CdS nanoparticles in soil induce metabolic reprogramming in broad bean (Vicia faba L.) roots and leaves†
Liyan
Tian
 a,
Huiling
Zhang
a,
Huiling
Zhang
 a,
Xiaopeng
Zhao
a,
Xueyuan
Gu
a,
Jason C.
White
a,
Xiaopeng
Zhao
a,
Xueyuan
Gu
a,
Jason C.
White
 b,
Lijuan
Zhao
a and
Rong
Ji
b,
Lijuan
Zhao
a and
Rong
Ji
 *a
*a
aState Key Laboratory of Pollution Control and Resource Reuse, School of the Environment, Nanjing University, Nanjing 210023, China. E-mail: ji@nju.edu.cn; Fax: +86 025 8968 0581; Tel: +86 025 8968 0581
bDepartment of Analytical Chemistry, The Connecticut Agricultural Experiment Station (CAES), New Haven, Connecticut 06504, USA
First published on 25th November 2019
Abstract
The rapid development of nanotechnology has raised concern regarding the environmental toxicity of nanoparticles (NPs). However, little is known about the molecular mechanisms underlying NP toxicity in plants. Broad bean (Vicia faba L.) plants were cultivated in soil amended with 0, 10, and 100 mg cadmium sulfide (CdS)-NPs per kg soil for 4 weeks, and then the phenotypic, biochemical, and metabolic responses of the plants to CdS-NP stress were evaluated. Metabolomics analysis revealed the significant up-regulation (1.2- to 39.2-fold) of several antioxidative metabolites, including N-acetyl-5-hydroxytryptamine, 2-hydroxybutanoic acid, putrescine, and flavone, upon CdS-NP exposure, but no negative phenotypic effects were visible (plant biomass, photosynthetic pigment contents, and lipid peroxidation). This observation was in accordance with the observed regulation of antioxidative-defense-related metabolic pathways (tyrosine pathway and phenylpropanoid biosynthesis) that were identified by biological pathway analysis. Importantly, twice as many metabolites were modulated in leaves than in roots, including three nitrogen-related (purine metabolism; alanine, aspartate, and glutamate metabolism; β-alanine metabolism) and two carbon-related (pantothenate and CoA biosynthesis and carbon fixation) metabolic pathways. These results indicate that to alleviate the toxicity of CdS-NP exposure in soil, plants significantly reprogram the metabolic profiles of leaves rather than of roots, which might subsequently impact both harvest and crop quality.
Environmental significanceThe rapid development of nanotechnology has raised concern regarding the environmental toxicity of nanoparticles (NPs). However, little is known about the molecular mechanisms underlying NP toxicity in plants. Understanding toxic mechanisms in organisms at the molecular level, especially in crop plants, is important for their sustainable use and development. In this study, although none of the phenotypic parameters of broad bean plants (photosynthetic pigments contents, biomass, and lipid peroxidation) were overtly impacted in response to CdS-NPs in soil during 28 d of exposure, metabolomics revealed marked and statistically significant alterations in the metabolite profiles of plant roots and leaves. The reprogramming of antioxidant metabolite production presumably reflected the molecular defense response of the plants to CdS-NP stress. The sensitive responses of flavone, putrescine and noradrenaline in the leaves suggest the use of these compounds in legumes as biomarkers of oxidative stress induced by the presence of CdS-NPs in soil. Metabolomics might thus be a suitable approach for the early detection of soil contamination by Cd. In the plants, the reprogramming of carbon and nitrogen metabolism (including sugars, organic acids, amino acids, and N-containing compounds) alleviated the toxicity of CdS-NPs, which may have been caused by free Cd2+ ions or perhaps by a particle-specific response. Importantly, this response might adversely impact crop yield and quality in plants under long-term exposure to CdS-NPs. |
Introduction
The wide-ranging applications of cadmium sulfide nanoparticles (CdS-NPs) include biological imaging, photovoltaic technology, glasses, plastics and ceramics.1–5 However, during the fabrication, use, storage and disposal of CdS-NPs, these materials are eventually released into the environment by a number of routes, including wastewater irrigation, biosolid fertilizer application and atmospheric deposition.6 Soil is the ultimate sink for most NPs and as such, terrestrial plants are likely to be highly exposed. Since plants are primary producers in terrestrial ecosystems, an understanding of the underlying mechanisms of NP toxicity to these plant species is critical to the accurate assessment risk to humans and other species.The uptake and toxicity of Cd-containing NPs (e.g., CdS, CdSe, and CdTe) in plants have been studied in hydroponic and sand cultures with ryegrass (Lolium perenne),7 onion (Allium cepa),7 rockcress (Arabidopsis thaliana),8,9 snow pea (Pisum sativum L.),10 soybean (Tohya variety),11 zucchini (Cucurbita pepo L.),12 and a woody plant (Kandelia obovata).13 In hydroponic cultures of A. thaliana, CdS-NP (80 mg kg−1) exposure strongly inhibited chlorophyll biosynthesis and the cellular respiration rate,8 and CdSe-NP (5.8 nM) exposure decreased the content ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG).9 In soil, the interaction of NPs with soil organic matter, clay minerals, microorganisms, and macrofauna often alters the fate and toxicity of these particles.14–18 Therefore, the effects of CdS-NPs on crop plants will likely differ between plants grown in soil vs. hydroponic or sand cultures.
Most previous studies examining the exposure of NPs to plants have used phenotypic parameters as end points,19 but this approach does not provide information on the underlying mechanisms leading to the toxicity or the detoxification of NPs. More recently, transcriptomics has been used to investigate the mechanisms by which A. thaliana is able to tolerate 80 mg CdS-NPs per kg under hydroponic culture. The results showed that of the 195 genes up-regulated, 32% were associated with detoxification, represented by reactive oxygen species (ROS) metabolism and the expression of lipid transfer proteins involved in defense.8 Further studies of the effects of CdS-NPs (size <10 nm) on soybean found that the plants were able to reduce the stress caused by CdS-NPs by increasing the amino acid content in their leaves.11 Importantly, plants contain >200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different low-molecular-weight (LMW) metabolites,20 including phenolic acids, carboxylic acids, carbohydrates and amino acids. These metabolites are the end products of gene expression and changes in their contents determine the ultimate biochemical phenotype of the plant.21–25 Plant metabolism is a highly dynamic series of processes and metabolomics can provide a snapshot of the in vivo metabolic profile.26 By undertaking a quantitative analysis of metabolites, metabolomics can yield accurate information on the physiological mechanisms induced by NP exposure. In previous studies, gas chromatography-mass spectrometry (GC-MS)-based metabolomics was used to explore plant responses under various NP exposures, including Cu(OH)2, CuO, CeO2, and Ag NPs.27–30 Different NPs were shown to induce distinctive metabolic changes in plants, mainly relating to pathways involved with lipids, energy and amino acids and the production of antioxidants.28–32
000 different low-molecular-weight (LMW) metabolites,20 including phenolic acids, carboxylic acids, carbohydrates and amino acids. These metabolites are the end products of gene expression and changes in their contents determine the ultimate biochemical phenotype of the plant.21–25 Plant metabolism is a highly dynamic series of processes and metabolomics can provide a snapshot of the in vivo metabolic profile.26 By undertaking a quantitative analysis of metabolites, metabolomics can yield accurate information on the physiological mechanisms induced by NP exposure. In previous studies, gas chromatography-mass spectrometry (GC-MS)-based metabolomics was used to explore plant responses under various NP exposures, including Cu(OH)2, CuO, CeO2, and Ag NPs.27–30 Different NPs were shown to induce distinctive metabolic changes in plants, mainly relating to pathways involved with lipids, energy and amino acids and the production of antioxidants.28–32
The objectives of the present study were (1) to investigate the phenotypic and biochemical responses of broad bean plants (Vicia faba L.) exposed to CdS-NPs in soil, (2) to analyze the changes in the metabolites of broad bean plants induced by CdS-NP exposure and (3) to explore the defense mechanisms of broad bean plants against CdS-NP stress at the molecular level. Broad bean (Vicia faba L.) is a leguminous plant and was chosen as it is easy to grow, has a low fertilizer requirement and serves as a forage crop for animals and a protein source for humans.33 Moreover, broad bean is an excellent model legume, as demonstrated with a number of studies of heavy metal pollution in the environment.34 Specifically, broad bean was grown in soil amended with 0, 10, and 100 mg CdS-NPs per kg and changes in several phenotypic parameters (plant biomass, photosynthetic pigments, lipid peroxidation and total phenolic compounds) and in the root and leaf metabolites were measured. The results provide a comprehensive picture of the effects of NPs on leguminous plants at both the physiological and the molecular level, which yields information critical to the accurate assessment of risk of these materials in the environment.
