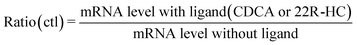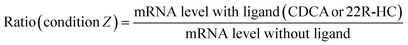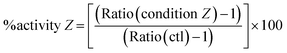Open Access Article
Open Access ArticleNuclear translocation of silver ions and hepatocyte nuclear receptor impairment upon exposure to silver nanoparticles†
Vanessa
Tardillo Suárez
 a,
Elizaveta
Karepina
bc,
Mireille
Chevallet
b,
Benoit
Gallet
a,
Elizaveta
Karepina
bc,
Mireille
Chevallet
b,
Benoit
Gallet
 d,
Cécile
Cottet-Rousselle
e,
Peggy
Charbonnier
b,
Christine
Moriscot
df,
Isabelle
Michaud-Soret
d,
Cécile
Cottet-Rousselle
e,
Peggy
Charbonnier
b,
Christine
Moriscot
df,
Isabelle
Michaud-Soret
 b,
Wojciech
Bal
g,
Alexandra
Fuchs
h,
Rémi
Tucoulou
a,
Pierre-Henri
Jouneau
c,
Giulia
Veronesi
b,
Wojciech
Bal
g,
Alexandra
Fuchs
h,
Rémi
Tucoulou
a,
Pierre-Henri
Jouneau
c,
Giulia
Veronesi
 ab and
Aurélien
Deniaud
ab and
Aurélien
Deniaud
 *b
*b
aESRF, The European Synchrotron, 71 Avenue des Martyrs, 38000 Grenoble, France
bUniv. Grenoble Alpes, CNRS, CEA, IRIG, Laboratoire de Chimie et Biologie des Métaux, F-38000 Grenoble, France. E-mail: aurelien.deniaud@cea.fr; Tel: + 33 (0)4 38 78 96 51
cUniv. Grenoble Alpes, CEA, IRIG, MEM, F-38000 Grenoble, France
dInstitut de Biologie Structurale, CEA, CNRS, Univ. Grenoble Alpes, 71 Avenue des Martyrs, F-38042 Grenoble, France
eUniv. Grenoble Alpes, INSERM U1055, Laboratory of Fundamental and Applied Bioenergetics (LBFA), and Environmental and System Biology (BEeSy), 38000 Grenoble, France
fIntegrated Structural Biology Grenoble (ISBG), CNRS, CEA, Université Grenoble Alpes, EMBL, 71 Avenue des Martyrs, F-38042, Grenoble, France
gInstitute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawinskiego 5a, 02-106 Warsaw, Poland
hDRF/BIG-INSERM U1160, Hôpital St. Louis, F-75010 Paris, France
First published on 9th April 2020
Abstract
The impact on human health of long-term exposure to low doses of silver nanoparticles (AgNPs) remains understudied. Cellular studies have shown the intracellular dissolution of AgNPs within endolysosomes followed by Ag(I) binding to biomolecular thiolate-containing molecules. However, the precise subcellular distribution of Ag(I) and the nature of the disrupted physiological pathways remained unknown. Novel imaging approaches enabled us to visualize the trafficking of AgNP-containing lysosomes towards a perinuclear location and a nuclear transfer of Ag(I) species with accumulation in the nucleoli. These Ag(I) species impaired nuclear receptor activity, disrupting critical mechanisms of liver physiology in low dose exposure scenarios, thus justifying further research into defining a framework for the safe use of AgNPs.
Environmental significanceSilver nanoparticles (AgNPs) are massively used as biocides in consumer products and medical devices. The biocidal activity arises from the long-lasting release of Ag(I) species that is due to AgNP oxidative dissolution. This process can occur under environmental and biological conditions leading to the exposure of various organisms, including humans, to Ag(I) species. The acute toxicity of AgNPs and Ag(I) has been extensively studied but the physiologic disruptions induced by chronic exposure to AgNPs remain understudied. Under conditions mimicking realistic exposures, an innovative elemental imaging technique enabled Ag(I) nuclear translocation to be visualized. Under these AgNP exposure conditions, specific nuclear receptors get inhibited, revealing an endocrine disruptor-like effect. Due to their tremendous affinity for thiolate, nuclear Ag(I) species most probably replace Zn(II) in Zn finger domains of nuclear receptors. |
1 Introduction
Silver nanoparticles (AgNPs) are widely used in everyday products for their biocidal activity and increasing production has led to environmental and human safety concerns over the past few years. Their biocidal properties stem from the durable release of Ag(I) ions from the NP's Ag(0) core due to surface oxidation,1,2 ions that are also toxic to mammalian cells.3In humans, the main and most extensively described exposure route for AgNPs is dietary.4,5 However, the impact of nanosilver-containing medical devices that can directly release Ag into lymphatic and blood vessels has been sorely disregarded. The leaching of AgNPs is a well-known drawback of nano-enabled products, which has been studied and quantified, for instance in fabrics and consumer products for children.6–8 AgNP-based medical devices, although designed to release biocidal Ag(I) ions, can also leach AgNPs. In order to prevent this undesired effect, synthetic strategies that allow adhesion of the NPs to the products are under investigation.9,10 Considering that the commercially available AgNP-based materials for clinical use are catheters and wound dressings,11 leaching from such products results in the release of AgNPs into the bloodstream.
The exposure route to AgNPs influences the biodistribution and body clearance of silver, as shown by an in vivo study where mice were exposed to a single dose of AgNPs either orally or intravenously.12 In both cases, Ag preferentially accumulates in the liver. However, in the case of parenteral exposure, the Ag concentration in the blood after 24 h is ten times higher, and its clearance is much slower than that after ingestion. This is most probably due to the direct introduction of Ag(0) from AgNPs into the bloodstream in the case of injection, whereas in the case of oral exposure AgNPs are readily transformed in the gastrointestinal tract, and Ag enters the bloodstream mainly in the form of various Ag(I) species (ref. 13 and for a recent review see ref. 4). Besides, several studies in burn patients revealed Ag blood levels in the low to high micromolar range proving the relevance of Ag exposure in the medical context.14–16 Therefore, it is of paramount importance to better understand the AgNP fate in the liver to predict the risk for patients during hospital care. In spite of its relevance, this exposure scenario has been as yet disregarded in nanotoxicology studies. In this context, we decided to address the impact of an exposure over several days to low and non-toxic concentrations of pristine AgNPs on critical hepatocyte functions.
Over the past few years, several studies have described the fate of AgNPs in various mammalian cell models.17–21 A consensus has emerged showing AgNP dissolution in endosomes and lysosomes and the subsequent distribution of Ag(I) species throughout the cell mainly in the form of Ag–thiolate complexes.18,20–22 This type of complexation comes as no surprise as Ag has a picomolar or higher affinity for thiolates depending on the coordination chemistry offered by the biomolecular ligands.23 However, recent imaging studies did not provide the subcellular distribution of Ag(I) species at the organelle level.3,21 The identification of cell compartments that can store Ag and/or that can be critically affected by this non-physiological and deleterious metal ion remains a critical question. Of particular importance is the long-standing controversy about AgNP entry into the nucleus. Indeed, in the literature, AgNPs have been shown in the nucleus only in few publications,24,25 while the majority of the studies did not. Moreover, the chemical composition of these NPs visualized in the nucleus was not proven. We described in detail the limits of those studies in a recent review.4
In the current study, we explored the interior of hepatic cells exposed to AgNPs by applying several high-resolution imaging methods to thin cell sections. The synergistic use of confocal microscopy, electron microscopy, elemental imaging by synchrotron X-ray fluorescence (XRF), and energy dispersive X-ray spectroscopy (EDX), complemented by chemical analysis through X-ray absorption spectroscopy (XAS), allowed us to infer the subcellular distribution of AgNPs and Ag–thiolate complexes. These experiments were carried out under long-term low dose exposure in order to be as realistic as possible. We thus highlighted that Ag(I) species but not AgNPs are translocated to the nucleus and we further analyzed their impact on nuclear functions. Hepatocyte nuclear receptors, such as the liver X receptor (LXR) and the farnesoid X receptor (FXR), also known as the bile acid receptor (BAR), were identified as molecular targets that were inhibited by these translocated metal ions even in a very low dose exposure scenario mimicking chronic exposure. AgNPs could thus trigger an endocrine disruptor-like effect.
2 Experimental
2.1 AgNP characterization
Highly homogenous 20 nm citrate-coated AgNPs were purchased from NanoComposix (20 nm Citrate BioPure™ Silver) and 90 nm PVP-coated AgNPs were purchased from Sigma-Aldrich. Both were previously characterized.21 Dynamic light scattering measurements were carried out in cell culture medium supplemented with 10% fetal calf serum at 25 °C in a UVette (Eppendorf) on a Nanostar instrument (Wyatt). Zeta potential was determined using a Malvern Nano ZS in 10 mM Hepes and 2 mM sodium citrate pH 7. In cell culture media, citrate- and PVP-coated AgNPs formed agglomerates of 160 nm and homogenous 90 nm particles, respectively (Table S1†).2.2 Cell culture
HepG2 cells were grown in MEM medium supplemented with 10% FBS and 1% antibiotics–antimycotic and exposed to cit-AgNPs or PVP-AgNPs at the indicated concentrations of total Ag and for the indicated times. The medium and AgNPs were renewed every 24 hours. When required, chenodeoxycholic acid (CDCA, Sigma-Aldrich) at 80 μM or the equivalent volume of DMSO was added for the last 17 hours of exposure or 22R-hydroxycholesterol (22R-HC, Sigma-Aldrich) at 10 μM or the equivalent volume of absolute ethanol was added for the last 20 hours of exposure.2.3 Spheroid preparation
Micropatterns were produced on glass slides by deep-UV photolithography by which a non-toxic cell repellent PEG-PLL coating was degraded locally by deep-UV light passing through a photomask (as described in ref. 26). Micropatterns were then coated with fibronectin (20 μg mL−1 in PBS) to form adhesive disks of 50 μm in diameter, surrounded by a cytophobic surface.HepG2/C3a cells were seeded on micropatterned surfaces at a density of 1.25 × 105 cells per cm2 and were allowed to attach. After 2 hours, unattached and floating cells were removed by extensively washing the surface and gentle flushing with a pipette. Cells on micropatterns were maintained in culture for up to one week, to allow spheroids to grow.
