Open Access Article
Open Access ArticleChemical and structural characterization of SeIV biotransformations by Stenotrophomonas bentonitica into Se0 nanostructures and volatiles Se species†
Miguel A.
Ruiz-Fresneda
 *a,
Abdurrahman S.
Eswayah
bc,
María
Romero-González‡
d,
Philip H. E.
Gardiner
*a,
Abdurrahman S.
Eswayah
bc,
María
Romero-González‡
d,
Philip H. E.
Gardiner
 b,
Pier L.
Solari
e and
Mohamed L.
Merroun
b,
Pier L.
Solari
e and
Mohamed L.
Merroun
 a
a
aDepartment of Microbiology, University of Granada, Granada, Spain. E-mail: mafres@ugr.es
bBiomolecular Sciences Research Centre, Sheffield Hallam University, Sheffield, UK
cBiotechnology Research Centre, Tripoli, Libya
dDepartment of Geography, University of Sheffield, Sheffield, UK
eMARS Beamline, Synchrotron SOLEIL, L'Orme des Merisiers, Saint-Aubin, Gif-sur-Yvette Cedex, France
First published on 18th June 2020
Abstract
The deep geological repository (DGR) system is widely accepted as the solution for the disposal of radioactive wastes in the future. This concept is based on several natural and engineered barriers such as bentonite clays, which will encase the metal containers holding the radioactive waste. Microorganisms living therein can influence the mobility of the radionuclides (e.g. selenium, uranium, etc.) present in such residues. In this work the bentonite isolate Stenotrophomonas bentonitica is shown to reduce selenite (SeIV) to elemental Se (Se0) nanostructures (amorphous and trigonal) and to volatile methylated Se−II species. Electron microscopy (HAADF-STEM) analysis of purified Se nanostructures supported the transformation process from amorphous to trigonal Se, proposed in previous studies. Infrared spectroscopy (ATR-FTIR) and X-ray photoelectron spectroscopy (XPS) revealed the presence of amine rich organic matter, covering the nanostructures, suggesting the role of proteins in their synthesis and transformation. In addition, X-ray absorption spectroscopy (XAS) of SeNPs associated to the cells confirmed the formation of different Se0 structures (amorphous and crystalline). Finally, the reduction of SeIV to volatile methylated species (DMDSe and DMSeS) was detected using a gas chromatography-mass spectrometry (GC-MS) system. The oxidation state and molecular coordination of Se in the purified Se nanostructures as well as the volatile Se species, by means of microscopic, spectroscopic, and gas chromatographic techniques, indicated their lower mobility and chemo-toxicity. This study thus highlights the potential environmental significance of microbial processes for the mobility and toxicity of selenium in future repositories, which in turn contribute to their safe implementation.
Environmental significanceThe safest option for the disposal of nuclear wastes is the deep geological repository system, which includes a variety of barriers composed by bentonites and other materials. Microorganisms can influence the mobility of radionuclides (e.g. selenium, uranium, etc.) present in such residues. The bentonite isolate Stenotrophomonas bentonitica is demonstrated herein to reduce SeIV to Se0 nanostructures (amorphous and trigonal) and volatile methylated Se−II. The identification of the oxidation state and the molecular coordination of Se in the purified Se nanostructures and the volatile Se species indicated their lower mobility and toxicity. This study highlights the potential environmental significance of microbial processes in the mobility and toxicity of selenium in future repositories, which in turn will help to support their implementation. |
1. Introduction
The impact of microbial processes on the safety of the future planned deep geological repositories (DGRs) for radioactive wastes is of current environmental concern. The repositories to be constructed underground at a depth of 500–1000 m, have been exhaustively studied from a physical, chemical, and geological point of view. However, few investigations have been conducted in order to study the impact of microbial biotransformations on their safety. It is well documented that microorganisms present in these geological disposal environments could play an important role in the corrosion of metal canisters containing radioactive wastes,1 thereby giving rise to the release of radionuclides. In addition, they can also lead to the alteration of geochemical and mineralogical properties of clays that will surround the containers in some DGR designs, in countries such as France, Switzerland, or Belgium.2 The gas production as a consequence of microbial activity (e.g. H2, CO2, CH4, etc.) could result in excess pressure build-up within the repositories thus compromising their integrity.3 Finally, microorganisms can cause changes in the speciation and hence the solubility of elements characteristic for these type of wastes, such as uranium (U), selenium (Se), curium (Cm), etc.4–6 Consequently, they may have a critical impact on the safety of DGRs in the event of radionuclides escape to the surrounding environment. The groundwater inflow has to be considered since it will definitely affect several aspects of the DGR system. The re-saturation of water after thousand years will probably result in the release of chemical compounds, including electron donors and acceptors, organic matter, and gases from concrete and bitumen materials, that will be employed in some repositories. Additionally, radionuclides could be dispersed through groundwater, allowing their mobilization.Se is considered as an important radionuclide of interest as it is present in radioactive wastes and spent nuclear fuel.7 Specifically, Se79 isotope formed by nuclear fission reactions is the main radioisotope of Se found in nuclear wastes.7 The solubility and mobility of this element depends to a great extent on its different oxidation states [selenate (SeVI), selenite (SeIV), elemental selenium (Se0) and selenide (Se−II)]. In contrast to toxic and highly reactive Se oxyanions (SeVI and SeIV), Se0 and methyl selenide forms are basically insoluble and less biologically available.8 Microbial reduction of toxic Se oxyanions to the insoluble Se0 in form of nanoparticles has been previously described by many authors.5,9,10 Some studies have demonstrated the presence of organic shells (proteins) that have been microbiologically produced and surround the Se nanospheres.11,12 The physico-chemical properties (size, morphology, crystallinity) and hence the mobility of Se nanospheres could be affected largely by the presence of these bioorganic layers (proteins, polysaccharides, etc.) associated with them.13,14 Thus, microorganisms could play an important role in the immobilization of Se within the DGR as well as the bioremediation of Se contaminated environments and the production of nanostructures.
The reduction of Se oxyanions (SeVI and SeIV) to Se−II has not been extensively investigated. However, Se0 can be reduced to Se−II by the action of few microorganisms such as Veillonella atypica and Bacillus selenitireducens.15,16 Therefore, the production of biogenic volatile methylated selenide compounds is a widespread process which has to considered when bacteria are present. Methylococcus capsulatus, Methylosinus trichosporium OB3b, Stenotrophomonas maltophilia, and Pseudomonas stutzeri NT-I have been shown to produce Se volatile species such as dimethyl selenide (DMSe) or dimethyl diselenide (DMDSe) as a consequence of a Se methylation process.17–19 Several possible mechanisms for Se biomethylation have been proposed. Chasteen20 suggested that methylation is preceded by the formation of Se0 from SeIV, after which, Se0 is reduced to a selenide form (H–Se–X) and subsequently methylated and converted to methane selenol and dimethyl selenide. Recently, Eswayah et al. 2019 (ref. 21) have proposed that methyl selenol is the precursor for the formation of Se0 and for methylated selenium species. The proposed mechanism by Challenger,22 does not proceed by the formation of Se0 suggesting that the methylation process from selenate involves several reductive and methylation steps leading eventually to the production of DMSe. The most frequently detected microbiologically produced Se methylated forms include DMSe, DMDSe and dimethyl selenyl sulphide (DMSeS). However, less commonly methylated forms including dimethyl selenone, dimethyl triselenide, dimethyl selenyl disulphide, etc. have been also detected.23,24 It is noteworthy that methylation of Se inorganic forms is considered a detoxification process, since methylated species are less toxic compared to Se oxyanions.25
In our recent publication we describe the ability of S. bentonitica, that has been isolated from Cabo de Gata's bentonite (Almeria, Spain), to reduce SeIV by producing amorphous and crystalline Se nanostructures.26 These bentonites are considered natural analogues of artificial barriers for future Spanish repositories.27 Individual amorphous Se (a-Se) nanospheres were initially synthesized intracellularly and after a period of 48–72 h they begin to coalesce forming aggregates within the extracellular space. After 144 h of incubation, different trigonal Se (t-Se) nanostructures with different morphologies (e.g. hexagons, polygons, and nanowires) were detected in addition to a-Se nanospheres. Therefore, a time-dependent transformation process from a-Se nanospheres to t-Se nanostructures was proposed. Extracellular flagella-like proteins produced by the cells could play an important role in the transformation process since nanospheres seem to coalesce on the axis of these proteins forming elongated shapes. Characterization of the organic matter surrounding the Se nanoparticles (SeNPs) produced by S. bentonitica is certainly of interest since they may be involved in the synthesis and transformation of the nanoparticles. However, no structural studies have yet been conducted to determine the local coordination of Se within the Se nanostructures. Therefore, the main objectives of the present work are: 1) to confirm the involvement of organic matter (e.g. proteins) in the transformation of Se nanostructures using a combination of X-ray photoelectron spectroscopy (XPS), attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR), and scanning transmission electron microscopy associated with a high-angle annular field detector (STEM-HAADF), 2) to characterize the local coordination of Se in the Se nanostructures with different morphology produced at different incubation times by means of X-ray absorption spectroscopy (XAS), and 3) to characterize the methylated Se volatile compounds (DMDSe and DMDSeS) produced by this strain using a combined thermal desorption and GC-MS (gas chromatography-mass spectrometry) system. This work will provide new molecular evidence on the impact of the bacterial strain S. bentonitica on the biotransformation of Se species and provide that is essential in determining the safety of future disposal of radioactive wastes.
