Effects of antibiotics on microbial community structure and microbial functions in constructed wetlands treated with artificial root exudates†
Xin-nan
Tong
 a,
Xin-ze
Wang
*a,
Xiao-juan
He
*a,
Zhe
Wang
b and
Wen-xuan
Li
c
a,
Xin-ze
Wang
*a,
Xiao-juan
He
*a,
Zhe
Wang
b and
Wen-xuan
Li
c
aSchool of Environmental Science and Engineering, Shanghai Jiaotong University, Shanghai 200240, PR China. E-mail: xinzewang@sjtu.edu.cn; hexiaojuan@sjtu.edu.cn
bState Key Laboratory of Eco-hydraulics in Northwest Arid Region, Xi'an University of Technology, Xi'an, Shaanxi 710048, PR China
cNUS Environmental Research Institute, National University of Singapore, 5A Engineering Drive 1, #02-01, Singapore 117411, Singapore
First published on 26th December 2019
Abstract
In the rhizosphere, plant root exudates can mediate the toxicity of antibiotics on microorganisms, yet the mechanisms are poorly understood. To simulate the antibiotic contamination of global rivers and lakes, the current study investigated the effects of two antibiotics (ofloxacin at 8.69 × 104 ng L−1 and tetracycline at 8.62 × 104 ng L−1) and their binary combination (8.24 × 104 ng L−1 ofloxacin and 7.11 × 104 ng L−1 tetracycline) on bacterial communities in micro-polluted constructed wetlands with and without artificial root exudates. The two antibiotics had no significant effects on the removal of excess carbon and nitrogen from the microcosms treated with and without exudates. Furthermore, with regard to bacterial community structure, antibiotic exposure increased the bacterial richness of bulk and exudate treated microcosms (P < 0.05). However, a significant increase (P < 0.05) in bacterial diversity elicited by ofloxacin and antibiotic mixture exposure was only observed in microcosms with exudates. In exudate treated microcosms, ofloxacin promoted the relative abundance of Arthrobacter spp., which are ofloxacin-resistant bacterial species, which significantly varied from what was observed in microcosms free of exudates. Moreover, tetracycline, ofloxacin and their combination all significantly increased the relative abundance of nitrogen cycling bacteria Rhizobacter spp. and Rhizobium spp., and decreased the relative abundance of antibiotic-resistant bacteria Pseudomonas spp. Simultaneously, with regard to bacterial community functions, the functional profiles (Kyoto Encyclopedia of Genes and Genomes) showed that the pathways of amino acid and carbohydrate metabolism were enhanced by antibiotics in microcosms with exudates. The findings illustrate that antibiotics not only alter the bacterial structure and composition but also change their functional properties in constructed wetlands, and these interruption effects could be affected by root exudates of plants, which may further reveal the ecological implication of plants in constructed wetlands.
Environmental significanceAntibiotics are considered to pose potential risks to microbial communities in constructed wetlands (CWs). In the rhizosphere of CWs, root exudates are proposed to play a primary role in regulating microbial community dynamics. A few studies showed that root exudates can stimulate or inhibit the impacts of antibiotics on microorganism growth and community tolerance. Nevertheless, no work has investigated how antibiotics in combination with root exudates influence the microbial community structure. Hence, the effects of two antibiotics and their combination on bacterial communities in bulk and root exudate added CWs were assessed. This work will provide comprehensive information on the response of bacteria to antibiotic exposure in the rhizosphere, which is important to gain further understanding of the protective effects of root exudates in antibiotic-contaminated wetlands. |
1. Introduction
As an efficient treatment technique, constructed wetlands (CWs) are often designed and built to remove the majority of nitrogen and carbon in micro-polluted surface water (typically nitrogen < 10 mg L−1).1,2 In CWs, biodegradation, substrate adsorption, and plant uptake all play specific roles in reducing the nitrogen and carbon loading, and biodegradation by microorganisms is the most predominant factor.3,4The rhizosphere, a micro-environment surrounding the plant root surface, is important in most wetland types. In the zone between the soil and living roots, root exudates stimulate or inhibit microbial populations and their activities.5 Exudates released by roots into the rhizosphere can be divided roughly into two main groups: low-molecular-weight carbon (LMWC) compounds such as sugars, amino acids, and organic acids, and water-insoluble materials such as cell walls, sloughed-off materials and other root debris and mucilage.5 LMWC compounds present in root exudates are readily assimilated by microorganisms and are proposed to play a primary role in regulating microbial community dynamics in the rhizosphere.6 Researchers investigated the effect of varying exudate profiles on microbial communities in the rhizosphere of Lupinus albus L. using DGGE analysis of 16S rRNA gene fragments.7 They observed that the strong differences in the bacterial community structure were attributed to aconitic, citric, and malic acids in root exudates. Wu et al.2 also showed that the abundance and community structure of nirK- and nirS-encoding bacteria in CWs were positively influenced by the concentrations of sucrose, glucose and oxalic acid in root exudates of wetland plants.
Micro-polluted surface water often contains chemicals, such as antibiotics, which have been increasingly recognized as emerging environmental contaminants in aquatic systems, due to their potential hazardous effects on ecosystems and human health.8 Antibiotics can kill or inhibit the growth of microorganisms through inhibition of synthesis of the cell wall, proteins, and/or nucleic acids, and the alteration of cell membranes.9,10 However, at sub-inhibitory (environmentally relevant) concentrations antibiotics can also stimulate the growth of some microbial populations.11–13 For instance, 100 mg kg−1 tetracycline, sulfamonomethoxine and ciprofloxacin can stimulate Proteobacteria, Actinobacteria, Arthrobacter, Firmicutes and Gemmatimonadetes phyla in the soil,14 which may result in the development of resistance.12,13 Meanwhile, higher numbers of sequences of Proteobacteria and Actinobacteria were found in substrate samples of a constructed wetland used to treat wastewater with 100 μg L−1 enrofloxacin and ceftiofur.15 Numerous published papers have found that antibiotics can also affect the microbial community structure, which might disturb microbial functions, resulting in notable ecological consequences.14,16–19
A few studies have been carried out on the effects or interactions of antibiotics on microbial communities from the rhizosphere of plants. Brandt et al.20 indicated that the bacterial community tolerance to sulfadiazine can be stimulated with pulse addition of artificial root exudates (i.e., the mixture of sugars, organic acids and amino acids). Similarly, researchers also demonstrated that natamycin (50–100 mg L−1) controls fungal growth in root free soils, whereas higher concentrations (up to 200 mg L−1) were needed to achieve the same effect in the rhizosphere.21 Therefore, root exudates can mediate the impacts of antibiotics on microorganisms, which may change the microbial community structure in the rhizosphere. Nevertheless, no work has investigated how antibiotics in combination with root exudates influence the microbial community structure in the rhizosphere. Therefore, the objective of the study was to pinpoint the effects of antibiotics in combination with root exudates on the microbial community structure and function in CWs. The role of root exudate compounds in the structure and activity of bacterial communities has been investigated by the application of artificial root exudates to substrates in microcosm studies.22 Therefore, we used artificial root exudates (composed of soluble sugars, low molecular weight organic acids and amino acids) to replace a single substrate or an undefined plant exudate. This study reports nutrient removal, and microbial diversity and composition in CWs following the exposure to two antibiotics (i.e., tetracycline and ofloxacin) in mono or binary combination along with bulk and artificial root exudate addition. Moreover, comparative analysis of the predicted functional diversity was performed to investigate the impacts of antibiotics on microbial community functions in CWs.