2.4 Subcellular fractionation and elemental quantification
HepG2 cells were cultured in 2D and exposed to cit-AgNPs (12.5 μM in Ag) for 24 to 72 hours. At the end of the exposure, cells were washed three times with PBS. About 100 million cells were subjected to subcellular fractionation using a protocol developed to purify nucleoli27 that we adapted to avoid the contamination of the nuclear fraction by endolysosomal vesicles rich in AgNPs. Indeed, at low centrifugation speed, the high density of trapped AgNPs pelleted the “light vesicles” containing AgNPs. Therefore, we purified nuclei through discontinuous sucrose/percoll gradient centrifugation in which nuclei are recovered at the interface between two layers and not at the bottom of the tubes where AgNP-containing vesicles can be found.After AgNP exposure for the indicated times, HepG2 cells were recovered by trypsinization. The cell pellet was resuspended in 7.5 mL of 10 mM Tris-HCl pH 7.4, 10 mM NaCl and 1 mM MgSO4 and incubated for 20 minutes on ice with regular mixing. NP40 was added at a final concentration of 0.3% and cell lysis was carried out with a Dounce homogenizer. The nuclei-containing pellet was collected by centrifugation at 1200g for 5 minutes. The pellet was resuspended in 2.5 mL of 10 mM Hepes pH 7.5, 10 mM MgSO4 and 0.25 M sucrose and loaded on top of a gradient of 10 mM Hepes pH 7.5, 0.25 M sucrose and 0.05 mM MgSO4 with a 2.5 mL layer of 20% percoll, a 2.5 mL layer of 50% percoll, a 2.5 mL layer of 60% percoll and a 1 mL layer of 70% percoll. The gradient was centrifuged for 1 hour at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C. The main fraction, the nuclei, was recovered at the interface between the 20 and 50% percoll layers and diluted in 10 mL of 10 mM Hepes pH 7.5, 0.25 M sucrose and 10 mM MgSO4. This fraction was centrifuged for 10 minutes at 1200g through a layer of 10 mL of 10 mM Hepes pH 7.5, 0.88 M sucrose and 0.05 mM MgSO4. The pellet contained pure nuclei that were resuspended in 10 mL of 10 mM Hepes pH 7.5, 0.34 M sucrose and 0.05 mM MgSO4. A fraction was kept for further analysis and the main part was sonicated 4 times for 30 seconds at 10% power with a microtip (Sonicator BioBlock Scientific Vibracell 72412). This solution was centrifuged for 20 minutes at 2000g through a layer of 10 mL of 10 mM Hepes pH 7.5, 0.88 M sucrose and 0.05 mM MgSO4. The nucleolar pellet was resuspended in 100 μL of 10 mM Hepes pH 7.5, 0.34 M sucrose and 0.05 mM MgSO4. The protein content of the nuclear and nucleolar fractions was quantified using the microBCA method (Pierce) following the manufacturer's instructions.
000g and 4 °C. The main fraction, the nuclei, was recovered at the interface between the 20 and 50% percoll layers and diluted in 10 mL of 10 mM Hepes pH 7.5, 0.25 M sucrose and 10 mM MgSO4. This fraction was centrifuged for 10 minutes at 1200g through a layer of 10 mL of 10 mM Hepes pH 7.5, 0.88 M sucrose and 0.05 mM MgSO4. The pellet contained pure nuclei that were resuspended in 10 mL of 10 mM Hepes pH 7.5, 0.34 M sucrose and 0.05 mM MgSO4. A fraction was kept for further analysis and the main part was sonicated 4 times for 30 seconds at 10% power with a microtip (Sonicator BioBlock Scientific Vibracell 72412). This solution was centrifuged for 20 minutes at 2000g through a layer of 10 mL of 10 mM Hepes pH 7.5, 0.88 M sucrose and 0.05 mM MgSO4. The nucleolar pellet was resuspended in 100 μL of 10 mM Hepes pH 7.5, 0.34 M sucrose and 0.05 mM MgSO4. The protein content of the nuclear and nucleolar fractions was quantified using the microBCA method (Pierce) following the manufacturer's instructions.
2.5 Silver quantification by ICP-AES
The amount of Ag present in the cells and in nuclei and nucleoli fractions was quantified by inductively coupled plasma atomic emission spectroscopy (ICP-AES) using a Shimadzu ICPE-9800 after overnight mineralization at 50 °C in 65% HNO3.2.6 RNA extraction and quantification
At the indicated timepoints, cells were harvested and mRNA was isolated using the Nucleospin RNA kit (Macherey-Nagel) for 2D cultures or the NucleoSpin RNA Plus XS kit (Macherey-Nagel) for the spheroid samples. The RNA concentration was determined using a NanoDrop spectrophotometer (ND-1000). RNA was reverse transcribed with the Affinity script qPCR cDNA synthesis kit (Agilent), according to the manufacturer's instructions using random primers. Quantitative PCR was performed as described in ref. 21, qPCRs were run in triplicate, and quantification was performed using comparative regression (Cq determination mode) using Cfx (Bio-Rad Cfx manager) with GAPDH and HPRT amplification signals as housekeeping genes to correct for total RNA content, while the untreated sample was used as the “calibrator”. At least three biological replicates were performed for the presented data. The primers used are described in Table S2, ESI.†2.7 Nuclear receptor activity
The activity of selected nuclear receptors was determined by measuring the amount of one of their target transcripts upon addition of a specific ligand of the receptor. This activity was compared in hepatocytes exposed or not to AgNPs in order to measure the impact of these NPs on nuclear receptor activity. We followed the activity of two nuclear receptors: FXR and LXR. For the former, CDCA was used to induce the expression of bsep (coding for Bile Salt Export Pump, BSEP),28 and for the latter, 22R-HC29 was added and abcg5 (coding for the ATP-binding cassette subfamily G member 5) expression was monitored. In this case, the control (ctl) sample (unexposed to AgNPs) was set to 100% activity of FXR or LXR and it corresponds to the ratio:For all conditions of AgNP exposure, we similarly calculated the ratio:
In order to determine the percentage of activity under condition Z, we applied the following equation:
Subtracting 1 takes into account that the value of a control in mRNA normalized relative expression is 1.
2.8 Sample preparation for EM and nano-XRF
At the desired timepoint, monolayers of HepG2 cells were fixed overnight at room temperature in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio mixture of 4% paraformaldehyde, 0.4% glutaraldehyde in 0.2 M PHEM pH 7.2 and culture medium, washed in 0.1 M PHEM pH 7.2, and fixed for 30 minutes in 2% paraformaldehyde, 0.2% glutaraldehyde in 0.1 M PHEM pH 7.2, washed in 0.1 M PHEM pH 7.2 and post-fixed in 1% OsO4 in 0.1 M PHEM buffer for 1 h at room temperature. Cells were then dehydrated in graded ethanol series, and flat-embedded using an Epoxy Embedding Medium kit (Sigma-Aldrich). Sections (200 or 400 nm) were cut on a Leica UC7 ultra-microtome using a DiATOME 35° diamond knife and collected on 50 nm-thick Si3N4 grids (Oxford Instruments). The same preparation protocol was used for spheroids, where some sections were collected on formvar carbon coated 100 mesh copper grids for scanning transmission electron microscopy coupled with energy-dispersive X-ray spectroscopy (STEM-EDX) and the block was then used for focused ion beam scanning electron microscopy (FIB-SEM).
1 ratio mixture of 4% paraformaldehyde, 0.4% glutaraldehyde in 0.2 M PHEM pH 7.2 and culture medium, washed in 0.1 M PHEM pH 7.2, and fixed for 30 minutes in 2% paraformaldehyde, 0.2% glutaraldehyde in 0.1 M PHEM pH 7.2, washed in 0.1 M PHEM pH 7.2 and post-fixed in 1% OsO4 in 0.1 M PHEM buffer for 1 h at room temperature. Cells were then dehydrated in graded ethanol series, and flat-embedded using an Epoxy Embedding Medium kit (Sigma-Aldrich). Sections (200 or 400 nm) were cut on a Leica UC7 ultra-microtome using a DiATOME 35° diamond knife and collected on 50 nm-thick Si3N4 grids (Oxford Instruments). The same preparation protocol was used for spheroids, where some sections were collected on formvar carbon coated 100 mesh copper grids for scanning transmission electron microscopy coupled with energy-dispersive X-ray spectroscopy (STEM-EDX) and the block was then used for focused ion beam scanning electron microscopy (FIB-SEM).
2.9 Nano-XRF data acquisition and analysis
X-ray fluorescence (XRF) experiments were carried out at the state-of-the-art hard X-ray nanoprobe beamline ID16B at the ESRF.30 The incoming photon energy was set at 29.6 keV and the beam was focused using Kirkpatrick–Baez mirrors down to 50 × 50 nm2. The fluorescence emission from the sample was recorded using two 3-element silicon drift detector (SDD) arrays positioned at 13° from the sample. The photon flux on the sample was ∼1011 photons per s. High resolution maps were recorded at room temperature, raster scanning the sample in the X-ray focal plane with 100 × 100 nm2 step size and 500 ms dwell time/pixel.Hyperspectral images were analyzed using the PyMCA software package (http://pymca.sourceforge.net/).31 The detector response was calibrated over a thin film multilayer sample from AXO (RF8-200-S2453). XRF data were energy calibrated, normalized by the incoming photon flux, and batch-fitted in order to extract spatially resolved elemental concentrations, assuming a biological matrix of light elements and density 1 g cm−3 according to NIST standards (https://physics.nist.gov/cgi-bin/Star/compos.pl?matno=261). At least three different cells per exposure condition were analyzed, and their averages and standard deviations are reported in Fig. 2A.