2. Experimental
2.1. Quantification of biological SeIV reduction rate
The SeIV reduction rate of S. bentonitica was determined by growing the cells aerobically in LB (Luria-Bertani) liquid medium (30 ml) supplemented with different initial SeIV concentration (0.1, 0.25 and 2 mM). SeIV-treated cells were incubated at 28 °C by shaking at 180 rpm. After different times of incubation (0, 6, 24, 48, 120 and 144 h) 1 ml of the samples were taken and centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g; 15 min; room temperature) to remove the cells and other debris. The SeIV concentration in the resultant supernatant was determined by using a Perkin Elmer Flexar HPLC (high performance liquid chromatography) pump attached to a Hamilton PRP-X100 column (4.6 × 250 mm) and coupled with a Perkin Elmer ICP-MS (inductively coupled plasma mass spectrometry) NexION350X. Ammonium citrate (5 mM; pH 5.2) containing methanol (2% v/v) was employed as mobile phase to achieve the separation at a flow rate of 1 ml min−1. SeIV amended-media without bacterial cells were used as controls. All samples were prepared in triplicate. In addition, a standard calibration curve with different solutions of known SeIV concentrations was prepared (0.02–1 mM).
000 × g; 15 min; room temperature) to remove the cells and other debris. The SeIV concentration in the resultant supernatant was determined by using a Perkin Elmer Flexar HPLC (high performance liquid chromatography) pump attached to a Hamilton PRP-X100 column (4.6 × 250 mm) and coupled with a Perkin Elmer ICP-MS (inductively coupled plasma mass spectrometry) NexION350X. Ammonium citrate (5 mM; pH 5.2) containing methanol (2% v/v) was employed as mobile phase to achieve the separation at a flow rate of 1 ml min−1. SeIV amended-media without bacterial cells were used as controls. All samples were prepared in triplicate. In addition, a standard calibration curve with different solutions of known SeIV concentrations was prepared (0.02–1 mM).
2.2. Effect of SeIV on the bacterial growth
The potential of S. bentonitica to tolerate SeIV was established by growing the cells under the same conditions described above (section 3.1.). Cell growth was evaluated by quantifying the total protein content in the bacterial cell extracts, using a modification of the method described by Dhanjal and Cameotra.28 The total protein content was correlated with the increase in the cell growth. A 1 ml aliquot of bacterial culture was taken at different time intervals in order to measure the growth rate based on the protein content of the cells determined by using Bradford reagent (Bio-Rad®). Bovine serum albumin (BSA) was used as a standard. All measurements were performed in triplicate.2.3. Flow cytometry
The cell viability and the metabolic activity of S. bentonitica in the presence of SeIV under oxic conditions were determined by using flow cytometry technique. For this purpose, the bacterial species S. bentonitica was amended with 2 mM of SeIV and incubated at 28 °C and 180 rpm on a rotary shaker. All experiments were done in triplicate. After 24 and 144 h incubation, the cells were harvested after centrifugation at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and 4 °C for 10 min. The resultant pellet was washed four times in phosphate buffered saline (PBS) pH 7 and then, the cells were dispersed in PBS, and the cell density was adjusted to approximately 106 cells per ml. For cell viability test, fluorescein diacetate (FDA) and propidium iodide (PI) were added into each sample to a final concentration of 20 μl ml−1 and 2 μl ml−1, respectively. For the metabolic activity test, 3,30-dihexyloxacarbocyanine iodide (DiOC6) was employed to give a final concentration of 20 μl ml−1. Finally, the samples were analysed by Forward Scatter using a FACSCantoII™ cytometer (Becton Dickinson). SeIV-free cultures and dead cells used as controls were obtained by heating the biomass at 90 °C.
000 × g and 4 °C for 10 min. The resultant pellet was washed four times in phosphate buffered saline (PBS) pH 7 and then, the cells were dispersed in PBS, and the cell density was adjusted to approximately 106 cells per ml. For cell viability test, fluorescein diacetate (FDA) and propidium iodide (PI) were added into each sample to a final concentration of 20 μl ml−1 and 2 μl ml−1, respectively. For the metabolic activity test, 3,30-dihexyloxacarbocyanine iodide (DiOC6) was employed to give a final concentration of 20 μl ml−1. Finally, the samples were analysed by Forward Scatter using a FACSCantoII™ cytometer (Becton Dickinson). SeIV-free cultures and dead cells used as controls were obtained by heating the biomass at 90 °C.
2.4. Bacterial synthesis, extraction, and purification of selenium nanoparticles
SeNPs were synthesized aerobically by S. bentonitica under 2 mM SeIV stress as previously described (section 3.1.). After periods of 48 and 144 h incubation, the SeNPs were extracted and purified following the method described by Sonkusre et al.29 Finally, purified SeNPs were spread onto LB agar plates to determine the efficiency of the extraction process in removing the organic material surrounding the particles.2.5. Attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR)
The samples of bacterial cells supplemented with 2 mM SeIV were harvested after 48 and 144 h incubation and subjected to centrifugation (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 10 min). The precipitates were washed 3 times with distilled water and dried at 28 °C. Then the samples were powdered in an agate mortar and kept in a desiccator. Finally, 1 mg of the powder was mixed with 100 mg of dried potassium bromide (KBr) and pressed forming disks. Bacterial cells solutions without SeIV were used as a control. In addition, purified SeNPs extracted after 48 and 144 h incubation, respectively were analysed by using the same technique.
000 × g, 10 min). The precipitates were washed 3 times with distilled water and dried at 28 °C. Then the samples were powdered in an agate mortar and kept in a desiccator. Finally, 1 mg of the powder was mixed with 100 mg of dried potassium bromide (KBr) and pressed forming disks. Bacterial cells solutions without SeIV were used as a control. In addition, purified SeNPs extracted after 48 and 144 h incubation, respectively were analysed by using the same technique.
ATR-FTIR measurements were performed on a Perkin Elmer Spectrum One FT-IR spectrometer, equipped with a silver gate evolution ATR accessory, consisting of a germanium crystal. A total of 200 scans for the purified SeNPs and 32 for the other samples were collected for each sample in wavenumber range from 4000 to 600 cm−1. Spectra obtained were baseline corrected and normalised to 1.5 absorbance (arbitrary units) using the 1650 cm−1 amide I absorption band from the control sample (S. bentonitica biomass without Se). All data acquisition and processing were performed using the Perkin Elmer Spectrum version 3.3 to obtain peak positions and relative peak intensities.
2.6. X-ray photoelectron spectroscopy (XPS)
Both purified and cell associated SeNPs obtained as indicated for ATR-FTIR experiments under the same conditions (2 mM SeIV, 48 and 144 h of incubation) were analysed by XPS. In addition, a cell biomass powder was used as control. The powder samples were all mounted by pressing them into indium foil, which was adhered using double sided carbon tape to a paper label stuck to the XPS sample mount.The analyses were carried out using a Kratos Supra instrument with a monochromated aluminium source, and two analysis points per sample. Survey scans were collected between 1200 to 0 eV binding energy, at 160 eV pass energy, 1 eV intervals, and 300 seconds per sweep. High-resolution O 1s, N 1s, C 1s, S 2p, P 2p and Se 3d XPS spectra were also collected at 20 eV pass energy and 0.1 eV intervals for each analysis point over an appropriate energy range, with one 300 second sweep for all elements except N 1s and S 2p for which two sweeps were used. The data collected was calibrated in intensity using a transmission function characteristic of the instrument (determined using software from NPL) to make the values instrument independent. The data were analysed using the theoretical Schofield relative sensitivity factors. All peaks positions were calibrated with respect to the C–C/C–H peak position at 285.0 eV.
2.7. HAADF-STEM analysis
The morphology and elemental composition analysis of the purified SeNPs were analysed by using Transmission electron microscopy (TEM) equipped with energy dispersive X-ray (EDX). EDX analysis was performed at 300 kV using a spot size of 4 Å and a live counting time of 50 s. The structural characterization of the SeNPs were analysed by using selected-area electron diffraction (SAED).TEM samples consisting of purified SeNPs produced by S. bentonitica after 48 and 144 hours of incubation were prepared using the conventional negative staining. Finally, the samples were examined under a HAADF-STEM microscope FEI TITAN G2 80-300. STEM specimen holders were cleaned by plasma prior to STEM analysis in order to minimize contamination.
2.8. X-ray absorption spectroscopy (XAS)
S. bentonitica cells were grown in LB media amended with 2 mM SeIV. After 24, 72 and 144 h of incubation the cells in contact with SeIV were harvested by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g; 10 min). Then, the samples were prepared as previously described in Merroun et al. (2005)30 with some modifications. The resultant pellets were washed twice with sterile double distilled water to remove the cell debris and the interfering ingredients of the growth medium before being dried at 28 °C for 24–48 h. Then the samples were powdered in an agate mortar and kept in a desiccator. Finally, the powdered samples were prepared by spreading and pressing them onto 10 mm × 3 mm area of Kapton tape, which was sealed between additional layers. Selenium references (sodium selenate, sodium selenite, Se0 foil and selenium sulfide) XAS data were collected in transmission mode. Each standard was prepared by mixing the powder with dried cellulose and pressed forming disks (13 mm of diameter).