Materials and methods
CdS-NP characterization
CdS-NPs (Ksp = 7.94 × 10−27) were purchased from Kela Co., Ltd., China. The hydrodynamic size and zeta potential of the particles were determined by mixing 1 and 10 mg CdS-NPs in 100 mL of nanopure water and then sonicating the suspensions for 10 min at 25 °C using a flat-tip probe sonicator (Ymnl-1000Y, Nanjing Immanuel Instrument Equipment Co. Ltd., Nanjing, PR China) at a power of 65 W in cycles of 5 s of sonication and a 5 s pause. The hydrodynamic diameter of CdS-NPs in nanopure water was 140–615 nm with an average of 294 nm, and the zeta potential was −26.3 ± 0.7 mV, as measured by dynamic light scattering (Malvern series ZEN 3500, Malvern Instruments Ltd., Worcester, UK). The morphology of CdS-NPs was examined by scanning electron microscopy (S-3400N II, Hitachi, Tokyo, Japan) (Fig. S1A in the ESI†). The CdS-NPs ranged in size from 10 to 100 nm as determined by transmission electron microscopy (JEM-200CX, JEOL, Japan) (ESI† Fig. S1B and C). This kind of CdS-NPs is sold for industrial use and could represent the contamination of CdS-NPs in the environment.Broad bean exposure
Field soil was collected from Harbin City, Heilongjiang Province, China. The soil samples were air-dried for 2 weeks and then sieved through a 2 mm mesh. The physiochemical properties of the soil, including total organic matter, total phosphorous and cation exchange capacities (CECs), are described in the ESI† Table S1. Broad bean (Vicia faba L.) seeds were purchased from Luwang Seed Co. Ltd. (Sichuan Province, China). Three treatments were established: control (no CdS-NPs), 10 mg CdS-NPs per kg, and 100 mg CdS-NPs per kg. A treatment with free Cd2+ was not established because the concentration of dissolved Cd2+ ions from CdS-NPs in deionized water at 25 °C was low (1.28 × 10−8 mg L−1) according to the solubility product (Ksp) of CdS (7.94 × 10−27).35 Black plastic boxes (each 7 cm × 5 cm × 8 cm) containing 30 holes with a diameter of 0.2 cm at the bottom were filled with 100 g of soil containing 0, 10, or 100 mg CdS-NPs per kg soil dry weight, hereafter expressed as mg CdS-NPs per kg soil. After 24 h, broad bean seeds were sown at a depth of 1 cm. Six replicate plants (one plant per pot) were grown for each treatment. The plants were cultivated for 28 days in a greenhouse under a 25 °C/20 °C day/night cycle. The relative humidity in the greenhouse was 65–70%. During the 16 h photoperiod, active radiation was set at 180 μmol m−2 s−1. The pots were watered three times (each 4 mL) daily to maintain 60% of field water capacity.Physiological responses
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min, 0.1 mL of the supernatant was mixed with 2 mL of 20% TCA and 2 mL of 0.5% thiobarbituric acid. This mixture was heated in a water bath at 95 °C for 30 min and then allowed to cool to room temperature. The UV absorbance was measured at 450, 532 and 600 nm. Lipid peroxidation was expressed as μmol malondialdehyde (MDA) equivalents per g fresh weight.37
000g for 15 min, 0.1 mL of the supernatant was mixed with 2 mL of 20% TCA and 2 mL of 0.5% thiobarbituric acid. This mixture was heated in a water bath at 95 °C for 30 min and then allowed to cool to room temperature. The UV absorbance was measured at 450, 532 and 600 nm. Lipid peroxidation was expressed as μmol malondialdehyde (MDA) equivalents per g fresh weight.37
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min, 0.1 mL of the supernatant was mixed with Folin–Ciocalteu phenol reagent (0.25 mL) and then after 1 min, with 3 mL of 5% Na2CO3 solution. The sample was allowed to stand for 1 h in the dark, after which the absorbance at 760 nm was measured. The total phenolic content of the sample was expressed as mg of gallic acid equivalents (GAE) per g fresh weight.
000g for 15 min, 0.1 mL of the supernatant was mixed with Folin–Ciocalteu phenol reagent (0.25 mL) and then after 1 min, with 3 mL of 5% Na2CO3 solution. The sample was allowed to stand for 1 h in the dark, after which the absorbance at 760 nm was measured. The total phenolic content of the sample was expressed as mg of gallic acid equivalents (GAE) per g fresh weight.
Elemental analyses
At harvest, the plants were washed first with water and then with deionized water to remove residual particles. After removal of the surface water, the leaves, stems and roots were weighed. A portion of the tissues was dried at 80 °C for 72 h and ground into powder for metal content analysis. Briefly, 0.01 g of dried tissue was digested in a mixture of 6 mL H2O2 and 4 mL HNO3 at 160 °C for 40 min using a microwave digester (Milestone Ethos Up, Italy). The resulting suspension was filtered through a membrane with 0.22 μm pore size. Inductively coupled plasma-mass spectrometry (ICP-MS, NexION-300, PerkinElmer, USA) was used to measure the Cd and Zn concentrations of the extracts. K, Ca, Mg, and Fe concentrations were quantified using ICP with optical emission spectrometry (ICP-OES, Optima 5300, PerkinElmer, USA). The standard reference material (SRM 1573a, tomato leaves purchased from NIST, Gaithersburg, USA) was digested and measured using the same procedures. The recoveries of Cd and Zn were in the range of 93–106%. To determine the carbon, nitrogen, and sulfur concentrations in plant biomass, dried plant samples were ground in an agate mortar and passed through a 35-mesh (0.5 mm) sieve. Two mg of the ground plant tissue was analyzed using an elemental analyzer (Vario EL II elemental analyzer, Elementar, Germany) (ESI† Table S2). After removal of the plants, the soil remaining in the pot was collected, mixed thoroughly and water-extracted. The levels of water-extractable Cd and micronutrients (Zn, K, Ca, Mg, and Fe) in the soil were quantified using ICP-MS and ICP-OES (ESI† Table S3).Metabolite analysis in leaves and roots
At harvest, a portion of fresh tissues was ground into powder in liquid N2 and stored at −80 °C for metabolite analysis by GC-MS.29 Briefly, 60 mg of ground roots and leaves were extracted with 360 μL of cold methanol. Forty μL of 2-chloro-L-phenylalanine in methanol (0.3 mg mL−1) was added as an internal standard and the mixture was then ultrasonicated for 30 min at room temperature. After the addition of 200 μL of chloroform, the mixture was vortexed and 400 μL of water was added. The mixture was again vortexed, ultrasonicated at room temperature for 30 min and centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C. A quality control (QC) sample was prepared by mixing aliquots of all treatment samples based on the assumption that the QC sample contained a mean concentration of all components present in the samples under investigation. An aliquot (300 μL) of the supernatant was transferred to a glass vial and dried using a centrifugal vacuum concentrator at room temperature. Eighty μL of methoxyamine hydrochloride in pyridine solution (15 mg mL−1) was subsequently added. The resultant mixture was vortexed for 2 min and incubated at 37 °C for 90 min. Eighty μL of trifluoroacetamide containing 1% trimethylchlorosilane and 20 μL of n-hexane were added to the mixture, which was then vortexed for 2 min and heated at 70 °C for 60 min. The mixture was allowed to stand at room temperature for 30 min before analysis by GC-MS (for details, see the ESI†).
000g for 10 min at 4 °C. A quality control (QC) sample was prepared by mixing aliquots of all treatment samples based on the assumption that the QC sample contained a mean concentration of all components present in the samples under investigation. An aliquot (300 μL) of the supernatant was transferred to a glass vial and dried using a centrifugal vacuum concentrator at room temperature. Eighty μL of methoxyamine hydrochloride in pyridine solution (15 mg mL−1) was subsequently added. The resultant mixture was vortexed for 2 min and incubated at 37 °C for 90 min. Eighty μL of trifluoroacetamide containing 1% trimethylchlorosilane and 20 μL of n-hexane were added to the mixture, which was then vortexed for 2 min and heated at 70 °C for 60 min. The mixture was allowed to stand at room temperature for 30 min before analysis by GC-MS (for details, see the ESI†).