2.10 X-ray absorption spectroscopy (XAS) data acquisition and analysis
Ag K-edge X-ray absorption spectra were acquired on the bending-magnet beamline BM23 of the ESRF.32 HepG2 cells were exposed to AgNPs for the indicated times, pelleted and resuspended in PBS containing 20% glycerol (20 μL). Drops of cell suspensions or reference solutions (50 μL) were deposited in the XAS sample holder equipped with Kapton windows and immediately frozen in liquid N2, then transferred into the He cryostat and measured at 10 K. The incoming photon energy was scanned around the Ag K edge (25.514 keV) with a fixed exit double crystal Si(311) monochromator (Kohzu, Japan) cooled with liquid N2. Harmonic rejection was achieved with Pt coated mirrors. Slits were opened to provide a beam size at sample of ∼3 × 1 mm2 and minimize the photon density, in order to avoid radiation damage. The energy was calibrated over an Ag foil mounted downstream to the sample. XAS spectra were acquired in fluorescence mode, with a 13-element Ge detector from Canberra. Data processing and linear combination analysis were performed with the Athena software;33 the chosen fitting range for linear combination analysis was [−20; 80] eV with respect to the edge energy.2.11 STEM-EDX
For the chemical identification of AgNPs, thin sections were observed in scanning/transmission electron microscopy (STEM) mode and analyzed by EDX using either a Zeiss Merlin microscope operated at 30 kV and fitted with two Bruker SDD detectors or a FEI/Tecnai Osiris microscope operated at 80 kV.2.12 FIB-SEM acquisition and data analysis
Focused ion beam (FIB) tomography was performed with a Zeiss NVision 40 dual-beam microscope. In a first step, the surface of the sample was protected by the deposition in the microscope of a 2 μm thick Pt or C layer, on a surface of typically 20 × 30 μm2. The resin-embedded HepG2/C3a spheroids were then cut in cross-sections, slice by slice, with a Ga+ ion beam (typically with a current of 300 or 700 pA at a beam acceleration of 30 kV). After a thin slice was removed with the ion beam, the newly exposed surface was imaged in the SEM at 1.5 kV using the in-column EsB backscatter detector. For each slice, a thickness of 8 nm was removed, and the SEM images were recorded with a pixel size of 4 × 4 nm2. The image stack was then registered by cross-correlation using the StackReg plugin in the Fiji software.34 Manual segmentation was performed in Ilastik35 and projected in three-dimension using AVIZO (FEI, USA).2.13 Reflection confocal microscopy
HepG2 cells were seeded on a two-well Thermo Scientific™ Nunc™ Lab-Tek™ chambered cover glass suitable for confocal microscopy. At 60% confluency, cells were incubated with 1 μM Hoechst 33342 for 30 minutes at room temperature then washed with complete medium. Confocal microscopy was carried out in a perfusion chamber (POC Chamber, LaCom®, Erbach, Germany) and incubation system (O2–CO2–0C, PeCom®, Erbach, Germany) that was mounted on a Leica TCS SP2 AOBS inverted laser scanning confocal microscope using a 63× oil immersion objective (HCX PL APO 63.0 × 1.40 OIL). Cells were exposed to cit-AgNPs or PVP-AgNPs at the indicated concentrations of total Ag just before the first image (t0) then images were acquired on the confocal microscope every 30 minutes and up to 8 hours for cit-AgNPs and 4 h for PVP-AgNPs, respectively. For each timepoint, a z-stack of 26 pictures with a 1 μm z-step was captured. The laser excitation was set at 351–364 nm for Hoechst, and 488 nm (25%) for the reflection settings. The fluorescence emission was set at 410–485 nm for Hoechst. The detector for reflected light was set at 480–500 nm. For co-localization with Lamp1, after the incubation with AgNPs, cells were fixed with formalin (Sigma) for 30 minutes at room temperature. Fixed cells were permeabilized with 0.5% Triton X-100 in Tris-buffered saline containing 0.05% Tween 20 for 60 minutes at room temperature and then blocked with 0.1% Triton X-100/3% BSA in PBS containing 0.05% Tween 20 for 30 minutes at room temperature. Blocked cells were incubated with antibody against Lamp1 (Santa-Cruz, sc20011) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 in antibody buffer (0.1% Triton X-100/1% BSA/0.05% Tween 20 in PBS) for 60 minutes at room temperature. Cells were then incubated with Alexa Fluor 488 goat anti-mouse secondary antibody diluted 1
50 in antibody buffer (0.1% Triton X-100/1% BSA/0.05% Tween 20 in PBS) for 60 minutes at room temperature. Cells were then incubated with Alexa Fluor 488 goat anti-mouse secondary antibody diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 (Life Technologies) in antibody buffer for 60 minutes at room temperature. For all the above-mentioned experimental conditions, cells were washed in PBS in between subsequent steps. Cells were finally imaged using a Leica TCS SP2 AOBS inverted laser scanning confocal microscope using a 63× oil immersion objective (HCX PL APO 63.0 × 1.40 OIL) as for live imaging.
1000 (Life Technologies) in antibody buffer for 60 minutes at room temperature. For all the above-mentioned experimental conditions, cells were washed in PBS in between subsequent steps. Cells were finally imaged using a Leica TCS SP2 AOBS inverted laser scanning confocal microscope using a 63× oil immersion objective (HCX PL APO 63.0 × 1.40 OIL) as for live imaging.
2.14 Statistical analysis
All the quantitative data presented with a standard deviation are based on at least three biological replicates except for XRF experiments that are based on at least three different cells. The statistical comparison of qPCR quantification is based on a standard Student t-test. The method of quantification for FXR and LXR activation is described above in this experimental section.3 Results and discussion
3.1 Hepatocyte exposure and AgNP transformations
In this study, we used 20 nm citrate-coated AgNPs (cit-AgNPs) and 90 nm PVP-coated AgNPs (PVP-AgNPs) that formed objects of 160 and 90 nm size, respectively, in cell culture media21 (Table S1, ESI†). We had previously identified concentrations of 50 and 100 μM Ag as subtoxic in HepG2 cells for citrate- and PVP-coated AgNPs, respectively.21 In the current study, the main part of the work was performed at concentrations 4-fold lower, i.e. 12.5 μM (cit-AgNP) and 25 μM (PVP-AgNP) that led to a similar Ag uptake in 24 hours. These are also non-toxic concentrations and for all the experiments performed in this study, cellular viability was between 85 and 100% based on the trypan blue assay (Fig. S1, ESI†). Besides, these conditions did not show significant stress based on heme oxygenase (HMOX) expression (Fig. S2A, ESI†). Indeed, HMOX is regulated by Nrf2, a redox stress sensor, usually activated by metal NPs.36,37 Under the chosen conditions, the only marker activated by these low AgNP concentrations was metallothionein (MT), a classical marker of metal ions release into the cytosol (Fig. S2B, ESI†). In our previous study, at subtoxic AgNP concentration, we made use of XAS analysis to quantify Ag ion release from AgNPs in hepatocytes.21 After 24 hours of exposure, between 21 and 46% of Ag species in cells were Ag–thiolate. Herein, a similar XAS analysis was performed on HepG2 cells exposed for 24 or 48 hours to 12.5 μM cit-AgNPs and for 24 hours to 25 μM PVP-AgNPs. The proportion of Ag–thiolate and AgNPs in hepatocytes was determined using the spectra of a AgNP suspension and of a Ag–GSH complex as standards (Fig. S3, ESI†). The spectra of cells showed similar features, suggesting similar dissolution fractions under the explored exposure conditions. The main features of the AgNP reference spectrum were present in the spectra of exposed cells, but heavily smoothed as a consequence of the predominance of Ag–thiolate species (represented by the Ag–GSH reference compound), as observed previously.20,21 The semi-quantitative evaluation of the different Ag species in cells obtained by linear combination fitting (Table S3, Fig. S3 red curves, ESI†) confirmed an extensive dissolution for both cit-AgNPs (12.5 μM) and PVP-AgNPs (25 μM) after only 24 hours of exposure leading to 70 and 76% of Ag(I)–thiolate complexes, respectively. This dissolution process was maintained over time because 77% of uptaken cit-AgNPs were dissolved after 48 hours of exposure. It is therefore interesting to notice that lower exposure concentrations lead to higher dissolution of AgNPs into Ag(I) ions since we previously showed that not more than 50% of internalized AgNPs were dissolved after 24 h of exposure to 100 μM AgNPs.21 Altogether, these data showed that AgNPs rapidly dissolved upon uptake and continuously over time under exposure to low and non-toxic concentrations of AgNPs where Ag(I)–thiolate is the main form of Ag in hepatocytes.3.2 Distribution of Ag species within hepatocytes upon exposure to low AgNP concentrations
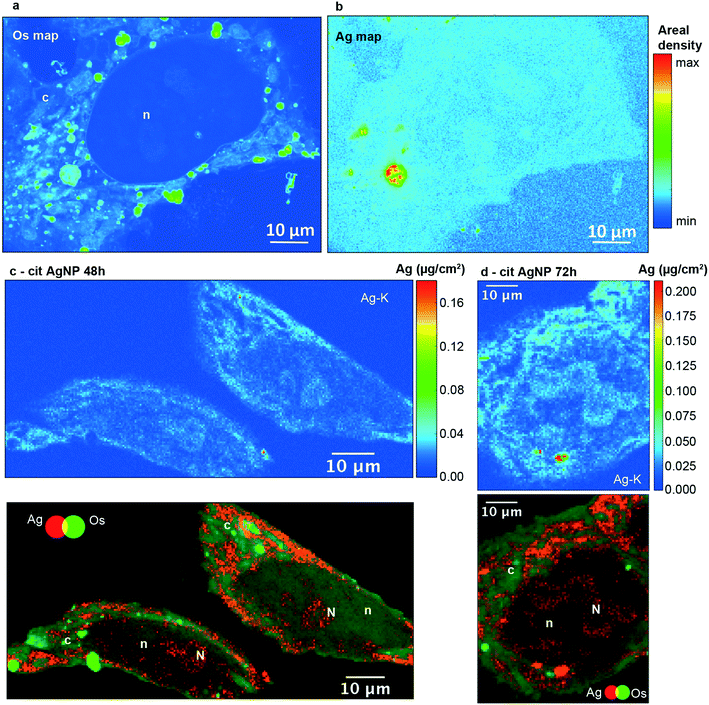