000 × g; 10 min). Then, the samples were prepared as previously described in Merroun et al. (2005)30 with some modifications. The resultant pellets were washed twice with sterile double distilled water to remove the cell debris and the interfering ingredients of the growth medium before being dried at 28 °C for 24–48 h. Then the samples were powdered in an agate mortar and kept in a desiccator. Finally, the powdered samples were prepared by spreading and pressing them onto 10 mm × 3 mm area of Kapton tape, which was sealed between additional layers. Selenium references (sodium selenate, sodium selenite, Se0 foil and selenium sulfide) XAS data were collected in transmission mode. Each standard was prepared by mixing the powder with dried cellulose and pressed forming disks (13 mm of diameter).
Selenium K-edge X-ray absorption spectra were collected at the MARS beamline at the SOLEIL synchrotron facility, which is the French bending magnet beamline dedicated to the study of radioactive materials.31 For the experiment, the ring was operated at 2.75 GeV in 8-bunch mode (100 mA). The optics consisted on a double-crystal monochromator with horizontal dynamical focusing set to the Si(220) crystals, and -coated mirrors for vertical focusing and rejection of higher harmonics set to the Si strips.32 Spectra were collected in transmission mode using a 13-element Ge detector (EG & G ORTEC, USA). Data were processed following standard procedures by using the Athena/Artemis codes.33 Background removal was performed by means of a pre-edge linear function. Atomic absorption was simulated with a square-spline function. The theoretical phase and amplitude functions used in data analysis were calculated with FEFF8 (ref. 34) using Se foil as model. The amplitude reduction factor (S02) was held constant at 1.0 for the FEFF8 calculation and extended x-ray absorption fine structure (EXAFS) fits. The shift in threshold energy, ΔE0, was varied as a global parameter in the fits.
2.9. Detection and identification of volatile Se compounds
A combined thermal desorption GC-MS system was employed for the detection and identification of Se volatile species produced by the bacterial strain S. bentonitica. Cultures of S. bentonitica were grown in the presence of SeIV (0.1 and 2 mM) in 250 ml conical Quickfit® flask capped with Suba-Seals (Sigma®) to avoid the loss of gases during gas extraction with hypodermic syringes. SeIV-free cultures (biotic) and SeIV-amended media (abiotic) were used as controls. The gaseous samples were taken and analysed as described previously by Eswayah et al.17 after 24 and 144 h of incubation. The National Institute of Standards and Technology (NIST) MS search program (version 2011) was employed to identify compounds based on their mass spectrum.3. Results
3.1. Time course experiments for SeIV reduction
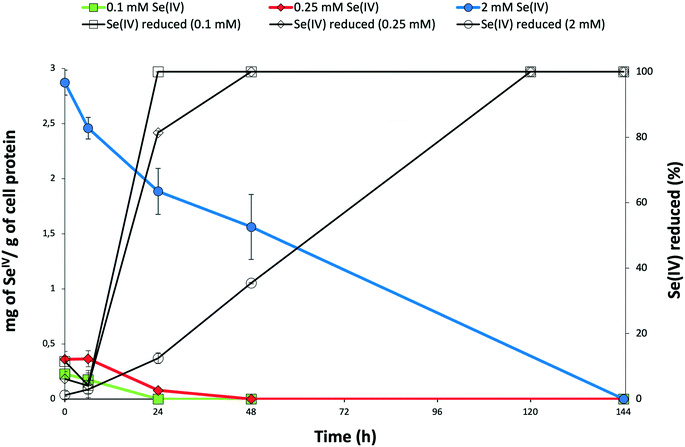
The time course of SeIV reduction by the strain BII-R7 of S. bentonitica was studied in the presence of different initial SeIV concentration. SeIV was completely reduced after 24, 48 and 120 h, if the cells were incubated with 0.1, 0.25 and 2 mM, respectively (Fig. 1). Therefore, SeIV removal depends on the initial concentration, where SeIV is removed faster at low initial concentration (0.1 mM) than at high concentration (2 mM). These results clearly show the efficiency with which the strain S. bentonitica BII-R7 reduces SeIV. No variation in SeIV concentration in the abiotic controls (SeIV amended-media without bacterial cells) confirmed that the bacterial strain is involved in the reduction of SeIV concentration (data not shown). The results displayed in Fig. 1 also show the decrease of the amount of SeIV per g of cell protein with time at all that concentrations assayed, which mirror the increase in the percentage of reduced SeIV.The decrease in the amount of SeIV at a given initial concentration matches with the growth profile of the cells as measured by the total protein content. The presence of SeIV has an inhibitory effect at the early stage of the bacterial growth as shown by the initial long lag phases in the first 48 h (Fig. 2). As we have previously reported, this stress is most probably due to the presence of SeIV.26 The optimum growth of SeIV-treated cells was reached at 96 h (for 0.1 mM SeIV) and around 144 h (for 0.25 and 2 mM SeIV) once the SeIV has been completely reduced.
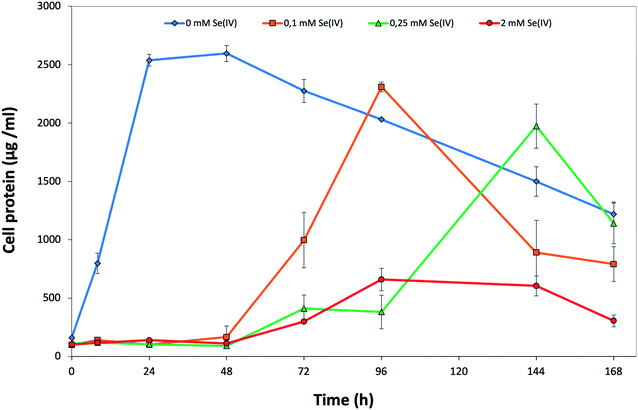 | ||
| Fig. 2 Growth profile of S. bentonitica in the absence and the presence of 0.1, 0.25, and 2 mM SeIV. Selenite was added at time zero. Each curve shows means based on the results of triplicates. | ||
The effect of SeIV on cell viability of S. bentonitica was studied by the live-dead staining approach conducted with propidium iodide (PI) and fluorescein diacetate (FDA). PI enters cells with damaged membranes staining nucleic acids,35 while FDA stains viable cells.36 With regard to the metabolic activity test, the fluorescent dye DiOC6 was used in order to bind polarized membranes of active cells.37 The percentages of viable and active cells of S. bentonitica population untreated and treated with an initial SeIV concentration of 2 mM and incubated for 24 and 144 h are displayed in Fig. S1.† Cell viability was affected negatively by SeIV. Specifically, 46.3 and 36.4% of cell populations were found to be viable after 24 and 144 h, respectively. In contrast, a greater number of viable cell population (100%) was observed in the untreated samples. On the other hand, the metabolic activity test showed a lower oxidative response under SeIV stress. Specifically, 59 and 72.2% were found to be active after 24 and 144 h respectively, comparing with untreated cells (97 and 97.8% of active cells). This increase on cell activity from 59 to 72.2% could be explained by the fact that at 24 h cells are still in the lag phase (see Fig. 2), probably adapting their metabolic processes to respond to Se stress. As soon as they are able to tolerate Se they grow as observed at 144 h. The results of the flow cytometry measurements concur with the growth curve suggesting a toxic effect of SeIV on S. bentonitica cells.
The reduction of SeIV could be also observed by the appearance of a red colour in SeIV-amended cultures. However, under 0.1 mM SeIV stress, the red colour changed to light brown colour after 48 h incubation. This observation would suggest that other mechanisms in addition to reduction seem to be involved. The absence of SeIV at 48 h (Fig. 1) discards an oxidation of Se compounds as possible mechanism. However, a volatilization process to methylated Se products it is possible. In fact, a characteristic garlic-like odor generated in the SeIV-treated cultures suggested the formation of volatile Se species.
3.2. Synthesis and purification of the selenium nanoparticles
The SeNPs were biogenically produced by S. bentonitica by reducing the oxyanion SeIV to non-toxic Se0 as described in Ruiz-Fresneda et al.26 The study revealed the formation of amorphous Se nanospheres and different crystalline Se nanostructures were found both intracellularly and extracellularly. The nanostructures were isolated using an improved method reported by Sonkusre et al.29 for the recovery of the SeNPs. This procedure includes cell lysis using lysozyme (Sigma®) and French press. After which, the separation of the nanoparticles from the cell debris was conducted by a two-phase water–octanol extraction (Fig. 3A). Finally, the water phase containing the SeNPs was subsequently washed with chloroform, absolute ethanol, ethanol (70%) and resuspended in water, obtaining the solution of purified SeNPs (Fig. 3B). No cell growth was detected when the harvested and purified SeNPs were incubated on LB agar plates, thus indicating the high efficiency of the purification steps in removing bacterial cells.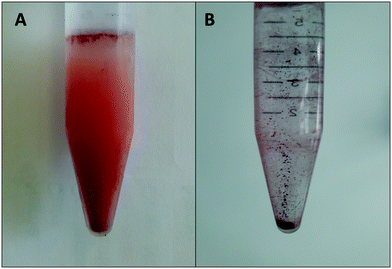 | ||
| Fig. 3 Separation of SeNPs from cell debris by a two-phase water–octanol extraction system (A) and final solution of purified SeNPs in water (B). | ||
3.3. Microscopic characterization of the purified selenium nanoparticles
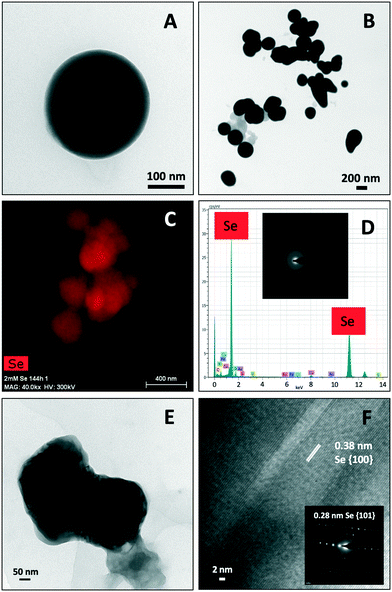
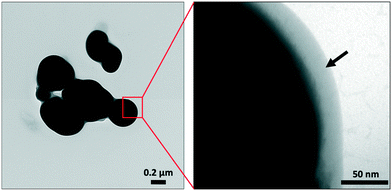
STEM-HAADF micrographs of purified SeNPs produced by S. bentonitica after 48 and 144 h of incubation showed the presence of spherical electron-dense structures with sizes ranging from 100 to 400 nm (Fig. 4A and B). These structures frequently appeared forming aggregates (Fig. 4B). EDX analysis and EDX elemental-distribution maps confirmed that they consisted of Se (Fig. 4C and D). Additionally, the ED pattern indicated their amorphous nature (Fig. 4D inset). Changes in the spherical morphology have been shown to occur in some Se nanospheres (Fig. 4B), probably as an intermediate step in the transformation from amorphous nanospheres to trigonal Se crystals as reported in Ruiz-Fresneda et al.26 The presence of Se crystals in the 144 h sample also supports the proposed transformation process (Fig. 4E). The HRTEM image of an individual Se nanostructure confirmed its crystallinity as indicated by a d-spacing of 0.38 nm (Fig. 4F). An additional d-spacing of 0.28 nm was calculated from the SAED pattern (Fig. 4F inset). These d-spacings (0.38 and 0.28 nm) could correspond to (100) and (101) planes of t-Se, respectively suggesting the trigonal crystalline phase of these nanostructures.Negative staining of the samples allowed the detection of an organic layer coating the Se nanostructures (Fig. 5); suggesting the presence of amine rich organic material. The results show the efficiency of the purification method since no cell debris was detected, and only the SeNPs was evident.