2. Materials and methods
2.1 Selection of antibiotics
Numerical studies have shown that tetracyclines and fluoroquinolones are the commonly detected antibiotics in global lakes and rivers.23,24 Moreover, tetracycline (TET) and ofloxacin (OFL) have been found at significantly high concentrations across the globe, reflecting high usage of these antibiotics as human and veterinary medicine.24 Among them, high levels of OFL were detected in the Isakavagu–Nakkavagu stream (1 × 104 ng L−1), Hai River system (1.17 × 104 ng L−1), Wangyang River (1 × 104 ng L−1), and Pearl River (1.56 × 103 ng L−1) (Table S2†).24 In the case of TET, high levels were seen in the Hai River (2.5 × 104 ng L−1), Wangyang River (2.5 × 104 ng L−1), and Nanming River (6.8 × 103 ng L−1) (Table S2†).24 Besides the high detection frequency and concentration, the ecological risks of TET and OFL were also remarkable. For example, based on the risk quotient values, tetracycline and ofloxacin may present a high ecological risk to organisms living in Taihu Lake.25 Therefore, TET and OFL were selected as representative antibiotics and the different combinations (mono and binary) were set.2.2 Experimental set-up
Twenty-four wetland microcosms were set up in PVC containers (diameter 14 cm, height 20 cm) and kept under greenhouse (25 °C) and dark conditions. The containers were wrapped in aluminum foil to avoid light penetration, mimicking real environmental conditions. All microcosms were operated in batch mode, having only a tap at the base of the plastic containers for sample collection. All microcosms had a 0.18 m layer of gravel (particle size: 5–10 mm). Before the experiment, all microcosms were cultivated using synthetic wastewater for 15 days (replaced every three days). Synthetic wastewater was prepared with tap water and nutrients, including KH2PO4, glucose, (NH4)2SO4, and KNO3 (1.46 mg L−1 to 2.21 mg L−1 NH4+–N, 8.65 mg L−1 to 11.28 mg L−1 NO3−–N, 0.55 mg L−1 to 0.92 mg L−1 PO43−–P, and 22.32 mg L−1 to 20.54 mg L−1 total organic carbon).2.3 Experimental design
After this cultivation phase, all series of microcosms were designed as 4 × 2 factorial with four antibiotic treatments and two concentrations of root exudates (Fig. 1). All treatments were incubated in triplicate. Among them, 12 microcosms were fed with simulated wastewater and antibiotics (Control, OFL, TET, and OT), and another set of 12 microcosms received simulated wastewater, antibiotics and simplified exudates (Control–R, OFL–R, TET–R, and OT–R). The case of exposure to two antibiotics contained mono and binary combinations (Control: no antibiotic, OFL: 8.62 × 104 ng L−1 ofloxacin, TET: 8.69 × 104 ng L−1 tetracycline, OT: mixture of 8.24 × 104 ng L−1 ofloxacin and 7.11 × 104 ng L−1 tetracycline) (Text S1†). The exudates were prepared as described in previous research,26 and following the concentration of organic carbon (22.5 mg L−1) in root exudates measured by Wu et al.2 The composition and concentration of artificial root exudates used were as follows: D-fructose 0.0625 nmol L−1, D-glucose 0.0625 nmol L−1, sucrose 0.0625 nmol L−1, succinic acid 0.0313 nmol L−1, tartaric acid 0.0313 nmol L−1, glutamate 0.0156 nmol L−1 and serine 0.0156 nmol L−1.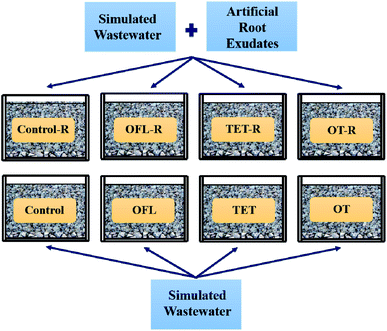 | ||
| Fig. 1 Schematic diagram of the experimental design (OFL = ofloxacin, TET = tetracycline, OT = the mixture of tetracycline and ofloxacin, R indicates the microcosms treated with exudates). | ||
The wastewater in the microcosms was replaced every three days with new antibiotic-supplemented wastewater, and NH4+, NO3−, TOC, pH and DO of the influent and effluent were measured. The nutrient concentrations of the microcosm influent were 1.71 mg L−1 to 2.00 mg L−1 NH4+–N, 9.99 mg L−1 to 10.61 mg L−1 NO3−–N, and 0.69 mg L−1 to 0.78 mg L−1 PO43−–P. The influent’s dissolved oxygen (DO) was 8.1 mg L−1 to 8.3 mg L−1, pH was 7.00 to 7.24, and total organic carbon (TOC) was 48.69 mg L−1 to 51.49 mg L−1 with exudates and 20.54 mg L−1 to 21.74 mg L−1 without exudates. On the 30th day of the experiment samples of gravel (30 ± 2 g) were retrieved from microcosms and stored at −80 °C for DNA analyses.
