Open Access Article
Open Access ArticleThe longer the worse: a combined proteomic and targeted study of the long-term versus short-term effects of silver nanoparticles on macrophages†
Bastien
Dalzon
a,
Catherine
Aude-Garcia‡
a,
Hélène
Diemer
b,
Joanna
Bons
b,
Caroline
Marie-Desvergne
c,
Julien
Pérard
a,
Muriel
Dubosson
c,
Véronique
Collin-Faure
a,
Christine
Carapito
b,
Sarah
Cianférani
b,
Marie
Carrière
 d and
Thierry
Rabilloud
d and
Thierry
Rabilloud
 *a
*a
aChemistry and Biology of Metals, Univ. Grenoble Alpes, CNRS UMR5249, CEA, IRIG-DIESE-CBM, F-38054 Grenoble, France. E-mail: thierry.rabilloud@cnrs.fr
bLaboratoire de Spectrométrie de Masse BioOrganique (LSMBO), Université de Strasbourg, CNRS, IPHC UMR 7178, 67000 Strasbourg, France
cUniversité Grenoble-Alpes, CEA, Nanosafety Platform, Medical Biology Laboratory (LBM), 17 rue des Martyrs, F-38054 Grenoble, France
dUniv. Grenoble-Alpes, CEA, CNRS UMR 5819, INAC-SyMMES, Chimie Interface Biologie pour l'Environnement, la Santé et la Toxicologie (CIBEST), F-38054 Grenoble, France
First published on 27th May 2020
Abstract
Despite considerable research effort devoted to the study of the effects of silver nanoparticles on mammalian cells in recent years, data on the potential long term effects of this nanomaterial remain scarce, and centered on epithelial cells. The aim of this study was to explore the effects of silver nanoparticles on macrophages. To this end, RAW 264.7 murine macrophages were exposed to either 1 μg ml−1 silver nanoparticles for 20 days, i.e. a chronic exposure scheme, or to 20 μg ml−1 silver nanoparticles for 24 hours, i.e. an acute exposure scheme. A proteomic study was then conducted to study and compare the cellular responses to both exposure schemes. They proved to be essentially different, and stronger for the chronic exposure scheme. Targeted validation studies showed effects of chronic exposure to silver nanoparticles on detoxifying enzymes such as flavin reductase, which was increased, and on central metabolism enzymes such as triose phosphate isomerase, the activity of which decreased under chronic exposure to silver nanoparticles. Chronic exposure to silver nanoparticles also induced a decrease of reduced glutathione content, a decreased phagocytic activity and reduced macrophage responses to lipopolysaccharide, as exemplified by nitric oxide and interleukin 6 production. Overall, chronic exposure to silver nanoparticles induced stronger effects than acute exposure on macrophages in the metabolic (glutathione level, mitochondrial potential) and functional (phagocytosis, cytokine production) parameters tested.
Environmental significanceSilver nanoparticles are known to have profound effects on living cells. Because of their widespread use, contamination is almost unavoidable. However, most toxicological studies to date use an acute exposure to subtoxic doses of silver nanoparticles and read the biological effects immediately after exposure. While this is standard toxicological practice, this exposure scheme does not reflect the most likely exposure, i.e. a repeated exposure to low doses. To address this concern, we have performed a parallel study comparing the effects of a unique high dose and the same cumulative dose delivered over 20 days. We have used macrophages as the target cell type, as they are a primary scavenger cell type in many organisms. To get a better appraisal of the biological responses, we have used a combination of proteomic and targeted experiments. The main output of this study is that a repeated exposure to silver nanoparticles induces stronger biological effects than a single exposure, although less silver is internalized upon repeated exposure. This result shows that the exposure regime is a very important variable to take into account and that toxicological studies using repeated exposure to low doses should be performed. |
1. Introduction
Silver nanoparticles are more and more frequently used as biocides in many consumer products and in healthcare products such as wound dressing, or as a surface biocide in catheters and prostheses.1 The biocidal activity is mainly due to liberation of toxic silver ions, the particles themselves showing weak intrinsic toxicity.2 While well tolerated, products containing silver nanoparticles are not without effects. For example, a localized argyria has been demonstrated to occur when using silver coated healthcare products.3,4 It can be seen as a repeated exposure of proximal tissue to silver doses that are higher than the occupational doses, as the latter do not induce any argyria.5 In parallel with the local argyria, repeated exposure to moderate dose of silver nanoparticles has been shown to alter physiological parameters, while being non lethal.6–10 In the frame of occupational and food-derived exposures, in vitro long term studies of the effects of silver nanoparticles on cells have concentrated on the barrier cell types, i.e. epithelial cells.11–15For all exposure routes, including liberation in the bloodstream,16 which is a frequent route for healthcare products, resident macrophages represent the common scavenger cell type that internalizes the particular material. In the case of silver nanoparticles, reported effects of a chronic exposure to silver nanoparticles appear to be generally deleterious and often show altered inflammatory responses,6,9,10 although the internalization of silver nanoparticles by macrophages can help them in killing otherwise resistant bacteria, and thus be helpful.17
The deleterious effects of silver nanoparticles on macrophages have been previously reported in several papers (e.g.18–20). However, these studies use a single dose of silver nanoparticles at relatively high concentrations to investigate the cellular responses.
As this exposure scenario is quite different from the in vivo situations, e.g. in case of use of silver nanoparticles-containing healthcare products, we therefore decided to investigate in more details the responses of macrophage to a repeated exposure to silver nanoparticles, and to compare the results to those obtained with a single high dose, which is the standard practice in nanotoxicology. To this purpose, we used a proteomic approach, which has given interesting insights for the study of cellular responses to acute exposures to several nanoparticles,21–27 including silver nanoparticles.28–30
2. Material and methods
All experiments were carried out on at least three independent biological replicates. Unless specified otherwise, all chemicals were purchased from Sigma-Aldrich and were at least 99% pure.2.1. Nanoparticles
PVP-coated silver nanoparticles (Ag-NP) were purchased from Sigma, directly as a concentrated suspension (catalog number #758329). The zeta potential, size distribution and agglomeration state of Ag-NP suspensions were characterized by dynamic light scattering on a ZetaSizer nanoZS (Malvern Instrument). TEM images of Ag-NPs were obtained on a JEOL 1200EX TEM operating at 80 kV (Grenoble Institut des Neurosciences, Grenoble, France).2.2. Cell culture
The mouse macrophage cell line RAW264.7 was obtained from the European Cell Culture Collection (Salisbury, UK). The cells were cultured in RPMI 1640 medium + 10% fetal bovine serum (FBS). For routine culture, cells were seeded on non-adherent flasks (e.g. suspension culture flasks from Greiner) at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml and harvested 48 hours later, at 100
000 cells per ml and harvested 48 hours later, at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0000 cells per ml. Cell viability was measured by a dye exclusion assay, either with eosin (1 mg ml−1) under the microscope31 or with propidium iodide (1 μg ml−1) in a flow cytometry mode.32
0000 cells per ml. Cell viability was measured by a dye exclusion assay, either with eosin (1 mg ml−1) under the microscope31 or with propidium iodide (1 μg ml−1) in a flow cytometry mode.32
For determination of the useful dose, cells were seeded at 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. They were exposed to nanoparticles on the following day and harvested after a further 24 hours in culture.
000 cells per ml. They were exposed to nanoparticles on the following day and harvested after a further 24 hours in culture.