3.4. Spectroscopic characterization of biogenic purified and cell associated Se nanostructures
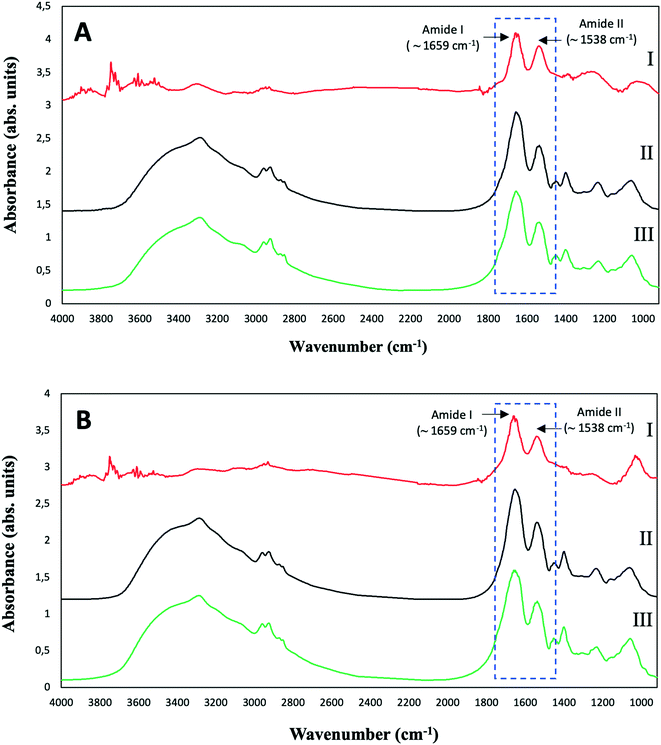 | ||
| Fig. 6 ATR-FTIR spectra of purified SeNPs (I), and S. bentonitica biomass in the absence (II) and presence (III) of 2 mM SeIV after 48 (A) and 144 h (B) of incubation. | ||
The control spectra showed the typical ATR-FTIR spectral regions of bacterial biomass with absorption peaks within the regions dominated by stretching vibrations of functional groups in proteins, lipids and polysaccharides (Table S1†). The spectra of the bacterial cells grown in the presence of Se show no remarkable shifts in the absorption bands in comparison with controls. This suggests that the amount of SeNPs in the bacterial culture was too low to give any detectable spectroscopic contribution specifically from SeNPs.12 In the purified SeNPs samples, the absorption bands at around 1659 and 1538 cm−1 mainly due to C-O vibrations corresponding to Amide I as well as N–H and C–N vibrations of peptide band attributed to Amide II of proteins were detected. The non-presence of complementary absorption bands characteristic of stretching in functional groups of proteins suggests that amide I and II bands correspond to amino acid residues coating the SeNPs. These results confirmed the presence of organic layers composed by amine rich matter surrounding the SeNPs as suggested by STEM-HAADF analysis. The detection of typical protein bands of isolated SeNPs biogically produced by the bacteria Azospirillum brasilense Sp7 and Stenotrophomonas maltophilia SeITE02 have been reported in other studies.38,39
It was found that 4 peaks gave the best fit for the C 1s spectra, most probably due to C–C and C–H bonds, C–O and C–N which have not been resolved, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and O–C
O and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O type carbon environments (Fig. S3†). The relative concentrations of the different components are most similar to the biomass control and the cell associated nanoparticles. However, the deconvoluted carbon peak for the purified nanoparticles showed components at different positions from the other samples, suggesting a slightly different composition of the organic material (Fig. S3†). The oxygen high resolution spectra show a similar result with peak areas and binding energies slightly different for the purified nanoparticles than the rest of the samples (Fig. S3†). The high nitrogen concentrations recorded at a binding energy of approximately 400 eV are consistent with the high concentrations of amine groups (Table S3; Fig. S3†). All samples appeared to show a second much smaller component at a slightly higher binding energy that most likely corresponds to protonated amines (Table S3; Fig. S3†).
O type carbon environments (Fig. S3†). The relative concentrations of the different components are most similar to the biomass control and the cell associated nanoparticles. However, the deconvoluted carbon peak for the purified nanoparticles showed components at different positions from the other samples, suggesting a slightly different composition of the organic material (Fig. S3†). The oxygen high resolution spectra show a similar result with peak areas and binding energies slightly different for the purified nanoparticles than the rest of the samples (Fig. S3†). The high nitrogen concentrations recorded at a binding energy of approximately 400 eV are consistent with the high concentrations of amine groups (Table S3; Fig. S3†). All samples appeared to show a second much smaller component at a slightly higher binding energy that most likely corresponds to protonated amines (Table S3; Fig. S3†).
The Se 3d peak is made up of two components: the Se 3d3/2 and the Se 3d5/2. Values from the literature (https://xpssimplified.com/elements/selenium.php) suggest these are separated by 0.86 eV with a fixed area ratio of 0.735![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The curve fits used to fit the data, have defined these two parameters. It was found that two pairs of peaks were necessary to fit each Se peak, and the widths of these pairs were allowed to vary independently. From the literature metallic selenium Se0 is expected 3d5/2 at 55.1 eV, whereas SeIV is expected at approximately 59.4 eV, SeVI at approximately 61 eV, SeII at approximately 57.7 eV and Se−II at binding energies <55 eV.40 It has also been seen that Se0 can be polarised leading to slightly higher binding energies.41 The binding energy at 55.2 eV in the purified nanoparticles (Fig. 7A) (Table S3†) confirmed the presence of Se0. Indeed, for these samples it is suggested that all the selenium is Se0, but approximately 50% of the signal could be due to Se influenced by the outer organic layer wrapping the SeNPs, which has polarised the Se atoms, while the remainder is from a true metallic Se0 core. The narrow peak width of the metallic Se0 peak is typical of a metal sample. For the cell associated nanoparticles (Fig. 7B) it should be noted that the intensity of the selenium signal is initially much lower, so the data are much more noisy. It is suggested for these samples the selenium is a mix of polarised Se0 and SeII.
1. The curve fits used to fit the data, have defined these two parameters. It was found that two pairs of peaks were necessary to fit each Se peak, and the widths of these pairs were allowed to vary independently. From the literature metallic selenium Se0 is expected 3d5/2 at 55.1 eV, whereas SeIV is expected at approximately 59.4 eV, SeVI at approximately 61 eV, SeII at approximately 57.7 eV and Se−II at binding energies <55 eV.40 It has also been seen that Se0 can be polarised leading to slightly higher binding energies.41 The binding energy at 55.2 eV in the purified nanoparticles (Fig. 7A) (Table S3†) confirmed the presence of Se0. Indeed, for these samples it is suggested that all the selenium is Se0, but approximately 50% of the signal could be due to Se influenced by the outer organic layer wrapping the SeNPs, which has polarised the Se atoms, while the remainder is from a true metallic Se0 core. The narrow peak width of the metallic Se0 peak is typical of a metal sample. For the cell associated nanoparticles (Fig. 7B) it should be noted that the intensity of the selenium signal is initially much lower, so the data are much more noisy. It is suggested for these samples the selenium is a mix of polarised Se0 and SeII.