2.4 DNA extraction, amplification, and sequencing
DNA was extracted from the membranes on gravel samples using a protocol modified from Wu et al. (2017).2 Briefly, 20 g of gravel was extracted with 1× PBS buffer. The samples were homogenized for 2 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 25 °C (Pico-21, Thermo Fisher, USA). Approximately 0.2 g of biofilm was collected, and DNA samples in the precipitate were further purified using the E.Z.N.A™ Mag-Bind Soil DNA Kit (OMEGA, USA). DNA integrity and purity were monitored on 1% agarose gels. DNA concentration and purity were measured using a spectrophotometer (UVP, USA). To determine the impact of antibiotic treatment on microbial diversity and composition, the 16S rRNA gene targeting the V3–V4 region was amplified from DNA extracts from gravel using PCR with the 341F/805R primer pair. Then, the PCR products were detected using 1% agarose gel electrophoresis and purified. Sequencing libraries were generated using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA) following manufacturer's recommendations, and index codes were added. Finally, the library was sequenced, and the microbial communities were analyzed using the Illumina MiSeq300 platform (Shanghai Sunggong Technology Co., Ltd.; Shanghai, China). Detailed information is provided in the ESI (Text S2).†
000 rpm, 25 °C (Pico-21, Thermo Fisher, USA). Approximately 0.2 g of biofilm was collected, and DNA samples in the precipitate were further purified using the E.Z.N.A™ Mag-Bind Soil DNA Kit (OMEGA, USA). DNA integrity and purity were monitored on 1% agarose gels. DNA concentration and purity were measured using a spectrophotometer (UVP, USA). To determine the impact of antibiotic treatment on microbial diversity and composition, the 16S rRNA gene targeting the V3–V4 region was amplified from DNA extracts from gravel using PCR with the 341F/805R primer pair. Then, the PCR products were detected using 1% agarose gel electrophoresis and purified. Sequencing libraries were generated using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA) following manufacturer's recommendations, and index codes were added. Finally, the library was sequenced, and the microbial communities were analyzed using the Illumina MiSeq300 platform (Shanghai Sunggong Technology Co., Ltd.; Shanghai, China). Detailed information is provided in the ESI (Text S2).†
2.5 Sequence processing and data analysis
Sequences that were shorter than 200 base pairs (bp) and with a quality score below 20 were removed from the data sets. Then, the normalized samples were individually classified and analyzed using the Ribosomal Database Project (RDP) database (http://rdp.cme.msu.edu/). High-quality reads related to 16S rDNA were clustered into operational taxonomic units (OTUs) using UPARSE software (v8.1.1756) at a sequence identity of 97%, using the UCHIME algorithm (v4.2.40).2 Abundance-based coverage estimator (ACE) richness and Shannon diversity were calculated via the Mothur analysis (http://www.mothur.org) at a distance level of 3%.2 Bacterial community structures and compositions were compared using nonmetric multidimensional scaling (NMDS) analysis by means of Bray–Curtis similarities in R using the metaMDS function.27 Permutational multivariate analyses of variance were compared using “Anosim” in the vegan R package.28 Functional inferences were made from the Kyoto Encyclopedia of Genes and Genomes (KEGG) catalog using the Tax4Fun program, which is connected to the SILVA database.28,292.6 Statistical analysis
For excess nutrients, water chemistry parameters, bacterial richness and diversity, significant differences between samples were evaluated using parametric one-way analysis of variance (ANOVA). Significant (p < 0.05) differences were evaluated using Tukey comparison tests. All statistical analyses were performed using commercial software SPSS version 19.0 (SPSS, Inc., Chicago, IL, USA).3. Results and discussion
3.1 Pollutant removal from wetland microcosms
To determine the influence of different antibiotics on the pollutant removal from the microcosms, nutrient levels (NH4+–N, NO3−–N and TOC) and dissolved oxygen in the microcosms and their corresponding removal efficiencies of contaminants are presented in Table 1. Exudate-treated microcosms had noticeably higher NO3−–N and TOC removal efficiency and lower DO concentration than microcosms without exudates (p < 0.01). However, in the microcosms with and without exudates, tetracycline, ofloxacin and their mixture did not significantly affect NH4+–N, NO3−–N and TOC removal efficiencies (p > 0.05). Interestingly, DO concentrations in all treatments were always higher than 2 mg L−1, which was essential for nitrification.30 In microcosms without exudates, exposure to both tetracycline, and the mixture of tetracycline and ofloxacin reduced the DO significantly (p < 0.05). In contrast, the DO was stable (1.9 mg L−1 to 2.2 mg L−1) and was not influenced by antibiotic exposure in exudate treated microcosms. Different from NH4+–N, NO3−–N, TOC and DO, the effluent pH was comparable across all microcosms over the course of this study (Table S3†).| NH4+–N (%) | NO3−–N (%) | TOC (%) | DO (mg L−1) | |
|---|---|---|---|---|
| a OFL = ofloxacin, TET = tetracycline, OT = the mixture of tetracycline and ofloxacin, R indicates the microcosms treated with root exudates; * indicates that the value decreased or increased significantly (p < 0.05) in comparison with the control treatment. | ||||
| Control | 93.7 ± 3.8 | 13.4 ± 7.7 | 33.6 ± 4.2 | 4.0 ± 1.2 |
| OFL | 89.5 ± 6.1 | 19.3 ± 6.4 | 35.4 ± 3.2 | 3.5 ± 0.9 |
| TET | 83.1 ± 4.8 | 15.0 ± 5.4 | 25.5 ± 4.7 | 2.9 ± 0.3* |
| OT | 95.8 ± 1.4 | 18.4 ± 3.4 | 25.7 ± 5.2 | 3.2 ± 0.5* |
| Control–R | 89.0 ± 2.4 | 71.2 ± 14.4 | 67.2 ± 4.5 | 2.2 ± 0.2 |
| OFL–R | 78.8 ± 6.0 | 70.4 ± 16.6 | 67.6 ± 6.3 | 2.2 ± 0.4 |
| TET–R | 82.9 ± 3.3 | 73.8 ± 18.6 | 76.0 ± 3.5 | 1.9 ± 0.9 |
| OT–R | 83.8 ± 5.5 | 59.1 ± 14.7 | 62.3 ± 8.9 | 2.0 ± 0.3 |
Although the inhibitory or even toxic effects of antibiotics on nitrogen removal have attracted the attention of many researchers,31,32 in this study, no significant differences were observed between control and antibiotic treated microcosms in terms of carbon and nitrogen removal, demonstrating the stability of constructed wetlands for contaminant removal from antibiotic polluted water.
3.2 Taxonomic annotation and alpha diversity
After quality filtering, 388![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 039 sequences were detected and assigned to 5347 OTUs from all the biofilm samples, and then 362
039 sequences were detected and assigned to 5347 OTUs from all the biofilm samples, and then 362![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 657 sequences were subsampled to compare community structures and compositions. The rarefaction plots (Fig. S1†) plateaued with the current sampling effort, and the coverage was high (>97%) for all biofilm samples, indicating that the OTUs of each bacterial library were adequately sampled.
657 sequences were subsampled to compare community structures and compositions. The rarefaction plots (Fig. S1†) plateaued with the current sampling effort, and the coverage was high (>97%) for all biofilm samples, indicating that the OTUs of each bacterial library were adequately sampled.