Statistical analysis
The relative abundances of the metabolites were analyzed using a supervised partial least squares discriminant analysis (PLS-DA) and a multivariate analysis, both run using MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/). These analyses were conducted to normalize (by sum) the data by log transformation, such that individual features were more comparable.39 A variable importance in projection (VIP) value was obtained by an unbiased estimate of the classification error of the results of the PLS-DA analysis. A VIP value >1.5 was considered to indicate a significant impact on the metabolite of interest.40 Biological pathways were analyzed on the basis of GC-MS data using MetaboAnalyst 4.0. Significant differences in mean plant biomass, stem length, number of leaves, photosynthetic pigments, MDA, and total phenolic compounds as well as Cd and mineral nutrient concentrations, in broad bean and soil between treatments, were evaluated in a one-way ANOVA followed by a Tukey–Kramer post hoc test, performed using SPSS 24.0 software (SPSS, Chicago, USA) and plotted using GraphPad® Prism 8.0.1 (GraphPad Software Inc., USA). Differences were considered statistically significant at p < 0.05.Results and discussion
Cd uptake in broad bean
Exposure to CdS-NPs increased the Cd content of all broad bean tissues in a dose-dependent fashion (p < 0.05) (ESI† Fig. S2A), with roots > stems > leaves. The Cd content in leaves (1.0 ± 0.1 mg kg−1), stems (2.3 ± 0.4 mg kg−1), and roots (35.1 ± 7.0 mg kg−1) in the high-dose (100 mg kg−1) CdS-NP treatment was nearly 27-, 30-, and 100-fold higher than in the control, respectively. Since CdSe-NPs with the size used in previous study could not be translocated to the leaves of plants,9 as such, Cd detected in the leaves in this study was mostly from dissolved Cd2+ from CdS particles, which could be changed by the environment of the rhizosphere, such as pH and root exudates,41 whereas Cd in the roots could be attributed to both Cd2+ and CdS particles. Previous study has shown that CdS-NPs (<10 nm) in aqueous medium could be translocated to the xylem/phloem of the roots of woody plants by endocytosis.13 The concentration of water-extractable Cd in soil was significantly higher in the 100 mg kg−1 dose of CdS-NPs than in the control soil (p < 0.05) (ESI† Table S3). Because water-soluble Cd represents the readily bioavailable proportion of Cd, the higher bioaccessibility of Cd indicated a greater accumulation of Cd in broad bean tissues of the 100 mg CdS-NPs per kg treatment.Photosynthetic pigments and biomass of broad bean
The Chl a, Chl b and carotenoid content of plants exposed to different doses of CdS-NPs are shown in the ESI† Fig. S4A. Chl b levels were unchanged by either dose of CdS-NPs. Chl a and carotenoid concentrations were significantly increased in plants exposed to 10 mg CdS-NPs per kg (p < 0.05), whereas 100 mg CdS-NPs per kg had no significant effect on either parameter (ESI† Fig. S4A). A previous study reported the complete inhibition of Chl biosynthesis in A. thaliana growing in hydroponic culture with exposure to 80 mg L−1 CdS-NPs.8 The mechanism by which the lower (10 mg kg−1) but not the higher (100 mg kg−1) dose of CdS-NPs increased the Chl a and carotenoid contents of the plants is unclear but it may involve hormesis; this phenomenon was previously demonstrated for wheat plants (Triticum aestivum L.) grown in soil contaminated with Cd at 3.3 mg kg−1.42 However, the broad bean phenotype was unaffected by CdS-NPs in the current study; neither tissue biomass nor stem length was changed by particle exposure (ESI† Fig. S3).Lipid peroxidation and total phenolic compounds
MDA is a by-product of lipid peroxidation.43 As shown in the ESI† Fig. S4B, the MDA content in leaves and roots was not significantly affected by exposure to CdS-NPs, indicating that neither dose induced measurable lipid peroxidation after 28 d of exposure. Phenolic compounds are non-enzymatic antioxidants in plants that play a key role in antioxidative defense.44 CdS-NP exposure had no effect on the content of total phenolic compounds in plant roots relative to controls (ESI† Fig. S4C). However, exposure of leaves to 100 mg CdS-NPs per kg resulted in a significant decrease in phenolic content as compared to the 10 mg CdS-NPs per kg treatment (p < 0.05) but no significant difference as compared to the control. The latter could be attributed to hormetic effects of plant responses to pollutants,45 which is a concentration response phenomenon characterized by low-dose stimulation and has been observed in a wide range of taxonomic groups (including microbes, plants, and animals), i.e., in our study, lower dose of CdS-NPs had a positive effect to induce production of phenolic compounds in leaves of broad bean; the induction of oxidative stress by the higher dose could be due to Cd2+ ions released from the particles, as supported by the Cd tissue content data above, because a decrease in the content of phenolic compounds at the Cd2+ concentration of 7.0 mg L−1 was also observed in tea leaf calli.46 A long-term exposure to CdS-NPs would have visible effects on lipid peroxidation and total phenolic compounds because of the continuous dissolution of Cd2+ from CdS-NPs, which needs further investigation.Macro- and micro-nutrients in broad bean
Mineral nutrients are essential for plant metabolism. While CdS-NPs did not significantly affect the content of K, Ca, Mg, and Fe in the leaf, stem or root tissues (ESI† Table S2), the Zn content was significantly increased in roots and leaves by 12.5% and 16.2%, respectively (p < 0.05), upon exposure to 100 mg kg−1 CdS-NPs (ESI† Fig. S2B). Zn and Cd exert antagonistic effects in terms of their accumulation in plants,47 and plants have been shown to alleviate Cd-induced toxicity by preferentially taking up Zn.48 This may explain the accumulation of Zn in the broad bean plants, particularly given that the amount of water-extractable Zn in the soil of plants exposed to 10 and 100 mg CdS-NPs per kg decreased significantly (ESI† Table S3) by 26.2% and 57.8%, respectively. In addition, Zn has a role in stabilizing and protecting biomembranes against oxidative and peroxidative damage.49,50 In fact, the accumulation of Zn in plant tissues to alleviate Cd-induced stress may explain why plant biomass was unchanged (ESI† Fig. S3) and lipid peroxidation was not induced (ESI† Fig. S4B), in spite of the increase in the Cd content of the plants' tissues (ESI† Fig. S2A).Metabolomics of broad bean roots
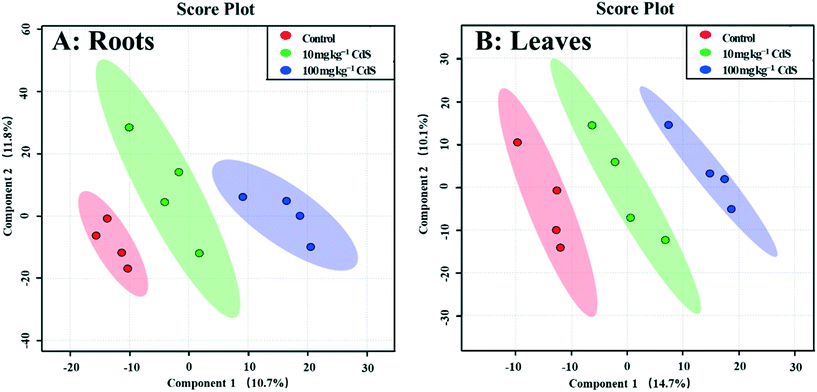
A GC-MS-based non-target metabolomics analysis identified and semi-quantified 242 metabolites in the roots of broad bean plants. The PLS-DA loading plot shows that the metabolites of plants under the two CdS-NP treatments were clearly separated from those of the control (Fig. 1), indicating that CdS-NPs altered the plant root metabolite profile. The PLS-DA model provides a VIP score for all metabolites, with a VIP >1.5 indicative of compounds most responsible for the separation.51 ESI† Fig. S5A lists the 31 metabolites with a VIP score >1.5. The levels of metabolites in roots that were significantly changed by CdS-NPs were identified with a univariate (independent t-test) and subsequent multivariate analysis (Table 1).| Metabolite | VIP | CdS-NP concentration (mg per kg soil) | |||
|---|---|---|---|---|---|