In order to visualize the subcellular distribution of both AgNPs and Ag–thiolates, we performed X-ray fluorescence (XRF) nano-imaging on thin cell sections. XRF imaging is an elemental analytical technique that cannot distinguish between different Ag species. However, thanks to the combination with XAS and electron microscopy, and to the comparison with Ag distribution after exposure to Ag-salts, we could previously assign the XRF signals in entire cells to different Ag species: intense, confined Ag signals were assigned to AgNPs, whereas a weak signal diffused throughout the cell was assigned to Ag–thiolates.21 In the present work, XAS showed that AgNPs and Ag(I)–thiolates are the main existing species, and we made use of different microscopy techniques to localize these species within the cell. XRF images of 200 nm sections yielded an extremely detailed picture of the inside of the cell, with the clear identification of various vesicles and compartments by OsO4 staining (Fig. 1A). In HepG2 cells exposed to low concentrations of AgNPs over three days, we observed Ag hot spots inside vesicles, most probably corresponding to pristine or partially dissolved NPs (Fig. 1B, red-yellow pixels). Using confocal microscopy, both citrate and PVP AgNPs can be detected in reflection mode (Fig. S4, ESI,† red) and they were often observed to be co-localized with Lamp1 stained vesicles, thus identifying such vesicles as lysosomes (Fig. S4, ESI,† green, co-localization red-green highlighted with white arrows). In addition, under the same experimental conditions, similar perinuclear lysosome vesicles containing dark particles were observed by STEM (Fig. S5 and S6, ESI†). EDX showed that they contained Ag as well as sulfur in the case of both cit-AgNPs and PVP-AgNPs (Fig. S5 and S6, ESI†). A variable proportion of the detected elements was observed within the lysosome, with some areas of stronger co-localization (for instance in Fig. S5B, ESI,† lower panels comparing Ag and S EDX maps). This lets us hypothesize the formation in lysosomes of transient species in-between the pristine AgNPs and the Ag(I)–thiolate complexes revealed by XAS, which would represent the first step of surface oxidation necessary for Ag(I) ion release.38,39 However, considering that XAS proved that Ag(I)–thiolate species represent 70–75% of all Ag atoms in cells, we must exclude that all the thiolates are simply adsorbed at the surface of AgNPs, in particular if we consider that the fraction of surface Ag atoms in a 20 nm NP is about 5%. Such a high proportion of Ag–thiolate complexes can form only if Ag(I) ions are released from AgNPs. X-ray fluorescence images are consistent with this hypothesis, since different levels of intensity in the Ag signal appear within lysosomes. Our data suggest that AgNP dissolution and Ag(I) release is initiated in lysosomes, consistent with XRF images showing a much higher Ag level in lysosomes compared to the cytosol, where it was homogeneously low (Fig. 1B, green compared to light blue pixels). Therefore, Ag(I) species released from AgNPs inside lysosomes are not free to leave these vesicles, but are under the control of limiting transport mechanisms that may involve metal-ion transporters.40 Besides, the low intensity Ag signal visualized all over the cell and in particular in the cytosol (Fig. 1B, light blue pixels) can be assigned to Ag(I)–thiolate molecular complexes. This conclusion is supported by the evidence that the low intensity XRF signal is undetectable by EDX (Fig. S5 and S6, ESI†) and the XAS data show that more than 70% of total intracellular Ag is Ag(I)–thiolate (Table S3 and Fig. S3, ESI†).The cell nucleus was easily identified by its low retention of Os staining (Fig. 1A, denoted as n) and a low but significant signal for Ag was observed in this organelle (Fig. 1B). In order to perform an accurate elemental quantification, 400 nm thick sections were used. Ag was observed in the nuclei of cells exposed for 48, 72 and 96 hours to cit-AgNPs (Fig. 1C and D and S7A and B, ESI†) and 72 hours to PVP-AgNPs (Fig. S7C, ESI†). Moreover, the superimposition of Os (green) and Ag (red) maps revealed areas in the nucleus with higher Ag content (Fig. 1C and D and S7A–C, denoted as N, ESI†). These regions were putatively identified as nucleoli, which are usually observed as darker areas by electron microscopy.41 The presence of Ag in these areas was confirmed by a specific XRF Ag-Kα peak (Fig. S8, ESI†).
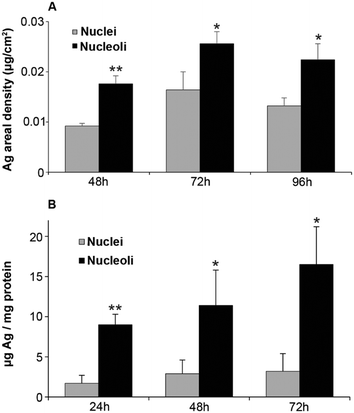
Between 48 and 72 hours of exposure (Fig. 2A and S9, ESI†), Ag concentrations increased in the nuclei and in the putative nucleoli structures, while a slight decrease occurred after 72 hours. In the case of cit-AgNPs, the amount of Ag per million cells increased at a slower rate than with PVP-AgNPs, and both reached a plateau around 0.7–0.8 μg Ag per 106 cells that corresponds to about 4 ×109 Ag atoms per cell (Fig. S10, ESI†). The uptake of cit-AgNPs between 72 and 96 hours of exposure is thus limited. It is therefore possible that internalized silver is distributed to other organelles between 72 and 96 hours explaining this slight decrease in the nucleus.
In order to confirm these data, we quantified Ag in nuclei and bound to nucleoli following subcellular fractionation after 24, 48 and 72 hours of exposure. In order to avoid contamination of the nuclei fraction by endosomes and lysosomes loaded with dense AgNPs, sucrose/percoll density gradient centrifugation was performed. A continuous uptake of Ag in nuclei was observed over 72 hours (Fig. 2B). Nucleoli clearly accumulated Ag since elemental quantification normalized on the protein amount showed about five times higher Ag concentrations compared to the total nuclei fraction with the same kinetic trend (Fig. 2B). This quantification is not absolute and the ratio of nucleoli/nuclei is higher than that based on XRF quantification but both methods show consistently a higher amount of Ag in nucleoli.
Altogether, our study is the first to show that Ag(I) species enter nuclei and are preferentially accumulated in nucleoli. These novel results are supported by the identification of proteins from the nucleoli possessing Ag-affinity in vitro, the so-called AgNOR (Nucleolar Organizer Region)42,43 that most probably bind Ag via stretches of glutamate or aspartate residues.44 However, carboxylates do not possess a high affinity for Ag(I), making Ag-rich nucleoli a reservoir of Ag(I) that can be eventually released in the nucleus and bind higher affinity protein sites.
3.3 Impact of Ag(I) species on nuclear functions
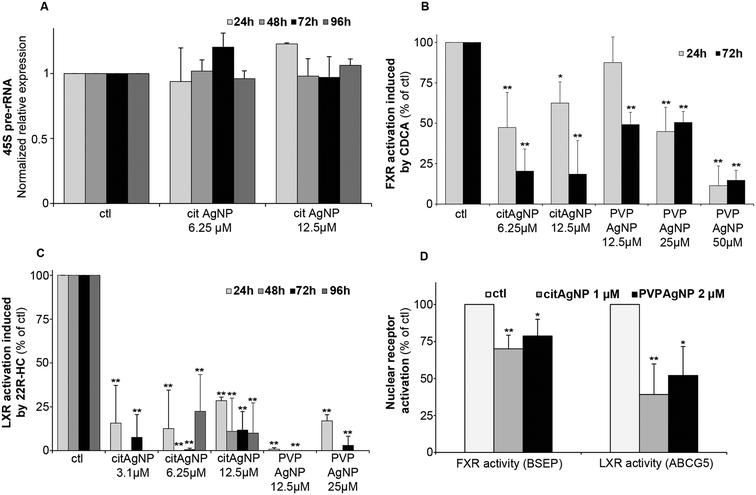
The initial phase of ribosome biogenesis takes place in nucleoli,43,45 in particular with the biosynthesis of the 45S pre-ribosomal RNA (pre-rRNA). We assessed a possible impact of Ag binding to nucleolar proteins on this function. The amount of 45S pre-rRNA was determined by quantitative real-time PCR (qPCR) in HepG2 cells exposed or not to cit-AgNPs at 6.25 and 12.5 μM for up to 4 days (Fig. 3A). 45S pre-rRNA production remained almost constant under the tested conditions. We only observed a slight overexpression after 24 hours of exposure to cit-AgNPs at 12.5 μM. It is possible that Ag binding to AgNOR slightly disturbs nucleolar functions but as ribosomes are essential, cells have no doubt developed ways to efficiently correct for alteration in their biogenesis pathway. In addition, Ag binding to AgNOR occurs in domains with unknown functions for these proteins, which could explain a limited impact on ribosome biogenesis.The nucleus carries out many critical functions and any disruption can play a role in the development of non-transmissible pathologies such as cancers. Environmental pollutants, those classified as endocrine disruptors for instance, impair the activity of some nuclear receptors (for review see ref. 46). These are ligand-activated transcription factors with finely tuned activity. They contain zinc finger DNA binding domains that are structured by zinc ions coordinated by four cysteines (Cys). This amino-acid possesses thiol functions that are very good ligands for Cu(I) or Ag(I) ions and it has been observed in vitro that Cu(I) can displace Zn(II) from zinc finger domains.47–49 Recently, it was shown that an excess of intracellular Cu(I) in Wilson disease impairs the function of nuclear receptors in the liver.28,50,51 To assess the activity of crucial liver nuclear receptors, we determined transcript levels of target genes under the control of these receptors upon addition of their specific ligands under standard growth conditions (100% activation) versus those in cells exposed to AgNPs (Fig. 3B–D). Farnesyl X receptor (FXR) is critical in maintaining bile salt homeostasis, and the addition of chenodeoxycholic acid (CDCA) into the culture medium triggers the expression of bile salt export protein (BSEP) mRNA.28 The exposure of HepG2 cells to cit-AgNPs at 6.25 or 12.5 μM for 24 hours inhibited about half of FXR's activation and a prolonged exposure to 72 hours led to 80% inhibition (Fig. 3B). PVP-AgNPs possessed a lower inhibitory capacity leading to only about 50% inhibition after 72 hours at 12.5 and 25 μM, and 50 μM was required to inhibit more than 85% of FXR's activity (Fig. 3B). The level of FXR mRNA expression was also determined by qPCR under the different exposure conditions (Fig. S11A, ESI†). It was observed that CDCA itself induced a slight down-regulation of FXR expression most probably due to a feedback loop avoiding hyper-activation of FXR. The CDCA-induced down-regulation was observed under all tested conditions except at 50 μM PVP-AgNPs. Moreover, cit-AgNPs at 12.5 μM and PVP-AgNPs at 50 μM induced a moderate and high down-regulation of FXR, respectively, most probably because of the stress induced to hepatocytes by these concentrations of AgNP exposure as shown by HMOX low and moderate mRNA overexpression, respectively (Fig. S2A, ESI†). Overall, for an unknown reason, FXR is more sensitive to cit-AgNPs than to PVP-AgNPs.