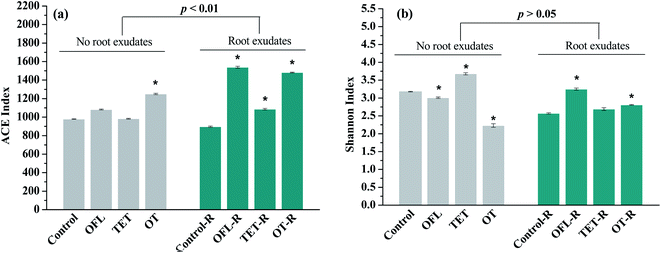
Bacterial community complexity was evaluated using the alpha components including the species richness (ACE index) and diversity (Shannon index) in both microcosms with and without exudates (Fig. 2). Diverse microbial assemblages were observed in Control, OFL, TET, OT, Control–R, OFL–R, TET–R, and OT–R, with average observed OTUs of 532, 697, 691, 674, 583, 790, 726, and 654, respectively, suggesting different amounts of unique species. The ACE and Shannon indices in control microcosms were similar to those in other constructed wetlands,33,34 and the smaller values may be attributed to the low nutritional level. For bacterial richness, the extent of ACE in microcosms with exudates (896 to 1539, p < 0.01) was significantly larger than that in microcosms without exudates (979 to 1250). In general, the microcosms with antibiotics showed greater species richness than their corresponding controls. In contrast to bacterial richness, the difference of Shannon indices between microcosms with (2.57 to 3.25) and without exudates (2.23 to 3.68) was not significant (p > 0.05). The effects of antibiotics on diversity varied. In microcosms without exudates, ofloxacin (p < 0.05) and the mixture of tetracycline and ofloxacin (p < 0.01) significantly decreased bacterial diversity, but not in the microcosms exposed to tetracycline. However, in microcosms treated with exudates, bacterial diversity was increased by antibiotics, particularly the effects of ofloxacin and the mixture of tetracycline and ofloxacin were significant (p < 0.01, p < 0.05). In conclusion, antibiotics increased bacterial richness in microcosms with and without exudates. Meanwhile ofloxacin and the mixture of tetracycline and ofloxacin decreased bacterial diversity in microcosms without exudates, but significantly improved it in exudate treated microcosms.
This study for the first time provided experimental evidence for a significantly increased bacterial community diversity associated with antibiotic exposure in microcosms treated with pulse additions of exudates. However, in microcosms without exudates, significantly lower diversity indexes were observed in treatments with ofloxacin and the mixture of antibiotics when compared with the control. There is ample evidence showing that contamination of soils with antibiotics can elicit a reduction in microbial diversity.35,36 Furthermore, in a batch reactor, Lin et al.14 observed that the microbial diversity index decreased with application of 100 mg kg−1 tetracycline. However, in bulk and planted soils, changes in bacterial diversity elicited by antibiotic exposure were only observed in bulk soil.37,38 Diversity is considered as an important feature of ecosystem integrity, and it acts as a genetic reservoir and implies functional redundancy, which increases community resilience to disturbance.39 Therefore, in the present study, exudates might protect the microbial community from antibiotic stress. Furthermore, researchers demonstrated that exudates can increase the bacterial community tolerance to antibiotics, which may be the result of enhanced microbial diversity.40
3.3 Bacterial community composition and structure
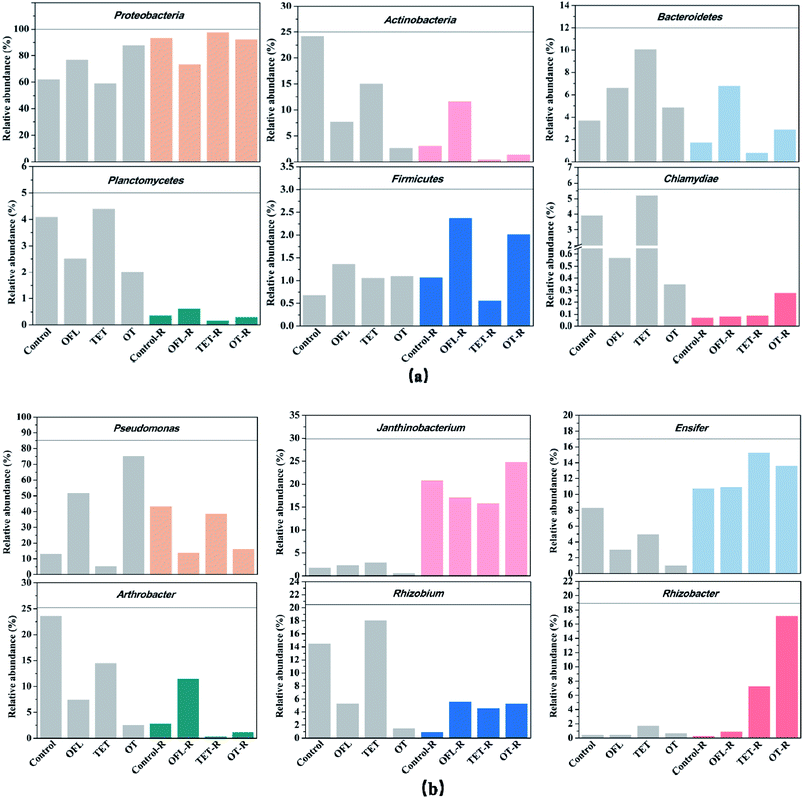
In the present study, a clear differentiation between the bacterial communities in the antibiotic treated and antibiotic free microcosms was observed in terms of taxonomic composition and community structure. The 16S rRNA gene encoding bacteria were assigned to 15 different phyla, which accounted for a total of 99.6% to 99.9% of the species (Fig. S2a and Table S4†). Among the 15 phyla, 6 were found in all 8 treatments: Proteobacteria, Actinobacteria, Bacteroidetes, Planctomycetes, Firmicutes, and Chlamydiae (Fig. 3a). These 6 phyla accounted for 95.1% to 99.7% of the bacterial communities in all microcosms. Antibiotic treatment had different effects when exudates were added to one set of microcosms. When exudates were not added, dosing with ofloxacin and the mixture of tetracycline and ofloxacin increased the relative abundance of Proteobacteria (p < 0.01, p < 0.01), whereas tetracycline decreased its relative abundance (p < 0.01, Table S5†). Meanwhile, the relative abundance of Actinobacteria was reduced by antibiotics in microcosms without exudates (p < 0.01). If exudates were added, exposure to ofloxacin and the mixture of antibiotics decreased the relative abundance of Proteobacteria (p < 0.01, p < 0.01), but exposure to tetracycline increased it (p < 0.01).In all 8 microcosms, 37 different genera were identified (Fig. S2a and Table S6†) and Fig. 3b shows the 6 most abundant genera. Tetracycline and ofloxacin impacted unique genera including both antibiotic resistant and sensitive species of bacteria. In microcosms without exudates, ofloxacin significantly increased the percentage of Pseudomonas spp. (Table S5†), which are known to harbor antibiotic resistance plasmids and exhibit resistance to several antibiotics.41–45 Ofloxacin also significantly stimulated the relative abundance of Parasegetibacter spp. (Table 2), which belong to the phylum Bacteroidetes in which some species have quinolone antibiotic resistance.46 However, Arthrobacter spp., which demonstrate resistance to ofloxacin,14,47,48 decreased with ofloxacin in microcosms without exudates. Researchers found that Pseudomonas spp. increased in relative abundance with tetracycline.49,50 However, in microcosms without exudates, tetracycline decreased the percentage of Pseudomonas spp., which is different from that for ofloxacin and the mixture of antibiotics.