| 10 | 100 | 10 | 100 | ||
| p | Fold changea | ||||
| Amino acids and derivatives | |||||
| O-Acetylserine | 0.26 | 0.004 | 0.186 | 1.6 | 1.2 |
| Cysteinylglycine | 1.91 | 0.751 | 0.620 | 0.8 | 1.2 |
| N-Ethylglycine | 1.60 | 0.046 | 0.112 | 0.7 | 0.6 |
| Glycine-proline | 3.31 | 0.941 | 0.192 | 0.9 | 0.5 |
| Glutamine | 0.08 | 0.014 | 0.799 | 1.5 | 1.1 |
| L-Glutamic acid | 1.95 | 0.986 | 0.252 | 1.0 | 2.6 |
| N-Methyl-DL-alanine | 1.90 | 0.258 | 0.243 | 0.8 | 0.7 |
| N-Methyl-L-glutamic acid | 1.52 | 0.524 | 0.469 | 2.0 | 2.4 |
| N-Methyltryptophan | 2.20 | 0.799 | — | 1.3 | NDb |
| N-containing compounds | |||||
|---|---|---|---|---|---|
| Adipamide | 3.54 | 0.643 | 0.035 | 0.6 | 3.2 |
| N-Acetyl-5-hydroxytryptamine | 2.79 | 0.039 | 0.141 | 2.7 | 1.5 |
| 2′-Deoxyadenosine | 2.55 | 0.519 | 0.043 | 1.1 | 0.7 |
| 2,4-Diaminobutanoic acid | 1.87 | 0.909 | — | 0.9 | ND |
| DL-Dihydrosphingosine | 1.55 | — | — | NMc | NM |
| 3-Hydroxypyridine | 3.26 | 0.268 | 0.071 | 0.7 | 0.4 |
| Isoxanthopterin | 0.43 | 0.371 | 0.036 | 0.8 | 0.6 |
| Maleimide | 0.26 | 0.055 | 0.002 | 0.8 | 1.4 |
| 6-Methylmercaptopurine | 0.82 | 0.015 | 0.467 | 0.3 | 1.1 |
| Noradrenaline | 2.16 | 0.020 | 0.008 | 0.5 | 0.3 |
| Thioctamide | 3.69 | 0.707 | 0.017 | 1.5 | 4.4 |
| Uracil | 0.34 | 0.998 | 0.008 | 1.0 | 1.6 |
| Urea | 1.70 | 0.806 | 0.654 | 1.2 | 1.3 |
| Sugars and sugar alcohols | |||||
|---|---|---|---|---|---|
| Dihydrocarvone | 0.42 | 0.694 | 0.037 | 0.8 | 1.7 |
| Glucose | 1.79 | 0.589 | 0.259 | 1.1 | 0.6 |
| Palatinitol | 0.49 | 0.066 | 0.008 | 1.3 | 1.9 |
| Ribose | 0.35 | 0.256 | 0.046 | 0.8 | 0.6 |
| Sedoheptulose | 2.08 | 0.536 | 0.209 | 1.4 | 2.2 |
| Threitol | 5.00 | 0.709 | 0.018 | 1.0 | 0.2 |
| Turanose | 1.63 | 0.514 | 0.804 | 1.4 | 1.1 |
| Organic acids | |||||
|---|---|---|---|---|---|
| Benzoylformic acid | 1.84 | — | — | NM | NM |
| Digalacturonic acid | 0.15 | 0.022 | 0.246 | 1.2 | 1.2 |
| 2-Hydroxybutanoic acid | 1.44 | — | — | NM | NM |
| 3-Hydroxypropionic acid | 1.51 | 0.380 | 0.273 | 1.4 | 1.5 |
| 2-Ketovaleric acid | 1.49 | 0.175 | 0.108 | 1.2 | 0.6 |
| Toluenesulfonic acid | 1.84 | — | 0.724 | ND | 1.5 |
| Succinic acid | 0.19 | 0.021 | 0.054 | 1.5 | 1.2 |
| Others | |||||
|---|---|---|---|---|---|
| a Relative abundance of metabolites in roots in the 10 and 100 mg CdS-NP per kg treatments compared with the control. b New metabolites appeared in both CdS-NP treatments. c Not detected. | |||||
| Benzoin | 1.61 | 0.944 | 0.464 | 1.0 | 1.3 |
| 1-Hydroxyanthraquinone | 0.09 | 0.022 | 0.607 | 0.6 | 0.9 |
| 21-Hydroxypregnenolone | 2.33 | — | — | NM | NM |
| 3-Methylamino-1,2-propandiol | 0.05 | 0.024 | 0.018 | 0.8 | 0.8 |
| Naphthalene | 2.44 | 0.299 | 0.131 | 4.0 | 0.2 |
| Octanal | 1.72 | — | — | NM | NM |
| Phosphate | 2.21 | 0.273 | 0.568 | 0.9 | 0.8 |
| 2,4,6-Trihydroxybenzophenone | 3.31 | 0.261 | 0.173 | 4.6 | 4.4 |
The levels of a number of ROS scavenging root metabolites were significantly affected by both doses of CdS-NPs, including N-acetyl-5-hydroxytryptamine, noradrenaline and 2-hydroxybutanoic acid. N-Acetyl-5-hydroxytryptamine levels increased 2.7-fold and 1.5-fold in the roots of broad bean plants exposed to 10 and 100 mg CdS-NPs per kg, respectively, compared to the control (Table 1). N-Acetyl-5-hydroxytryptamine is the precursor of N-acetyl-5-methoxytryptamine, a compound that directly scavenges the ROS induced by various stress conditions.52,53 Therefore, the up-regulation of N-acetyl-5-hydroxytryptamine suggests that CdS-NPs triggered the overproduction of ROS in broad bean roots. By contrast, 10 and 100 mg CdS-NPs per kg decreased the noradrenaline content of the roots significantly by 50% and 70%, respectively (Table 1). Noradrenaline, the downstream product of dopamine, is a scavenger of free radicals in plants and may thus cause a decrease in the photosynthetic reduction of oxygen.54 A down-regulation of noradrenaline may therefore reflect an imbalance of ROS production and scavenging. 2-Hydroxybutanoic acid stimulates the production of 4-diaminobutanoic acid (GABA), a powerful antioxidant that also scavenges ROS.55,56 While 2-hydroxybutanoic acid was not detected in the control, its levels in the roots of broad bean plants were markedly increased (Table 1). Taken together, the up-regulation of N-acetyl-5-hydroxytryptamine and 2-hydroxybutanoic acid and the down-regulation of noradrenaline appear to be part of a systematic defense strategy employed by broad bean against CdS-NP-induced oxidative stress. The up-regulation of antioxidants may have contributed to the unchanged MDA content of the roots as well as the overall lack of effect on biomass.
High-dose CdS-NP exposure also caused significant changes in the levels of a number of amino acids and their derivatives. For example, O-acetylserine, cysteinylglycine, L-glutamic acid, N-methyl-L-glutamic acid and glutamine increased by 10–160%, and N-ethylglycine, N-methyltryptophan, glycine-proline, and N-methyl-DL-alanine were decreased by 30–100% (Table 1). Amino acids are not only the building blocks of proteins but also play key roles in a number of physiological pathways. Thus, the up- and down-regulation of amino acids and their derivatives may signal the alteration of protein biosynthesis and catabolism. Cysteinylglycine is a degradation product of glutathione (GSH), a soluble antioxidant that removes ROS in plants.57 Thus, an increase in this amino acid derivative in the exposed plants suggests that CdS-NPs induced oxidative stress and therefore, degradation of GSH. Glutamic acid and glutamine are related to primary nitrogen metabolism. Their up-regulation in broad bean treated with high-dose CdS-NPs suggests that primary nitrogen metabolism in the plants was not inhibited by the particles. In addition to amino acids and their derivatives, the N-containing compounds urea, uracil, maleimide, adipamide and thioctamide were significantly increased in broad bean roots exposed to a high dose of CdS-NPs (Table 1). Together, these results indicate that CdS-NPs disrupted nitrogen metabolism in the exposed plants. Urea is a nitrogen source for bacteria, algae, fungi, and plants and is involved in nitrogen fixation in legumes.58 Accordingly, the impact of CdS-NPs on nitrogen fixation merits further study.
The levels of metabolites involved in carbohydrate metabolism were also significantly changed by CdS-NP exposure. In plants exposed to 100 mg CdS-NPs per kg (Table 1), glucose, ribose, and threitol levels decreased by 40–80%, whereas palatinitol, sedoheptulose, and turanose levels were significantly up-regulated (Table 1). Sugars play an important role in signal transduction, stress defenses, and energy metabolism,59,60 while sugar alcohols serve as energy conservation compounds.61 Thus, the down-regulation of sugar levels and the up-regulation of sugar alcohol levels indicate that the plants use the analyzed sugars as an energy source in their stress defense responses, resulting in an accumulation of the respective high-energy compounds. Similarly, the levels of succinic acid, an intermediate of the TCA cycle that provides the carbon skeleton for carbohydrate biosynthesis, increased significantly by 50% and 20% in plants exposed to 10 and 100 mg kg−1 CdS-NPs, respectively (Table 1). In summary, CdS exposure impacted both nitrogen and carbon metabolisms in the roots of broad bean plants cultivated in soil.