Liver X receptor (LXR) is a key nuclear receptor for liver metabolism as it is a major regulator of glucose, lipid and cholesterol metabolisms. LXR can be activated by various ligands including cholesterol derivatives such as 22R-hydroxycholesterol (22R-HC)29 that induces the expression of ABCG5, an ABC transporter involved in biliary cholesterol excretion. Just like FXR, the exposure of HepG2 cells to cit-AgNPs for only 24 hours at 6.25 or 12.5 μM inhibited 70–85% of LXR's activity (Fig. 3C). Moreover, a 48 hour exposure to AgNPs was enough to completely abolish LXR activity (Fig. 3C) and a slightly lower inhibition was observed for a 96 hour exposure to 6.25 μM, most probably because of the redistribution of Ag throughout the cell as shown by nano-XRF (Fig. 2A). Similar results were observed with PVP-AgNPs with a complete inhibition of LXR at 12.5 μM (Fig. 3C). In order to exclude a direct effect on LXR expression, we quantified LXR mRNA by qPCR. For LXR, we were able to detect both LXRα and LXRβ mRNAs in HepG2 cells and we choose to follow LXRβ expression under our various exposure conditions (Fig. S11B, ESI†). In contrast to FXR, the ligand 22R-HC did not induce a down-regulation of LXRβ expression. Cit-AgNPs at 3.1 μM did not affect LXRβ expression, while concentrations of 6.25 and 12.5 μM induced only a slight down-regulation between 24 and 72 hours of exposure that was lost for 96 hours of exposure (Fig. S11B, ESI†). In the case of PVP-AgNP exposure, only 12.5 μM for 72 hours induced a slight down-regulation of LXRβ, while the level of mRNA was quite stable under all other conditions.
The concentration of cit-AgNPs was then decreased down to 3.1 μM Ag in order to observe effects on LXR activity that could not be related to metal stress in hepatocytes. Under these conditions, MTs were only minimally overexpressed (Fig. S2B, ESI†) and LXRβ was very stable (Fig. S11B, ESI†). Interestingly, such a low concentration of AgNPs led to about 80 and 90% inhibition of LXR after 24 and 72 hours of exposure (Fig. 3C), respectively. Thus, we prove that the exposure to low and non-toxic concentrations of AgNPs impairs nuclear receptor activity in hepatocytes with an extent of inhibition that is receptor-dependent. Indeed, we observed a complete inhibition of LXR and only a partial inhibition of FXR that could be due to different subcellular localizations of these two nuclear receptors under basal conditions.
Finally, in order to very closely mimic chronic exposure to low but constant levels of Ag, we studied exposure to 1 μM cit-AgNPs or 2 μM PVP-AgNPs that led to a low but detectable intracellular Ag concentration. Its level stabilized after 7 to 10 days around 45 and 70 ng Ag per 106 cells (between 2.5 and 4 × 108 Ag atoms per cell) for cit-AgNPs and PVP-AgNPs, respectively (Fig. S12, ESI†), while lower and higher concentrations delivered undetectable and time-dependent increasing amounts of Ag in hepatocytes, respectively. In the latter case, critical Ag levels were reached at one point making it impossible to work under chronic-mimicking conditions as AgNP exposure concentrations ended up being too high, while 1 μM cit-AgNPs and 2 μM PVP-AgNPs were ideal to approach this objective. Similar effects were observed with 1 μM cit-AgNP and 2 μM PVP-AgNP exposure with only a moderate inhibition of FXR, 30 and 20%, respectively, and a significant inhibition of LXR, 60 and 50%, respectively (Fig. 3D). These results confirmed the lower sensitivity of FXR to Ag(I) and also proves that chronic exposure to AgNPs delivered sufficient amounts of Ag(I) to disrupt nuclear receptor activity. Overall, the experiments on nuclear functions did not show major differences between AgNPs with different coatings except a slightly higher concentration required for PVP to reach the same level of toxicity.
3.4 From AgNP endocytosis to Ag(I) nuclear entry, a rapid process
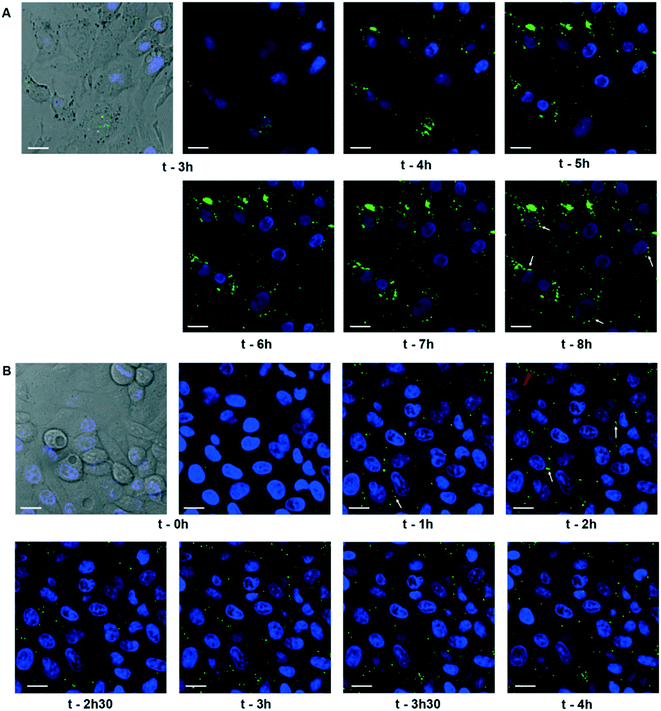
Our data have clearly demonstrated the rapid uptake by nuclei of a significant amount of Ag(I) ions that accumulate in nucleoli and induce dose-dependent effects on nuclear functions (Fig. 1–3). A debate is still ongoing concerning the possible uptake of the nanoparticulate form of AgNPs in the nuclei, but the methods used to demonstrate this hypothesis cannot be considered as conclusive (for detailed explanations, see the recent review in ref. 4). In order to deepen our understanding of AgNP trafficking towards nuclei, we performed combined confocal reflected light microscopy and 3D-EM analysis using FIB-SEM. The particle nature of AgNPs makes it possible to visualize their dynamics by reflectance confocal microscopy (Fig. 4 and S13–S15, ESI†).52 Cit-AgNPs could only be detected in cells three hours after exposure, while PVP-AgNPs were observed as clear single spots immediately following their addition (Fig. 4A and S13, ESI†versus4B and S14, ESI†). This is consistent with our previous observations of isolated PVP-AgNPs as opposed to agglomerated cit-AgNPs during endocytosis21 and with the kinetics of uptake of both types of AgNPs (Fig. S10, ESI†). We also recently published electron microscopy micrographs revealing PVP-AgNPs bound to the cell membrane at the interface between cells.4 This observation was made on thin cell sections and was specific to PVP-AgNPs. Similarly, confocal reflectance microscopy images showed particles evenly distributed on lines or curves between cells (Fig. 4B and S14, red arrows, ESI†) in addition to intracellular particles.Therefore, for an unknown reason, we confirmed that a fraction of the PVP-AgNPs but not of the cit-AgNPs binds to membranes between cells as described in Marchioni et al.4 For cit-AgNPs, the reflection signal in the cell increased over time with a tendency to accumulate in the perinuclear region (Fig. 4A and S13, white arrows, ESI†). In the case of PVP-AgNPs, the dynamics were faster and NPs that came in the vicinity of nuclei disappeared rapidly (Fig. 4B and S14, white arrows, ESI†). Therefore, within just a few hours, AgNPs reached the vicinity of the nucleus where they promptly dissolved. In order to go a step further, the PVP-AgNP distribution within hepatocytes grown as spheroids was also obtained at nanometer resolution using FIB-SEM. This method provided the unique capability to observe individual NPs without any preparation-dependent artifacts that might lead to erroneous localization of NPs within the cell. Indeed, the biological sample is embedded in a resin and imaged 8 nm layer by 8 nm layer thanks to Ga gun nanomachining (for review see ref. 53) thus avoiding diamond knife alterations of the AgNP location. Moreover, for this experiment, a derivative of the HepG2 cell line, named HepG2/C3a,54 was used to grow hepatocytes in 3D as spheroids, as this cell line is capable of forming functional bile canaliculi in 3D culture, providing a model closer to functional tissue. Similar to our data on 2D cell culture, FXR activity was readily inhibited in hepatocytes grown as spheroids after 48 hours of exposure to 12.5 μM cit-AgNPs or 25 μM PVP-AgNPs (Fig. S16, ESI†) as observed in 2D culture (Fig. 3B). These results increase the relevance of our data on nuclear receptor inhibition upon AgNP exposure. FIB-SEM analysis revealed that PVP-AgNPs were only observed in vesicles, most probably endosomes and/or lysosomes (Fig. 5A white arrows and Video S1, ESI†). Moreover, in accordance with our previous data (Fig. S5 and S6, ESI†), AgNPs were frequently found in lysosomes close to the nucleus, as shown in the 3D reconstruction of a typical hepatocyte (Fig. 5B and C and Video S1, ESI†) as well as in thin sections analyzed by STEM-EDX that confirmed the dark particles to be transformed AgNPs, since Ag and S were detected (Fig. S17, ESI†). These results confirmed the reflection confocal microscopy data adding nanometer resolution. Thorough analysis of the original SEM micrographs used to make the 3D reconstruction confirmed the proximity between AgNP-containing vesicles and the nucleus, showing close contacts between both membranes (Fig. 5D, red arrow). However, AgNPs were never observed inside nuclei.