| Ofloxacin | Tetracycline | Mixture | Ofloxacin–R | Tetracycline–R | Mixture–R | |
|---|---|---|---|---|---|---|
| a (1) Inhibition or stimulation percentage = 100 × (genus relative abundance in microcosms exposed to antibiotics − control genus relative abundance)/control genus relative abundance. Stimulation occurs when this calculation is positive and inhibition when negative. (2) This table excludes microcosm genera accounting for less than 1% of the total composition in all libraries of microcosm samples. Activation: the sum of the activation percentages. Inhibition: the sum of the inhibition percentage. *The three genera in each microcosm most affected by ofloxacin, tetracycline, or the mixture of ofloxacin and tetracycline. | ||||||
| Pseudomonas | 3.98* | −0.59 | 4.42* | −0.71 | −0.10 | −0.57 |
| Janthinobacterium | 0.60 | 0.58 | −0.68 | −0.26 | −0.23 | 0.35 |
| Ensifer | −0.54 | −0.39 | −0.88 | −0.09 | 0.43 | 0.43 |
| Arthrobacter | −0.60 | −0.38 | −0.90 | 2.56* | −0.86 | −0.52 |
| Rhizobium | −0.54 | 0.25 | −0.90 | 4.33* | 3.90* | 5.35* |
| Unclassified | 0.19 | 0.75 | −0.42 | 0.03 | −0.30 | 0.43 |
| Rhizobacter | 0.30 | 2.56* | 0.34 | 1.67 | 22.70* | 61.61* |
| Gemmobacter | 0.50 | 2.66* | 0.16 | 0.68 | 0.64 | −0.18 |
| Parasegetibacter | 7.08* | 0.07 | 5.63* | 6.02* | −0.99* | 1.24 |
| Sediminibacterium | 0.04 | 2.06* | −0.48 | −0.55 | −0.53 | 0.68 |
| Parachlamydia | −0.87 | 0.33 | −0.92 | −0.03 | 0.22 | 3.50* |
| Massilia | −0.92* | −0.67 | −0.95* | 0.31 | −0.68 | −0.31 |
| Bradyrhizobium | −0.67 | −0.22 | −0.79 | −0.09 | −0.45 | −0.37 |
| Inhibition | −4.14 | −2.25 | −6.92 | −1.73 | −4.14 | −1.95 |
| Activation | 12.69 | 7.20 | 10.55 | 15.60 | 27.89 | 73.59 |
The rhizosphere is considered to be a hotspot for antibiotic resistance transfer, where the effects of antibiotics on microorganisms may be influenced by exudation of nutrients,35 and distinct microbial communities might exhibit various microbial functions in microcosms with and without exudates. In exudate treated microcosms, ofloxacin, tetracycline and their mixture all decreased the relative abundance of Pseudomonas spp. This showed that association of antibiotics and exudates could have a synergistic antibacterial activity against some antibiotic-resistant bacteria, such as Pseudomonas spp. However, ofloxacin increased the relative abundance of Arthrobacter spp., which did not appear in microcosms free of exudates. Lin et al.14 thought that the genus Arthrobacter spp. can be able to dissipate quinolone as an additional source of carbon and energy. Meanwhile, the degradation of polycyclic aromatic hydrocarbons by Arthrobacter spp. has been proved to be strengthened by root exudates.51 Therefore, we hypothesize that the growth of Arthrobacter spp. was stimulated by root exudates when ofloxacin was added to the microcosms. In exudate treated microcosms, tetracycline, ofloxacin and their mixture significantly increased the relative abundance of Rhizobacter spp. and Rhizobium spp., which are responsible for nitrification and nitrogen fixation.52–54 However, the NH4+–N and NO3−–N removal efficiencies in exudate treated microcosms were stable, which showed that the stimulation of Rhizobacter spp. and Rhizobium spp. by antibiotics did not affect the nitrogen removal ability in microcosms.
Comparison of the relative abundance of bacteria at the genus level was carried out between the antibiotic-treated and control microcosms. Taxa with relatively high percentages (>1%) at the genus level (with definitive taxonomic information) are shown in Table 2. In the absence of exudates, the inhibitory degree of ofloxacin and the mixture of antibiotics was stronger than in exudate treated microcosms. Meanwhile, the degree of activation caused by antibiotics was significantly lower in microcosms without exudates than in exudate treated microcosms. In exudate treated microcosms, the degree of activation in the treatments with the mixture of antibiotics was much higher than in tetracycline or ofloxacin treatments. Differences in microbial response observed in the case of the mixture of antibiotics (compared to single antibiotic and control treatments) indicated that this mixture may have a different mode of action than the single compounds. For example, in microcosms with and without exudates, tetracycline and ofloxacin both decreased the relative abundance of Janthinobacterium spp. and Sediminibacterium spp., but the effects of the mixture were opposite.
3.4 Similarity analysis of community structures
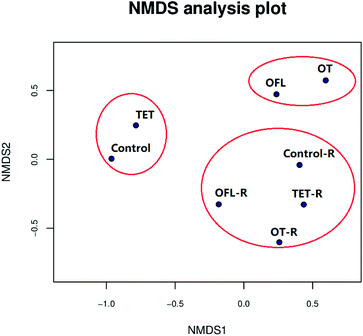
The NMDS was performed based on a Bray–Curtis distance matrix. In these plots, microbial communities that are similar are in closer proximity than dissimilar communities.55 As shown in Fig. 4, NMDS demonstrated that all samples were obviously clustered into three groups based on operational taxonomic unit tables. These three groups were significantly different as confirmed by the Anosim analyses (P = 0.004, R = 0.93) (Table S7†), consistent with the heatmap analysis (Fig. S3 and S4†). In comparison to the ofloxacin and ofloxacin–exudate treated microcosms, the tetracycline and tetracycline–exudate treated microcosms had more similar bacterial communities to the controls and control–exudate microcosms, suggesting greater similarity between the bacterial communities in the tetracycline and control microcosms. Meanwhile, in microcosms without exudates, similar microbial structures were observed between the microcosms exposed to the binary mixture and ofloxacin, which might indicate that ofloxacin affects the microbial structure more significantly than tetracycline, particularly at higher taxonomic levels (Fig. S4†). In general, microcosms treated with exudates clustered together, which confirmed an obvious role of exudates in shaping bacterial community structures.Collectively, the results from the analysis of community structures indicated that antibiotics significantly affected the bacterial community structure, but root exudates could weaken this effect, which meant that in CWs, exudates released by plant roots may protect bacterial communities from antibiotic stress.