For treatment with nanoparticles, cells were seeded at 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml on classical, adherent cell culture flasks and left for 24 hours at 37 °C for cell adhesion. The medium volume was adjusted to keep a constant medium height for all the culture supports used (2 ml for 6-well plates, 5 ml for 25 cm2 flasks, 15 ml for 75 cm2 flasks and 35 ml for 175 cm2 flasks). The cells were grown to confluence for 48 hours. From then on, the culture medium was changed every 48 hours. For the acute exposure, the cells were kept post-confluence for 2 days. They were then treated with silver nanoparticles for 24 hours. After this period of time, the cell culture medium was removed and the cells were collected by scraping in PBS, then rinsed twice in PBS and processed. In the chronic exposure scheme, the treatment started just after confluence was reached. The cells were treated daily with 1 μg ml−1 silver nanoparticles for 20 days, with a culture medium change every two days. The cells were then collected as described above. Thus, four conditions were eventually analyzed:
000 cells per ml on classical, adherent cell culture flasks and left for 24 hours at 37 °C for cell adhesion. The medium volume was adjusted to keep a constant medium height for all the culture supports used (2 ml for 6-well plates, 5 ml for 25 cm2 flasks, 15 ml for 75 cm2 flasks and 35 ml for 175 cm2 flasks). The cells were grown to confluence for 48 hours. From then on, the culture medium was changed every 48 hours. For the acute exposure, the cells were kept post-confluence for 2 days. They were then treated with silver nanoparticles for 24 hours. After this period of time, the cell culture medium was removed and the cells were collected by scraping in PBS, then rinsed twice in PBS and processed. In the chronic exposure scheme, the treatment started just after confluence was reached. The cells were treated daily with 1 μg ml−1 silver nanoparticles for 20 days, with a culture medium change every two days. The cells were then collected as described above. Thus, four conditions were eventually analyzed:
| Acute control: cells let at confluence for 8 days |
| Acute treated: cells let at confluence for 7 days, then treated with silver nanoparticles (20 μg ml−1) for 1 day |
| Chronic control: cells let at confluence for 20 days |
| Chronic treated: cells treated at confluence for 20 days (1 μg ml−1 per day silver nanoparticles) |
2.3. Phagocytosis and particle internalization assay
The phagocytic activity was measured using fluorescent latex beads (1 μm diameter, green labelled, catalog number L4655 from Sigma), as described previously.33 After a 2 h 30 treatment with the fluorescent beads in culture medium at 37 °C, the cells were harvested in PBS with propidium iodide (1 μg mL−1). Viability and phagocytic activity were measured simultaneously by flow cytometry on a FacsCalibur instrument (Beckton Dickinson). The dead cells (propidium positive) were excluded from the analysis.2.4. Mitochondrial transmembrane potential measurement
The mitochondrial transmembrane potential was assessed by rhodamine 123 uptake,27 using a low rhodamine concentration (80 nM) to avoid intramitochondrial fluorescence quenching that would result in a poor estimation of the mitochondrial potential.342.5. Enzyme assays
The enzymes were assayed according to published procedures (see below).The cell extracts for enzyme assays were prepared as described previously.27 The dehydrogenases or dehydrogenases-coupled activities were assayed at 500 nm using the phenazine methosulfate/iodonitrotetrazolium coupled assay.35 The enzyme assay buffer contained 25 mM Hepes NaOH pH 7.5, 5 mM magnesium acetate, 100 mM potassium nitrate and 1% Triton X-100. It also contained 30 μM phenazine methosulfate, 200 μM iodonitrotetrazolium chloride, 250 μM of the adequate cofactor (NAD or NADP) and 1–5 mM of the organic substrate, which was used to start the reaction. Triose phosphate isomerase was assayed with dihydroxyacetone phosphate and a glyceraldehyde dehydrogenase-coupled assay.36 Glutathione reductase was assayed through NADPH consumption.37
2.6. Reduced glutathione assay
Classical glutathione assays are difficult to apply to macrophages, as the oxidizing activities present in their phagolysosomes oxidize the cytosolic reduced glutathione when the cells are homogeneized. To alleviate this problem, we used a spectrophotometric assay utilizing the cellular glutathione S-transferases and the chlorodinitrobenzene substrate.38,39 After their exposure to silver in 6 well plates, cells were finally treated for 30 minutes with 25 μM chlorodinitrobenzene in the culture medium, resulting in the conjugation of glutathione to the substrate and consumption of the free reduced glutathione.40,41 The culture medium was removed and the cell layer rinsed twice in Hepes NaOH buffer (10 mM pH 7.5) containing 2 mM magnesium acetate and 0.25 M sucrose. The cell layer was then lysed on a rocking table for 15 minutes in 800 μl of Hepes NaOH buffer (10 mM pH 7.5) containing 2 mM magnesium acetate and 1 mg ml−1 tetradecyldimethylammonio propane sulfonate (SB 3-14). The suspension was collected and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 minutes to pellet particulate material. The concentration of glutathione was determined by reading the absorbance at 340 nm, using an extinction coefficient of 9600 M−1 cm−1.38 To normalize the results, the protein concentration of the extracts was determined by a modified dye-binding assay,42 and the final results were expressed in mM glutathione/mg protein.
000g for 15 minutes to pellet particulate material. The concentration of glutathione was determined by reading the absorbance at 340 nm, using an extinction coefficient of 9600 M−1 cm−1.38 To normalize the results, the protein concentration of the extracts was determined by a modified dye-binding assay,42 and the final results were expressed in mM glutathione/mg protein.
2.7. NO production and cytokines production
The cells were grown to confluence in a 6 well plate and pre-treated with nanoparticles as described above for flasks. For the final 18 hours of culture, half of the wells were treated with 100 ng ml−1 LPS (from Salmonella, purchased from Sigma), and arginine monohydrochloride was added to all the wells (5 mM final concentration) to give a high concentration of substrate for the nitric oxide synthase. After 18 hours of incubation, the cell culture medium was recovered, centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes to remove cells and debris, and the nitrite concentration in the supernatants was read at 540 nm after addition of an equal volume of Griess reagent and incubation at room temperature for 30 minutes.
000g for 10 minutes to remove cells and debris, and the nitrite concentration in the supernatants was read at 540 nm after addition of an equal volume of Griess reagent and incubation at room temperature for 30 minutes.
For cytokine production, a commercial kit (BD cytometric bead array, catalog number 552364 from BD Biosciences) was used.
2.8. Silver quantification
For measuring the Ag-NP uptake and dissolution in cells, 5 ml cell cultures in 25 cm2 culture flasks were used, with the exposure scheme described above. At the end of the exposure period, the NP-containing medium was removed and the cell layer was gently rinsed twice with complete culture medium.The cells were then harvested by scraping in buffer A (Hepes 50 mM pH 7.5, sorbitol 200 mM, magnesium acetate 2 mM), and collected by centrifugation (500 g, 5 min). After a further rinse in buffer A, the cells were lysed by incubation in 10-cell pellet volume of lysis buffer (Hepes 50 mM pH 7.5, magnesium acetate 2 mM, tetradecyldimethylammonio propane sulfonate (SB 3-14), 0.15% (w/v)) for 20 minutes on ice. Non sedimentable (soluble) silver is obtained by centrifugation of an aliquot of the extracts (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 30 minutes, 4 °C) and collection of the upper half of the supernatant.
000g, 30 minutes, 4 °C) and collection of the upper half of the supernatant.
The extracts were mineralized by the addition of one volume of suprapure 65% HNO3 and incubation on a rotating wheel at room temperature for 18 h.
Mineralized samples were then diluted in ultrapure grade HNO3 (1% v/v) and analyzed on a Nexion 300X ICP-MS (Perkin Elmer, Waltham, MA) equipped with a concentric nebulizer and operated in standard mode. External calibration was performed using a certified ionic Ag ICP-MS standard and Yttrium was used as an internal standard.
The technic was validated based on linearity, repeatability, reproducibility, accuracy, inter-sample contamination criterions, and the analytical limit of detection determined as the sum of the mean and three standard deviations of 20 ultrapure water samples was 0.035 μg L−1 of Ag in injected sample. Multielemental quality controls were used to validate each run of analysis.
In some experiments, inductively coupled plasma atomic emission spectroscopy (ICP-AES) (Shimadzu ICP 9000 with mini plasma torch in axial reading mode) was used to measure the metal content. Standard solutions of Ag for atomic absorption spectroscopy (Sigma Aldrich) were used for quantification (calibration curve between 1.9 to 1000 μg L−1 with 1% HNO3 (Fluka)). Mineralized samples were diluted with pure water to a final concentration of 1% of HNO3 prior to analysis.
To normalize the results, the protein concentration of the extracts was determined by a modified dye-binding assay.42
2.9. Proteomics
The 2D gel based proteomic experiments were essentially carried out as previously described,26,27 on independent biological triplicates, except for the acutely exposed cells for which a quadruplicate was obtained. The isoelectric focusing dimension used a linear 4–8 pH gradient, and the second dimension a homogeneous 10% acrylamide gel spanning a molecular mass window of 14–200 kDa. Protein detection was carried out by silver staining. Two-dimensional gel images were analyzed by the Delta2D software, and the spot intensities are expressed as a percentage of the sum of all detected spots, thereby normalizing the intensities between gels. Statistical analysis was carried out by a heteroschedastic Student T-test (also known as Welch T test). For the general analysis a first pass filter of p < 0.25 was carried out. This allowed decreasing the noise brought by proteins that do not show a consistent variation in the biological phenomena under investigation. For the detailed analysis only proteins showing a p ≤ 0.05 in one T-test (acutely exposed cells vs. acute control or chronically-exposed cells vs. chronic control) were selected.Protein identification was carried out by mass spectrometry as described previously.27 The MS/MS data were interpreted using a local Mascot server with MASCOT 2.5.1 algorithm (Matrix Science, London, UK) against UniProtKB/SwissProt (version 2017_09, 555594 sequences). The research was carried out in all species. Spectra were searched with a mass tolerance of 15 ppm for MS and 0.05 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Protein identifications were validated with at least two peptides with Mascot ion score above 30.
3. Results
3.1. Silver nanoparticles characterization and viability effects
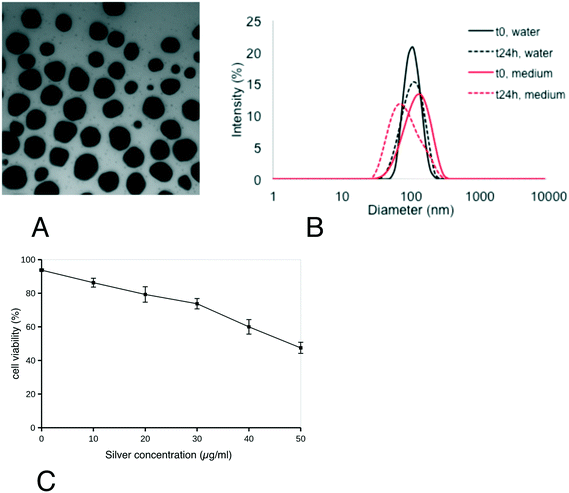
First, the characteristics of the silver nanoparticles were verified and their effect on cell viability was assayed. From the results shown in Fig. 1, the silver nanoparticles showed a spherical shape with a diameter of 49.3 ± 12.6 nm. Their hydrodynamic diameter (Z-average) in water was 103 ± 2 nm (polydispersity index: 0.15 ± 0.04) (B). This value did not change significantly upon dilution of Ag-NP suspension in exposure medium, where it was 110 ± 2 nm (polydispersity index: 0.11 ± 0.01). After 24 h at 37 °C, the size distribution of Ag-NPs in water and medium slightly decreased (Z-average: 98 ± 1 nm/Pdi: 0,08 ± 0,01 and Z-average: 94 ± 1 nm/Pdi: 0,23 ± 0,01 in water and medium, respectively). Their zeta potential, when diluted in water, was −11.4 ± 1,0 mV. From the viability curve, a dose of 20 μg ml−1 was selected for all the subsequent experiments, corresponding to the LD20. This dose ensures strong effects on cells while keeping cellular mortality at an acceptable level for subsequent experiments. As previous experiments had shown that the cellular differentiated features were kept for up to 20 days of confluence culture, the dose was split at 1 μg ml−1 per day silver nanoparticles for the chronic exposure scheme.Experiments were then conducted to determine the actual silver dose reaching the cells after a 24 hours exposure at the two concentrations used (1 and 20 μg ml−1). The results, shown in Table S1,† indicated that ca. 75% of the silver input was internalized by the cells, regardless of the input concentration.