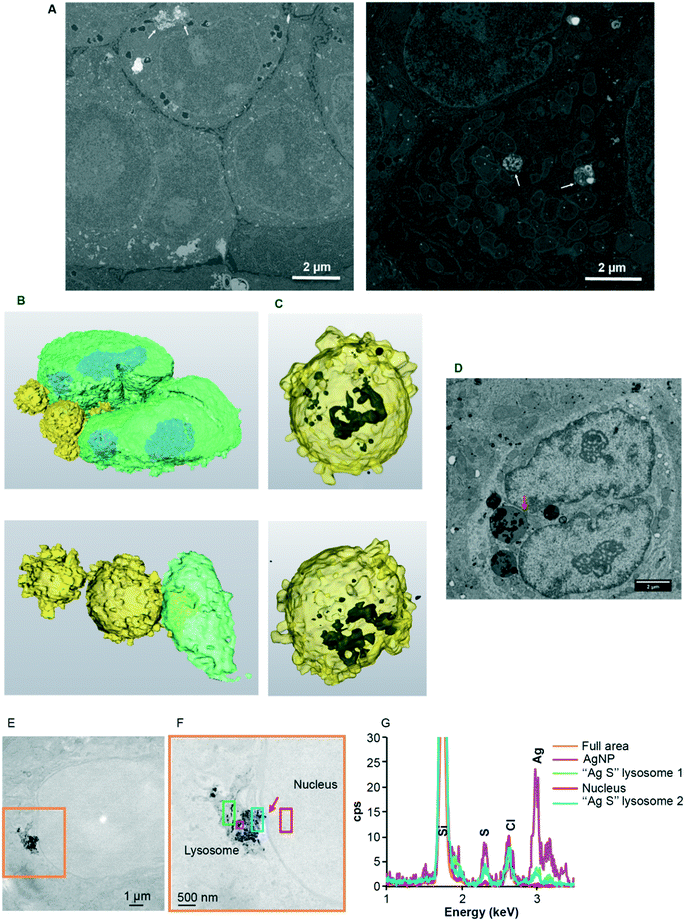 | ||
| Fig. 5 EM analysis of the subcellular distribution of AgNP-containing vesicles. (A) Original micrographs extracted from two different FIB-SEM stacks of images (see also Video S1† for stack 1 with the corresponding segmentation). Endolysosomes containing AgNPs close to the nucleus (left) and in the middle of the mitochondrial network (right) are indicated with white arrows. (B) 3D reconstruction of a nucleus (green) with bound vesicles (yellow) containing AgNPs (black) seen by transparency (C). (D) SEM micrograph from the stack used to make the 3D reconstruction (B and C). This micrograph shows a direct contact between the AgNP-containing vesicle and the nucleus (red arrow). Scale bars represent 2 μm. (E) Large field of view STEM micrograph showing the presence of a lysosome containing NPs in contact with a nucleus in HepG2 cells exposed for 72 hours to 12.5 μM cit-AgNPs. (F) STEM micrograph of the orange area in (E) at higher magnification. The red arrow pinpoints a region of close contact between the lysosome and the nuclei. The different colored boxes correspond to areas scanned by EDX with the spectra presented in (G). (G) EDX spectra of different regions in the energy range 1–3.5 keV. The positions of Si, S, Cl and Ag peaks are highlighted. | ||
It must be considered that XAS analysis (section 3.1) proved that AgNPs represent only ∼30% of the total Ag species encountered in cells 24 h after exposure, whereas Ag–thiolate complexes represent ∼70% of species. This extent of dissolution is consistent with the observed perinuclear localization of AgNP-containing endolysosomes. Indeed, the closer the lysosomes are to the nucleus, the lower their internal pH (ref. 55) and lower pH values favor AgNP dissolution. Nonetheless, the combination of all our data prompted us to reconsider our vision on AgNP dissolution. Indeed, different forms of species containing Ag and S exist within the cell. XRF microscopy showed intense Ag signals within endosomes and lysosomes and low Ag signals in the cytosol and other organelles such as nuclei. However, STEM-EDX experiments revealed that lysosomes contain both AgNPs close to the pristine form and highly transformed AgNPs (Fig. 5 and S5 and S6, ESI†). This is particularly clear in Fig. 5E–G where darker particles have a high signal of Ag (pink square in Fig. 5F), while grey particle-like areas have a low Ag signal with the presence of S in a similar intensity (blue and green squares in Fig. 5F). Interestingly, in Fig. 5F, we can almost see a connection between a lysosome and the nucleus (red arrow) but no particle nor Ag signal was observed in the nucleus even in areas that look darker in STEM. Similarly, Ag was never detected in the cytosol by EDX, while it was by XRF both in the cytosol and nuclei. Species containing Ag and S thus exist in two forms, particle-like in lysosomes and soluble elsewhere. Therefore, our results constitute the first evidence of a rapid trafficking of endosomes maturing into lysosomes that contain AgNPs under conditions favoring their rapid transformation into Ag(I) species, which are then partly transferred as soluble complexes into the nucleus where they bind and inhibit nuclear receptors. Mechanisms governing the shuttling of nuclear receptors between the cytoplasm and nucleus are complex and not fully understood,56,57 and are partly receptor-specific. In the case of LXR, the metabolic status of the cell (i.e. glucose level) can favor LXR nuclear translocation.58 Therefore, the fast kinetics of Ag(I) nuclear transfer provides one possible explanation for the difference in sensitivity to AgNPs between FXR and LXR. Indeed, Ag(I) directly delivered to the nucleus could rapidly and efficiently bind and inhibit nuclear LXR.
4 Conclusions
In the current study, we developed an innovative elemental imaging methodology that we combined with complementary imaging approaches, altogether enabling unprecedented localization of both AgNPs and Ag(I) species at the organelle level. AgNPs were only observed in endosomal and lysosomal vesicles, which move towards a perinuclear localization. The combination of methods revealed the transformation of AgNPs into dense species containing Ag and S within lysosomes. Highly sensitive elemental imaging revealed a direct and rapid entry of soluble Ag(I) species into the nucleus where it accumulates, weakly bound to nucleoli, and impairs nuclear receptor activity. Indeed, one day of exposure is sufficient to partially or totally inhibit a set of nuclear receptors under conditions very close to medical device exposure.14–16 Besides, extremely low doses leading to less than 50 fg of silver per cell are sufficient to inhibit more than half of LXR activity, a key regulator of liver metabolism.Based on previously published in vitro data47–49 and on our recent study59 we can ascribe the observed effects on nuclear receptor activity to a displacement of Zn(II) by Ag(I) in the zinc finger DNA binding domains, as already proposed with Cu(I) in a mouse model of Wilson disease.28,50,51 Zinc fingers require a tetra-coordination geometry to be able to bind their specific DNA-binding domains upon ligand binding. We showed that Ag(I) ions displace Zn(II) from a model zinc finger triggering the formation of an Ag4S4 cluster, which alters the finger structure.59 This in vitro data provides the basis to explain the inhibition of nuclear receptor activity observed upon AgNP exposure. The impairment of this activity is highly detrimental to liver metabolism as it will affect lipid, cholesterol and bile salt homeostasis, among others. Moreover, released Ag(I) ions could also interact with other nuclear receptors, possibly leading to developmental diseases46 such as those observed with endocrine disruptors. It could therefore be of high relevance to study the impact of the combined exposure to AgNPs and endocrine disruptors that target the same protein family. In addition, the binding of released Ag(I) cannot be limited to nuclear receptors. Zinc finger proteins account for 3% of the human genome60 and many sites with two or three exposed thiols exist in proteins. It would therefore be of high interest to determine the whole Ag metalloproteome upon realistic exposure to dietary or medical AgNPs.