3.5 Microbial functional potential of biofilm communities
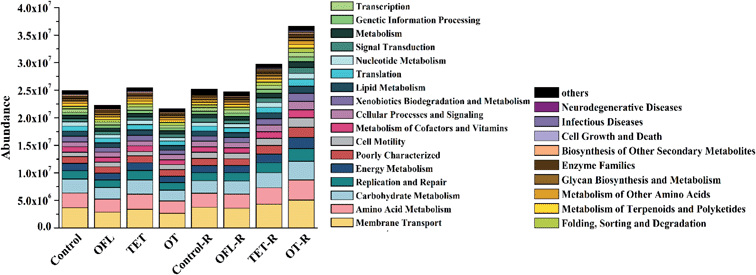
To investigate the effects of antibiotics and root exudates on microbial functional diversity, the sequences obtained from 16S data were annotated to the KEGG database (Fig. 5).56 Forty-one functional categories were detected with metabolism having the highest abundance, which mainly consisted of membrane transport (12.1% to 14.8%), amino acid metabolism (9.9% to 10.8%), carbohydrate metabolism (8.9% to 10.2%), and replication and repair (6.0% to 6.6%) (Table S8†). Similar to the community structure, all samples were also obviously clustered into the same three groups based on microbial functions (Anosim, P = 0.002, R = 0.913), which meant that antibiotics and root exudates could not only change the community structure but also affect microbial functions.There were significant differences in the functional composition among the different treatments, although the magnitude of difference among antibiotic-treated and control treatments was relatively low (generally less than 1%, Table S9†). Probably because of the additional amino acids and sugars, amino acid and carbohydrate metabolism were more enriched in exudate treated microcosms than in microcosms without exudates. Specifically, in microcosms without exudates, ofloxacin and the mixture of tetracycline and ofloxacin both decreased the abundances of amino acid metabolism, carbohydrate metabolism, energy metabolism, and xenobiotic biodegradation and metabolism. However, in microcosms treated with exudates, amino acid metabolism and carbohydrate metabolism were significantly enriched by antibiotics. These two metabolic pathways are strongly linked to the degradation of alanine, aspartate, glutamate and other carbohydrates,57 and the alterations of these functional properties might be associated with the stability of nitrogen cycling in the microcosms. Thus, these results showed that antibiotics influence the metabolic performance and potentially the metabolism of nutrients of specific communities in the rhizosphere which contains exudation of nutrients.
4. Conclusion
The responses of microbial communities to tetracycline, ofloxacin and their mixture were evaluated in microcosms treated with and without exudates. The carbon and nitrogen removal were not significantly affected by antibiotics in microcosms treated with and without exudates. Furthermore, a significant increase in bacterial diversity elicited by ofloxacin and antibiotic mixture exposure was only observed in microcosms with exudates. In exudate treated microcosms, ofloxacin promoted the relative abundance of Arthrobacter spp., which significantly varied from what was observed in microcosms free of exudates. Moreover, tetracycline, ofloxacin and their combination all significantly altered the relative abundances of antibiotic resistant bacteria (Rhizobacter spp., Rhizobium spp., and Pseudomonas spp.). Simultaneously, the functional profiles (KEGG) showed that the pathways of amino acid and carbohydrate metabolism were enhanced by antibiotics in microcosms with exudates. Further investigation is needed regarding the increase of antibiotic resistant organisms in CWs. Although the increased loading of carbon and nitrogen substrates is beneficial for improving microbial ecosystem functions in antibiotic-contaminated wetlands, the potential disturbance of the water bacterial communities may have the potential risk of spreading antibiotic resistance in CWs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support by the National Key Project on Prevention and Control of Water Pollution of the Ministry of Environmental Protection of China (2017ZX07203-005) (2012ZX07105-003).Notes and references
- L. Huang, X. Gao, M. Liu, G. Du, J. Guo and T. Ntakirutimana, Correlation among soil microorganisms, soil enzyme activities, and removal rates of pollutants in three constructed wetlands purifying micro-polluted river water, Ecol. Eng., 2012, 46, 98–106 CrossRef.
- H. Wu, X. Wang, X. He, S. Zhang, R. Liang and J. Shen, Effects of root exudates on denitrifier gene abundance, community structure and activity in a micro-polluted constructed wetland, Sci. Total Environ., 2017, 598, 697–703 CrossRef CAS PubMed.
- A. Garcia-Rodriguez, V. Matamoros, C. Fontas and V. Salvado, The ability of biologically based wastewater treatment systems to remove emerging organic contaminants–a review, Environ. Sci. Pollut. Res. Int., 2014, 21, 11708–11728 CrossRef PubMed.
- J. Chen, H. Xu, Y. Sun, L. Huang, P. Zhang, C. Zou, B. Yu, G. Zhu and C. Zhao, Interspecific differences in growth response and tolerance to the antibiotic sulfadiazine in ten clonal wetland plants in South China, Sci. Total Environ., 2016, 543, 197–205 CrossRef CAS PubMed.
- J. Lynch, The rhizosphere, Wiley, London, UK, 1990, p. 458 Search PubMed.
- H. P. Bais, T. L. Weir, L. G. Perry, S. Gilroy and J. M. Vivanco, The role of root exudates in rhizosphere interactions with plants and other organisms, Annu. Rev. Plant Biol., 2006, 57, 233–266 CrossRef CAS PubMed.
- P. Marschner, D. Crowley and C. H. Yang, Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type, Plant Soil, 2004, 261, 199–208 CrossRef CAS.
- M. Patel, R. Kumar, K. Kishor, T. Mlsna, C. U. Pittman Jr and D. Mohan, Pharmaceuticals of Emerging Concern in Aquatic Systems: Chemistry, Occurrence, Effects, and Removal Methods, Chem. Rev., 2019, 119, 3510–3673 CrossRef CAS PubMed.