3.2. Global analysis of the proteomic results
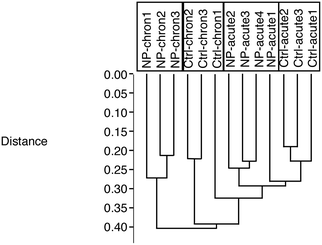
From the total protein abundances data obtained from the gel analysis software, a subset of putatively variable proteins (T-test p < 0.25 in either the acute exposure vs. control or the chronic exposure vs. control comparisons) was extracted. This subset was then tested using the PAST statistical suite.43 As a first test, a hierarchical clustering was performed, and the results are shown in Fig. 2. This test clearly showed that the cells chronically exposed to silver stood apart from all the other groups, and that the second most important variable was the cell aging, as the chronic control group also stood apart from the two acute groups (control and treated).3.3. Detailed analysis of the proteomic results
We extracted the intensity values for the spots that were significantly altered (T-test p ≤ 0.05) between one of the silver-treated groups and its corresponding control. We thus obtained two subsets, one for the spots changed in the chronic exposure scheme and one for the spots changed in the acute exposure scheme. The two subsets, accounting in total for 152 spots, were then merged into a single table of 85 variable and identified proteins (Table 1). The corresponding 2D gel images are shown on Fig. 3 and Fig. S1 and S2.† The mass spectrometry identification parameters are detailed in Table S2† and the mass spectrometry data are deposited in Pride under the identifier PXD016379.44| Acc number | Protein name | Ratio/T-test | Ratio/T-test |
|---|---|---|---|
| Acute/ctrl A | Chron/ctrl C | ||
| a Proteins that do not change significantly but are included as controls. | |||
| O09172 | Glutamate–cysteine ligase regulatory subunit | 0.91/0.39 | 1.91/0.01 |
| O35345 | Importin subunit alpha-7 | 1.22/0.27 | 0.58/0.04 |
| O35639 | Annexin A3 | 0.87/0.30 | 1.63/0.01 |
| O55023 | Inositol monophosphatase 1 | 0.94/0.52 | 2.18/0.0001 |
| O70370 | Cathepsin S | 0.81/0.28 | 0.73/0.02 |
| O88456 | Calpain small subunit 1 | 1.48/0.02 | 1.67/0.13 |
| O88487 | Cytoplasmic dynein 1 intermediate chain 2 | 0.81/0.04 | 1.64/0.05 |
| O88544 | COP9 signalosome complex subunit 4 | 0.87/0.03 | 1.22/0.05 |
| P09528 | Ferritin heavy chain | 0.53/0.08 | 2.19/0.01 |
| P10605 | Cathepsin B | 0.91/0.31 | 1.82/0.04 |
| P14824 | Annexin A6 | 0.88/0.61 | 2.62/0.001 |
| P17751/aa | Triose phosphate isomerase | 0.99/0.93 | 0.99/0.96 |
| P17751/ba | Triose phosphate isomerase | 0.92/0.25 | 0.82/0.09 |
| P17751/c | Triose phosphate isomerase | 0.98/0.91 | 0.67/0.04 |
| P17751/da | Triose phosphate isomerase | 1.11/0.46 | 0.73/0.39 |
| P17751 ∑ | Triose phosphate isomerase | 1.01/0.76 | 0.82/0.04 |
| P18242 | Cathepsin D | 1.12/0.67 | 3.46/0.04 |
| P18760 | Cofilin 1 | 1.50/0.09 | 0.51/0.05 |
| P23591 | GDP-L-fucose synthase | 0.62/0.06 | 0.37/0.01 |
| P26516 | Proteasome non-ATPase regulatory subunit 7 | 0.98/0.92 | 1.36/0/02 |
| P31938 | Dual specificity mitogen-activated protein kinase kinase 1 | 0.73/0.21 | 0.75/0.05 |
| P45376 | Aldo-keto reductase family 1 member B1 | 0.75/0.04 | 0.94/0.49 |
| P46061 | Ran GTPase-activating protein 1 | 0.97/0.54 | 1.33/0.02 |
| P47738 | aldehyde dehydrogenase, mitochondrial | 1.52/0.05 | 1.87/0.18 |
| P47753 | F-Actin-capping protein subunit alpha-1 | 1.06/0.21 | 1.31/0.02 |
| P47791 | Glutathione reductase, mitochondrial | 1.24/0.18 | 1.82/0.02 |
| P50171 | Estradiol 17-beta-dehydrogenase 8 | 0.61/0.02 | 1.58/0.03 |
| P54227 | Stathmin | 1.41/0.02 | 0.54/0.16 |
| P57759 | Endoplasmic reticulum resident protein 29 | 0.80/0.05 | 0.68/0.17 |
| P61982 | 14-3-3 gamma protein | 0.9/0.66 | 0.76/0.04 |
| P62137 | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 0.75/0.02 | 0.99/0.97 |
| P62748 | Hippocalcin-like protein 1 | 0.93/0.73 | 0.61/0.01 |
| P67778 | prohibitin | 0.88/0.09 | 1.26/0.01 |
| P70202 | Latexin | 0.89/0.51 | 1.42/0.03 |
| Q3SXD3 | HD domain-containing protein 2 | 1.44/0.04 | 0.35/0.02 |
| Q3U1J4 | DNA damage-binding protein 1 | 0.45/0.04 | 1.26/0.27 |
| Q3UM45 | Protein phosphatase 1 regulatory subunit 7 | 0.60/0.05 | 0.76/0.19 |
| Q60854 ∑ | Serpin B6 | 0.99/0.72 | 1.40/0.01 |
| Q60854/a | Serpin B6 | 1.07/0.54 | 1.50/0.04 |
| Q60854/b | Serpin B6 | 0.96/0.76 | 1.23/0.04 |
| Q60854/c | Serpin B6 | 0.98/0.61 | 1.46/0.02 |
| Q61205 | Platelet-activating factor acetylhydrolase IB subunit gamma | 0.84/0.42 | 0.60/0.01 |
| Q64105 | Sepiapterin reductase | 0.98/0.90 | 0.57/0.002 |
| Q64152/a | Transcription factor btf3 | 1.04/0.88 | 0.62/0.02 |
| Q64152/b | Transcription factor btf3 | 1.11/0.67 | 0.85/0.01 |
| Q64152 ∑ | Transcription factor btf3 | 1.07/0.77 | 0.72/0 .01 |
| Q64442 | sorbitol dehydrogenase | 1.74/0.03 | 1.44/0.15 |
| Q6P3D0 | U8 snoRNA-decapping enzyme | 0.41/0.01 | 1.18/0.34 |
| Q6ZQI3 | malectin | 0.78/0.08 | 1.57/0.03 |
| Q7TQI3 | Ubiquitin thioesterase OTUB1 | 1.10/0.51 | 0.66/0.04 |
| Q80UW8 | DNA-directed RNA polymerases I, II, and III subunit RPABC1 | 1.36/0.01 | 0.81/0.41 |
| Q8BH04 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | 0.84/0.05 | 1.36/0.13 |
| Q8BMF4 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | 0.80/0.23 | 1.43/0.03 |
| Q8BV13 | COP9 signalosome complex subunit 7b | 0.88/0.68 | 0.34/0.05 |
| Q8CAY6 | Acetyl-CoA acetyltransferase, cytosolic | 0.58/0.02 | 0.98/0.94 |
| Q8CDN6 | Thioredoxin-like protein 1 | 0.76/0.07 | 0.40/0.03 |
| Q8K2B3 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit | 1.07/0.41 | 1.08/0.02 |
| Q91WN1 | DnaJ homolog subfamily C member 9 | 0.71/0.09 | 0.56/0.04 |
| Q91ZJ5 | UTP–glucose-1-phosphate uridylyltransferase | 1.66/0.12 | 0.52/0.03 |
| Q91ZR2 | Sorting nexin-18 | 0.79/0.34 | 3.21/0.01 |
| Q921I9 | Exosome complex component RRP41 | 1.55/0.03 | 0.78/0.30 |
| Q923D2 | Flavin reductase | 1.05/0.72 | 1.89/0.04 |
| Q99K51/a | Plastin-3 | 0.74/0.03 | 2.45/0.01 |
| Q99K51/b | Plastin-3 | 0.63/0.09 | 2.79/0.01 |
| Q99KR3 | Endoribonuclease LACTB2 | 0.56/0.04 | 1.27/0.56 |
| Q99LC5 | Electron transfer flavoprotein subunit alpha | 0.75/0.10 | 1.70/0.03 |
| Q9CQ02 | COMM domain-containing protein 4 | 0.53/0.005 | 1.86/0.10 |
| Q9CQ65 | S-Methyl-5′-thioadenosine phosphorylase | 0.68/0.03 | 1.49/0.03 |
| Q9CQU0 | Thioredoxin domain-containing protein 12 | 1.42/0.17 | 0.57/0.01 |
| Q9CXW3 | Calcyclin binding protein | 0.68/0.02 | 0.87/0.25 |
| Q9CYZ2 | Tumor protein D54 | 0.89/0.49 | 1.78/0.004 |
| Q9CZ13 | Cytochrome b–c1 complex subunit 1 | 0.91/0.22 | 1.36/0.03 |
| Q9D2G2 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | 0.78/0.27 | 1.22/0.04 |
| Q9D6J6 | NADH dehydrogenase [ubiquinone] flavoprotein 2 | 0.68/0.03 | 1.36/0.07 |
| Q9D8C4 | Interferon-induced 35 kDa protein homolog | 0.89/0.41 | 0.45/0.02 |
| Q9D964 | Glycine amidinotransferase, mitochondrial | 1.39/0.06 | 1.58/0.02 |
| Q9DB05 | Alpha-soluble NSF attachment protein | 1.23/0.07 | 0.66/0.01 |
| Q9DBP5 | UMP-CMP kinase | 1.02/0.94 | 0.55/0.03 |
| Q9DCX2 | ATP synthase subunit d | 0.85/0.23 | 1.29/0.02 |
| Q9ER38 | Torsin-3A | 0.56/0.03 | 2.34/0.02 |
| Q9ERF3 | WD repeat-containing protein 61 | 1.41/0.04 | 1.26/0.33 |
| Q9JHW2 | Omega-amidase NIT2 | 0.90/0.50 | 0.69/0.02 |
| Q9QXK7 | Cleavage and polyadenylation specificity factor subunit 3 | 1.23/0.29 | 1.49/0.01 |
| Q9QYB1 | Chloride intracellular channel protein 4 | 1.37/0.17 | 1.97/0.002 |