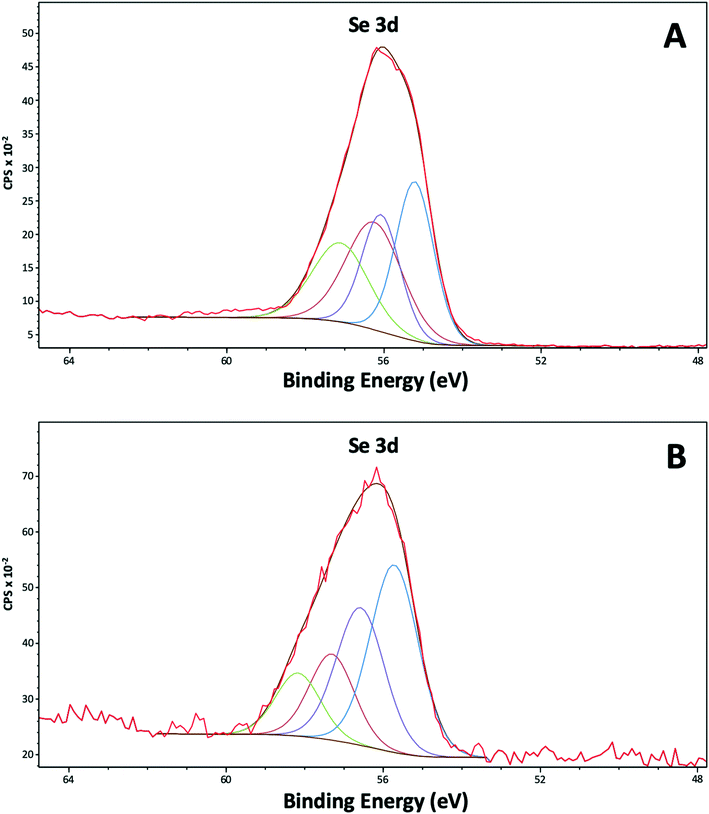 | ||
| Fig. 7 XPS high resolution spectra of selenium (Se 3d) from the purified SeNPs (A) and the cell associated SeNPs (B) after 144 h of incubation. | ||
The XPS results confirmed the presence of outer layer of amine rich organic material capping the SeNPs as previously indicated by HAADF-STEM and ATR-FTIR analysis. In addition, the Se reduction products appear to be present in the zero-valent state (Se0). The lack of binding energies corresponding to SeIV is consistent with the results of the time course experiments of SeIV reduction, where SeIV was completely reduced.
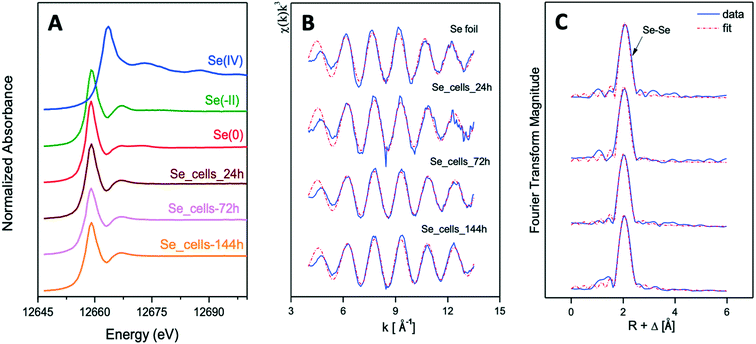
The Se K-edge EXAFS spectra of a Se foil and of biogenic SeNPs samples produced by S. bentonitica BII-R7 at 24, 72, and 144 h along with their corresponding Fourier transforms (FT) are presented in Fig. 8B and C. The fit parameters of the calculated spectra are summarized in Table 1.
| Sample | Shell | N | R [Å] | σ 2 [Å2] | ΔE [eV] |
|---|---|---|---|---|---|
| a Errors in coordination numbers are ±25% and standard deviations as estimated by EXAFSPAK. b Errors in distance are ±0.02 Å. c Debye–Waller factor. | |||||
| Se foil | Se–Se | 3.2 ± 0.2 | 2.37 | 0.0040 | −10.1 |
| SeIV–cells-24 h | Se–Se | 3.3 ± 0.2 | 2.34 | 0.0045 | −10.6 |
| SeIV–cells-72 h | Se–Se | 2.5 ± 0.1 | 2.33 | 0.0032 | −12.5 |
| SeIV–cells-144 h | Se–Se | 2.5 ± 0.1 | 2.34 | 0.0030 | −9.47 |
FT peak distances are reported in units of Å and are uncorrected for scattering phase shift, i.e., R + ΔR. The fit of the EXAFS spectra of the 3 experimental samples showed the presence of one Se–Se coordination shell at a bond distance of about 2.33–2.34 ± 0.02 Å. No differences were observed for the bond distance of this shell within the 3 samples. Similar values were reported for the Se–Se coordination shell of SeNPs produced by M. capsulatus and M. trichosporium17 and A. brasilense,9 with bond distances at 2.35 and 2.34 Å, respectively. According to these studies, the values obtained indicated the amorphous nature of the SeNPs. The coordination number of this coordination sphere is ranging from 2 to 3. The Debye–Waller factor (σ2) of this shell, ranging from 0.0030 to 0.0045 Å2 in all samples, decrease with reaction time, suggesting an increase of structural order with time (see Table 1). This could explain, as reported by Eswayah et al.,17 the biotransformation of amorphous SeNPs (24 h) to trigonal Se nanostructures (72 h and 144 h). Attempts to fit the second Se–Se coordination shell at about 3.63–3.70 ± 0.02 Å failed due to the small amplitude of this peak in the 3 studied samples. The bond distance of this shell would give insights on the crystalline Se, as was indicated by Eswayah et al.17 and Scheinost and Charlet.42 Thus, due to the fact that EXAFS is a bulk technique, no strong structural evidence on the transformation of amorphous Se to trigonal Se was demonstrated, as it was suggested by STEM/HAADF analysis.
3.5. Volatilization of selenium species by S. bentonitica
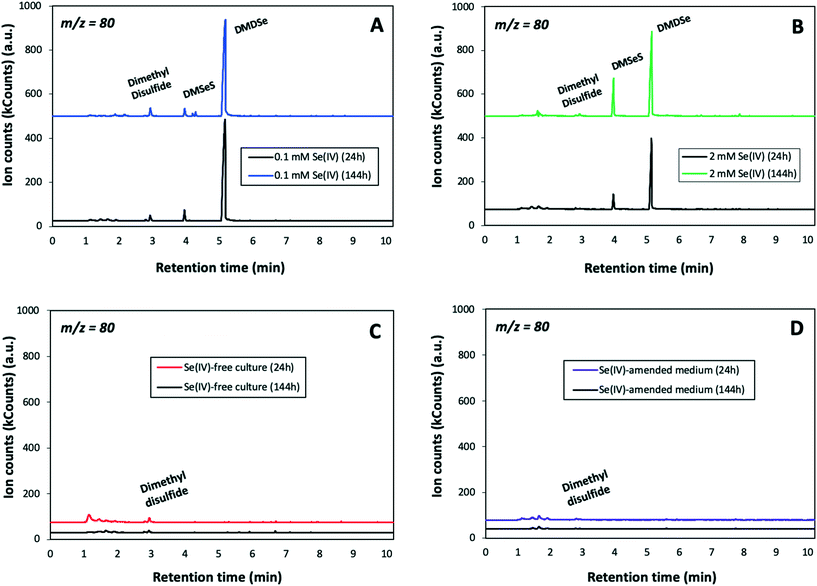
The headspace gas above the cultures was sampled (200 ml) and analyzed by GC–MS system in order to identify volatile Se species. GC-MS extracted ion chromatograms for 80 m/z (specific for Se) of the samples amended with 0.1 and 2 mM SeIV concentration showed the production of dimethyl diselenide (DMDSe) and dimethyl selenenyl sulphide (DMSeS) after 24 and 144 h (Fig. 9A and B). No differences in the production of volatile Se species were observed with different initial SeIV concentration and incubation time. The absence of Se volatile compounds in biotic and abiotic controls (Fig. 9C and D) suggested that they are produced by the bacterial cells. In addition, it is noteworthy that dimethyl disulfide (DMDS) was detected in both samples and controls since this compound could be involved in the formation of Se volatile species.20 Chasteen20 previously reported that a disproportionation reaction between DMDSe and DMDS to produce DMSeS is quite probable. The biotic and abiotic production of DMDS has been previously reported in the literature.43,44A number of microorganisms including phylogenetically-related species to S. bentonitica have been reported for their ability to produce volatile Se compounds largely in form of DMSe and DMDSe. Dungan et al. and Kagami et al.18,19 demonstrated the formation of DMSe, DMDSe and DMSeS by the bacterial species Stenotrophomonas maltophilia and Pseudomonas stutzeri NT-I, respectively. Bacteria were found to mainly generate DMSe as the most common form, while DMDSe were produced in a lesser extent.8 In our case, S. bentonitica produces DMDSe and not DMSe. Less common methylated Se compounds such as DMSeS, dimethyl selenyl disulphide (DMSeDS), dimethyl triselenide (DMTSe), and dimethyl selenone [(CH3)2SeO2] have also been reported to be generated by several microbes.23,24 However, the specific mechanism involved in their production still remains unclear.