In conclusion, the current work is a pioneer study in understanding physiological disruptions induced by exposure to very low doses of AgNPs mimicking chronic exposure. Further work is now required to precisely determine our exposure levels in order to clearly define an AgNP safety framework. In this context, we recently developed robust, versatile and safer biocides based on assemblies of AgNPs.61 Moreover, the idea of using AgNPs in cancer therapy recently emerged62 and this work on a detailed analysis of AgNP trafficking and transformation in cells can be highly valuable for these types of development. Once again, deciphering toxicity mechanisms of nanomaterials could lead to nanomedicine applications as described recently by Peng et al. who made use of nanomaterial-induced endothelial leakiness to favor cancer therapy.63
Author contributions
AD designed the study and interpreted the data. AD, AF, GV and WB wrote the paper. AD, MC, WB and GV contributed to data interpretation. AD and MC performed cellular experiments including sample preparation for imaging. BG and CM optimized XRF sample preparation and prepared samples for imaging. GV, VTS and RT participated in synchrotron nanoprobe experiments, data acquisition and analysis, and interpretation of the results. GV performed XAS experimental design, data acquisition and analysis. PHJ, AD and EK performed STEM-EDX and FIB-SEM experimental design, data acquisition and analysis. CCR and MC performed confocal microscopy and image analysis. AD, PC and MC performed qPCR experiments and analysis. AF and MC implemented spheroid preparation. IMS contributed to nanomaterial and nanotoxicology expertise.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the European Synchrotron Radiation Facility for providing beam time (in-house research on ID16B-NA and experiment LS-2639 on BM23), and O. Mathon for the excellent support during XAS data acquisition. This work used the platforms of the Grenoble Instruct-ERIC Center (ISBG: UMS 3518 CNRS-CEA-UGA-EMBL) with support from the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-05-02) and GRAL, a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003) within the Grenoble Partnership for Structural Biology (PSB). The IBS Electron Microscopy facility is headed by Guy Schoehn and supported by the Auvergne Rhône-Alpes Region, the Fondation Recherche Medicale (FRM), the fonds FEDER and the GIS-Infrastructures en Biologie Sante et Agronomie (IBISA). This work is also a contribution to the Labex Serenade (ANR-11-LABX-0064) funded by the French Government’s “Investissements d’Avenir” ANR program, through the A*MIDEX project (ANR-11-IDEX-0001-02). This research is part of the LabEx ARCANE and CBH-EUR-GS (grant ANR-17-EURE-0003). This work was supported by the CEA Transversal Programs Toxicology and Nanoscience through the NanoTox-RX grant and the CEA DRF-Impulsion grant FIB-Bio. This work was also funded by the Université Grenoble Alpes – AGIR grant NanoSilverSafe and IRS NanoBIS grant and by the RHU program ANR-16-RHUS-0005. EK was supported by a CMIRA Accueil Sup 2016 fellowship.References
- J. Liu, D. A. Sonshine, S. Shervani and R. H. Hurt, Controlled release of biologically active silver from nanosilver surfaces, ACS Nano, 2010, 4, 6903–6913 CrossRef CAS PubMed.
- Z. M. Xiu, Q. B. Zhang, H. L. Puppala, V. L. Colvin and P. J. Alvarez, Negligible particle-specific antibacterial activity of silver nanoparticles, Nano Lett., 2012, 12, 4271–4275 CrossRef CAS PubMed.
- V. De Matteis, M. A. Malvindi, A. Galeone, V. Brunetti, E. De Luca, S. Kote, P. Kshirsagar, S. Sabella, G. Bardi and P. P. Pompa, Negligible particle-specific toxicity mechanism of silver nanoparticles: the role of Ag+ ion release in the cytosol, Nanomedicine, 2015, 11, 731–739 CrossRef CAS PubMed.
- M. Marchioni, P.-H. Jouneau, M. Chevallet, I. Michaud-Soret and A. Deniaud, Silver nanoparticle fate in mammals: Bridging in vitro and in vivo studies, Coord. Chem. Rev., 2018, 364, 118–136 CrossRef CAS.
- S. W. P. Wijnhoven, W. J. G. M. Peijnenburg, C. A. Herberts, W. I. Hagens, A. G. Oomen, E. H. W. Heugens, B. Roszek, J. Bisschops, I. Gosens, D. Van De Meent, S. Dekkers, W. H. De Jong, M. van Zijverden, A. J. A. M. Sips and R. E. Geertsma, Nano-silver – a review of available data and knowledge gaps in human and environmental risk assessment, Nanotoxicology, 2009, 3, 109–138 CrossRef CAS.
- K. Kulthong, S. Srisung, K. Boonpavanitchakul, W. Kangwansupamonkon and R. Maniratanachote, Determination of silver nanoparticle release from antibacterial fabrics into artificial sweat, Part. Fibre Toxicol., 2010, 7, 8 CrossRef PubMed.
- M. E. Quadros, R. T. Pierson, N. S. Tulve, R. Willis, K. Rogers, T. A. Thomas and L. C. Marr, Release of silver from nanotechnology-based consumer products for children, Environ. Sci. Technol., 2013, 47, 8894–8901 CrossRef CAS PubMed.
- B. Reidy, A. Haase, A. Luch, K. A. Dawson and I. Lynch, Mechanisms of Silver Nanoparticle Release, Transformation and Toxicity: A Critical Review of Current Knowledge and Recommendations for Future Studies and Applications, Materials, 2013, 6, 2295–2350 CrossRef CAS PubMed.
- S. Agnihotri, S. Mukherji and S. Mukherji, Immobilized silver nanoparticles enhance contact killing and show highest efficacy: elucidation of the mechanism of bactericidal action of silver, Nanoscale, 2013, 5, 7328–7340 RSC.
- K. Wu, Y. Yang, Y. Zhang, J. Deng and C. Lin, Antimicrobial activity and cytocompatibility of silver nanoparticles coated catheters via a biomimetic surface functionalization strategy, Int. J. Nanomed., 2015, 10, 7241–7252 CAS.
- S. Nakamura, M. Sato, Y. Sato, N. Ando, T. Takayama, M. Fujita and M. Ishihara, Synthesis and Application of Silver Nanoparticles (Ag NPs) for the Prevention of Infection in Healthcare Workers, Int. J. Mol. Sci., 2019, 20, 3620 CrossRef CAS PubMed.
- K. Park, E. J. Park, I. K. Chun, K. Choi, S. H. Lee, J. Yoon and B. C. Lee, Bioavailability and toxicokinetics of citrate-coated silver nanoparticles in rats, Arch. Pharmacal Res., 2011, 34, 153–158 CrossRef CAS PubMed.
- P. Bove, M. A. Malvindi, S. S. Kote, R. Bertorelli, M. Summa and S. Sabella, Dissolution test for risk assessment of nanoparticles: a pilot study, Nanoscale, 2017, 9, 6315–6326 RSC.
- E. C. M. Mimura, J. P. M. Favoreto, M. E. Favero, K. L. Bonifacio, T. S. Peixe, A. A. Morita, D. S. Barbosa, M. J. S. Yabe and A. J. F. Carrilho, Silver serum levels in burned patients treated with silver sulfadiazine and its toxicity on inflammatory cells, Burns, 2019 DOI:10.1016/j.burns.2019.11.012.
- M. Trop, M. Novak, S. Rodl, B. Hellbom, W. Kroell and W. Goessler, Silver-coated dressing acticoat caused raised liver enzymes and argyria-like symptoms in burn patient, J. Trauma: Inj., Infect., Crit. Care, 2006, 60, 648–652 CrossRef PubMed.
- A. T. Wan, R. A. Conyers, C. J. Coombs and J. P. Masterton, Determination of silver in blood, urine, and tissues of volunteers and burn patients, Clin. Chem., 1991, 37, 1683–1687 CrossRef CAS.
- A. R. Gliga, S. Skoglund, I. O. Wallinder, B. Fadeel and H. L. Karlsson, Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- X. Jiang, T. Miclăuş, L. Wang, R. Foldbjerg, D. S. Sutherland, H. Autrup, C. Chen and C. Beer, Fast intracellular dissolution and persistent cellular uptake of silver nanoparticles in CHO-K1 cells: implication for cytotoxicity, Nanotoxicology, 2014, 9, 181–189 CrossRef PubMed.
- R. P. Singh and P. Ramarao, Cellular uptake, intracellular trafficking and cytotoxicity of silver nanoparticles, Toxicol. Lett., 2012, 213, 249–259 CrossRef CAS PubMed.
- G. Veronesi, C. Aude-Garcia, I. Kieffer, T. Gallon, P. Delangle, N. Herlin-Boime, T. Rabilloud and M. Carriere, Exposure-dependent Ag+ release from silver nanoparticles and its complexation in AgS2 sites in primary murine macrophages, Nanoscale, 2015, 7, 7323–7330 RSC.
- G. Veronesi, A. Deniaud, T. Gallon, P. H. Jouneau, J. Villanova, P. Delangle, M. Carriere, I. Kieffer, P. Charbonnier, E. Mintz and I. Michaud-Soret, Visualization, quantification and coordination of Ag+ ions released from silver nanoparticles in hepatocytes, Nanoscale, 2016, 8, 17012–17021 RSC.
- L. Wang, T. Zhang, P. Li, W. Huang, J. Tang, P. Wang, J. Liu, Q. Yuan, R. Bai, B. Li, K. Zhang, Y. Zhao and C. Chen, Use of Synchrotron Radiation-Analytical Techniques To Reveal Chemical Origin of Silver-Nanoparticle Cytotoxicity, ACS Nano, 2015, 9, 6532–6547 CrossRef CAS PubMed.
- N. W. H. Adams and J. R. Kramer, Potentiometric Determination of Silver Thiolate Formation Constants Using a Ag2S Electrode, Aquat. Geochem., 1999, 5, 1–11 CrossRef CAS.
- P. V. AshaRani, G. Low Kah Mun, M. P. Hande and S. Valiyaveettil, Cytotoxicity and genotoxicity of silver nanoparticles in human cells, ACS Nano, 2009, 3, 279–290 CrossRef CAS PubMed.
- H. R. Kim, M. J. Kim, S. Y. Lee, S. M. Oh and K. H. Chung, Genotoxic effects of silver nanoparticles stimulated by oxidative stress in human normal bronchial epithelial (BEAS-2B) cells, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2011, 726, 129–135 CrossRef CAS PubMed.
- A. Azioune, N. Carpi, Q. Tseng, M. Théry and M. Piel, in Methods in Cell Biology, ed. L. Cassimeris and P. Tran, Academic Press, 2010, vol. 97, pp. 133–146 Search PubMed.
- A. Scherl, Y. Couté, C. Déon, A. Callé, K. Kindbeiter, J.-C. Sanchez, A. Greco, D. Hochstrasser and J.-J. Diaz, Functional Proteomic Analysis of Human Nucleolus, Mol. Biol. Cell, 2002, 13, 4100–4109 CrossRef CAS PubMed.
- C. R. Wooton-Kee, A. K. Jain, M. Wagner, M. A. Grusak, M. J. Finegold, S. Lutsenko and D. D. Moore, Elevated copper impairs hepatic nuclear receptor function in Wilson's disease, J. Clin. Invest., 2015, 125, 3449–3460 CrossRef PubMed.
- E. Kaneko, M. Matsuda, Y. Yamada, Y. Tachibana, I. Shimomura and M. Makishima, Induction of intestinal ATP-binding cassette transporters by a phytosterol-derived liver X receptor agonist, J. Biol. Chem., 2003, 278, 36091–36098 CrossRef CAS PubMed.