| Q9QYJ0 | DnaJ homolog subfamily A member 2 | 1.04/0.87 | 0.42/0.02 |
| Q9QZ06 | Toll-interacting protein | 0.96/0.68 | 1.27/0.02 |
| Q9QZ88 | Vacuolar protein sorting-associated protein 29 | 0.87/0.55 | 0.46/0.001 |
| Q9WUU7 | Cathepsin Z | 1.33/0.36 | 4.23/0.01 |
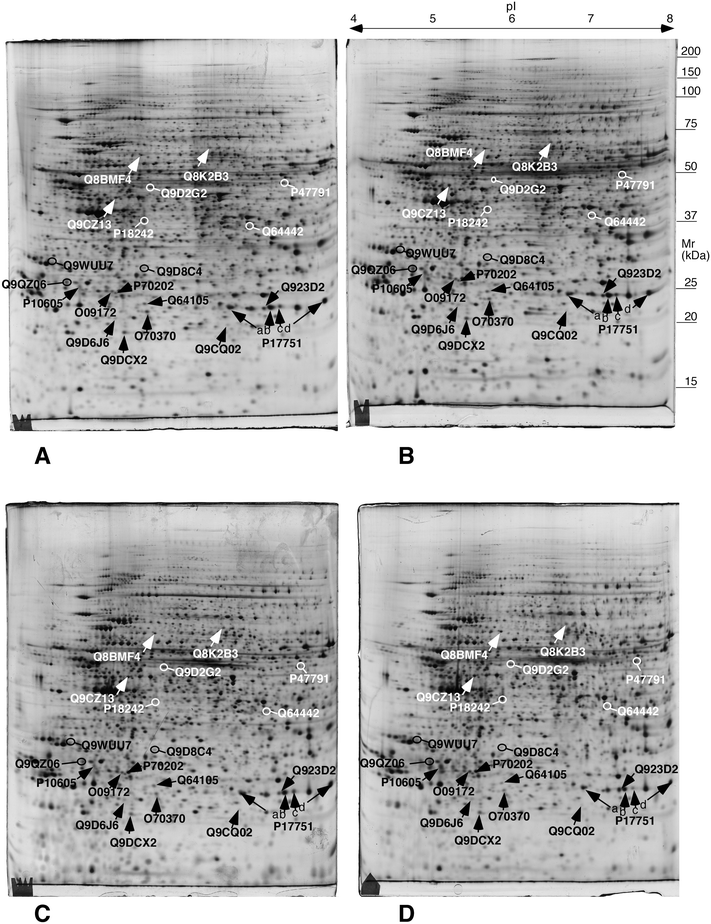 | ||
| Fig. 3 Representative 2D gels of the different conditions. The proteins highlighted in this figure are mentioned in the targeted experiments described hereafter. A: control acute; B: AgNP-treated, acute scheme; C: control chronic; D: AgNP-treated, chronic scheme. The white and black arrows and protein names have been chosen only for legibility purposes. They point to proteins whose abundances change significantly (p < 0.05 by Student T test) between either the acute exposure vs. control or the chronic exposure vs. control comparisons. These proteins are listed in Table 1. | ||
In order to cope with the multiple testing issue, we used several approaches such as the Benjamini–Hochberg approach,45 the sequential goodness of fit approach46 and the sequential Fisher test approach.47 All selected spots showed a very low q-value for the condition for which they were variable compared to the corresponding control (Table S3†).
In order to further analyze the proteomic results, we applied a pathway analysis through the DAVID tool48,49 (publicly available at https://david.ncifcrf.gov/), and the results of these analyses are shown in Tables S4 and S5.†
Several important cellular pathways were highlighted by this analysis, such as antioxidant pathways (e.g. glutathione reductase, glutamate cysteine ligase, flavin reductase), mitochondrial proteins (e.g. ATP synthase, respiratory complexes I–III, pyruvate dehydrogenase, oxoglutarate dehydrogenase), lysosomal proteins (e.g. cathepsins B, D, S and Z), or central metabolism enzymes (e.g. sorbitol dehydrogenase, triose phosphate isomerase). We also used knowledge obtained for other nanoparticles using a proteomic screen, as detailed in the following sections.
3.4. Enzymatic activities
For some of the modulated proteins, we first checked whether their enzyme activities (Table 2), which is the relevant parameter in terms of cell physiology, followed or not the proteomic results. For triose phosphate isomerase, sorbitol dehydrogenase and flavin reductase the enzymatic activity followed the trend indicated by proteomics, i.e. a decrease for triose phosphate isomerase and an increase for sorbitol dehydrogenase and flavin reductase. The results were different for glutathione reductase, for which the enzymatic activity was constant, while proteomics indicated an increase in the protein amount upon cell exposure to silver in both modes. In this case, however, we showed that the enzymatic activity is inhibited by silver ion. Thus, the decrease in the relative enzymatic activity observed upon silver treatment (compared to the protein amount) may be explained by the inhibition of the enzyme by the silver ion present in the cells in this case (as shown by the ICP-MS results).| Acute control | Ag NP acute | Chronic control | Ag NP chronic | Proteomic fold change acute | Proteomic fold change chronic | |
|---|---|---|---|---|---|---|
| Enzymes assayed: TPIS: triose phosphate isomerase; DHSO: sorbitol dehydrogenase; GSR: glutathione reductase; BLVRB: flavin reductase. All the activities are expressed in nmole substrate converted/min mg−1 total protein. Statistical significance of the results in the Student T test (compared to the matched control). a p < 0.05. b p < 0.01. c p < 0.1 (Student T-test and Mann–Whitney U test). d Proteomic fold change reported for the only triose phosphate isomerase spot that shows a significant abundance change. e The GSR activity is sensitive to silver ion. Activity of the acute control samples measured in the presence of 1 μM Ag+ : 35.3 ± 2.7 nmole substrate converted/min mg−1 total protein. | ||||||
| TPIS | 65 ± 8.4 | 68.8 ± 8.6 | 67.99 ± 4.6 | 43 ± 0.63b | 0.98d | 0.67d |
| DHSO | 4.56 ± 0.44 | 6.31 ± 0.98a | 5.54 + 0.97 | 6.02 ± 0.36c | 1.74 | 1.44 |
| GSRe | 65.48 ± 4.17 | 66.26 ± 4.32 | 39.95 ± 4.78 | 41.8 ± 3.2 | 1.24 | 1.82 |
| BLVRB | 4.24 ± 0.33 | 4.27 ± 0.54 | 5.15 ± 0.36 | 6.91 ± 0.18b | 1.05 | 1.89 |
3.5. Mitochondrial potential
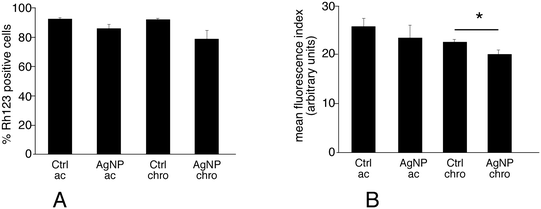
The keyword mitochondrion appeared in pathway analyses. Correspondingly, several mitochondrial proteins appeared modulated upon exposure of macrophages to silver nanoparticles, among which a few subunits of the respiratory complexes, some components of the mitochondrial matrix involved in electron transfer (electron transfer flavoprotein) and some proteins involved in the general energy metabolism (pyruvate dehydrogenase, oxoglutarate dehydrogenase). As this may suggest perturbations in the mitochondrial functions, we investigated the mitochondrial transmembrane potential. The results, shown on Fig. 4, indicated that nearly all viable cells accumulated rhodamine 123 and thus had a strong mitochondrial transmembrane potential. Nonetheless, the amount of accumulated rhodamine 123 was lower in cells chronically exposed to silver nanoparticles compared to the corresponding control, and this effect, albeit of low magnitude, was statistically significant. In cells acutely exposed to silver, no significant change in the mitochondrial membrane potential was observed. In addition, the proportion of cells with a high mitochondrial potential is also a good indicator of viability. The results shown in Fig. 4 indicated that both treatment schemes with silver nanoparticles induced a decrease in cell viability (−15% for the acute exposure, −21% for the chronic exposure) that is in line with the LD20.3.6. Glutathione level
Beside glutathione reductase, which has been studied in section 3.4, one of the proteins increased after exposure to silver nanoparticles is glutamate cysteine ligase subunit M, i.e. the regulatory subunit of the enzyme involved in the first step of glutathione biosynthesis, which is the limiting step of the pathway.50 In a previous work on copper nanoparticles,33 a change in the amount of this protein correlated with a change in the level of free reduced glutathione. Thus, this induction could be a response of cells to decreased intracellular glutathione levels due to exposure to Ag-NPs. Moreover, we previously reported that the levels of free glutathione was reduced in primary macrophages exposed to silver nanoparticles,51 and that intracellular dissolution of silver nanoparticles led to the formation of silver–glutathione complexes.52 In the present exposure conditions, a statistically significant reduction (p ≤ 0.05, Mann–Whitney U test) of the intracellular free, reduced glutathione content was measured:| Acute control: 5.91 ± 1.13 nmoles GSH/mg protein |
| Acute AgNP: 4.60 ± 0.87 nmoles GSH/mg protein |
| Chronic control: 5.90 ± 1.40 nmoles GSH/mg protein |
| Chronic AgNP: 3.63 ± 0.49 nmoles GSH/mg protein |
3.7. Phagocytosis
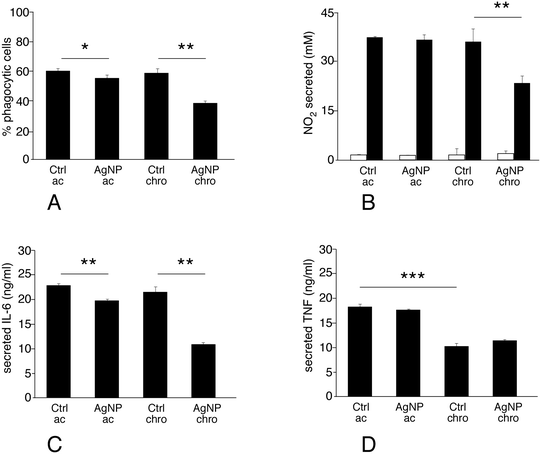
Phagocytosis is one of the most important functions of the macrophages. The proteomic analysis highlighted important changes in the levels of several cathepsins, which may indicate alterations in the phagolysosomal pathway. Moreover, actin binding appeared as a modulated pathway, and is important for phagocytosis. Thus, the phagocytic activity of macrophages exposed to silver nanoparticles was assessed. The proportion of phagocytic cells did not change appreciably upon treatment with silver nanoparticles in the acute exposure mode. Conversely, chronic exposure to silver nanoparticles significantly decreased their phagocytic capacity (Fig. 5A).3.8. NO and cytokine production