4. Discussion
Se biotransformation has been well-described as a possible metal-microorganism interaction for bioremediation purposes.45,46 However, few studies have been conducted about its influence in the safety of the future DGR systems. The ability of the bacterial species S. bentonitica to reduce SeIV to Se0 forming Se nanostructures with different morphology (e.g. nanospheres, hexagons, polygons, and nanowires) has been described in a recently published work.26 The time-dependent transformation of a-Se nanospheres to crystalline t-Se nanostructures was proposed as indicated by microscopy and spectroscopy results. Extracellular flagella-like proteins and organic matter produced by the cells seem to be involved in the transformation process serving as template for a-Se nanospheres aggregation.In the present work a combination of spectroscopic (XPS, ATR-FTIR, XAS) and microscopic (STEM-HAADF) techniques were employed to confirm the involvement of biogenic proteins in the transformation of Se nanostructures. The employment of a broad range of techniques allowed us to address this issue and to give realistic and accurate insights into the chemical and structural properties of these nanostructures, which would help to understand the impact of this bacterium in the mobility of Se under DGR relevant conditions. The STEM-HAADF results of the purified SeNPs showed the presence of crystalline nanostructures attached to extensive a-Se nanospheres aggregations, what is in good agreement with the proposed transformation process from amorphous to crystalline Se. The change in the shape of the nanospheres also agrees with the proposed transformation since the final crystalline products present different morphologies (e.g. hexagonal, polygonal, and nanowires). The conventional negative staining of these samples showed organics layers capping the purified SeNPs suggesting the existence of proteins surrounding them. Finally, the presence of amine rich organic material coating the SeNPs was confirmed by means of ATR-FTIR and XPS. The identification of amine rich proteins from these organic layers in the present study suggests the role of proteins on the nanospheres aggregation and transformation. These results are in agreement with the studies of Bao et al. and Debieux et al.47,48 who reported the critical role of proteins in the nucleation and self-assembly of SeNPs produced by Thauera selenatis and Bacillus oryziterrae, respectively. To the best of our knowledge, the aggregation of Se nanospheres would allow their precipitation and settlement and would be beneficial for the surroundings of the repository. The qualitative identification of proteins associated to the SeNPs could contribute to a better understanding of the mechanisms of their synthesis. However, the use of quantitative proteomic analysis would further improve our understanding about the involvement of specific proteins in the bioreduction process (results will be published soon). This would indeed be helpful to improve the production of SeNPs on an industrial scale and thus enable their applications (e.g. for bioremediation purposes). Several studies have recently included proteomic analysis to investigate the synthesis of SeNPs. Xu et al.49 reported that the organic layer of the SeNPs produced by the bacterial strain Comamonas testosteroni S44 are composed mainly of proteins as indicated by quantitative analysis. Minor amounts of carbohydrates and lipids were detected, suggesting the major role of proteins in the formation of SeNPs. They reported that binding proteins to nanoparticles were nonspecific and could be a result of physicochemical processes, in contrast to previous studies indicating the role of specific proteins such as SefA.47 However, they demonstrated that the charged amino acid rich proteins have a higher binding ability to the surface of the SeNPs. These results could be useful for the production of SeNPs by biologically produced proteins for industrial and medical applications. Future proteomic studies on the biogenic SeNPs produced by S. bentonitica are in progress in order to further investigate the possible role of proteins on the synthesis of SeNPs and hence on the mobilization of Se present within the repository system.
The local structure of the reduced Se was characterized by XAS. The spectrum in the XANES region identified Se0 as the oxidation state of the Se reduction products as previously suggested due to the colour change observed in the cultures. On the other hand, the EXAFS region showed the presence of two coordination shells at a bond distance of about 2.34–2.35 ± 0.02 Å for the first Se–Se1 shell and about 3.63–3.70 ± 0.02 Å for the second Se–Se2 shell. It is well documented that the bond distance of the first Se–Se shell (Se–Se1) is intimately related to the structure of Se nanostructures. The shortest bond length of this shell (2.32–2.34 ± 0.02 Å) is associated to red a-Se.50 The largest Se–Se1 bond distance (2.37 ± 0.02 Å) is related to t-Se.51 In the case of monoclinic Se (m-Se), the Se–Se1 bond distance is calculated to be ranging from 2.32 to 2.34 ± 0.02 Å.52 Therefore, the obtained Se–Se1 bond distance for the studied samples (2.34–2.35 ± 0.02 Å) could correspond to the average distance of Se–Se1 shell from different Se structures (amorphous, monoclinic and trigonal). The identification of a-Se and t-Se nanoparticles previously detected by STEM-HAADF and VP-FESEM26 supported this hypothesis. The bond distances obtained for the second Se–Se2 shell correspond to crystalline Se according to Eswayah et al.17 They showed via EXAFS measurements of SeNPs, produced by M. capsulatus and M. Trichosporium, a Se–Se2 of 3.69 Å. According to Scheinost and Charlet,42 the Se–Se shell at 3.67–3.69 Å of Se reduced by Mackinawite minerals is in line with the structure of m-Se. Definitely, EXAFS analysis suggested that the SeNPs produced by S. bentonitica includes amorphous, monoclinic and trigonal structures, according to the calculated bond distance values. The higher Debye–Waller factor (σ2) of Se–Se1 shell in the 24 h sample indicated the higher structural disorder of this sample in comparison with 72 and 144 h samples. This is observation is in good agreement with the microscopy results obtained in Ruiz-Fresneda et al.,26 where amorphous SeNPs were predominantly observed at 24 h and higher structurally ordered (crystalline) Se were formed subsequently.
The Se reduction efficiency of S. bentonitica under aerobic conditions could be tested with a HPLC system coupled to ICP-MS. The results showed the complete SeIV reduction under different initial concentrations assayed. The time course of SeIV reduction by S. bentonitica is similar to other bacterial strains belonging to the same genus such as S. maltophilia SeITE02.39 Under 2 mM SeIV stress SeITE02 strain is able to reduce up to 86% after 192 h of incubation. In this study, S. bentonitica showed a higher reduction efficiency with a 100% of reduced SeIV after 120 h of incubation at the same SeIV concentration. The high Se reduction efficiency of the bentonite isolate S. bentonitica would be beneficial within the radioactive disposal system, where SeIV will be present.53 In addition, the obtained data would be indispensable for the geochemical models used to predict the influence of biogeochemical processes on Se mobility within the DGR system. Aerobic conditions are expected to dominate the DGRs after their closure, as oxygen will enter during the construction and operational periods. However, after that, anoxic conditions are supposed to be established relatively fast. These results show that S. bentonitica may maintain cell viability and activity under SeIV stress, and are able to reduce SeIV aerobically; similar to our results obtained under anaerobic conditions, reported in a previous work.54 This suggests that the reduction of SeIV could occur both aerobically and anaerobically within the DGR system. In the presence of oxygen, this facultative aerobic bacterium will use oxygen rather than other electron acceptors for respiration. Nitrate and acetate are used as electron acceptor and donor, respectively by this bacterium under anaerobic conditions.54 Both are compounds that will exist in the geodisposal system mainly as degradation products from radiolysis and chemolysis.55,56 In addition, it is assumed that different geochemical processes such as degradation of cellulose, from several materials present in radioactive waste, could lead to the generation of nutrients and growth substrates that microbes can use to support growth.
To further investigate the reduction mechanism involved, the production of volatile Se species was studied. The biomethylation to volatile Se products results in the removal of SeVI and SeIV and the transfer of Se from aquatic and terrestrial sites to the atmosphere in form of methyl selenides.8,57 The insolubility and limited bioavailability of methyl selenides in contrast to the highly reactive Se oxyanions is well known.58 For this reason, methylation is considered one of the most important transformation processes in relation to bioremediation of Se. Losi and Frankenberger59 demonstrated that oxidation and resolubilization of Se0 to SeIV and SeVI microbially induced in soil occurs at relatively low rates. Hence, the reduction of Se oxyanions to Se0 could be only a temporary solution for bioremediation of contaminated environments.25,59 This fact makes biomethylation of Se an effective approach for the bioremediation of contaminated environments because it would allow the complete removal of Se from soil and water.25,57 The production of methylated Se compounds (DMDSe and DMSeS) by S. bentonitica confirmed the transformation to Se−II oxidation state of the Se reduction products since methylation involve reduction to Se−II when the initial oxidation states are SeVI, SeIV and Se0.57 Both reduction and methylation of Se oxyanions (SeVI and SeIV) are considered a detoxification process for microorganisms since volatile compounds DMSe and DMDSe are 500 to 700 times less toxic than Se oxyanions and other derivatives.8 This fact suggests the potential positive influence that S. bentonitica would have in the planned repositories of radioactive wastes by converting toxic Se oxyanions to methylated Se species with lower toxicity. However, some Se volatile species such as DMSe could be rapidly demethylated in anoxic sediments to methane (CH4), carbon dioxide (CO2), and hydrogen selenide (H2Se) by methanogens and sulphate-reducing bacteria.60 It is important to note that gas production within the DGR system can affect the integrity and safety of clay barriers.61 Despite the lower toxicity of the Se volatile species formed, the risk potential associated to radioactive substances must be considered. The isotope 79Se with a long half- life is a significant emitter and contributes to the radioactivity generated in the repository.
The presence of dimethyl disulphide (DMDS) in the samples suggested its role in the formation of Se volatile species by a disproportionation reaction between DMDSe and DMDS to produce DMSeS as previously suggested by Chasteen and Bentley.62 However, more studies are required to determine the specific mechanism involved.