- K. Kummerer, Antibiotics in the aquatic environment–a review–part I, Chemosphere, 2009, 75, 417–434 CrossRef PubMed.
- K. K. Brandt, A. Amezquita, T. Backhaus, A. Boxall, A. Coors, T. Heberer, J. R. Lawrence, J. Lazorchak, J. Schonfeld, J. R. Snape, Y. G. Zhu and E. Topp, Ecotoxicological assessment of antibiotics: a call for improved consideration of microorganisms, Environ. Int., 2015, 85, 189–205 CrossRef CAS PubMed.
- G. L. French, Bactericidal agents in the treatment of MRSA infections–the potential role of daptomycin, J. Antimicrob. Chemother., 2006, 58, 1107–1117 CrossRef CAS PubMed.
- C. Ding and J. He, Effect of antibiotics in the environment on microbial populations, Appl. Microbiol. Biotechnol., 2010, 87, 925–941 CrossRef CAS PubMed.
- P. Grenni, V. Ancona and A. Barra Caracciolo, Ecological effects of antibiotics on natural ecosystems: a review, Microchem. J., 2018, 136, 25–39 CrossRef CAS.
- H. Lin, D. Jin, T. E. Freitag, W. Sun, Q. Yu, J. Fu and J. Ma, A compositional shift in the soil microbiome induced by tetracycline, sulfamonomethoxine and ciprofloxacin entering a plant-soil system, Environ. Pollut., 2016, 212, 440–448 CrossRef CAS PubMed.
- F. Santos, C. M. R. Almeida, I. Ribeiro, A. C. Ferreira and A. P. Mucha, Removal of veterinary antibiotics in constructed wetland microcosms – response of bacterial communities, Ecotoxicol. Environ. Saf., 2019, 169, 894–901 CrossRef CAS PubMed.
- H. Heuer and K. Smalla, Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months, Environ. Microbiol., 2007, 9, 657–666 CrossRef CAS PubMed.
- G. C. Ding, V. Radl, B. Schloter-Hai, S. Jechalke, H. Heuer, K. Smalla and M. Schloter, Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine, PLoS One, 2014, 9, e92958 CrossRef PubMed.
- J. Ma, H. Lin, W. Sun, Q. Wang, Q. Yu, Y. Zhao and J. Fu, Soil microbial systems respond differentially to tetracycline, sulfamonomethoxine, and ciprofloxacin entering soil under pot experimental conditions alone and in combination, Environ. Sci. Pollut. Res. Int., 2014, 21, 7436–7448 CrossRef CAS PubMed.
- X. Tong, X. Wang, X. He, K. Xu and F. Mao, Effects of ofloxacin on nitrogen removal and microbial community structure in constructed wetland, Sci. Total Environ., 2019, 656, 503–511 CrossRef CAS PubMed.
- K. K. Brandt, O. R. Sjøholm, K. A. Krogh, B. Halling-Sørensen and O. Nybroe, Increased pollution-induced bacterial community tolerance to sulfadiazine in soil hotspots amended with artificial root exudates, Environ. Sci. Technol., 2009, 43, 2963–2968 CrossRef CAS PubMed.
- M. A. Mohamed, L. Ranjard, C. Catroux, G. Catroux and A. Hartmann, Effect of natamycin on the enumeration, genetic structure and composition of bacterial community isolated from soils and soybean rhizosphere, J. Microbiol. Methods, 2005, 60, 31–40 CrossRef CAS PubMed.
- S. Henry, S. Texier, S. Hallet, D. Bru, C. Dambreville, D. Cheneby, F. Bizouard, J. C. Germon and L. Philippot, Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates, Environ. Microbiol., 2008, 10, 3082–3092 CrossRef CAS PubMed.
- Y. Yang, W. Song, H. Lin, W. Wang, L. Du and W. Xing, Antibiotics and antibiotic resistance genes in global lakes: a review and meta-analysis, Environ. Int., 2018, 116, 60–73 CrossRef CAS PubMed.
- R. Singh, A. P. Singh, S. Kumar, B. S. Giri and K.-H. Kim, Antibiotic resistance in major rivers in the world: a systematic review on occurrence, emergence, and management strategies, J. Cleaner Prod., 2019, 234, 1484–1505 CrossRef CAS.
- M. Nkoom, G. Lu and J. Liu, Occurrence and ecological risk assessment of pharmaceuticals and personal care products in Taihu Lake, China: a review, Environ. Sci.: Processes Impacts, 2018, 20, 1640–1648 RSC.
- B. Griffiths, K. Ritz, N. Ebblewhite and G. Dobson, Soil microbial community structure: effects of substrate loading rates, Soil Biol. Biochem., 1999, 31, 145–153 CrossRef CAS.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, J. Huntley, N. Fierer, S. M. Owens, J. Betley, L. Fraser, M. Bauer, N. Gormley, J. A. Gilbert, G. Smith and R. Knight, Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms, ISME J., 2012, 6, 1621–1624 CrossRef CAS PubMed.
- L. Miao, P. Wang, J. Hou, Y. Yao, Z. Liu, S. Liu and T. Li, Distinct community structure and microbial functions of biofilms colonizing microplastics, Sci. Total Environ., 2019, 650, 2395–2402 CrossRef CAS PubMed.
- K. P. Asshauer, B. Wemheuer, R. Daniel and P. Meinicke, Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data, Bioinformatics, 2015, 31, 2882–2884 CrossRef CAS PubMed.
- F. Ye and Y. Li, Enhancement of nitrogen removal in towery hybrid constructed wetland to treat domestic wastewater for small rural communities, Ecol. Eng., 2009, 35, 1043–1050 CrossRef.
- A. Rico, M. R. Dimitrov, R. P. Van Wijngaarden, K. Satapornvanit, H. Smidt and P. J. Van den Brink, Effects of the antibiotic enrofloxacin on the ecology of tropical eutrophic freshwater microcosms, Aquat. Toxicol., 2014, 147, 92–104 CrossRef CAS PubMed.
- L. Hou, G. Yin, M. Liu, J. Zhou, Y. Zheng, J. Gao, H. Zong, Y. Yang, L. Gao and C. Tong, Effects of sulfamethazine on denitrification and the associated N2O release in estuarine and coastal sediments, Environ. Sci. Technol., 2015, 49, 326–333 CrossRef CAS PubMed.
- Y. Long, Z. Zhang, X. Pan, B. Li, S. Xie and Q. Guo, Substrate influences on archaeal and bacterial assemblages in constructed wetland microcosms, Ecol. Eng., 2016, 94, 437–442 CrossRef.
- T. He, W. Guan, Z. Luan and S. Xie, Spatiotemporal variation of bacterial and archaeal communities in a pilot-scale constructed wetland for surface water treatment, Appl. Microbiol. Biotechnol., 2016, 100, 1479–1488 CrossRef CAS PubMed.