Production of nitric oxide and of pro-inflammatory cytokines such as IL-6 and TNF upon stimulation is one of the specialized functions of macrophages. In addition, the proteomic screen highlighted proteins that may be related, directly or indirectly, with these processes. One interesting example is represented by sepiapterin reductase, which catalyses the production of tetrahydrobiopterin, an obligatory cofactor of NO synthase. Other examples are represented by potential regulators of transduction of the pro-inflammatory signal in the nucleus (e.g. COMM domain-containing protein 4 and interferon-induced 35 kDa protein) or more upstream in the cytosol (e.g. toll-interacting protein), or proteins known to regulate inflammation by indirect mechanisms (e.g. latexin). For some of these proteins, such as sepiapterin reductase and interferon-induced 35 kDa protein, changes observed at the proteomic level have been correlated to changes in the nitric oxide and cytokines production.26,27 It was thus legitimate to evaluate the inflammatory status in cells exposed to silver nanoparticles. To this purpose IL6, TNF and NO production were measured after treatment of macrophages with silver nanoparticles under two schemes: i) treatment with nanoparticles only, or ii) treatment with nanoparticles and lipopolysaccharide (LPS) for the last 18 hours of exposure. The first scheme investigated the intrinsic pro-inflammatory action of the nanoparticles, while the second one investigated the interference of nanoparticles with a standard pro-inflammatory response induced by a bacterial stimulus.Silver nanoparticles did not show an intrinsic pro-inflammatory effect, as detected from NO production (Fig. 5B, grey bars). The situation was very different for the combined nanoparticles-LPS treatment (Fig. 5B–D, black bars). In this case, the response to acute exposure did not differ significantly from that of the control cells (exposed to LPS only). Oppositely, the NO production was significantly lower in chronically-exposed cells compared to control cells. This trend was also observed for interleukin-6 production (where a slight but statistically significant was however observed upon acute exposure, Fig. 5C), while TNF-alpha production was not changed significantly by exposure to silver in either mode. It is worth noting, however, that TNF-alpha production is sensitive to cell aging (Fig. 5D), with a 50% reduced production under LPS stimulation after 20 days of culture.
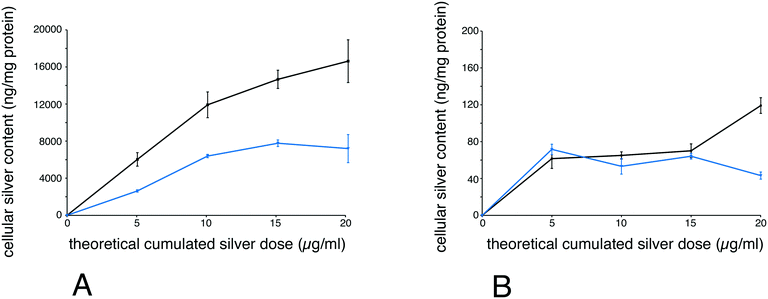
Finally, to better understand why chronically-exposed cells were more affected than acutely-exposed cells, we performed silver accumulation measurements, both for total silver and ionic silver. To this purpose, we exposed cells chronically to 1 μg ml−1 per day for 5, 10, 15 and 20 days. In parallel, cells were exposed to the same cumulated dose (i.e. 5, 10, 15 and 20 μg ml−1) for 24 hours. After exposure, the total and soluble silver contents were determined for each point. The results, shown in Fig. 6, indicate that total accumulated silver was lower for chronically-exposed cells than for acutely exposed cells. For soluble silver, the concentrations were similar and relatively constant for chronically-exposed cells and for acutely exposed cells for up to 15 days, and were then lower for chronically-exposed cells at 20 μg ml−1 cumulated dose.
4. Discussion
Although completely irreplaceable on the level of relevance, animal experimentation in toxicology poses important challenges in terms of speed, cost and also ethical issues, as exemplified by studies on the long term effects of ammunition.54 Furthermore, new regulations such as the REACH regulation in EU dramatically increase the amount of toxicological data to be produced. There is thus a general trend to decrease as much as possible the use of animal experimentation, following the 3R (refine, reduce, replace) guidelines. This however requires mandatorily to increase the relevance of data obtained on in vitro models and thus to refine these models and their use, with encouraging recent results regarding silver nanomaterials.11–15,55In in vitro nanotoxicological studies, the cell type, exposure scheme and nanoparticles used are critical features. Regarding silver nanoparticles, we tried to use an industrial product to maximise the relevance of our study. As such, PVP-coated silver nanoparticles have been used as antibacterial agents,56 in conductive inks,57–59 in electromechanics60 or in sensors.61and thus appear a relevant choice.
Regarding the exposure schemes, most in vitro nanotoxicology studies use a single exposure to relatively high doses of materials (see for example ref. 62–64). This strategy mimics an accidental exposure to a high dose, but not a repeated exposure to low doses, which is more realistic in mimicking occupational scenarios. Very few in vitro studies using a repeated exposure scheme have been reported in the literature. Interestingly, these studies report different outcomes than those observed for a single acute dose. Moreover, chronic exposure leads sometimes to stronger cell response compared to acute exposure, but the opposite also occurs.11,65,66 Thus, cell responses to a repeated exposure scenario are still not well understood, and more data need to be generated.
Until now, macrophages have not been used in repeated exposure scenarios, despite the fact that they are the primary scavenger cells for all particulate material in the body and are relevant for many exposure routes.16,67 To fill this gap of knowledge, we report here the cellular responses of macrophages to either acute or repeated exposure to silver nanoparticles, using an in vitro model. Primary macrophages cannot be maintained in vitro for a long period of time, which limits their use to a few days of exposure.51 In the present study, the response of a macrophage cell line to silver nanoparticles was analysed, in such a way that cells were exposed at confluence, thereby limiting the effect of proliferation on cellular responses. This system is different from those used previously in other cell types, which are regularly split during the exposure period.12–15 This difference is however in line with the in vivo physiology, as resident macrophages have a long life span68 and self-renew slowly,69 a feature that is mimicked by our confluent culture system.
Besides the culture scheme, the exposure dose used is also an important factor. We have used a dose (1 ppm per day) that is much higher than the dose previously used on keratinocytes (0.4 ppb),11 but it is similar to doses used in recent papers on bronchial15 and intestinal14 cells. The latter doses have been calculated based on exposure data, so we are confident that the dose that we used should not be irrelevant as compared to real in vivo doses. Furthermore, the existence of localized argyria under some clinical circumstances34 calls for the possibility of relatively high doses released under a chronic exposure scheme.