5. Conclusions
The results of the present study have shown the reduction of the toxic selenium oxyanion SeIV to less toxic species (Se0 and volatile Se−II) by the bacterium S. bentonitica, isolated from Spanish bentonites. The findings clearly demonstrated the high efficiency and tolerance of S. bentonitica in reducing SeIV and producing SeNPs and volatile methylated Se. The STEM-HAADF results support the transformation process from amorphous to crystalline Se proposed in a previous work. The local structure characterization of the Se0 reduction products by means of XAS suggested the presence of amorphous, monoclinic, and trigonal Se and evidenced the biotransformation of amorphous Se nanospheres to trigonal Se crystals. Additionally, the detection of volatile methylated Se species such as DMDSe and DMSeS confirmed the ability of S. bentonitica to reduce SeIV to the chemical species Se−II. The bioproduction of methylated Se is interesting as a promising tool for bioremediation purposes due to its lower toxicity comparing with Se oxyanions. Indeed, the Se reduction products in form of Se0 and Se−II may play an important role in ensuring the safety of DGR because of the biotransformation products are of lower solubility and toxicity.The identification of organic layers mainly composed by amine rich material coating the biogenic SeNPs suggested the involvement of proteins in their synthesis and transformation. Consequently, proteins could influence their mobility and hence their fate in the environment. However, additional investigations are necessary to better understand the specific role of protein coating in the solubility and mobility of the Se reduction products for the deep disposal of radioactive wastes. Definitely, this investigation has provided a new perspective on the potential impact of microorganisms on the Se mobility through planned DGRs of radioactive wastes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Euratom research and training programme 2014–2018 under grant agreement no. 661880 and the Mobility Grant awarded by the University of Granada in the framework of the Official Call for International PhD Students launched by the Vice-Rectorate for Internationalization.. The authors acknowledge the assistance of Maria del Mar Abad Ortega, Concepción Hernández Castillo, and Jaime Lazuen Alcón (Centro de Instrumentación Científica, University of Granada, Spain) for their help with microscopy and flow cytometry measurements. The authors also thank the assistance of Deborah Hammond (Sheffield Surface Analysis Centre, University of Sheffield, UK), Joseph Hufton, and Jaime Gomez-Bolivar with the XPS, ATR-FTIR analysis, and XAS measurements, respectively.References
- N. R. Smart, A. P. Rance, B. Reddy, L. Hallbeck, K. Pedersen and A. J. Johansson, In situ evaluation of model copper-cast iron canisters for spent nuclear fuel: a case of microbiologically influenced corrosion(MIC), Corros. Eng., Sci. Technol., 2014, 49(6), 548–553 CrossRef CAS.
- P. Delage, Y. J. Cui and A. M. Tang, Clays in radioactive waste disposal, J. Rock Mech. Geotech. Eng., 2010, 2(2), 111–123 CrossRef.
- A. Bagnoud, K. Chourey, R. L. Hettich, I. De Bruijn, A. F. Andersson and O. X. Leupin, et al. Reconstructing a hydrogen-driven microbial metabolic network in Opalinus Clay rock, Nat. Commun., 2016, 7, 12770 CrossRef CAS PubMed.
- E. Krawczyk-Bärsch, U. Gerber, K. Müller, H. Moll, A. Rossberg and R. Steudtner, et al. Multidisciplinary characterization of U(VI) sequestration by Acidovorax facilis for bioremediation purposes, J. Hazard. Mater., 2018, 347, 233–241 CrossRef.
- D. Song, X. Li, Y. Cheng, X. Xiao, Z. Lu and Y. Wang, et al. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction, Sci. Rep., 2017, 7(1), 3239 CrossRef PubMed.
- M. Lopez-Fernandez, H. Moll and M. L. Merroun, Reversible pH-dependent curium(III) biosorption by the bentonite yeast isolate Rhodotorula mucilaginosa BII-R8, J. Hazard. Mater., 2018, 370, 156–163 CrossRef PubMed.
- J. Ikonen, M. Voutilainen, M. Söderlund, L. Jokelainen, M. Siitari-Kauppi and A. Martin, Sorption and diffusion of selenium oxyanions in granitic rock, J. Contam. Hydrol., 2016, 192, 203–211 CrossRef CAS.
- L. Ranjard, S. Nazaret and B. Cournoyer, Freshwater bacteria can methylate selenium through the thiopurine methyltransferase pathway, Appl. Environ. Microbiol., 2003, 69(7), 3784–3790 CrossRef CAS PubMed.
- M. Vogel, S. Fischer, A. Maffert, R. Hübner, A. C. Scheinost and C. Franzen, et al. Biotransformation and detoxification of selenite by microbial biogenesis of selenium-sulfur nanoparticles, J. Hazard. Mater., 2018, 344, 749–757 CrossRef CAS PubMed.
- L. C. Staicu, C. J. Ackerson, P. Cornelis, L. Ye, R. L. Berendsen and W. J. Hunter, et al., Pseudomonas moraviensis subsp. stanleyae, a bacterial endophyte of hyperaccumulator Stanleya pinnata, is capable of efficient selenite reduction to elemental selenium under aerobic conditions, J. Appl. Microbiol., 2015, 119(2), 400–410 CrossRef CAS PubMed.
- J. Dobias, E. I. Suvorova and R. Bernier-Latmani, Role of proteins in controlling selenium nanoparticle size, Nanotechnology, 2011, 22(19), 195605 CrossRef CAS PubMed.
- A. A. Kamnev, P. V. Mamchenkova, Y. A. Dyatlova and A. V. Tugarova, FTIR spectroscopic studies of selenite reduction by cells of the rhizobacterium Azospirillum brasilense Sp7 and the formation of selenium nanoparticles, J. Mol. Struct., 2017, 1140, 106–112 CrossRef CAS.
- R. Jain, N. Jordan, S. Tsushima, R. Hübner, S. Weiss and P. N. L. Lens, Shape change of biogenic elemental selenium nanomaterials from nanospheres to nanorods decreases their colloidal stability, Environ. Sci.: Nano, 2017, 4(5), 1054–1063 RSC.
- B. Buchs, M. W. H. Evangelou, L. H. E. Winkel and M. Lenz, Colloidal properties of nanoparticular biogenic selenium govern environmental fate and bioremediation effectiveness, Environ. Sci. Technol., 2013, 47(5), 2401–2407 CrossRef CAS PubMed.
- C. I. Pearce, V. S. Coker, J. M. Charnock, R. A. D. Pattrick, J. F. W. Mosselmans and N. Law, et al. Microbial manufacture of chalcogenide-based nanoparticles via the reduction of selenite using Veillonella atypica: An in situ EXAFS study, Nanotechnology, 2008, 19(15), 155603 CrossRef.
- M. J. Herbel, J. S. Blum, R. S. Oremland and S. E. Borglin, Reduction of elemental selenium to selenide: Experiments with anoxic sediments and bacteria that respire Se-oxyanions, Geomicrobiol. J., 2003, 20(6), 587–602 CrossRef CAS.
- A. S. Eswayah, T. J. Smith, A. C. Scheinost, N. Hondow and P. H. E. Gardiner, Microbial transformations of selenite by methane-oxidizing bacteria, Appl. Microbiol. Biotechnol., 2017, 101(17), 6713–6724 CrossRef CAS PubMed.
- R. S. Dungan, S. R. Yates and W. T. Frankenberger, Transformations of selenate and selenite by Stenotrophomonas maltophilia isolated from a seleniferous agricultural drainage pond sediment, Environ. Microbiol., 2003, 5(4), 287–295 CrossRef CAS.
- T. Kagami, T. Narita, M. Kuroda, E. Notaguchi, M. Yamashita and K. Sei, et al. Effective selenium volatilization under aerobic conditions and recovery from the aqueous phase by Pseudomonas stutzeri NT-I, Water Res., 2013, 47(3), 1361–1368 CrossRef CAS.
- T. G. Chasteen, Confusion between dimethyl selenenyl sulfide and dimethyl selenone released by bacteria, Appl. Organomet. Chem., 1993, 7(5), 335–342 CrossRef CAS.
- A. S. Eswayah, N. Hondow, A. C. Scheinost, M. Merroun, M. Romero-González and T. J. Smith, et al. Methyl Selenol as a Precursor in Selenite Reduction to Se/S Species by Methane-Oxidizing Bacteria. Kelly RM, editor, Appl. Environ. Microbiol., 2019, 85(22), e01379-19 CrossRef PubMed.
- F. Challenger, Biological Methylation, Chem. Rev., 1945, 36(3), 315–361, DOI:10.1021/cr 60115a003.
- D. C. Reamer and W. H. Zoller, Selenium biomethylation products from soil and sewage sludge, Science, 1980, 208(4443), 500–502 CrossRef CAS PubMed.
- R. Burra, G. A. Pradenas, R. A. Montes, C. C. Vásquez and T. G. Chasteen, Production of dimethyl triselenide and dimethyl diselenenyl sulfide in the headspace of metalloid-resistant Bacillus species grown in the presence of selenium oxyanions, Anal. Biochem., 2010, 396(2), 217–222 CrossRef CAS PubMed.
- W. T. Frankenberger and M. Arshad, Bioremediation of selenium-contaminated sediments and water, BioFactors, 2001, 14(1–4), 241–254 CrossRef CAS PubMed.
- M. A. Ruiz Fresneda, J. Delgado Martín, J. Gómez Bolívar, M. V. Fernández Cantos, G. Bosch-Estévez and M. F. Martínez Moreno, et al. Green synthesis and biotransformation of amorphous Se nanospheres to trigonal 1D Se nanostructures: Impact on Se mobility within the concept of radioactive waste disposal, Environ. Sci.: Nano, 2018, 5(9), 2103–2116 RSC.
- M. V. Villar, Pérez del Villar L, Martín PL, Pelayo M, Fernández AM, Garralon A, et al. The study of Spanish clays for their use as sealing materials in nuclear waste repositories: 20 years of progress, J. Iber. Geol., 2006, 32(1), 15–36 Search PubMed.