Metabolomics of broad bean leaves
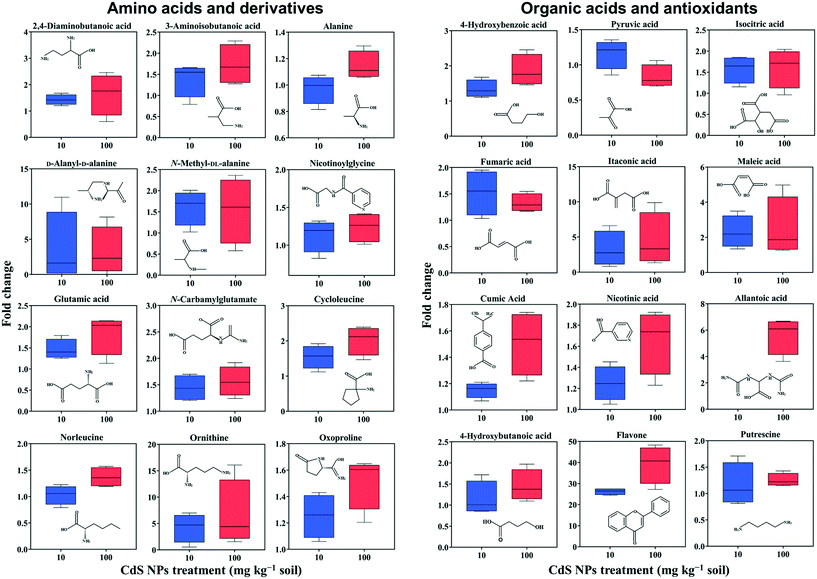
In total, 275 metabolites in broad bean leaves were identified and semi-quantified by GC-MS. Similar to the roots, the PLS-DA loading plot shows that both CdS-NP treatments were clearly separated from the control along PC1, which explained 14.7% of the total variance (Fig. 1). Interestingly, this result suggests that although the leaves were not directly exposed to CdS-NPs, the particles significantly altered the metabolite profile of bean leaves. The 87 metabolites that changed significantly are listed in the ESI† Table S4, and the 28 metabolites with a VIP score >1.5 are shown in the ESI† Fig. S5B. Flavone, putrescine, and noradrenaline are three metabolites associated with ROS removal and antioxidative defense and their levels increased significantly in response to both doses of CdS-NPs (Fig. 2). Levels of the flavonoid flavone, which has a high ROS-scavenging ability,62 were increased 13- and 39-fold, respectively, following exposure of the plants to 10 and 100 mg CdS-NPs per kg (Fig. 2). Flavonoids play different roles in the ecology of plants, including as stress protectants and signaling molecules.17,63–65 In addition, the levels of the common polyamine putrescine increased 30% in leaves exposed to the high dose of CdS-NPs compared to the control (p > 0.05) (Fig. 2). Putrescine scavenges ROS, activates antioxidants and protects biomembranes and biomolecules by inducing metal chelation.66 Noradrenaline levels in leaves were also significantly increased (ESI† Table S4). As discussed above, noradrenaline exhibits antioxidative activity in addition to its participation in many cellular processes.54 Taken together, the up-regulation of three antioxidants in broad bean leaves indicates active detoxification by the plants to achieve oxidative equilibrium and thus alleviate the oxidative stress induced by CdS-NPs. This high level of response explains the absence of oxidative damage and phenotypic changes in the leaves of exposed plants.Similar to roots, significant changes in amino acids and their derivatives were also observed in the leaves. The levels of alanine, glutamic acid, norleucine, cycloleucine, ornithine and oxoproline increased significantly in response to 100 mg CdS-NPs per kg as compared to the control (p < 0.05) (Fig. 2). An increase in amino acid levels in soybean leaves exposed to CdS-NPs (size <10 nm) as determined by liquid chromatography coupled to MS was previously reported.11 The amino acid glutamic acid is involved in ammonia assimilation,67 and its levels increased 1.8-fold under high-dose CdS-NP exposure (Fig. 2). The levels of ornithine, the major nitrogenous compound in nodulated and non-nodulated plants,68 increased 6.6-fold under high-dose CdS-NP exposure (Fig. 2). Allantoic acid is a component of the soluble nitrogen pool of nodulated plants,68 and its levels increased 4.2-fold under high-dose CdS-NP exposure (Fig. 2). These data suggested a positive impact of CdS-NPs on plant nitrogen metabolism, and although the mechanisms are unclear, the increased synthesis of storage proteins may provide a means of detoxifying these particles.
A high-dose CdS-NPs also significantly increased the levels of 2,4-diaminobutanoic acid and 3-aminobutanoic acid in broad bean leaves (Fig. 2). These two compounds are derivatives of GABA, a non-protein amino acid. In addition, the levels of both a GABA degradation product (4-hydroxybutanoic acid) and a GABA precursor (putrescine) increased significantly (p < 0.05) in leaves exposed to CdS-NPs (Fig. 2). The up-regulation of these four GABA-related compounds has previously been shown to be part of an active plant response to biotic and abiotic stresses.64,65,69 Cysteinylglycine in leaves (ESI† Table S4) increased by 12% in response to 100 mg CdS-NPs per kg, very similar to the results in the roots (Table 1). As mentioned above, cysteinylglycine is the degradation product of the antioxidant GSH. Thus, its up-regulation suggests that CdS-NPs activated GSH to remove excess amounts of ROS in both the roots and the leaves of broad bean plants.
Nitrogen and carbon metabolism are linked through the TCA cycle. High-dose CdS-NPs up-regulated several TCA cycle intermediates in the leaves, including isocitric acid, fumaric acid, itaconic acid and maleic acid (Fig. 2),70–72 indicating enhanced activation of the TCA cycle in the exposed plants.
Nicotinic acid is a key component of plant pathways involved in redox homeostasis and stress signaling.73 The up-regulation of nicotinic acid in broad bean plants exposed to 100 mg CdS-NPs per kg (Fig. 2) suggests a defense response of the leaves to oxidative stress induced by the particles. This conclusion is supported by the significant increase of the antioxidant 4-hydroxybenzoic acid in the leaves of exposed plants (Fig. 2).