In addition to the chronic exposure scheme, we added a control experiment with the same cumulative dose delivered acutely, i.e. over 24 hours. This control experiment has been omitted for some studies on epithelial cells,13–15 but included in other studies.11,12,65,70 As these previous studies on epithelial cells have shown that short- and long-term responses are very different, such a control may have been seen as superfluous. However, macrophages are highly phagocytic cells that store the material that they cannot degrade (e.g. mineral material) as exemplified in.67 Thus, it can be argued that a single exposure to the total dose may recapitulate what is observed upon repeated exposure, and this is why we included this control experiment. As cell aging has been shown to be an important parameter,11 the acute exposure control could be performed on aged cells or on fresh cells. In line with a scheme validated by previous publications,11,12 we decided to perform the control on fresh cells, as it allows the investigation of the molecular effects of cell aging, especially when using “omic” approaches (e.g. in ref. 11). More importantly, this scheme also allows the comparison of the data obtained in the acute exposure scheme with those present in the nanotoxicology literature, where fresh cells are always used.
Using this in vitro system, we showed that a repeated low dose exposure produced a stronger impact on cells compared to a single dose exposure at the same theoretical cumulative dose. Targeted experiments confirmed that a repeated exposure produces stronger effects on cellular metabolism and on specialized macrophages functions (e.g. phagocytosis and cytokine production) than a single high dose exposure. This suggests that chronic exposure to silver nanoparticles may produce stronger disturbances in the function of the innate immune system than acute exposure. Moreover, the set of proteins with modified intracellular content differs in the repeated exposure scheme compared to the acute one. In response to the chronic exposure scheme, the highlighted proteins belong essentially to antioxidant responses (e.g. glutathione reductase, glutamate cysteine ligase, flavin reductase), cytoskeletal proteins (e.g. plastin, cofilin, actin capping protein) energy metabolism (e.g. ATP synthase, respiratory complexes I–III, pyruvate dehydrogenase, oxoglutarate dehydrogenase, triose phosphate isomerase) and phagolysosomal proteins (e.g. cathepsins B, D, S and Z). Quite interestingly, these proteins did not respond to the acute exposure scheme. This suggests that the innate immune response may be affected differently by chronic and acute exposures to silver nanoparticles. Therefore, effort should be put in generating data from chronic exposure scenarios, which are more realistic.
The next step was then to correlate the intensity of the impact on cells with the amount of internalized silver. There is ample evidence in the literature that particulate silver is very weakly toxic,2,20 and that the toxicity of silver nanoparticles is mediated by the intracellular release of silver ions.2,20,71,72 We thus measured total and soluble silver levels for both exposure schemes.
Measurements of total silver levels showed that they were lower for the repeated exposure scheme. This could be due to cell renewal over time, a decrease in silver uptake upon cell aging or a progressive effect of silver internalization on the phagocytic process itself, as supported by the lower phagocytic capacity observed for chronically-exposed cells.
It could be argued, however, that repeated exposure could result in a higher dissolution of silver because of longer dissolution time, thus explaining the stronger effects of a repeated exposure. However, we showed that dissolved silver intracellular content was constant for nontoxic doses. This suggests a sustained release of silver from cells, as documented in primary macrophages,51 probably via a complexation to glutathione.52 This hypothesis is further supported by the decrease in the free glutathione levels that we observed in repeatedly-exposed cells, a mechanism that may contribute to the deleterious effects of silver.73 When toxicity begins to appear, i.e. at 20 μg ml−1 cumulated dose, the soluble silver content diverged between chronically-exposed cells and acutely-exposed cells. In acutely-exposed cells, the soluble silver level increased, suggesting that the systems controlling intracellular silver ion content became overwhelmed, leading to toxicity. In chronically-exposed cells, the soluble silver level was close to the levels observed for earlier time points, suggesting a different toxicity mechanism, based on progressive attrition of cellular functions at this low but sustained silver ion concentration.
Conclusions
Thus, the overall and major conclusion of this work is that even with a lower intracellular release of silver ions, chronic exposure to silver nanoparticles, if sustained for enough time (e.g. in comparison with a four-day exposure51) induces a progressive “wearing-down” of macrophage functions, which was not observed for an acute exposure to the same cumulative dose. This phenomenon may be part of the picture explaining the silver chronic toxicity observed in several in vivo models.6–10 Indeed, body barriers are the combination of an epithelium that forms a physical barrier and of scavenging cells that either remove particulate material that has crossed the epithelial barrier (e.g. in skin or intestine), or keep the epithelial surface clean for exchanges (e.g. in lung alveolae). Moreover, it is now known that both cell types (epithelial and innate immune cells) can trigger an immune response. Thus, our study on macrophages complements previous studies on epithelial cells,13–15 and shows that very different cell types share a deleterious response to chronic exposure to silver nanoparticles, even at sub-toxic doses. Put in the broader context of long term silver toxicity, the in vitro studies using repeated exposure to low doses shall be seen as a first step to bridge the current gap existing between classical in vitro toxicology using single exposures and results obtained with chronic exposures on animals.Authors contributions
Experimental contributions
BD and CAG performed the mitochondrial potential, phagocytosis, nitric oxide and cytokine production experiments, and the cell cultures required for these experiments. VCF performed the 2D gel experiments. HD and SC performed the mass spectrometry identifications of the proteins, while TR, JB and CC performed the computerized analysis of the proteomic experiments. CMD, JP and MD performed the ICP measurements. MC performed the nanoparticles characterization. TR provided the cell cultures needed for proteomics, and performed the glutathione assay experiments and the enzyme assays.Redactional contributions
TR and MC drafted the initial manuscript, for which all the coauthors in charge of some experiments drafted the section corresponding to their experiments. The manuscript was critically reviewed, amended and approved by all co-authors.Conflicts of interest
The authors report no conflicts of interest.Acknowledgements
This work was funded by the CNRS, The University of Grenoble, the University of Strasbourg Unistra, the Région Alsace, the French National Research Program for Environmental and Occupational Health of ANSES (PNREST 2011/025, Innimunotox Grant) and the toxicology project of the CEA (Nanostress grant).This work is a contribution to the Labex Serenade (no. ANR-11-LABX-0064) funded by the “Investissements d'Avenir” French Government program of the French National Research Agency (ANR) through the A*MIDEX project (no. ANR-11-IDEX-0001-02).
This work used the platforms of the French Proteomic Infrastructure (ProFI) project (grant ANR-10-INBS-08-03).
The authors would like to thank K. Pernet-Gallay for help in TEM image acquisition.
Lastly, BD thanks the CNRS for a handicap PhD fellowship.
References
- C. A. Dos Santos, M. M. Seckler, A. P. Ingle, I. Gupta, S. Galdiero, M. Galdiero, A. Gade and M. Rai, Silver nanoparticles: therapeutical uses, toxicity, and safety issues, J. Pharm. Sci., 2014, 103, 1931–1944 CrossRef CAS PubMed.
- V. De Matteis, M. A. Malvindi, A. Galeone, V. Brunetti, E. De Luca, S. Kote, P. Kshirsagar, S. Sabella, G. Bardi and P. P. Pompa, Negligible particle-specific toxicity mechanism of silver nanoparticles: the role of Ag+ ion release in the cytosol, Nanomedicine, 2015, 11, 731–739 CrossRef CAS PubMed.
- X. Q. Wang, H. E. Chang, R. Francis, H. Olszowy, P. Y. Liu, M. Kempf, L. Cuttle, O. Kravchuk, G. E. Phillips and R. M. Kimble, Silver deposits in cutaneous burn scar tissue is a common phenomenon following application of a silver dressing, J. Cutaneous Pathol., 2009, 36, 788–792 CrossRef PubMed.
- P. Berger, J. B. Ricco, P. Liqui Lung and F. L. Moll, Localized argyria caused by metallic silver aortic grafts: a unique adverse effect, J. Geophys. Res. Biogeosci, 2013, 46, 565–568 CAS.
- B. A. Weldon, E. M. Faustman, G. Oberdorster, T. Workman, W. C. Griffith, C. Kneuer and I. J. Yu, Occupational exposure limit for silver nanoparticles: considerations on the derivation of a general health-based value, Nanotoxicology, 2016, 10, 945–956 CrossRef CAS PubMed.
- J. H. Sung, J. H. Ji, J. U. Yoon, D. S. Kim, M. Y. Song, J. Jeong, B. S. Han, J. H. Han, Y. H. Chung, J. Kim, T. S. Kim, H. K. Chang, E. J. Lee, J. H. Lee and I. J. Yu, Lung function changes in Sprague-Dawley rats after prolonged inhalation exposure to silver nanoparticles, Inhalation Toxicol., 2008, 20, 567–574 CrossRef CAS PubMed.
- J. H. Sung, J. H. Ji, J. D. Park, J. U. Yoon, D. S. Kim, K. S. Jeon, M. Y. Song, J. Jeong, B. S. Han, J. H. Han, Y. H. Chung, H. K. Chang, J. H. Lee, M. H. Cho, B. J. Kelman and I. J. Yu, Subchronic inhalation toxicity of silver nanoparticles, Toxicol. Sci., 2009, 108, 452–461 CrossRef CAS PubMed.
- E. J. Park, E. Bae, J. Yi, Y. Kim, K. Choi, S. H. Lee, J. Yoon, B. C. Lee and K. Park, Repeated-dose toxicity and inflammatory responses in mice by oral administration of silver nanoparticles, Environ. Toxicol. Pharmacol., 2010, 30, 162–168 CrossRef CAS PubMed.
- W. H. De Jong, L. T. Van Der Ven, A. Sleijffers, M. V. Park, E. H. Jansen, H. Van Loveren and R. J. Vandebriel, Systemic and immunotoxicity of silver nanoparticles in an intravenous 28 days repeated dose toxicity study in rats, Biomaterials, 2013, 34, 8333–8343 CrossRef CAS PubMed.
- R. Ebabe Elle, S. Gaillet, J. Vide, C. Romain, C. Lauret, N. Rugani, J. P. Cristol and J. M. Rouanet, Dietary exposure to silver nanoparticles in Sprague-Dawley rats: effects on oxidative stress and inflammation, Food Chem. Toxicol., 2013, 60, 297–301 CrossRef CAS.
