Sun and J. F. Gui, Fish viperin exerts a conserved antiviral function through RLR-triggered IFN signaling pathway, Dev. Comp. Immunol., 2014, 47, 140–149 CrossRef CAS PubMed.
- L. Canesi, C. Ciacci, E. Bergami, M. P. Monopoli, K. A. Dawson, S. Papa, B. Canonico and I. Corsi, Evidence for immunomodulation and apoptotic processes induced by cationic polystyrene nanoparticles in the hemocytes of the marine bivalve Mytilus, Mar. Environ. Res., 2015, 111, 34–40 CrossRef CAS PubMed.
- J. Neefjes, M. M. L. Jongsma and I. Berlin, Stop or Go? Endosome positioning in the establishment of compartment architecture, dynamics and F¡function, Trends Cell Biol., 2017, 27, 580–594 CrossRef CAS PubMed.
- J. Martínez-Fábregas, A. Prescott, S. van Kasteren, D. L. Pedrioli, I. McLean, A. Moles, T. Reinheckel, V. Poli and C. Watts, Lysosomal protease deficiency or substrate overload induces an oxidative-stress mediated STAT3-dependent pathway of lysosomal homeostasis, Nat. Commun., 2018, 9, 1–16 CrossRef PubMed.
- P. A. Gleeson, The role of endosomes in innate and adaptive immunity, Semin. Cell Dev. Biol., 2014, 31, 64–72 CrossRef CAS PubMed.
- M. Teles, C. Fierro-Castro, P. Na-Phatthalung, A. Tvarijonaviciute, T. Trindade, A. M. V. M. Soares, L. Tort and M. Oliveira, Assessment of gold nanoparticle effects in a marine teleost (Sparus aurata) using molecular and biochemical biomarkers, Aquat. Toxicol., 2016, 177, 125–135 CrossRef CAS PubMed.
- I. Brandts, M. Teles, A. P. Gonçalves, A. Barreto, L. Franco-martinez and A. Tvarijonaviciute, Effects of nanoplastics on Mytilus galloprovincialis after individual and combined exposure with carbamazepine, Sci. Total Environ., 2018, 643, 775–784 CrossRef CAS PubMed.
- L. Gnügge, D. Meyer and W. Driever, Pancreas development in zebrafish, Methods Cell Biol., 2004, 2004, 531–551 CrossRef.
- W. Z. Stephens, A. R. Burns, K. Stagaman, S. Wong, J. F. Rawls, K. Guillemin and B. J. M. Bohannan, The composition of the zebrafish intestinal microbial community varies across development, ISME J., 2016, 10, 644–654 CrossRef PubMed.
- V. H. Quinlivan and S. A. Farber, Lipid uptake, metabolism, and transport in the larval zebrafish, Front. Endocrinol., 2017, 8, 1–11 Search PubMed.
- M. van Pomeren, N. R. Brun, W. J. G. M. Peijnenburg and M. G. Vijver, Exploring uptake and biodistribution of polystyrene (nano)particles in zebrafish embryos at different developmental stages, Aquat. Toxicol., 2017, 190, 40–45 CrossRef CAS PubMed.
- L. M. Skjolding, G. Ašmonaitė, R. I. Jølck, T. L. Andresen, H. Selck, A. Baun and J. Sturve, An assessment of the importance of exposure routes to the uptake and internal localisation of fluorescent nanoparticles in zebrafish (Danio rerio), using light sheet microscopy, Nanotoxicology, 2017, 11, 351–359 CrossRef CAS PubMed.
- M. Cole and T. S. Galloway, Ingestion of nanoplastics and microplastics by Pacific oyster larvae, Environ. Sci. Technol., 2015, 49, 14625–14632 CrossRef CAS PubMed.
- C. Della Torre, E. Bergami, A. Salvati, C. Faleri, P. Cirino, K. A. Dawson and I. Corsi, Accumulation and embryotoxicity of polystyrene nanoparticles at early stage of development of sea urchin embryos Paracentrotus lividus, Environ. Sci. Technol., 2014, 48, 12302–12311 CrossRef CAS PubMed.
- T. Kögel, Ø. Bjorøy, B. Toto, A. M. Bienfait and M. Sanden, Micro- and nanoplastic toxicity on aquatic life: Determining factors, Sci. Total Environ., 2020, 709, 5817 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0en00553c |
| This journal is © The Royal Society of Chemistry 2020 |