- S. Jechalke, H. Heuer, J. Siemens, W. Amelung and K. Smalla, Fate and effects of veterinary antibiotics in soil, Trends Microbiol., 2014, 22, 536–545 CrossRef CAS PubMed.
- M. Uddin, J. Chen, X. Qiao, R. Tian, Y. Arafat and X. Yang, Bacterial community variations in paddy soils induced by application of veterinary antibiotics in plant-soil systems, Ecotoxicol. Environ. Saf., 2019, 167, 44–53 CrossRef CAS PubMed.
- J. P. Fernandes, C. M. Almeida, M. C. Basto and A. P. Mucha, Response of a salt marsh microbial community to antibiotic contamination, Sci. Total Environ., 2015, 532, 301–308 CrossRef CAS PubMed.
- J. P. Fernandes, C. M. R. Almeida, A. C. Pereira, I. L. Ribeiro, I. Reis, P. Carvalho, M. C. P. Basto and A. P. Mucha, Microbial community dynamics associated with veterinary antibiotics removal in constructed wetlands microcosms, Bioresour. Technol., 2015, 182, 26–33 CrossRef CAS PubMed.
- A. Bissett, C. Burke, P. L. Cook and J. P. Bowman, Bacterial community shifts in organically perturbed sediments, Environ. Microbiol., 2007, 9, 46–60 CrossRef CAS PubMed.
- K. K. Brandt, O. R. Sjøholm, K. A. Krogh, B. Halling-Sørensen and O. Nybroe, Increased pollution-induced bacterial community tolerance to sulfadiazine in soil hotspots amended with artificial root exudates, Environ. Sci. Technol., 2009, 43, 2963–2968 CrossRef CAS PubMed.
- J. L. Martinez, Environmental pollution by antibiotics and by antibiotic resistance determinants, Environ. Pollut., 2009, 157, 2893–2902 CrossRef CAS PubMed.
- A. Malik and A. Aleem, Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater, Environ. Monit. Assess., 2011, 178, 293–308 CrossRef CAS PubMed.
- C. L. Amorim, A. S. Maia, R. B. Mesquita, A. O. Rangel, M. C. van Loosdrecht, M. E. Tiritan and P. M. Castro, Performance of aerobic granular sludge in a sequencing batch bioreactor exposed to ofloxacin, norfloxacin and ciprofloxacin, Water Res., 2014, 50, 101–113 CrossRef CAS PubMed.
- X. L. Gao, M. F. Shao, Q. Wang, L. T. Wang, W. Y. Fang, F. Ouyang and J. Li, Airborne microbial communities in the atmospheric environment of urban hospitals in China, J. Hazard. Mater., 2018, 349, 10–17 CrossRef CAS PubMed.
- H. Zhou, X. Wang, Z. Li, Y. Kuang, D. Mao and Y. Luo, Occurrence and Distribution of Urban Dust-Associated Bacterial Antibiotic Resistance in Northern China, Environ. Sci. Technol. Lett., 2018, 5, 50–55 CrossRef CAS.
- Y. Xu, W. Yu, Q. Ma and H. Zhou, Occurrence of (fluoro)quinolones and (fluoro)quinolone resistance in soil receiving swine manure for 11 years, Sci. Total Environ., 2015, 530–531, 191–197 CrossRef CAS PubMed.
- K. J. Forsberg, S. Patel, M. K. Gibson, C. L. Lauber, R. Knight, N. Fierer and G. Dantas, Bacterial phylogeny structures soil resistomes across habitats, Nature, 2014, 509, 612–616 CrossRef CAS PubMed.
- Q. Kong, X. He, Y. Feng, M. S. Miao, Q. Wang, Y. D. Du and F. Xu, Pollutant removal and microorganism evolution of activated sludge under ofloxacin selection pressure, Bioresour. Technol., 2017, 241, 849–856 CrossRef CAS PubMed.
- N. F. F. Moreira, C. Narciso-da-Rocha, M. I. Polo-Lopez, L. M. Pastrana-Martinez, J. L. Faria, C. M. Manaia, P. Fernandez-Ibanez, O. C. Nunes and A. M. T. Silva, Solar treatment (H2O2, TiO2-P25 and GO-TiO2 photocatalysis, photo-fenton) of organic micropollutants, human pathogen indicators, antibiotic resistant bacteria and related genes in urban wastewater, Water Res., 2018, 135, 195–206 CrossRef CAS PubMed.
- Q. Wang, X. Li, Q. Yang, Y. Chen and B. Du, Evolution of microbial community and drug resistance during enrichment of tetracycline-degrading bacteria, Ecotoxicol. Environ. Saf., 2019, 171, 746–752 CrossRef CAS PubMed.
- J. C. Thomas, E. Cable, R. T. Dabkowski, S. Gargala, D. McCall, G. Pangrazzi, A. Pierson, M. Ripper, D. K. Russell and C. L. Rugh, Native Michigan plants stimulate soil microbial species changes and PAH remediation at a legacy steel mill, Int. J. Phytorem., 2012, 15, 5–23 CrossRef CAS PubMed.
- M. I. Khan, J. Yang, B. Yoo and J. Park, Improved RDX detoxification with starch addition using a novel nitrogen-fixing aerobic microbial consortium from soil contaminated with explosives, J. Hazard. Mater., 2015, 287, 243–251 CrossRef CAS PubMed.
- J. Naamala, S. K. Jaiswal and F. D. Dakora, Antibiotics Resistance in Rhizobium: Type, Process, Mechanism and Benefit for Agriculture, Curr. Microbiol., 2016, 72, 804–816 CrossRef CAS PubMed.
- T. Liu, Y. J. Mao, Y. P. Shi and X. Quan, Start-up and bacterial community compositions of partial nitrification in moving bed biofilm reactor, Appl. Microbiol. Biotechnol., 2017, 101, 2563–2574 CrossRef CAS PubMed.
- A. Ramette, Multivariate analyses in microbial ecology, FEMS Microbiol. Ecol., 2007, 62, 142–160 CrossRef CAS PubMed.
- L. Miao, P. Wang, J. Hou, Y. Yao, Z. Liu, S. Liu and T. Li, Distinct community structure and microbial functions of biofilms colonizing microplastics, Sci. Total Environ., 2019, 650, 2395–2402 CrossRef CAS PubMed.
- E. Neis, C. Dejong and S. Rensen, The role of microbial amino acid metabolism in host metabolism, Nutrients, 2015, 7, 2930–2946 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9em00458k |
| This journal is © The Royal Society of Chemistry 2020 |