- K. K. Comfort, L. K. Braydich-Stolle, E. I. Maurer and S. M. Hussain, Less is more: long-term in vitro exposure to low levels of silver nanoparticles provides new insights for nanomaterial evaluation, ACS Nano, 2014, 8, 3260–3271 CrossRef CAS.
- N. Chen, Z. M. Song, H. Tang, W. S. Xi, A. Cao, Y. Liu and H. Wang, Toxicological Effects of Caco-2 Cells Following Short-Term and Long-Term Exposure to Ag Nanoparticles, Int. J. Mol. Sci., 2016, 17, E974 CrossRef PubMed.
- W. H. Choo, C. H. Park, S. E. Jung, B. Moon, H. Ahn, J. S. Ryu, K. S. Kim, Y. H. Lee, I. J. Yu and S. M. Oh, Long-term exposures to low doses of silver nanoparticles enhanced in vitro malignant cell transformation in non-tumorigenic BEAS-2B cells, Toxicol. In Vitro, 2016, 37, 41–49 CrossRef CAS PubMed.
- L. Vila, R. Marcos and A. Hernandez, Long-term effects of silver nanoparticles in caco-2 cells, Nanotoxicology, 2017, 11, 771–780 CrossRef CAS PubMed.
- A. R. Gliga, S. Di Bucchianico, J. Lindvall, B. Fadeel and H. L. Karlsson, RNA-sequencing reveals long-term effects of silver nanoparticles on human lung cells, Sci. Rep., 2018, 8, 6668 CrossRef PubMed.
- A. Kermanizadeh, C. Chauche, D. Balharry, D. M. Brown, N. Kanase, J. Boczkowski, S. Lanone and V. Stone, The role of Kupffer cells in the hepatic response to silver nanoparticles, Nanotoxicology, 2014, 8, 149–154 CrossRef CAS.
- A. N. Yilma, S. R. Singh, S. Dixit and V. A. Dennis, Anti-inflammatory effects of silver-polyvinyl pyrrolidone (Ag-PVP) nanoparticles in mouse macrophages infected with live Chlamydia trachomatis, Int. J. Nanomed., 2013, 8, 2421–2432 Search PubMed.
- E.-J. Park, J. Yi, Y. Kim, K. Choi and K. Park, Silver nanoparticles induce cytotoxicity by a Trojan-horse type mechanism, Toxicol. In Vitro, 2010, 24, 872–878 CrossRef CAS PubMed.
- K. C. Nguyen, L. Richards, A. Massarsky, T. W. Moon and A. F. Tayabali, Toxicological evaluation of representative silver nanoparticles in macrophages and epithelial cells, Toxicol. In Vitro, 2016, 33, 163–173 CrossRef CAS.
- A. Pratsinis, P. Hervella, J. C. Leroux, S. E. Pratsinis and G. A. Sotiriou, Toxicity of Silver Nanoparticles in Macrophages, Small, 2013, 9, 2576–2584 CrossRef CAS PubMed.
- M.-H. Cha, T. Rhim, K. H. Kim, A.-S. Jang, Y.-K. Paik and C.-S. Park, Proteomic identification of macrophage migration-inhibitory factor upon exposure to TiO2 particles, Mol. Cell. Proteomics, 2007, 6, 56–63 CrossRef CAS PubMed.
- Y. Y. Tsai, Y. H. Huang, Y. L. Chao, K. Y. Hu, L. T. Chin, S. H. Chou, A. L. Hour, Y. D. Yao, C. S. Tu, Y. J. Liang, C. Y. Tsai, H. Y. Wu, S. W. Tan and H. M. Chen, Identification of the Nanogold Particle-Induced Endoplasmic Reticulum Stress by Omic Techniques and Systems Biology Analysis, ACS Nano, 2011, 5, 9354–9369 CrossRef CAS PubMed.
- J. G. Teeguarden, B. J. Webb-Robertson, K. M. Waters, A. R. Murray, E. R. Kisin, S. M. Varnum, J. M. Jacobs, J. G. Pounds, R. C. Zanger and A. A. Shvedova, Comparative Proteomics and Pulmonary Toxicity of Instilled Single-Walled Carbon Nanotubes, Crocidolite Asbestos, and Ultrafine Carbon Black in Mice, Toxicol. Sci., 2011, 120, 123–135 CrossRef CAS PubMed.
- H. Karlsson, J. Lindbom, B. Ghafouri, M. Lindahl, C. Tagesson, M. Gustafsson and A. G. Ljungman, Wear Particles from Studded Tires and Granite Pavement Induce Pro-inflammatory Alterations in Human Monocyte-Derived Macrophages: A Proteomic Study, Chem. Res. Toxicol., 2011, 24, 45–53 Search PubMed.
- O. Okoturo-Evans, A. Dybowska, E. Valsami-Jones, J. Cupitt, M. Gierula, A. R. Boobis and R. J. Edwards, Elucidation of Toxicity Pathways in Lung Epithelial Cells Induced by Silicon Dioxide Nanoparticles, PLoS One, 2013, 8, e72363 CrossRef CAS PubMed.
- S. Triboulet, C. Aude-Garcia, L. Armand, V. Collin-Faure, M. Chevallet, H. Diemer, A. Gerdil, F. Proamer, J. M. Strub, A. Habert, N. Herlin, A. Van Dorsselaer, M. Carriere and T. Rabilloud, Comparative proteomic analysis of the molecular responses of mouse macrophages to titanium dioxide and copper oxide nanoparticles unravels some toxic mechanisms for copper oxide nanoparticles in macrophages, PLoS One, 2015, 10, e0124496 CrossRef PubMed.
- B. Dalzon, C. Aude-Garcia, V. Collin-Faure, H. Diemer, D. Beal, F. Dussert, D. Fenel, G. Schoehn, S. Cianferani, M. Carriere and T. Rabilloud, Differential proteomics highlights macrophage-specific responses to amorphous silica nanoparticles, Nanoscale, 2017, 9, 9641–9658 RSC.
- T. Verano-Braga, R. Miethling-Graff, K. Wojdyla, A. Rogowska-Wrzesinska, J. R. Brewer, H. Erdmann and F. Kjeldsen, Insights into the cellular response triggered by silver nanoparticles using quantitative proteomics, ACS Nano, 2014, 8, 2161–2175 CrossRef CAS PubMed.
- A. Oberemm, U. Hansen, L. Bohmert, C. Meckert, A. Braeuning, A. F. Thunemann and A. Lampen, Proteomic responses of human intestinal Caco-2 cells exposed to silver nanoparticles and ionic silver, J. Appl. Toxicol., 2016, 36, 404–413 CrossRef CAS.
- A. Braeuning, A. Oberemm, J. Gorte, L. Bohmert, S. Juling and A. Lampen, Comparative proteomic analysis of silver nanoparticle effects in human liver and intestinal cells, J. Appl. Toxicol., 2018, 38, 638–648 CrossRef CAS.
- W. E. Hathaway, L. A. Newby and J. H. Githens, The Acridine Orange Viability Test Applied to Bone Marrow Cells. I. Correlation with Trypan Blue and Eosin Dye Exclusion and Tissue Culture Transformation, Blood, 1964, 23, 517–525 CrossRef CAS PubMed.
- A. Moore, C. J. Donahue, K. D. Bauer and J. P. Mather, Simultaneous measurement of cell cycle and apoptotic cell death, Methods Cell Biol., 1998, 57, 265–278 CrossRef CAS PubMed.
- S. Triboulet, C. Aude-Garcia, M. Carriere, H. Diemer, F. Proamer, A. Habert, M. Chevallet, V. Collin-Faure, J. M. Strub, D. Hanau, A. Van Dorsselaer, N. Herlin-Boime and T. Rabilloud, Molecular responses of mouse macrophages to copper and copper oxide nanoparticles inferred from proteomic analyses, Mol. Cell. Proteomics, 2013, 12, 3108–3122 CrossRef CAS PubMed.
- S. W. Perry, J. P. Norman, J. Barbieri, E. B. Brown and H. A. Gelbard, Mitochondrial membrane potential probes and the proton gradient: a practical usage guide, BioTechniques, 2011, 50, 98–115 CrossRef CAS PubMed.
- K. M. Mayer and F. H. Arnold, A colorimetric assay to quantify dehydrogenase activity in crude cell lysates, J. Biomol. Screening, 2002, 7, 135–140 CrossRef CAS PubMed.
- B. Plaut and J. R. Knowles, pH-dependence of the triose phosphate isomerase reaction, Biochem. J., 1972, 129, 311–320 CrossRef CAS PubMed.
- I. Carlberg and B. Mannervik, Glutathione reductase, Methods Enzymol., 1985, 113, 484–490 CAS.
- M. Warholm, C. Guthenberg, C. von Bahr and B. Mannervik, Glutathione transferases from human liver, Methods Enzymol., 1985, 113, 499–504 CAS.
- B. Dalzon, J. Bons, H. Diemer, V. Collin-Faure, C. Marie-Desvergne, M. Dubosson, S. Cianferani, C. Carapito and T. Rabilloud, A Proteomic View of Cellular Responses to Anticancer Quinoline-Copper Complexes, Proteomes, 2019, 7, E26 CrossRef PubMed.
- B. A. Arrick, C. F. Nathan, O. W. Griffith and Z. A. Cohn, Glutathione depletion sensitizes tumor cells to oxidative cytolysis, J. Biol. Chem., 1982, 257, 1231–1237 CAS.