- S. Dhanjal and S. S. Cameotra, Aerobic biogenesis of selenium nanospheres by Bacillus cereus isolated from coalmine soil, Microb. Cell Fact., 2010, 9, 52 CrossRef PubMed.
- P. Sonkusre, R. Nanduri, P. Gupta and S. Cameotra, Improved Extraction of Intracellular Biogenic Selenium Nanoparticles and their Specificity for Cancer Chemoprevention, J. Nanomed. Nanotechnol., 2014, 05(2), 194 Search PubMed.
- M. L. Merroun, J. Raff, A. Rossberg, C. Hennig, T. Reich and S. Selenska-Pobell, Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12, Appl. Environ. Microbiol., 2005, 71(9), 5532–5543 CrossRef CAS PubMed.
- B. Sitaud, P. L. Solari, S. Schlutig, I. Llorens and H. Hermange, Characterization of radioactive materials using the MARS beamline at the synchrotron SOLEIL, J. Nucl. Mater., 2012, 425(1–3), 238–243 CrossRef CAS.
- P. L. Solari, S. Schlutig, H. Hermange and B. Sitaud, MARS, a new beamline for radioactive matter studies at SOLEIL, J. Phys.: Conf. Ser., 2009, 190(1), 012042 CrossRef.
- B. Ravel and M. Newville, ATHENA, ARTEMIS, HEPHAESTUS: Data analysis for X-ray absorption spectroscopy using IFEFFIT, J. Synchrotron Radiat., 2005, 12(4), 537–541 CrossRef CAS PubMed.
- A. Ankudinov and B. Ravel, Real-space multiple-scattering calculation and interpretation of x-ray-absorption near-edge structure, Phys. Rev. B: Condens. Matter Mater. Phys., 1998, 58(12), 7565–7576 CrossRef CAS.
- A. L. Givan, Flow Cytometry: An Introduction, in Flow Cytometry Protocols, ed. T. S. Hawley and R. G. Hawley, 2011, pp. 1–29 Search PubMed.
- L. C. F. Stubberfield and P. J. A. Shaw, A comparison of tetrazolium reduction and FDA hydrolysis with other measures of microbial activity, J. Microbiol. Methods, 1990, 12(3–4), 151–162 CrossRef CAS.
- F. David, A. Berger, R. Hänsch, M. Rohde and E. Franco-Lara, Single cell analysis applied to antibody fragment production with Bacillus megaterium: Development of advanced physiology and bioprocess state estimation tools, Microb. Cell Fact., 2011, 10, 23 CrossRef CAS PubMed.
- A. A. Kamnev, FTIR spectroscopic studies of bacterial cellular responses to environmental factors, plant-bacterial interactions and signalling, Spectroscopy, 2008, 22(2–3), 83–95 CrossRef CAS.
- S. Lampis, E. Zonaro, C. Bertolini, D. Cecconi, F. Monti and M. Micaroni, et al. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles, J. Hazard. Mater., 2017, 324, 3–14 CrossRef CAS PubMed.
- D. Briggs and M. P. Seah, Practical Surface Analysis by Auger and X-ray photoelectron spectroscopy, John Wiley & Sons, New York, 1983, p. 533, DOI:10.1002/sia.740060611.
- H. Rupp and U. Weser, X-ray photoelectron spectroscopy of some selenium containing amino acids, Bioinorg. Chem., 1975, 5(1), 21–32 CrossRef CAS PubMed.
- A. C. Scheinost and L. Charlet, Selenite reduction by mackinawite, magnetite and siderite: XAS characterization of nanosized redox products, Environ. Sci. Technol., 2008, 42(6), 1984–1989 CrossRef CAS PubMed.
- O. Carrión, J. Pratscher, A. R. J. Curson, B. T. Williams, W. G. Rostant and J. Colin Murrell, et al. Methanethiol-dependent dimethylsulfide production in soil environments, ISME J., 2017, 11(10), 2379–2390 CrossRef PubMed.
- M. J. Higgins, Y.-C. Chen, D. P. Yarosz, S. N. Murthy, N. A. Maas and D. Glindemann, et al. Cycling of Volatile Organic Sulfur Compounds in Anaerobically Digested Biosolids and its Implications for Odors, Water Environ. Res., 2006, 78(3), 243–252 CrossRef CAS PubMed.
- S. P. W. Hageman, R. D. van der Weijden, A. J. M. Stams and C. J. N. Buisman, Bio-production of selenium nanoparticles with diverse physical properties for recovery from water, Int. J. Miner. Process., 2017, 169, 7–15 CrossRef CAS.
- L. Che, Y. Dong, M. Wu, Y. Zhao, L. Liu and H. Zhou, Characterization of Selenite Reduction by Lysinibacillus sp. ZYM-1 and Photocatalytic Performance of Biogenic Selenium Nanospheres, ACS Sustainable Chem. Eng., 2017, 5(3), 2535–2543 CrossRef CAS.
- C. M. Debieux, E. J. Dridge, C. M. Mueller, P. Splatt, K. Paszkiewicz and I. Knight, et al. A bacterial process for selenium nanosphere assembly, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(33), 13480–13485 CrossRef CAS PubMed.
- P. Bao, K. Q. Xiao, H. J. Wang, H. Xu, P. P. Xu and Y. Jia, et al. Characterization and potential applications of a selenium nanoparticle producing and nitrate reducing bacterium Bacillus oryziterrae sp. nov., Sci. Rep., 2016, 6, 34054 CrossRef CAS PubMed.
- D. Xu, L. Yang, Y. Wang, G. Wang, C. Rensing and S. Zheng, Proteins enriched in charged amino acids control the formation and stabilization of selenium nanoparticles in Comamonas testosteroni S44, Sci. Rep., 2018, 8, 4766 CrossRef PubMed.
- V. S. Minaev, S. P. Timoshenkov and V. V. Kalugin, Structural and Phase Transformation in Condensed Selenium, J. Optoelectron. Adv. Mater., 2005, 7(4), 1717–1741 CAS.
- A. Popov, Correlation between the molecular structure and properties of amorphous selenium, J. Phys. C: Solid State Phys, 1976, 9(24), 675 CrossRef.
- Y. H. Zhao, K. Lu and T. Liu, EXAFS study of structural characteristics of nanocrystalline selenium with different grain sizes, Phys. Rev. B: Condens. Matter Mater. Phys., 1999, 59(17), 11117–11120 CrossRef CAS.
- K. Dardenne, E. González-Robles, J. Rothe, N. Müller, G. Christill and D. Lemmer, et al. XAS and XRF investigation of an actual HAWC glass fragment obtained from the Karlsruhe vitrification plant(VEK), J. Nucl. Mater., 2015, 460, 209–215 CrossRef CAS.
- M. A. Ruiz-Fresneda, J. Gomez-Bolivar, J. Delgado-Martin, M. del Mar Abad-Ortega, I. Guerra-Tschuschke and M. L. Merroun, The bioreduction of selenite under anaerobic and alkaline conditions analogous to those expected for a deep geological repository system, Molecules, 2019, 24(21), 3868 CrossRef CAS.
- L. Abrahamsen, T. Arnold, H. Brinkmann, N. Leys, M. Merroun and K. Mijnendonckx, et al. A review of anthropogenic organic wastes and their degradation behaviour, MIND Proj [Internet], 2015, 97. Available from: http://www.mind15.eu/wp-content/uploads/2015/11/MIND-Deliverable-1.1-Review-of-anthropogenic-organic-wastes.pdf Search PubMed.
- A. Bertron, N. Jacquemet, B. Erable, C. Sablayrolles, G. Escadeillas and A. Albrecht, Reactivity of nitrate and organic acids at the concrete-bitumen interface of a nuclear waste repository cell, Nucl. Eng. Des., 2014, 268, 51–57 CrossRef CAS.
- A. S. Eswayah, T. J. Smith and P. H. E. Gardiner, Microbial transformations of selenium species of relevance to bioremediation, Appl. Environ. Microbiol., 2016, 82(16), 4848–4859 CrossRef CAS PubMed.
- J. W. Doran, Microorganisms and the Biological Cycling of Selenium, in Advances in Microbial Ecology Vol. 6 [Internet], ed. Marshall KC, Springer US, Boston, MA, 1982, pp. 1–32, Available from: DOI:10.1007/978-1-4615-8318-9_1.
- M. E. Losi and W. T. J. Frankenberger, Microbial oxidation and solubilization of precipitated elemental selenium in soil, J. Environ. Qual., 1998, 27(4), 836–843 CrossRef CAS.
- R. S. Oremland and J. P. Zehr, Formation of Methane and Carbon Dioxide from Dimethylselenide in Anoxic Sediments and by a Methanogenic Bacterium, Appl. Environ. Microbiol., 1986, 52(5), 1031–1036 CrossRef CAS.
- A. Bagnoud, I. de Bruijn, A. F. Andersson, N. Diomidis, O. X. Leupin, B. Schwyn and R. Bernier-Latmani, A minimalistic microbial food web in an excavated deep subsurface clay rock, FEMS Microbiol. Ecol., 2016, 92(1), fiv138 CrossRef PubMed.
- T. G. Chasteen and R. Bentley, Biomethylation of Selenium and Tellurium: Microorganisms and Plants, Chem. Rev., 2003, 103(1), 1–25 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0en00507j |
| ‡ Present address: School of Engineering and Materials Science, Queen Mary University of London, London, E1 4NS, UK. |
| This journal is © The Royal Society of Chemistry 2020 |