- G. Martinez-Criado, J. Villanova, R. Tucoulou, D. Salomon, J. P. Suuronen, S. Laboure, C. Guilloud, V. Valls, R. Barrett, E. Gagliardini, Y. Dabin, R. Baker, S. Bohic, C. Cohen and J. Morse, ID16B: a hard X-ray nanoprobe beamline at the ESRF for nano-analysis, J. Synchrotron Radiat., 2016, 23, 344–352 CrossRef CAS PubMed.
- V. A. Solé, E. Papillon, M. Cotte, P. Walter and J. Susini, A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra, Spectrochim. Acta, Part B, 2007, 62, 63–68 CrossRef.
- O. Mathon, A. Beteva, J. Borrel, D. Bugnazet, S. Gatla, R. Hino, I. Kantor, T. Mairs, M. Munoz, S. Pasternak, F. Perrin and S. Pascarelli, The time-resolved and extreme conditions XAS (TEXAS) facility at the European Synchrotron Radiation Facility: the general-purpose EXAFS bending-magnet beamline BM23, J. Synchrotron Radiat., 2015, 22, 1548–1554 CrossRef CAS PubMed.
- B. Ravel and M. Newville, ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT, J. Synchrotron Radiat., 2005, 12, 537–541 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji - an Open Source platform for biological image analysis, Nat. Methods, 2012, 9, 676–692 CrossRef CAS PubMed.
- C. Sommer, C. Straehle, U. Köthe and F. A. Hamprecht, Ilastik: Interactive learning and segmentation toolkit, 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, 2011, pp. 230–233 Search PubMed.
- M. Chevallet, B. Gallet, A. Fuchs, P. H. Jouneau, K. Um, E. Mintz and I. Michaud-Soret, Metal homeostasis disruption and mitochondrial dysfunction in hepatocytes exposed to sub-toxic doses of zinc oxide nanoparticles, Nanoscale, 2016, 8, 18495–18506 RSC.
- M. Cuillel, M. Chevallet, P. Charbonnier, C. Fauquant, I. Pignot-Paintrand, J. Arnaud, D. Cassio, I. Michaud-Soret and E. Mintz, Interference of CuO nanoparticles with metal homeostasis in hepatocytes under sub-toxic conditions, Nanoscale, 2014, 6, 1707–1715 RSC.
- A. P. Gondikas, A. Morris, B. C. Reinsch, S. M. Marinakos, G. V. Lowry and H. Hsu-Kim, Cysteine-induced modifications of zero-valent silver nanomaterials: implications for particle surface chemistry, aggregation, dissolution, and silver speciation, Environ. Sci. Technol., 2012, 46, 7037–7045 CrossRef CAS PubMed.
- M. Marchioni, T. Gallon, I. Worms, P.-H. Jouneau, C. Lebrun, G. Veronesi, D. Truffier-Boutry, E. Mintz, P. Delangle, A. Deniaud and I. Michaud-Soret, Insights into polythiol-assisted AgNP dissolution induced by bio-relevant molecules, Environ. Sci.: Nano, 2018, 5, 1911–1920 RSC.
- J. Lee, M. M. Pena, Y. Nose and D. J. Thiele, Biochemical characterization of the human copper transporter Ctr1, J. Biol. Chem., 2002, 277, 4380–4387 CrossRef CAS PubMed.
- K. I. Farley, Y. Surovtseva, J. Merkel and S. J. Baserga, Determinants of Mammalian Nucleolar Architecture, Chromosoma, 2015, 124, 323–331 CrossRef CAS PubMed.
- C. Goodpasture and S. E. Bloom, Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining, Chromosoma, 1975, 53, 37–50 CrossRef CAS PubMed.
- P. Roussel and D. Hernandez-Verdun, Identification of Ag-NOR Proteins, Markers of Proliferation Related to Ribosomal Gene Activity, Exp. Cell Res., 1994, 214, 465–472 CrossRef CAS PubMed.
- P. Roussel, P. Belenguer, F. Amalric and D. Hernandez-Verdun, Nucleolin is an Ag-NOR protein; This property is determined by its amino-terminal domain independently of its phosphorylation state, Exp. Cell Res., 1992, 203, 259–269 CrossRef CAS PubMed.
- J. L. Woolford and S. J. Baserga, Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae, Genetics, 2013, 195, 643–681 CrossRef CAS PubMed.
- C. E. Foulds, L. S. Trevino, B. York and C. L. Walker, Endocrine-disrupting chemicals and fatty liver disease, Nat. Rev. Endocrinol., 2017, 13, 445–457 CrossRef CAS PubMed.
- B. J. Deegan, A. M. Bona, V. Bhat, D. C. Mikles, C. B. McDonald, K. L. Seldeen and A. Farooq, Structural and Thermodynamic Consequences of the Replacement of Zinc with Environmental Metals on ERα-DNA Interactions, J. Mol. Recognit., 2011, 24, 1007–1017 CrossRef CAS PubMed.
- T. W. Hutchens, H. Allen Mark, M. Li Chee and T.-T. Yip, Occupancy of a C2-C2 type ‘Zinc-finger’ protein domain by copper Direct observation by electrospray ionization mass spectrometry, FEBS Lett., 1992, 309, 170–174 CrossRef CAS PubMed.
- P. F. Predki and B. Sarkar, Effect of replacement of "zinc finger" zinc on estrogen receptor DNA interactions, J. Biol. Chem., 1992, 267, 5842–5846 CAS.
- J. P. Hamilton, L. Koganti, A. Muchenditsi, V. S. Pendyala, D. Huso, J. Hankin, R. C. Murphy, D. Huster, U. Merle, C. Mangels, N. Yang, J. J. Potter, E. Mezey and S. Lutsenko, Activation of lxr/rxr pathway ameliorates liver disease in atp7b(−/−) (wilson disease) mice, Hepatology, 2016, 63, 1828–1841 CrossRef CAS PubMed.
- P. Wilmarth, K. Short, O. Fiehn, S. Lutsenko, L. David and J. L. Burkhead, A systems approach implicates nuclear receptor targeting in the Atp7b(−/−) mouse model of Wilson's Disease, Metallomics, 2012, 4, 660–668 RSC.
- C. S. Kim, X. Li, Y. Jiang, B. Yan, G. Y. Tonga, M. Ray, D. J. Solfiell and V. M. Rotello, Cellular imaging of endosome entrapped small gold nanoparticles, MethodsX, 2015, 2, 306–315 CrossRef PubMed.
- K. Narayan and S. Subramaniam, Focused ion beams in biology, Nat. Methods, 2015, 12, 1021–1031 CrossRef CAS PubMed.
- K. Wrzesinski, M. C. Magnone, L. V. Hansen, M. E. Kruse, T. Bergauer, M. Bobadilla, M. Gubler, J. Mizrahi, K. Zhang, C. M. Andreasen, K. E. Joensen, S. M. Andersen, J. B. Olesen, O. B. Schaffalitzky de Muckadell and S. J. Fey, HepG2/C3A 3D spheroids exhibit stable physiological functionality for at least 24 days after recovering from trypsinisation, Toxicol. Res., 2013, 2, 163–172 CrossRef CAS.
- D. E. Johnson, P. Ostrowski, V. Jaumouille and S. Grinstein, The position of lysosomes within the cell determines their luminal pH, J. Cell Biol., 2016, 212, 677–692 CrossRef CAS PubMed.
- B. E. Black, J. M. Holaska, F. Rastinejad and B. M. Paschal, DNA binding domains in diverse nuclear receptors function as nuclear export signals, Curr. Biol., 2001, 11, 1749–1758 CrossRef CAS PubMed.
- M. S. Mavinakere, J. M. Powers, K. S. Subramanian, V. R. Roggero and L. A. Allison, Multiple Novel Signals Mediate Thyroid Hormone Receptor Nuclear Import and Export, J. Biol. Chem., 2012, 287, 31280–31297 CrossRef CAS PubMed.
- A. Helleboid-Chapman, S. Helleboid, H. Jakel, C. Timmerman, C. Sergheraert, F. Pattou, J. Fruchart-Najib and J.-C. Fruchart, Glucose regulates LXRα subcellular localization and function in rat pancreatic β-cells, Cell Res., 2006, 16, 661 CrossRef CAS PubMed.
- K. Kluska, M. D. Peris-Diaz, D. Plonka, A. Moysa, M. Dadlez, A. Deniaud, W. Bal and A. Krezel, Formation of highly stable multinuclear AgnSn clusters in zinc fingers disrupts their structure and function, Chem. Commun., 2020, 56, 1329–1332 RSC.
- A. Klug, The discovery of zinc fingers and their applications in gene regulation and genome manipulation, Annu. Rev. Biochem., 2010, 79, 213–231 CrossRef CAS PubMed.
- M. Marchioni, G. Veronesi, I. Worms, W. L. Ling, T. Gallon, D. Leonard, C. Gateau, M. Chevallet, P. H. Jouneau, L. Carlini, C. Battocchio, P. Delangle, I. Michaud-Soret and A. Deniaud, Safer-by-design biocide made of tri-thiol bridged silver nanoparticle assemblies, Nanoscale Horiz., 2020, 5, 507–513 RSC.
- Y. G. Yuan, Q. L. Peng and S. Gurunathan, Silver nanoparticles enhance the apoptotic potential of gemcitabine in human ovarian cancer cells: combination therapy for effective cancer treatment, Int. J. Nanomed., 2017, 12, 6487–6502 CrossRef CAS PubMed.
- F. Peng, M. I. Setyawati, J. K. Tee, X. Ding, J. Wang, M. E. Nga, H. K. Ho and D. T. Leong, Nanoparticles promote in vivo breast cancer cell intravasation and extravasation by inducing endothelial leakiness, Nat. Nanotechnol., 2019, 14, 279–286 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9en01348b |
| This journal is © The Royal Society of Chemistry 2020 |