- I. M. Bruggeman, A. Spenkelink, J. H. Temmink and P. J. van Bladeren, Differential effects of raising and lowering intracellular glutathione levels on the cytotoxicity of allyl isothiocyanate, tert-butylhydroperoxide and chlorodinitrobenzene, Toxicol. In Vitro, 1988, 2, 31–35 CrossRef CAS.
- T. Rabilloud, Optimization of the cydex blue assay: A one-step colorimetric protein assay using cyclodextrins and compatible with detergents and reducers, PLoS One, 2018, 13, e0195755 CrossRef PubMed.
- O. Hammer, D. A. T. Harper and P. D. Ryan, Paleontological statistics software package for education and data analysis, Palaeontol. Electron., 2001, 4, XIX–XX Search PubMed.
- Y. Perez-Riverol, A. Csordas, J. Bai, M. Bernal-Llinares, S. Hewapathirana, D. J. Kundu, A. Inuganti, J. Griss, G. Mayer, M. Eisenacher, E. Perez, J. Uszkoreit, J. Pfeuffer, T. Sachsenberg, S. Yilmaz, S. Tiwary, J. Cox, E. Audain, M. Walzer, A. F. Jarnuczak, T. Ternent, A. Brazma and J. A. Vizcaino, The PRIDE database and related tools and resources in 2019: improving support for quantification data, Nucleic Acids Res., 2019, 47, D442–D450 CrossRef CAS PubMed.
- D. Yekutieli and Y. Benjamini, Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics, J. Stat. Plan. Inference, 1999, 82, 171–196 CrossRef.
- A. Carvajal-Rodriguez and J. de Una-Alvarez, Assessing significance in high-throughput experiments by sequential goodness of fit and q-value estimation, PLoS One, 2011, 6, e24700 CrossRef CAS PubMed.
- A. P. Diz, A. Carvajal-Rodriguez and D. O. Skibinski, Multiple hypothesis testing in proteomics: a strategy for experimental work, Mol. Cell. Proteomics, 2011, 10, M110 004374 CrossRef PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res., 2009, 37, 1–13 CrossRef CAS PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS PubMed.
- Y. Chen, H. G. Shertzer, S. N. Schneider, D. W. Nebert and T. P. Dalton, Glutamate cysteine ligase catalysis: dependence on ATP and modifier subunit for regulation of tissue glutathione levels, J. Biol. Chem., 2005, 280, 33766–33774 CrossRef CAS PubMed.
- C. Aude-Garcia, F. Villiers, V. Collin-Faure, K. Pernet-Gallay, P. H. Jouneau, S. Sorieul, G. Mure, A. Gerdil, N. Herlin-Boime, M. Carriere and T. Rabilloud, Different in vitro exposure regimens of murine primary macrophages to silver nanoparticles induce different fates of nanoparticles and different toxicological and functional consequences, Nanotoxicology, 2016, 10, 586–596 CrossRef CAS PubMed.
- G. Veronesi, C. Aude-Garcia, I. Kieffer, T. Gallon, P. Delangle, N. Herlin-Boime, T. Rabilloud and M. Carriere, Exposure-dependent Ag+ release from silver nanoparticles and its complexation in AgS2 sites in primary murine macrophages, Nanoscale, 2015, 7, 7323–7330 RSC.
- S. J. Berger, D. Gosky, E. Zborowska, J. K. Willson and N. A. Berger, Sensitive enzymatic cycling assay for glutathione: measurements of glutathione content and its modulation by buthionine sulfoximine in vivo and in vitro in human colon cancer, Cancer Res., 1994, 54, 4077–4083 CAS.
- J. F. Kalinich, C. A. Emond, T. K. Dalton, S. R. Mog, G. D. Coleman, J. E. Kordell, A. C. Miller and D. E. McClain, Embedded weapons-grade tungsten alloy shrapnel rapidly induces metastatic high-grade rhabdomyosarcomas in F344 rats, Environ. Health Perspect., 2005, 113, 729–734 CrossRef CAS.
- D. Toybou, C. Celle, C. Aude-Garcia, T. Rabilloud and J.-P. Simonato, A toxicology-informed, safer by design approach for the fabrication of transparent electrodes based on silver nanowires, Environ. Sci.: Nano, 2019, 6, 684–694 RSC.
- L. Kvitek, A. Panacek, J. Soukupova, M. Kolar, R. Vecerova, R. Prucek, M. Holecova and R. Zboril, Effect of surfactants and polymers on stability and antibacterial activity of silver nanoparticles (NPs), J. Phys. Chem. C, 2008, 112, 5825–5834 CrossRef CAS.
- M. A. H. Khondoker, S. C. Mun and J. Kim, Synthesis and characterization of conductive silver ink for electrode printing on cellulose film, Appl. Phys. A: Mater. Sci. Process., 2013, 112, 411–418 CrossRef CAS.
- D. Mau Chien, D. Thi My Dung and E. Fribourg-Blanc, Silver nanoparticles ink synthesis for conductive patterns fabrication using inkjet printing technology, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2015, 6, 015003 Search PubMed.
- M. Ismail and R. Jabra, Investigation the parameters affecting on the synthesis of silver nanoparticles by chemical reduction method and printing a conductive pattern, J. Mater. Environ. Sci., 2017, 8, 4152–4159 CAS.
- Q. Liu, L. Seveyrat, F. Belhora and D. Guyomar, Investigation of polymer-coated nano silver/polyurethane nanocomposites for electromechanical applications, J. Polym. Res., 2013, 20, 309 CrossRef.
- D. R. Raj, S. Prasanth, T. V. Vineeshkumar and C. Sudarsanakumar, Ammonia sensing properties of tapered plastic optical fiber coated with silver nanoparticles/PVP/PVA hybrid, Opt. Commun., 2015, 340, 86–92 CrossRef.
- S. Arora, J. M. Rajwade and K. M. Paknikar, Nanotoxicology and in vitro studies: the need of the hour, Toxicol. Appl. Pharmacol., 2012, 258, 151–165 CrossRef CAS PubMed.
- S. M. Hussain, D. B. Warheit, S. P. Ng, K. K. Comfort, C. M. Grabinski and L. K. Braydich-Stolle, At the Crossroads of Nanotoxicology in vitro: Past Achievements and Current Challenges, Toxicol. Sci., 2015, 147, 5–16 CrossRef CAS PubMed.
- A. Kermanizadeh, I. Gosens, L. MacCalman, H. Johnston, P. H. Danielsen, N. R. Jacobsen, A. G. Lenz, T. Fernandes, R. P. Schins, F. R. Cassee, H. Wallin, W. Kreyling, T. Stoeger, S. Loft, P. Moller, L. Tran and V. Stone, A Multilaboratory Toxicological Assessment of a Panel of 10 Engineered Nanomaterials to Human Health--ENPRA Project--The Highlights, Limitations, and Current and Future Challenges, J. Toxicol. Environ. Health, Part B, 2016, 19, 1–28 CAS.
- L. Armand, A. Tarantini, D. Beal, M. Biola-Clier, L. Bobyk, S. Sorieul, K. Pernet-Gallay, C. Marie-Desvergne, I. Lynch, N. Herlin-Boime and M. Carriere, Long-term exposure of A549 cells to titanium dioxide nanoparticles induces DNA damage and sensitizes cells towards genotoxic agents, Nanotoxicology, 2016, 10, 913–923 CrossRef CAS PubMed.
- M. Dorier, D. Beal, C. Marie-Desvergne, M. Dubosson, F. Barreau, E. Houdeau, N. Herlin-Boime and M. Carriere, Continuous in vitro exposure of intestinal epithelial cells to E171 food additive causes oxidative stress, inducing oxidation of DNA bases but no endoplasmic reticulum stress, Nanotoxicology, 2017, 11, 751–761 CAS.
- J. J. Powell, C. C. Ainley, R. S. Harvey, I. M. Mason, M. D. Kendall, E. A. Sankey, A. P. Dhillon and R. P. Thompson, Characterisation of inorganic microparticles in pigment cells of human gut associated lymphoid tissue, Gut, 1996, 38, 390–395 CrossRef CAS PubMed.
- J. Murphy, R. Summer, A. A. Wilson, D. N. Kotton and A. Fine, The prolonged life-span of alveolar macrophages, Am. J. Respir. Cell Mol. Biol., 2008, 38, 380–385 CrossRef CAS PubMed.
- F. Ginhoux and M. Guilliams, Tissue-Resident Macrophage Ontogeny and Homeostasis, Immunity, 2016, 44, 439–449 CrossRef CAS PubMed.
- L. Armand, M. Biola-Clier, L. Bobyk, V. Collin-Faure, H. Diemer, J. M. Strub, S. Cianferani, A. Van Dorsselaer, N. Herlin-Boime, T. Rabilloud and M. Carriere, Molecular responses of alveolar epithelial A549 cells to chronic exposure to titanium dioxide nanoparticles: A proteomic view, J. Proteomics, 2016, 134, 163–173 CrossRef CAS.
- A. R. Gliga, S. Skoglund, I. O. Wallinder, B. Fadeel and H. L. Karlsson, Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- R. F. Hamilton, S. Buckingham and A. Holian, The Effect of Size on Ag Nanosphere Toxicity in Macrophage Cell Models and Lung Epithelial Cell Lines Is Dependent on Particle Dissolution, Int. J. Mol. Sci., 2014, 15, 6815–6830 CrossRef PubMed.
- M. J. Piao, K. A. Kang, I. K. Lee, H. S. Kim, S. Kim, J. Y. Choi, J. Choi and J. W. Hyun, Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis, Toxicol. Lett., 2011, 201, 92–100 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9en01329f |
| ‡ Deceased Nov. 21st, 2018. This paper is dedicated to her memory. |
| This journal is © The Royal Society of Chemistry 2020 |