Pathway disruption in broad bean
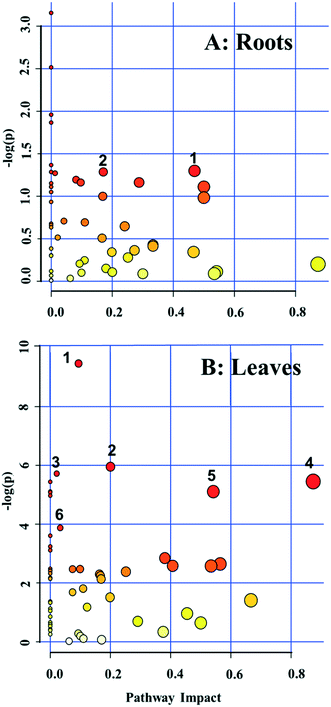
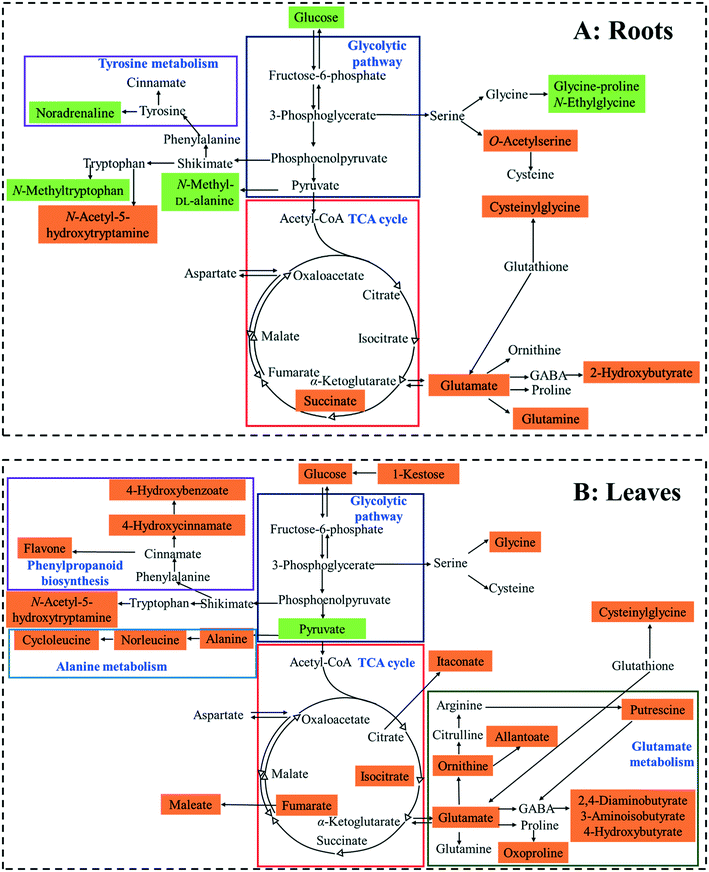
A pathway enrichment analysis was performed using the online tool MetaboAnalyst 4.0. The results are shown in Fig. 3. In the roots, two biological pathways, glyoxylate/dicarboxylate metabolism and tyrosine metabolism, were significantly changed in plants exposed to the higher dose of CdS-NPs (p < 0.05) (Fig. 3A). This finding is consistent with the above-discussed disruption of carbon and nitrogen metabolism in the roots of treated plants (summarized in Fig. 4A).In the leaves, pathway analysis revealed that six metabolic pathways were significantly (p < 0.05) disrupted (Fig. 3B): phenylpropanoid biosynthesis; pantothenate and CoA biosynthesis; purine metabolism; alanine, aspartate, and glutamate metabolism; β-alanine metabolism; and carbon fixation. The larger number of disturbed metabolic pathways in the leaves than in the roots was unexpected, given that the leaves were not directly exposed to the CdS-NPs. Thus, low-molecular-weight metabolites would seem to be a more sensitive endpoint than phenotypic parameters in toxicity assessments.28 Among these six biological pathways, the phenylpropanoid pathway is involved in all plant responses to biotic and abiotic stresses and in the activation of plant defense systems,74 which in this study included the production of a variety of antioxidative metabolites in leaves stressed by CdS-NP exposure (Fig. 4B). Three pathways related to nitrogen metabolism (purine metabolism, alanine, aspartate, and glutamate metabolism and β-alanine metabolism) and two related to carbon metabolism (pantothenate and CoA biosynthesis and carbon fixation) were also activated. Taken together, the analysis shows that CdS-NPs, especially at the higher dose, significantly activated the antioxidative defense system of broad bean plants and also impacted carbon and nitrogen metabolism in their leaves, which are markedly more sensitive than the roots to CdS-NP-induced stress.
Environmental implications
Although none of the phenotypic parameters of broad bean plants (photosynthetic pigment contents, biomass, and lipid peroxidation) were overtly impacted in response to CdS-NPs in soil during 28 d of exposure, metabolomics revealed marked and statistically significant alterations in the metabolite profiles of plant roots and leaves. The reprogramming of antioxidant metabolite production presumably reflected the molecular defense response of the plants to CdS-NP stress. The sensitive responses of flavone, putrescine and noradrenaline in the leaves suggest the use of these compounds in legumes as biomarkers of oxidative stress induced by the presence of CdS-NPs in soil. Metabolomics might thus be a suitable approach for the early detection of soil contamination by Cd.In the plants, the reprogramming of carbon and nitrogen metabolism (including sugars, organic acids, amino acids, and N-containing compounds) alleviated the toxicity of CdS-NPs, which may have been caused by free Cd2+ ions or perhaps by a particle-specific response. Importantly, this response might adversely impact crop yield and quality in plants under long-term exposure to CdS-NPs.
Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This study was financially supported by the National Key Research and Development Program of China (no. 2016YFD0800207), the National Natural Science Foundation of China (no. 31861133003, 41571130061), and the United States Department of Agriculture–National Institute of Food and Agriculture (USDA–NIFA CONH00147). We thank Dr. Xiang-dong Li for his suggestions to improve the manuscript.References
- L. Cheng, Q. J. Xiang, Y. L. Liao and H. W. Zhang, CdS-based photocatalysts, Energy Environ. Sci., 2018, 11, 1362–1391 RSC.
- NDRC, Medium and long-term development plan for renewable energy in China, Beijing, PR China: National Development and Reform Commission Website, 2007.
- D. W. Wakerley, M. F. Kuehnel, K. L. Orchard, K. H. Ly, T. E. Rosser and E. Reisner, Solar-driven reforming of lignocellulose to H2 with a CdS/CdOx photocatalyst, Nat. Energy, 2017, 2, 17021 CrossRef CAS.
- UNEP, Final review of scientific information on cadmium, Chemicals Branch DTIE, UN, 2010, pp. 87–89 Search PubMed.
- O. Chen, J. Zhao, V. P. Chauhan, J. Cui, C. Wong, D. K. Harris, H. Wei, H.-S. Han, D. Fukumura, R. K. Jain and M. G. Bawendi, Compact high-quality CdSe-CdS core-shell nanocrystals with narrow emission linewidths and suppressed blinking, Nat. Mater., 2013, 12, 445 CrossRef CAS PubMed.
- M. E. Vance, T. Kuiken, E. P. Vejerano, S. P. McGinnis, M. F. Hochella, Jr., D. Rejeski and M. S. Hull, Nanotechnology in the real world: redeveloping the nanomaterial consumer products inventory, Beilstein J. Nanotechnol., 2015, 6, 1769–1780 CrossRef CAS PubMed.
- N. Al-Salim, E. Barraclough, E. Burgess, B. Clothier, M. Deurer, S. Green, L. Malone and G. Weir, Quantum dot transport in soil, plants, and insects, Sci. Total Environ., 2011, 409, 3237–3248 CrossRef CAS PubMed.
- M. Marmiroli, L. Pagano, M. L. Savo Sardaro, M. Villani and N. Marmiroli, Genome-wide approach in Arabidopsis thaliana to assess the toxicity of cadmium sulfide quantum dots, Environ. Sci. Technol., 2014, 48, 5902–5909 CrossRef CAS PubMed.
- D. A. Navarro, M. A. Bisson and D. S. Aga, Investigating uptake of water-dispersible CdSe/ZnS quantum dot nanoparticles by Arabidopsis thaliana plants, J. Hazard. Mater., 2012, 211–212, 427–435 CrossRef CAS PubMed.
- S. Das, B. P. Wolfson, L. Tetard, J. Tharkur, J. Bazata and S. Santra, Effect of N-acetyl cysteine coated CdS:Mn/ZnS quantum dots on seed germination and seedling growth of snow pea (Pisum sativum L.): imaging and spectroscopic studies, Environ. Sci.: Nano, 2015, 2, 203–212 RSC.
- S. Majumdar, C. Ma, M. Villani, N. Zuverza-Mena, L. Pagano, Y. Huang, A. Zappettini, A. A. Keller, N. Marmiroli and O. P. Dhankher, Surface coating determines the response of soybean plants to cadmium sulfide quantum dots, NanoImpact, 2019, 100151 CrossRef.
- L. Pagano, F. Pasquali, S. Majumdar, R. De la Torre-Roche, N. Zuverza-Mena, M. Villani, A. Zappettini, R. E. Marra, S. M. Isch, M. Marmiroli, E. Maestri, O. P. Dhankher, J. C. White and N. Marmiroli, Exposure of Cucurbita pepo to binary combinations of engineered nanomaterials: physiological and molecular response, Environ. Sci.: Nano, 2017, 4, 1579–1590 RSC.
- R. L. Li, H. F. Sun, S. P. Wang, Y. H. Wang and K. F. Yu, Retention of CdS/ZnS quantum dots (QDs) on the Root Epidermis of woody plant and its implications by benzo[a]pyrene: evidence from the in situ synchronous nanosecond time-resolved fluorescence spectra method, J. Agric. Food Chem., 2018, 66, 814–821 CrossRef CAS PubMed.
- X. Gao, S. M. Rodrigues, E. Spielman-Sun, S. Lopes, S. Rodrigues, Y. L. Zhang, A. Avellan, R. M. B. O. Duarte, A. Duarte, E. A. Casman and G. V. Lowry, Effect of soil organic matter, soil pH, and moisture content on solubility and dissolution rate of CuO NPs in soil, Environ. Sci. Technol., 2019, 53, 4959–4967 CrossRef CAS PubMed.
- S. Rawat, V. L. R. Pullagurala, I. O. Adisa, Y. Wang, J. R. Peralta-Videa and J. L. Gardea-Torresdey, Factors affecting fate and transport of engineered nanomaterials in terrestrial environments, Current Opinion in Environmental Science & Health, 2018, 6, 47–53 Search PubMed.
- J. R. Lead, G. E. Batley, P. J. J. Alvarez, M. N. Croteau, R. D. Handy, M. J. McLaughlin, J. D. Judy and K. Schirmer, Nanomaterials in the environment: behavior, fate, bioavailability, and effects—an updated review, Environ. Toxicol. Chem., 2018, 37, 2029–2063 CrossRef CAS PubMed.
- K. Zhalnina, K. B. Louie, Z. Hao, N. Mansoori, U. N. da Rocha, S. Shi, H. Cho, U. Karaoz, D. Loqué, B. P. Bowen, M. K. Firestone, T. R. Northen and E. L. Brodie, Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly, Nat. Microbiol., 2018, 3, 470–480 CrossRef CAS PubMed.
- D. A. Navarro, S. Banerjee, D. F. Watson and D. S. Aga, Differences in soil mobility and degradability between water-dispersible CdSe and CdSe/ZnS quantum dots, Environ. Sci. Technol., 2011, 45, 6343–6349 CrossRef CAS PubMed.
- L. Pagano, E. Maestri, J. C. White, N. Marmiroli and M. Marmiroli, Quantum dots exposure in plants: minimizing the adverse response, Current Opinion in Environmental Science & Health, 2018, 6, 71–76 Search PubMed.
- C. Riedelsheimer, A. Czedik-Eysenberg, C. Grieder, J. Lisec, F. Technow, R. Sulpice, T. Altmann, M. Stitt, L. Willmitzer and A. E. Melchinger, Genomic and metabolic prediction of complex heterotic traits in hybrid maize, Nat. Genet., 2012, 44, 217 CrossRef CAS PubMed.
- O. Fiehn, J. Kopka, P. Dörmann, T. Altmann, R. N. Trethewey and L. Willmitzer, Metabolite profiling for plant functional genomics, Nat. Biotechnol., 2000, 18, 1157 CrossRef CAS PubMed.
- K. Stephan, S. Dirk, W. Lothar and G. Patrick, High-resolution plant metabolomics: from mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions, Plant J., 2012, 70, 39–50 CrossRef PubMed.
- C. H. Johnson, J. Ivanisevic and G. Siuzdak, Metabolomics: beyond biomarkers and towards mechanisms, Nat. Rev. Mol. Cell Biol., 2016, 17, 451 CrossRef CAS PubMed.
- L. W. Sumner, P. Mendes and R. A. Dixon, Plant metabolomics: large-scale phytochemistry in the functional genomics era, Phytochemistry, 2003, 62, 817–836 CrossRef CAS PubMed.
- N. Schauer and A. R. Fernie, Plant metabolomics: towards biological function and mechanism, Trends Plant Sci., 2006, 11, 508–516 CrossRef CAS PubMed.
- S. H. Guo, N. Hu, Q. S. Li, P. Yang, L. L. Wang, Z.-M. Xu, H. J. Chen, B. Y. He and E. Y. Zeng, Response of edible amaranth cultivar to salt stress led to Cd mobilization in rhizosphere soil: a metabolomic analysis, Environ. Pollut., 2018, 241, 422–431 CrossRef CAS PubMed.
- J. L. Zhang, Z. P. Zhou, Y. Pei, Q. Q. Xiang, X. X. Chang, J. Ling, D. Shea and L. Q. Chen, Metabolic profiling of silver nanoparticle toxicity in Microcystis aeruginosa, Environ. Sci.: Nano, 2018, 5, 2519–2530 RSC.
- H. Zhang, L. Lu, X. Zhao, S. Zhao, X. Gu, W. Du, H. Wei, R. Ji and L. Zhao, Metabolomics reveals the “invisible” responses of spinach plants exposed to CeO2 nanoparticles, Environ. Sci. Technol., 2019, 53, 6007–6017 CrossRef CAS PubMed.
- H. Zhang, W. Du, J. R. Peralta-Videa, J. L. Gardea-Torresdey, J. C. White, A. Keller, H. Guo, R. Ji and L. Zhao, Metabolomics reveals how cucumber (Cucumis sativus) reprograms metabolites to cope with silver ions and silver nanoparticle-induced oxidative stress, Environ. Sci. Technol., 2018, 52, 8016–8026 CrossRef CAS PubMed.
- L. Zhao, Y. Huang, A. S. Adeleye and A. A. Keller, Metabolomics reveals Cu(OH)2 nanopesticide-activated antioxidative pathways and decreased beneficial antioxidants in spinach leaves, Environ. Sci. Technol., 2017, 51, 10184–10194 CrossRef CAS PubMed.
- L. Pagano, A. D. Servin, R. De La Torre-Roche, A. Mukherjee, S. Majumdar, J. Hawthorne, M. Marmiroli, E. Maestri, R. E. Marra, S. M. Isch, O. P. Dhankher, J. C. White and N. Marmiroli, Molecular response of crop plants to engineered nanomaterials, Environ. Sci. Technol., 2016, 50, 7198–7207 CrossRef CAS PubMed.
- H. Salehi, A. Chehregani, L. Lucini, A. Majd and M. Gholami, Morphological, proteomic and metabolomic insight into the effect of cerium dioxide nanoparticles to Phaseolus vulgaris L. under soil or foliar application, Sci. Total Environ., 2018, 616–617, 1540–1551 CrossRef CAS PubMed.
- R. K. Singh, N. Bohra and L. Sharma, Valorizing faba bean for animal feed supplements via biotechnological approach: opinion, Biocatal. Agric. Biotechnol., 2019, 17, 366–368 CrossRef.
- M. Iqbal, Vicia faba bioassay for environmental toxicity monitoring: a review, Chemosphere, 2016, 144, 785–802 CrossRef CAS PubMed.
- M. I. Vučemilović, N. Vukelić and T. Rajh, Solubility and photocorrosion of small CdS particles, J. Photochem. Photobiol., A, 1988, 42, 157–167 CrossRef.
- H. K. Lichtenthaler, Chlorophylls and carotenoids: pigments of photosynthetic biomembranes, Methods Enzymol., 1987, 148, 350–382 CAS.
- H. Yang, C. Liu, D. F. Yang, H. S. Zhang and Z. G. Xi, Comparative study of cytotoxicity, oxidative stress and genotoxicity induced by four typical nanomaterials: the role of particle size, shape and composition, J. Appl. Toxicol., 2009, 29, 69–78 CrossRef CAS PubMed.
- K. Tawaha, F. Q. Alali, M. Gharaibeh, M. Mohammad and T. El-Elimat, Antioxidant activity and total phenolic content of selected Jordanian plant species, Food Chem., 2007, 104, 1372–1378 CrossRef CAS.
- F. Dieterle, A. Ross, G. Schlotterbeck and H. Senn, Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics, Anal. Chem., 2006, 78, 4281–4290 CrossRef CAS PubMed.
- J. G. Xia, N. Psychogios, N. Young and D. S. Wishart, MetaboAnalyst: a web server for metabolomic data analysis and interpretation, Nucleic Acids Res., 2009, 37, W652–W660 CrossRef CAS PubMed.
- K. A. Ubaid, X. Zhang, V. K. Sharma and L. Li, Fate and risk of metal sulfide nanoparticles in the environment, Environ. Chem. Lett., 2019 DOI:10.1007/s10311-019-00920-x.
- R. z. Lin, X. r. Wang, Y. Luo, W. c. Du, H. y. Guo and D. q. Yin, Effects of soil cadmium on growth, oxidative stress and antioxidant system in wheat seedlings (Triticum aestivum L.), Chemosphere, 2007, 69, 89–98 CrossRef CAS PubMed.
- S. S. Zhang, H. M. Zhang, R. Qin, W. S. Jiang and D. H. Liu, Cadmium induction of lipid peroxidation and effects on root tip cells and antioxidant enzyme activities in Vicia faba L, Ecotoxicology, 2009, 18, 814–823 CrossRef CAS PubMed.
- M. Shahid, B. Pourrut, C. Dumat, M. Nadeem, M. Aslam and E. Pinelli, Heavy-metal-induced reactive oxygen species: phytotoxicity and physicochemical changes in plants, Rev. Environ. Contam. Toxicol., 2014, 232, 1–44 CrossRef CAS PubMed.
- M. E. A. Carvalho, P. R. C. Castro and R. A. Azevedo, Hormesis in plants under Cd exposure: from toxic to beneficial element?, J. Hazard. Mater., 2019 DOI:10.1016/j.jhazmat.2019.121434, 121434.
- N. V. Zagoskina, E. A. Goncharuk and A. K. Alyavina, Effect of cadmium on the phenolic compounds formation in the callus cultures derived from various organs of the tea plant, Russ. J. Plant Physiol., 2007, 54, 237–243 CrossRef CAS.
- P. P. He, X. Z. Lv and G. Y. Wang, Effects of Se and Zn supplementation on the antagonism against Pb and Cd in vegetables, Environ. Int., 2004, 30, 167–172 CrossRef CAS PubMed.
- N. Sarwar, S. S. Malhi, M. H. Zia, A. Naeem, S. Bibi and G. Farid, Role of mineral nutrition in minimizing cadmium accumulation by plants, J. Sci. Food Agric., 2010, 90, 925–937 CAS.
- P. Aravind and M. N. V. Prasad, Zinc alleviates cadmium-induced oxidative stress in Ceratophyllum demersum L.: a free floating freshwater macrophyte, Plant Physiol. Biochem., 2003, 41, 391–397 CrossRef CAS.
- Y. T. Liu, Z. S. Chen and C. Y. Hong, Cadmium-induced physiological response and antioxidant enzyme changes in the novel cadmium accumulator, Tagetes patula, J. Hazard. Mater., 2011, 189, 724–731 CrossRef CAS PubMed.
- E. Saccenti, H. C. J. Hoefsloot, A. K. Smilde, J. A. Westerhuis and M. M. W. B. Hendriks, Reflections on univariate and multivariate analysis of metabolomics data, Metabolomics, 2014, 10, 361–374 CrossRef CAS.
- A. Kobylińska, S. Borek and M. M. Posmyk, Melatonin redirects carbohydrates metabolism during sugar starvation in plant cells, J. Pineal Res., 2018, 64, e12466 CrossRef PubMed.
- Y. Yu, Y. Lv, Y. Shi, T. Li, Y. C. Chen, D. Zhao and Z. W. Zhao, The role of phyto-melatonin and related metabolites in response to stress, Molecules, 2018, 23, 1887 CrossRef PubMed.
- A. Kulma and J. Szopa, Catecholamines are active compounds in plants, Plant Sci., 2007, 172, 433–440 CrossRef CAS.
- P. J. Allen, D. Wise, T. Greenway, L. Khoo, M. J. Griffin and M. Jablonsky, Using 1-D 1H and 2-D 1H J-resolved NMR metabolomics to understand the effects of anemia in channel catfish (Ictalurus punctatus), Metabolomics, 2015, 11, 1131–1143 CrossRef CAS.
- R. Wei, A. B. Ross, M. Su, J. Wang, S.-P. Guiraud, C. F. Draper, M. Beaumont, W. Jia and F.-P. Martin, Metabotypes related to meat and vegetable intake reflect microbial, lipid and amino acid metabolism in healthy people, Mol. Nutr. Food Res., 2018, 62, 1800583 CrossRef PubMed.
- B. Gong, S. S. Sun, Y. Y. Yan, X. Jing and Q. H. Shi, Glutathione metabolism and its function in higher plants adapting to stress, Antioxidants and Antioxidant Enzymes in Higher Plants, 2018, pp. 181–205 Search PubMed.
- A. Matiz, P. T. Mioto and H. Mercier, Urea in plants: metabolic aspects and ecological implications, Prog. Bot., 2019, 1–31 Search PubMed.
- I. Couee, C. Sulmon, G. Gouesbet and A. El Amrani, Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants, J. Exp. Bot., 2006, 57, 449–459 CrossRef CAS PubMed.
- M. Rosa, C. Prado, G. Podazza, R. Interdonato, J. A. González, M. Hilal and F. E. Prado, Soluble sugars, Plant Signaling Behav., 2009, 4, 388–393 CrossRef CAS PubMed.
- J. Sasse, E. Martinoia and T. Northen, Feed your friends: do plant exudates shape the root microbiome?, Trends Plant Sci., 2018, 23, 25–41 CrossRef CAS PubMed.
- K. Večeřová, Z. Večeřa, B. Dočekal, M. Oravec, A. Pompeiano, J. Tříska and O. Urban, Changes of primary and secondary metabolites in barley plants exposed to CdO nanoparticles, Environ. Pollut., 2016, 218, 207–218 CrossRef PubMed.
- P. G. Pietta, Flavonoids as antioxidants, J. Nat. Prod., 2000, 63, 1035–1042 CrossRef CAS PubMed.
- K. Nahar, M. Hasanuzzaman, M. M. Alam, A. Rahman, T. Suzuki and M. Fujita, Polyamine and nitric oxide crosstalk: antagonistic effects on cadmium toxicity in mung bean plants through upregulating the metal detoxification, antioxidant defense and methylglyoxal detoxification systems, Ecotoxicol. Environ. Saf., 2016, 126, 245–255 CrossRef CAS PubMed.
- F. M. Romero, S. J. Maiale, F. R. Rossi, M. Marina, O. A. Ruíz and A. Gárriz, in Polyamines: Methods and Protocols, ed. R. Alcázar and A. F. Tiburcio, Springer New York, New York, NY, 2018, pp. 37–49, DOI:10.1007/978-1-4939-7398-9_3.
- L. H. Chen, L. Wang, F. G. Chen, H. Korpelainen and C. Y. Li, The effects of exogenous putrescine on sex-specific responses of Populus cathayana to copper stress, Ecotoxicol. Environ. Saf., 2013, 97, 94–102 CrossRef CAS PubMed.
- B. G. Forde and P. J. Lea, Glutamate in plants: metabolism, regulation, and signalling, J. Exp. Bot., 2007, 58, 2339–2358 CrossRef CAS PubMed.
- A. J. Meijer, W. H. Lamers and R. A. Chamuleau, Nitrogen metabolism and ornithine cycle function, Physiol. Rev., 1990, 70, 701–748 CrossRef CAS PubMed.
- A. Fait, H. Fromm, D. Walter, G. Galili and A. R. Fernie, Highway or byway: the metabolic role of the GABA shunt in plants, Trends Plant Sci., 2008, 13, 14–19 CrossRef CAS PubMed.
- D. W. Chia, T. J. Yoder, W.-D. Reiter and S. I. Gibson, Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species, Planta, 2000, 211, 743–751 CrossRef CAS PubMed.
- M. G. Steiger, A. Rassinger, D. Mattanovich and M. Sauer, Engineering of the citrate exporter protein enables high citric acid production in Aspergillus niger, Metab. Eng., 2019, 52, 224–231 CrossRef CAS PubMed.
- G. Y. Wang, D. Huang, Y. Li, J. P. Wen and X. Q. Jia, A metabolic-based approach to improve xylose utilization for fumaric acid production from acid pretreated wheat bran by Rhizopus oryzae, Bioresour. Technol., 2015, 180, 119–127 CrossRef CAS PubMed.
- T. Berglund, A. Lindström, H. Aghelpasand, E. Stattin and A. B. Ohlsson, Protection of spruce seedlings against pine weevil attacks by treatment of seeds or seedlings with nicotinamide, nicotinic acid and jasmonic acid, Forestry, 2016, 89, 127–135 CrossRef.
- T. Vogt, Phenylpropanoid biosynthesis, Mol. Plant, 2010, 3, 2–20 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details on the methods and discussion. Tables showing the physiochemical properties and metal concentration of the experimental soil (Table S1), the concentration of mineral nutrients in plant tissues (Table S2), the concentration of water-extractable metals and nutrient elements in soil (Table S3), and the regulated metabolites in leaves in plants exposed to CdS-NP-amended soil for 28 days (Table S4). Figures showing the characterization of CdS-NPs (Fig. S1), tissue concentrations of Cd and Zn (Fig. S2), the fresh biomass of tissues, stem length, number of leaves (Fig. S3), the levels of photosynthetic pigments, MDA and total phenolic compounds in tissues (Fig. S4) and VIP scores from the PLS-DA analysis of metabolites in roots and leaves (Fig. S5). See DOI: 10.1039/c9en00933g |
| This journal is © The Royal Society of Chemistry 2020 |